Abstract
Background: A rapid molecular diagnostic test (MDT) is a test used to identify several different species of gram-negative bacteria and their genetic resistance markers. However, the impact of rapid MDT has not been established when combined with pharmacist involvement. Objective: To determine the impact of pharmacy involvement on patient outcomes when using rapid MDT. The primary outcome is the time from gram stain result to the first dose of the targeted antibiotic. Methods: This is a single-center, quasi-experimental, 1-group pretest-posttest design study of patients with gram-negative bacteremia in a community hospital. Hospitalized patients 18 years or older were included if they had a gram-negative blood culture. Patients were excluded if they were discharged or expired prior to culture results. Outcomes were compared between patients prior to and after implementation of the automated MDT. This research was determined to be exempt from institutional review board oversight consistent with West Florida Healthcare and in accordance with institutional policy. Results: The use of rapid MDT combined with pharmacist intervention resulted in a statistically significant decrease in the time to targeted antibiotic therapy (pre-intervention group, n = 77, 44.8 ± 17.8 hours versus post-intervention group, n= 80, 4.4 ± 5.8 hours; P ≤.001). There was no significant difference found between secondary outcomes. Limitations included small sample size as well as inconsistent documentation. Conclusions: The use of rapid MDT combined with pharmacist intervention resulted in a statistically significant decrease in the time to targeted antibiotic therapy.
Keywords: bacterial infections, diagnostic agents, antibiotics, clinical pharmacy, infectious diseases
Introduction
Bacteremia is a major cause of sepsis, being the source of 30%-40% of sepsis cases. Although bacteremia is often caused by gram-positive organisms, gram-negative organisms do play a role as well. 1 Delays in appropriate therapy and inappropriate empiric treatment have shown to increase patient mortality.2-4 Quick administration of appropriate antimicrobial therapy is crucial to the survival of patients. The increasing spread of multidrug-resistant (MDR) bacteria and the potential for multiple resistance mechanisms to simultaneously be present creates complex challenges and has led to the increased use of broad-spectrum antibiotics. 5 These unnecessarily broad regimens can not only increase selection of MDR strains in the hospital, thereby increasing mortality, but are also associated with higher costs, especially when utilizing carbapenems while awaiting final culture results. 6 Use of rapid molecular technology improves patient outcomes by providing the information to streamline patients from empiric antibiotic coverage to targeted treatment in a more efficient time frame than waiting for the traditional culture and sensitivity results. 7
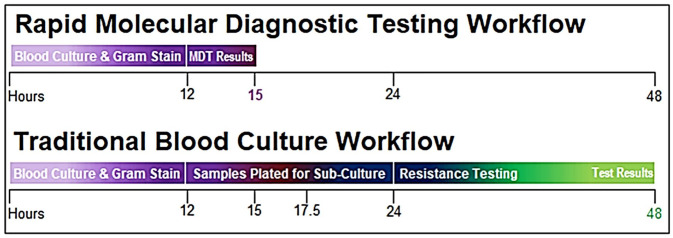
A rapid molecular diagnostic test (MDT) is used to identify several different organisms and their genetic resistance markers. The software used for this study identifies 4 different genera (Acinetobacter, Citrobacter, Enterobacter, and Proteus), 4 species (Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae, and Pseudomonas aeruginosa), and 6 different resistance genes (CTX-M, IMP, KPC, NDM, OXA, and VIM). The sensitivity is greater than 92% and specificity is greater than 99% for identification of gram-negative bacteria and genetic resistance markers. 8 Conventional methods require extended time periods due to the need for subculture, identification, and susceptibility testing while a rapid MDT provides the option for identification of the organisms and resistance within hours. 9 For reference, Figure 1 provides a timeline comparison of the rapid MDT assay versus traditional blood culture workflow.
Figure 1.
Rapid molecular diagnostic testing timeline.
Dodémont et al. evaluated the performance of the rapid MDT assay for accuracy and reliability. After assessing 125 positive blood cultures, 116 of the cultures were monomicrobial while only 9 were polymicrobial. The rapid MDT assay showed 99% and 83.3% accuracy for monomicrobial and polymicrobial, respectively. No misidentification occurred when assessing the organisms, and only 3 results were reported as false negatives. 10 Kim et al. also evaluated the performance of the rapid MDT assay. This study was able to assess 150 different gram-negative organism samples. Of these, 146 were monomicrobial and 138 were isolates that the rapid MDT assay was able to detect. When looking at the results from the organisms that the rapid MDT assay could potentially identify, all but one of the detected isolates were correctly identified (137 out of 138 isolates). 9 Hill et al. assessed 51 different gram-negative organisms, and the rapid MDT was able to correctly identify all 51 cultures. The assay was also able to correctly identify all 14 carbapenemase resistance mechanisms that were present. 11 The promising results from these studies prove that the rapid MDT results can provide opportunities for earlier interventions to promote improved patient outcomes. The purpose of this study was to assess the impact of a pharmacist-driven rapid MDT assay protocol.
Methods
Study Design
This was a single-center quasi-experimental, 1-group pretest-posttest design. It was conducted by chart review in a community hospital, evaluating the impact of pharmacists before and after an intervention was made. Data were collected from patient charts and de-identified, so informed consent was not required. This research was determined to be exempt from institutional review board oversight consistent with West Florida Healthcare and in accordance with institutional policy.
Patients
Hospitalized adult patients were included in this study if they had a gram-negative blood culture with one of the following organisms detected: Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter spp., Citrobacter spp., Enterobacter spp., or Proteus spp. Patients were excluded if they were discharged or expired prior to culture results or if the patient had a mixed infection. Patients were still included if they had mixed infections from a contaminant or if the blood culture grew a gram-positive organism prior to a gram-negative organism (rapid MDT assay could not be ran on the sample). Cultures were deemed contaminated based on clinical judgement and documentation of the provider. Mixed flora, patient presentation, and discordant growth between multiple cultures were specifically considered when determining whether a contaminant was present.
Study Design
The 2 cohorts included a pre-protocol implementation group (Group A) and a post-protocol implementation group (Group B). Group A patients were admitted between January 1, 2019, and June 30, 2019. Group B patients were admitted between September 7, 2020, and February 28, 2021. Data collected for each patient included patient demographics, length of hospital stay, intensive care unit (ICU) placement, Gram stain results, rapid MDT results, date of first empiric antibiotic received, date of targeted antibiotic received, mortality, antibiotic allergies, and acceptance rate of pharmacist interventions.
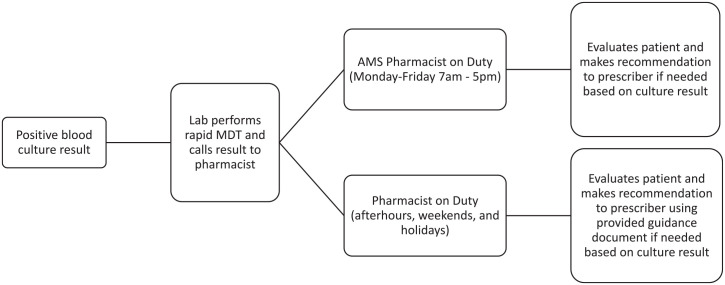
The rapid MDT for gram-negative organisms was implemented in November 2019. The rapid MDT assay is performed on all blood cultures that show gram-negative organism(s) identified on Gram stain review. Assay results, which are available in approximately 2 hours, are relayed to a pharmacist and documented. Once informed of the results, pharmacists communicate with the physician to ensure that the patient is on the most appropriate antibiotic. A specially trained Antimicrobial Stewardship (AMS) pharmacist is available during the weekdays to respond to results. If the AMS pharmacist is not available (afterhours, weekends, or holidays), the result is called to an alternative pharmacist on duty. Prior to implementation of the rapid MDT, an AMS pharmacist was responsible for responding to all positive blood cultures in Group A. Figure 2 shows the workflow of a positive blood culture.
Figure 2.
Blood culture result pathway. Abbreviation: AMS, Antimicrobial Stewardship.
Appropriate Antibiotic Choices
In order to train all pharmacists, several educational sessions were provided. These educational sessions explained the new RDT process, reviewed the guidance document, and provided patient case examples with potential interventions. The guidance document was developed using the genus, species, and resistance markers of different organisms detected by the rapid MDT as well as data from the hospital’s antibiogram. The patient case examples consisted of likely situations and interventions; for example, in a patient with gram-negative bacteremia on meropenem, when the laboratory calls the pharmacist and reports the MDT result of Escherichia coli with no resistance markers, the pharmacist will review the patient’s full history and if deemed appropriate (based on comorbidities, patient allergies, concurrent infections, likely source of bacteremia, etc.) will contact the provider to change from empiric coverage with a carbapenem to an appropriate antibiotic choice, which would be a third-generation cephalosporin based on our guidance document.
The definition of targeted antibiotic was different between the 2 cohorts. Targeted antibiotic in the pre-intervention cohort was defined as the narrowest antibiotic based on culture and sensitivity data as well as patient allergies, determined by pharmacist chart review. Targeted antibiotic in the post-intervention group was defined in the rapid MDT guidance document by using the results of local antibiogram data as well as resistance data provided by the rapid MDT result.
Endpoints
The primary endpoint was the time from first dose of empiric antibiotic to the first dose of the targeted antibiotic. Secondary endpoints include length of hospital stay, time to organism identification, and number of interventions made by pharmacists. For the purpose of this study, the time to organism identification was defined as the time from positive Gram stain to the time of final culture for the pre-implementation group or rapid diagnostic test for the post-implementation group, as documented in the electronic medical record.
Data Analysis
Demographic data were analyzed using the χ2 test. Results were analyzed using Kruskal-Willis and χ2 tests. A statistician analyzed the data using Statistical Analysis System (SAS) version 9.4 software. P values less than 0.05 were considered to be significant.
Results
The demographic data are shown in Table 1. There were no significant differences between the 2 groups. Although there was a greater percentage of females in the pre-implementation group, it was not statistically significant.
Table 1.
Patient Demographics.
| Demographic | Pre-group (n = 77) | Post-group (n = 80) | P |
|---|---|---|---|
| Female (%) | 48 (62.3) | 38 (47.5) | 0.0619 |
| Age ± SD (years) | 67.7 ± 18.1 | 72.6 ± 14.8 | 0.1621 |
| Weight ± SD (kg) | 82.3 ± 23.2 | 83.1 ± 28.7 | 0.8855 |
The primary outcome of this study was found to be statistically significant between the 2 cohorts as seen in Table 2. Patients in the post-implementation group were started on targeted antibiotics approximately 10 times faster than in the pre-implementation group.
Table 2.
Primary and Secondary Outcomes.
| Pre-group (n = 77) | Post-group (n = 80) | P | |
|---|---|---|---|
| Primary outcome | |||
| Average time to targeted antibiotic (hours ± SD) | 44.8 ± 17.8 | 4.4 ± 5.8 | <0.0001 |
| Secondary outcomes | |||
| Average length of stay (days ± SD) | 5.7 ± 3.1 | 6.7 ± 5.2 | 0.4244 |
| ICU admission (%) | 27 (35.1) | 22 (27.5) | 0.3065 |
| Average time to gram stain result (hours ± SD) | 16.9 ± 6.9 | 17.8 ± 5.8 | 0.0725 |
| Pharmacist interventions/opportunities (%) | 26/54 (48.2) | 15/21 (71.4) | 0.069 |
| Interventions accepted (%) | 23/26 (88.5) | 12/15 (80) | 0.5452 |
Abbreviation: ICU, intensive care unit.
The secondary outcomes can be found below the primary in Table 2. For secondary outcomes, average length of stay, ICU admission, time to Gram stain result, pharmacist interventions, and number of interventions accepted were similar in both groups.
Discussion
Multiple studies have evaluated the use of rapid MDT, but very few have assessed the impact that pharmacists can have on patient outcomes when using the results. Claeys et al. evaluated the use of a treatment algorithm developed by AMS pharmacists for appropriate antibiotic use based on the results of the rapid MDT assay results. This study was able to determine that the use of a treatment algorithm lead to 88.4% of patients receiving appropriate antibiotic care versus only 78.1% receiving appropriate antibiotics. 12 Bork et al. discovered a decreased use of the broad-spectrum antibiotics and an increased use of a more narrow option when analyzing the use of rapid MDT results on antibiotic usage. 3 This study provides data showing that the use of the rapid MDT gives pharmacists opportunities for de-escalation and more appropriate antibiotic usage. Cosgrove et al. and Holtzman et al. determined that rapid diagnostics can have little to no difference when implemented without an AMS program.13,14 Wenzler et al. also discussed the ability of rapid MDTs to be one the most powerful AMS interventions when used in conjunction with an AMS program. 15 These studies support the need for pharmacist involvement when using rapid diagnostic technology.
Based on our study results, the use of rapid MDT combined with pharmacist intervention improved average time to targeted antibiotic therapy by approximately 40 hours. This study agreed with Claeys et al. showing that the use of an AMS-derived treatment algorithm can lead to improved use of targeted antibiotics in a shorter time frame. 12 Additionally, this study confirmed the importance of utilizing an established algorithm plus pharmacist involvement when assessing rapid diagnostic results in gram-negative bacteremia. As mentioned earlier, in the studies by Cosgrove et al. and Holtzman et al., the absence of an algorithm and/or AMS guidance actually reduces the clinical benefits of using rapid MDT technology.13,14 Heyerly et al. also found that rapid identification combined with pharmacist intervention improved time to optimal antibiotic therapy. Although this study had a longer time to optimal antibiotic therapy compared with our study (8.4 hours vs 4.4 hours, respectively), both studies had a significantly shorter time to optimal therapy when utilizing rapid MDT results. 16 Gawrys et al. also found similar results in their study with a decrease in time to optimal therapy when utilizing rapid MDT in conjunction with pharmacist involvement. 17 Porter et al. specifically evaluated the use of the rapid MDT in a community hospital. Their findings were also in agreement with the result of this study and the multiple studies presented above show the importance of utilizing rapid MDT results along with pharmacist involvement. 18
Although not statistically significant (P = 0.069), there was a greater percentage of interventions in the post-implementation group than in the pre-implementation group (71.4% vs 48.2%, respectively). Of note, there were not as many opportunities for interventions in the post-implementation group as patients were already on the targeted antibiotic therapy. The rapid MDT process was new to all members during the assessment of the pre-intervention cohort, and data analysis proved that accurate notes were not provided on each patient with a rapid MDT result. Due to this, in some cases, it was difficult to track if interventions were caused by the pharmacist or the provider, which could lead to a falsely low intervention rate.
Strengths of this study include a standardized approach to antibiotic choices based on rapid MDT assay results and availability of pharmacists 24 hours a day for 7 days a week. Limitations of the study included small sample size (less than 100 patients in each group), inconsistent documentation on pharmacist interventions, and the process of documentation of interventions changed since the beginning of the study. Also, the cohorts were assessed at different times of year as the pre-implementation group was assessed from January to June, while the post-implementation group was assessed from September to February and could have led to seasonal differences. The gap in time that occurred between the pre-implementation group and the post-implementation group was simply due to logistics in the protocol and policy implementation, as well as staff education. At one point during the time frame for the post-intervention cohort, reagent for the rapid MDT ran out, which could have delayed some of the results of the rapid MDT. The delay in result could have led to increased time to targeted antibiotic therapy.
Conclusion
The use of rapid MDT combined with pharmacist intervention resulted in a statistically significant decrease in the time to targeted antibiotic therapy. Further studies need to be performed with accurate pharmacist documentation to determine the true pharmacist impact. Also, it would be useful to analyze the cost difference between the 2 cohorts to determine if there could be any cost benefit to the pharmacist interventions combined with rapid MDT.
Footnotes
Authors’ Note: This article was presented as a oral presentation at the Florida Residency Conference on May 14, 2021.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported (in whole or in part) by HCA Healthcare and/or an HCA Healthcare affiliated entity. The views expressed in this publication represent those of the author(s) and do not necessarily represent the official views of HCA Healthcare or any of its affiliated entities.
ORCID iD: Caitlin Bowman  https://orcid.org/0000-0002-9661-5960
https://orcid.org/0000-0002-9661-5960
References
- 1. Dekmezian M, Beal SG, Damashek MJ, Benavides R, Dhiman N. The SUCCESS model for laboratory performance and execution of rapid molecular diagnostics in patients with sepsis. Proc (Bayl Univ Med Cent). 2015;28:144-150. doi: 10.1080/08998280.2015.11929215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walker T, Dumadag S, Lee CJ, et al. Clinical impact of laboratory implementation of Verigene BC-GN microarray-based assay for detection of gram-negative bacteria in positive blood cultures. J Clin Microbiol. 2016;54:1789-1796. doi: 10.1128/JCM.00376-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bork JT, Leekha S, Heil EL, Zhao L, Badamas R, Johnson JK. Rapid testing using the Verigene gram-negative blood culture nucleic acid test in combination with antimicrobial stewardship intervention against gram-negative bacteremia. Antimicrob Agents Chemother. 2015;59:1588-1595. doi: 10.1128/AAC.04259-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kang CI, Kim SH, Park WB, et al. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother. 2005;49:760-766. doi: 10.1128/AAC.49.2.760-766.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pogue JM, Heil EL, Lephart P, et al. An antibiotic stewardship program blueprint for optimizing Verigene BC-GN within an institution: a tale of two cities. Antimicrob Agents Chemother. 2018;62:e02538-17. doi: 10.1128/AAC.02538-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rödel J, Karrasch M, Edel B, et al. Antibiotic treatment algorithm development based on a microarray nucleic acid assay for rapid bacterial identification and resistance determination from positive blood cultures. Diagn Microbiol Infect Dis. 2016;84:252-257. doi: 10.1016/j.diagmicrobio.2015.10.021 [DOI] [PubMed] [Google Scholar]
- 7. Belknap A, Grosser DS, Hale DA, et al. Clinical uptake of antimicrobial stewardship recommendations following nanosphere Verigene blood culture gram-negative reporting. Proc (Bayl Univ Med Cent). 2017;30:395-399. doi: 10.1080/08998280.2017.11930204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Verigene gram-negative blood culture nucleic acid test (BC-GN) [package insert]. Northbrook, IL: Nanosphere, Inc; 2015. [Google Scholar]
- 9. Kim JS, Kang GE, Kim HS, Kim HS, Song W, Lee KM. Evaluation of Verigene blood culture test systems for rapid identification of positive blood cultures. Biomed Res Int. 2016;2016:1081536. doi: 10.1155/2016/1081536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dodémont M, De Mendonça R, Nonhoff C, Roisin S, Denis O. Performance of the Verigene gram-negative blood culture assay for rapid detection of bacteria and resistance determinants. J Clin Microbiol. 2014;52:3085-3087. doi: 10.1128/JCM.01099-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hill JT, Tran KD, Barton KL, Labreche MJ, Sharp SE. Evaluation of the nanosphere Verigene BC-GN assay for direct identification of gram-negative bacilli and antibiotic resistance markers from positive blood cultures and potential impact for more-rapid antibiotic interventions. J Clin Microbiol. 2014;52:3805-3807. doi: 10.1128/JCM.01537-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Claeys KC, Schlaffer KE, Heil EL, Leekha S, Johnson JK. Validation of an antimicrobial stewardship-driven Verigene blood-culture gram-negative treatment algorithm to improve appropriateness of antibiotics. Open Forum Infect Dis. 2018;5:ofy233. doi: 10.1093/ofid/ofy233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cosgrove SE, Li DX, Tamma PD, et al. Use of PNA FISH for blood cultures growing gram-positive cocci in chains without a concomitant antibiotic stewardship intervention does not improve time to appropriate antibiotic therapy. Diagn Microbiol Infect Dis. 2016;86:86-92. doi: 10.1016/j.diagmicrobio.2016.06.016 [DOI] [PubMed] [Google Scholar]
- 14. Holtzman C, Whitney D, Barlam T, Miller NS. Assessment of impact of peptide nucleic acid fluorescence in situ hybridization for rapid identification of coagulase-negative staphylococci in the absence of antimicrobial stewardship intervention. J Clin Microbiol. 2011;49:1581-1582. doi: 10.1128/JCM.02461-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wenzler E, Timbrook TT, Wong JR, Hurst JM, MacVane SH. Implementation and optimization of molecular rapid diagnostic tests for bloodstream infections. Am J Health Syst Pharm. 2018;75:1191-1202. doi: 10.2146/ajhp170604 [DOI] [PubMed] [Google Scholar]
- 16. Heyerly A, Jones R, Bokhart G, Shoaff M, Fisher D. Implementation of a pharmacist-directed antimicrobial stewardship protocol utilizing rapid diagnostic testing. Hosp Pharm. 2016;51:815-822. doi: 10.1310/hpj5110-815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gawrys GW, Tun K, Jackson CB, et al. The impact of rapid diagnostic testing, surveillance software, and clinical pharmacist staffing at a large community hospital in the management of gram-negative bloodstream infections. Diagn Microbiol Infect Dis. 2020;98:115084. doi: 10.1016/j.diagmicrobio.2020.115084 [DOI] [PubMed] [Google Scholar]
- 18. Porter AM, Bland CM, Young HN, et al. Comparison of pharmacist-directed management of multiplex PCR blood culture results with conventional microbiology methods on effective and optimal therapy within a community hospital. Antimicrob Agents Chemother. 2018;63:e01575-18. doi: 10.1128/AAC.01575-18 [DOI] [PMC free article] [PubMed] [Google Scholar]