Abstract
Objectives
Recently, KPC-producing P. aeruginosa has rapidly emerged and expanded in East China. Here we described the clinical impact and characteristics of bloodstream infections (BSIs) from the dominant KPC-producing CRPA belonging to Sequence Type (ST) 463.
Methods
Retrospective cohort study was performed with CRPA BSI cases from 2019 to 2020 in a hospital in East China. Clinical characteristics, risk factors, and all-course mortality were evaluated. All CRPA isolates had whole-genome sequencing, antimicrobial susceptibility testing, and serum resistance assay. Representative isolates were tested for virulence in a Galleria mellonella infection model.
Results
Among the 50 CRPA BSI cases, ST463 predominated (48.0%). In multivariate analysis, we found three independent risk factors for fatal outcome: KPC carriage (OR 4.8; CI95% 1.0-23.7; P = 0.05), Pitt bacteremia score (OR 1.3; CI95% 1.0-1.6; P = 0.02), and underlying hematological disease (OR 8.5; CI95% 1.6-46.4; P = 0.01). The baseline clinical variables were not statistically different across STs, however the 28-day mortality was significantly higher in ST463 cases than that in non-ST463 cases (66.7% vs 33.3%, P = 0.03). ExoU and exoS virulence genes coexisted in all ST463 isolates, and the carbapenem resistant gene bla KPC were produced in almost all ST463 isolates, significantly higher than in the non-ST463 group(95.8% vs 7.7%, P<0.001). ST463 CRPA isolates also showed higher resistance rates to antipseudomonal cephalosporins, monobactam, and fluoroquinolones. And ST463 CRPA was confirmed hypervirulence in the larvae model. The genome of one ST463 CRPA strain showed that the bla KPC-2 gene was the sole resistance gene located on a 41,104bp plasmid pZYPA01, carried on a 7-kb composite transposon-like element flanked by two IS26 elements (IS26–Tn3-tnpA–ISKpn27–bla KPC-2–ISKpn6–IS26). Plasmid from various species presented core bla KPC-2 was franked by mobile genetic element ISKpn27 and ISKpn6.
Conclusions
In the ST463 CRPA BSI cohort, the mortality rates were higher than those in the non-ST463 CRPA BSI. The ST463 CRPA clone coharboring the bla KPC and exoU/exoS genes emerged and spread in East China, which might develop to a new threat in the clinic. Our results suggest that the surveillance of the new high-risk clone, ST463 CRPA, should be strengthened in China, even worldwide in the future.
Keywords: bloodstream infections, hypervirulence, carbapenem-resistant Pseudomonas aeruginosa , KPC, ST463
Introduction
Pseudomonas aeruginosa (PA) is a common and serious cause of nosocomial bloodstream infection; it is often severe when patients have compromised immunity. Despite improvements in medical care, the mortality of PA bacteremia remains considerably high (Thaden et al., 2017). Mortality in PA bloodstream infection is multifactorial, including the immune status of the host, the initial site of infection, the antibiotic treatment, and the microorganism (Hu et al., 2021a). Moreover, the presence of carbapenem-resistant Pseudomonas aeruginosa (CRPA) limits the treatment options and increases the risk of inadequate empirical therapy (Doi, 2019).
The common resistance mechanisms of P. aeruginosa to carbapenems are the loss of the outer membrane protein OprD and the overexpression of efflux pumps and/or the intrinsic chromosomally encoded AmpC β-lactamase (Li et al., 2012; Potron et al., 2015; Pang et al., 2019). Another mechanism for carbapenem resistance is the production of carbapenemases such as Metallo-β-lactamases. KPC-producing (class A β-lactamase) P. aeruginosa isolates have been occasionally reported in some countries since 2006 (Naas et al., 2013; Hu et al., 2015; Hagemann et al., 2018; Walkty et al., 2019). The bla KPC genes were frequently detected globally mainly among the species in the family Enterobacteriaceae (Feng et al., 2017). Some studies reported that bla KPC in CRPA might be transmitted from carbapenem-resistant Enterobacteriaceae. In China, KPC-2-producing Pseudomonas aeruginosa ST463 was first reported in 2015 and has rapidly spread in Zhejiang in recent years (Hu et al., 2015). However, clinical studies of BSIs by the emerging ST463 P. aeruginosa are still lacking.
In PA strains, the type III secretion system (T3SS) play a major role in their intrinsic virulence levels (Hauser, 2009). This secretion system injects potent cytotoxins into the cell and includes four effector proteins: exoU, a phospholipase; ExoS and ExoT, bifunctional proteins, and ExoY, an adenylate cyclase (Engel and Balachandran, 2009). In recent years, the cytotoxic exoU virulence gene of P. aeruginosa has been found to be associated with severe disease and poor outcomes. Furthermore, it is an important independent marker of early death in patients with P. aeruginosa BSI (Halavaty et al., 2012; Sawa et al., 2014; Peña et al., 2015). Almost all strains encode for exoT and exoY (Feltman et al., 2001), but the coexistence of exoU and exoS genes has rarely been reported due to their frequent mutual exclusivity (Hauser, 2009; Bradbury et al., 2010; Oliver et al., 2015). Previous studies indicated that the exoU genotype was also related to individual resistance to several antipseudomonal agents, including fluoroquinolones, carbapenems, and cephalosporins (Engel and Balachandran, 2009; Peña et al., 2015). However, one report illustrated that the co-presence of exoU and exoS was associated with a greater capacity for multi-drug resistance (Horna et al., 2019).
Herein, we described the occurrence of BSIs caused by KPC-producing CRPA that belonged to ST463. We investigate the high mortality rates associated with it in a tertiary hospital in East China in 2019–2020. We characterized the genomic alterations in the ST463 CRPA population and ascertained associated changes in phenotype and pathogenicity traits.
Materials And Methods
Study Population
We conducted a retrospective cohort study of CRPA BSI cases in a 2,500-bed public teaching hospital located in Zhejiang, China. The cases of CRPA BSIs were retrieved from the conventional microbiology laboratory database. The hospitalized patients with positive CRPA blood culture from January 2019 to December 2020 were selected. Five polymicrobial BSI cases were excluded from clinical analysis, but not from microbiological analysis. The study flowchart is shown in Figure 1 . Only the first isolate was selected for each patient. This study was approved by the institutional review board of the First Affiliated Hospital of Zhejiang University in China (approval no. IIT20210120B). Informed consent was waived by the review board due to the nature of the study. A standardized case record form was used to collect epidemiological and clinical data, as detailed in the Supplementary Materials.
Figure 1.
Study design. BSI, bloodstream infection; MLST, mulitilocus sequence typing; cgMLST, core genome MLST; SNPs, single nucleotide polymorphisms.
Microbiological Analysis
Frozen CRPA isolates were cultured and reidentified by VITEK 2 system (bioMérieux, France). We confirmed minimum inhibitory concentrations (MICs) of twelve antipseudomonal agents. Imipenem, meropenem, piperacillin-tazobactam, ceftazidime, cefepime, amikacin, gentamicin, ciprofloxacin, levofloxacin, aztreonam, and ceftazidime-avibactam were determined by the agar dilution method. The broth micro-dilution method was used to quantify antibacterial resistance against polymyxin. The results were interpreted according to Clinical and Laboratory Standards Institute (CLSI, 2021) standards (Papp-Wallace et al., 2010). All 50 CRPA isolates were sequenced using an Illumina Hiseq 2500 instrument (Qiagen, Valencia, CA, USA), and annotation was provided using the RAST server. Multi-locus sequence typing (MLST) and antibiotic resistance genes were conducted by the CGE server (https://cge.cbs.dtu.dk). Virulence factors were searched using the Virulence Factor DataBase (http://www.mgc.ac.cn/VFs/) (Liu et al., 2019). To determine the clonal relatedness, we performed a genome-wide gene-by-gene comparison by applying the SeqSphere+ software. Single nucleotide polymorphisms (SNPs) analysis was further conducted on CRPA isolates of the same Sequence Type clone. One isolate (ZYPA01) was carried out with long-read sequencing using the PacBio RSII platform. Sequence identity among the bla KPC-2 flanking genes of the isolate and KPC-encoding plasmids from P. aeruginosa reported previously, Escherichia coli or K. pneumoniae isolates were assessed by Blast analysis. Descriptions of detailed bioinformatics analysis are available in the Supplementary material.
Virulence Studies
We did a serum resistance assay with all 50 CRPA strains. We randomly selected five ST463 CRPA (ZYPA06, ZYPA10, ZYPA29, ZYPA32, and ZYPA36) and five non-ST463 CRPA (ZYPA08, ZYPA11, ZYPA23, ZYPA27, and ZYPA41) isolates as the representative for G. mellonella infection tests. The two assays were modified according to previously described protocols and are described in the Supplementary Methods . P. aeruginosa PAO1 was used as control.
Statistical Analysis
χ2 or Fisher exact test was used for categorical variables and t-test or Mann-Whitney U test for continuous variables to compare different groups in our study. Univariate analysis and multivariate analysis were performed to determine the risk factors for 28-day mortality. Variables with a P value of ≤ 0.05 in the univariate analysis were used in the binary logistic regression analysis to identify independent predictors. Kaplan-Meier survival curves (log-rank test) were obtained with the survminer R package. Kaplan-Meier survival curves were obtained in different groups. Analyses were two-tailed and the P-value of < 0.05 was considered significant. SPSS for Windows (IBM®, version 16.0) was used for the analysis.
Results
Overall Cohort Clinical Data
In this two-year analysis, we extracted and analyzed clinical data from the database for 45 patients with CRPA BSIs except for five polymicrobial cases. The epidemiological characteristics, clinical features, treatments, and clinical outcomes of the patients are listed in Table 1 . The median age of our cohort was 56 years and 37.8% were over 65 years. Most of the patients were male (71.1%). Majority of the patients had hematologic conditions (19, 42.2%). More than half (55.7%) of the patients had a prior hospitalization, with a median length of stay of 19 days, and 35.6% had been admitted to the intensive care unit. Most patients had invasive procedures prior to CRPA BSI: 66.7% of the patients underwent central venous catheterization, followed by mechanical ventilation (47.7%) and percutaneous catheterization (42.2%). Bacteremia was predominantly from respiratory tract infections (28.9%), followed by catheter-related and urinary tract infections (both 8.9%). Inflammatory indicators were elevated when bacteremia occurred. The median white blood cell (WBC) was 7.3*109/L, C-reactive protein (CRP) was 105.5 g/mL (increased), and procalcitonin (PCT) was 2.6 pg/mL (increased). Carbapenems combined with aminoglycosides were the most common antibiotic treatment (n = 15, 33.3%), followed by carbapenems combined with polymyxin B (n = 14, 31.1%). All-cause mortality rate at 7, 14 and 28 days was 44.4%, 48.9% and 51.1%, respectively. The clinical description of the 24 ST463 CRPA BSI cases is shown in Supplementary Table S1 .
Table 1.
Characteristics of patients with bloodstream infections caused by Carbapenem-Resistant P. aeruginosa according to sequence typing.
| Characteristic | Total (n = 45) | ST463 (n = 24) | Non-ST463 (n = 21) | P value |
|---|---|---|---|---|
| Demographic | ||||
| Sex (male) | 32 (71.11) | 18 (75.00) | 14 (66.67) | 0.54 |
| Age, years, median (IQR) | 56 (32.00-71.00) | 46.5 (25.25-66.00) | 65 (33.50-76.50) | 0.06 |
| Elderly (age>65y) | 17 (37.78) | 7 (29.17) | 10 (47.62) | 0.20 |
| BMI, kg/m2, mean (SD) | 21.71 (3.02) | 21.55 (2.30) | 21.97 (4.05) | 0.74 |
| Underlying medical conditions | ||||
| Diabetes | 6 (13.33) | 2 (8.33) | 4 (19.05) | 0.54 |
| Lung disease | 3 (6.67) | 2 (8.33) | 1 (4.76) | 1.00 |
| Kidney disease | 5 (11.11) | 2 (8.33) | 3 (14.29) | 0.87 |
| Hepatic disease | 8 (17.78) | 4 (16.67) | 4 (19.05) | 1.00 |
| Benign biliary diseases | 2 (4.44) | 0 (0) | 2 (9.52) | 0.21 |
| Cardiovascular diseases | 15 (33.33) | 6 (25.00) | 9 (42.86) | 0.20 |
| Hematological disease | 19 (42.22) | 12 (50.00) | 7 (33.33) | 0.26 |
| Solid malignant tumor | 3 (6.67) | 1 (4.17) | 2 (9.52) | 0.90 |
| Solid-organ transplant | 6 (13.33) | 4 (16.67) | 2 (9.52) | 0.79 |
| Hospital stay before BSI, median (IQR) | 19 (9.50-30.50) | 20 (9.25-36.50) | 18 (10.00-28.50) | 0.36 |
| Nosocomial | 38 (84.44) | 22 (91.67) | 16 (76.19) | 0.31 |
| Trauma | 3 (6.67) | 2 (8.33) | 1 (4.76) | 1.00 |
| Surgery | 22 (48.89) | 12 (50.00) | 10 (47.62) | 0.87 |
| Pitt bacteremia score, median (IQR)a | 3 (1.00-7.00) | 3.5 (1.00-9.25) | 2 (0.50-6.50) | 0.31 |
| Prior hospitalization (2 weeks) | 25 (55.56) | 13 (61.90) | 12 (50.00) | 0.42 |
| ICU (prior to bacteremia onset) | 16 (35.56) | 9 (37.50) | 7 (33.33) | 0.77 |
| CRPA BSI prior to hemodialysis | 11 (24.44) | 7 (29.17) | 4 (19.05) | 0.43 |
| Prior invasive procedure and/or devices | ||||
| Mechanical ventilation | 21 (47.73) | 12 (50.00) | 9 (45.00) | 0.74 |
| Urinary catheterization | 12 (27.91) | 7 (29.17) | 5 (26.32) | 0.84 |
| Central venous catheterization | 30 (66.67) | 16 (66.67) | 14 (66.67) | 1.00 |
| Percutaneous catheterization | 19 (42.22) | 13 (54.17) | 6 (28.57) | 0.08 |
| Laboratory examination | ||||
| White blood cell, median (IQR)b | 7.2 (0.33-15.30) | 2.9 (0.23-11.85) | 12.5 (0.70-15.60) | 0.16 |
| Neutrophilic granulocyte, median (IQR)c | 5.6 (0.06-13.30) | 1.8 (<0.01-10.40) | 10.9 (0.20-14.10) | 0.21 |
| Red blood cell, median (IQR)b | 2.31 (2.03-3.41) | 2.21 (1.94-2.66) | 2.39 (2.07-3.88) | 0.14 |
| Hemoglobin, median (IQR)b | 69 (62.50-105.50) | 67.5 (61.25-86) | 71 (63-117) | 0.23 |
| Platelet, median (IQR) a | 50 (11.00-247.50) | 20 (9-168) | 118 (25-252) | 0.04 |
| C-reactive protein, median (IQR)c | 105.50 (61.60-158.94) | 119.75 (84.67-160.17) | 86.9 (42.60-160.14) | 0.23 |
| Procalcitonin, median (IQR)c | 2.56 (0.42-15.05) | 2.68 (0.57-15.57) | 1.86 (0.21-17.67) | 0.51 |
| Albumin, median (IQR)d | 27.90 (24.30-29.93) | 27.10 (20.80-28.40) | 29.30 (25.40-31.90) | 0.03 |
| Agranulocytosis | 18 (40.00) | 11 (45.83) | 7 (33.33) | 0.39 |
| Source of bacteremia | ||||
| Lung | 13 (28.89) | 5 (20.83) | 8 (38.10) | 0.20 |
| Intra-abdominal infectione | 2 (4.44) | 1 (4.17) | 1 (4.76) | 1.00 |
| Skin and soft-tissue infection | 3 (6.67) | 2 (8.33) | 1 (4.76) | 1.00 |
| Surgical sites | 1 (2.22) | 1 (4.17) | 0 (0.00) | 1.00 |
| Catheter related | 4 (8.89) | 1 (4.17) | 3 (12.50) | 0.70 |
| Urinary tract infection | 4 (8.89) | 3 (12.50) | 1 (4.76) | 0.70 |
| Unknown | 19 (42.22) | 10 (41.67) | 9 (42.86) | 0.94 |
| Treatment | ||||
| Hormonal therapy | 29 (64.44) | 17 (70.83) | 12 (57.14) | 0.34 |
| Other immunotherapy | 20 (44.44) | 12 (50.00) | 8 (38.10) | 0.42 |
| Antibiotic After BSI | ||||
| Carbapenems + aminoglycosides | 15 (33.33) | 5 (20.83) | 10 (47.62) | 0.06 |
| Carbapenems + polymyxin B | 14 (31.11) | 4 (16.67) | 10 (47.62) | 0.03 |
| Ceftazidime-avibactam + aminoglycosides | 7 (15.56) | 1 (4.17) | 6 (28.57) | 0.07 |
| Ceftazidime-avibactam + carbapenems | 1 (2.22) | 0 (0.00) | 1 (4.76) | 0.47 |
| Ceftazidime-avibactam + polymyxin B | 4 (8.89) | 3 (12.50) | 1 (4.76) | 0.70 |
| Mortality | ||||
| All-cause death at 7 d postbacteremia | 20 (44.44) | 14 (58.33) | 6 (28.57) | 0.05 |
| All-cause death at 14 d postbacteremia | 22 (48.89) | 15 (62.50) | 7 (33.33) | 0.05 |
| All-cause death at 28 d postbacteremia | 23 (51.11) | 16 (66.67) | 7 (33.33) | 0.03 |
Data are presented as no. (%) unless otherwise indicated. Bolded numbers indicate that P < 0.05.
IQR, interquartile range; BMI, body mass index; SD, standard deviation; IAI, intra-abdominal infection; ICU, intensive care unit; BSI, bloodstream infection.
a. Evaluated 48 hours before or 24 hours after the first positive blood culture, whichever is the highest.
b. CRPA detected in the first blood culture.
c. The highest value before and after CRPA48 was detected in blood culture.
d. Lowest value at onset.
e. Including biliary tract and enteric canal.
Clinical Analysis According to Sequence Type
The 50 CRPA strains isolated from patients belonged to 24 different STs. As shown in Figure 2A (MLST plot), ST463 was the predominant type (48.0%, 24/50), and the other 26 strains showed a high degree of ST diversity. The rate of the bla KPC gene in ST463 CRPA was significantly higher than in non-ST463 CRPA (23/24, 95.8% vs 2/26, 6.7%; P < 0.001, Figure 2B ).
Figure 2.
(A) Pie chart showing sequence type distribution of 50 CRPA isolates; (B) Comparison of bla KPC carriage between ST463 and Non-ST463 strains (Fisher’s Exact Test). ST463 and bla KPC are highly associated (23 out of 24; 95.8%); (C) The Kaplan–Meier survival curves in patients infected with ST463 CRPA and with non-ST463 CRPA (P = 0.05); (D) The Kaplan–Meier survival curves between the two groups of patients infected with KPC-producing CRPA and non-KPC-producing CRPA (P = 0.008); (E) The K–M survival curves between KPC-producing ST463 CRPA and other STs CRPA that do not carry bla KPC (P = 0.018). The P value of Kaplan Meier’ analysis was calculated with log-rank test.
After the exclusion of five polymicrobial BSIs cases, patients were divided into two groups, ST463 CRPA-infected (n=24) and non-ST463 CRPA-infected (n=21), to determine whether there was specificity in the clinical features of ST463-infected patients. Notably, all-cause mortality at days 7, 14, and 28 was higher in ST463-infected than in non-ST463-infected patients (58.3% vs 28.6%, 62.5% vs 33.3%, and 66.7% vs 33.3%, respectively) ( Table 1 ). The 28-day survival curves based on different STs also showed a significant difference (P = 0.03) as detailed in Figure 2C . Furthermore, the ST463-infected group had a higher median Pitt bacteremia score of 3.5 compared with the non-ST463-infected group with a score of 2 (P = 0.3); this score is a predictor of early mortality risk in patients with BSI. There were no significant differences found in the clinical variables such as age, gender, underlying disease, or treatment received between the two groups; however, the amount of platelets was lower in the ST463-infected group (20 vs 118 *109/L, P=0.04) ( Table 2 ).
Table 2.
Analysis of risk factors for 28-Day mortality in 45 patients with Carbapenem-Resistant P. aeruginosa bloodstream infection.
| Covariate | Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|---|
| Survived (n = 22) | Died (n = 23) | P value | OR (95% CI) | P value | |
| Sex (male) | 14 (63.64) | 18 (78.26) | 0.28 | ||
| Age, years, mean (SD) | 54.86 (23.66) | 48 (22.54) | 0.32 | ||
| bla KPC | 7 (31.82) | 17 (73.91) | <0.01 | 4.87 (1.00-23.68) | 0.05 |
| Underlying medical conditions | |||||
| Hematological disease | 4 (18.18) | 15 (65.22) | <0.01 | 8.53 (1.57-46.35) | 0.01 |
| Solid malignant tumor | 2 (9.09) | 1 (4.35) | 0.97 | ||
| Solid-organ transplant | 1 (4.55) | 4 (17.39) | 0.37 | ||
| Pitt bacteremia score, median (IQR)a | 1.5 (0-3.25) | 6 (2-12) | 0.01 | 1.3 (1.04-1.64) | 0.02 |
| Source of bacteremia | |||||
| Respiratory tract infection | 7 (31.82) | 7 (30.43) | 0.92 | ||
| Intra-abdominal infectionb | 1 (4.55) | 1 (4.35) | 1 | ||
| Skin and soft-tissue infection | 0 (0) | 3 (13.04) | 0.25 | ||
| Surgical sites | 1 (4.55) | 0 (0) | 0.49 | ||
| Catheter related | 1 (4.55) | 1 (4.35) | 1 | ||
| Urinary tract infection | 3 (13.64) | 1 (4.35) | 0.57 | ||
| Unknown | 9 (40.91) | 10 (43.48) | 0.86 | ||
| Treatmentc | |||||
| Carbapenems + aminoglycosides | 12 (54.55) | 11 (47.83) | 0.65 | ||
| Carbapenems + polymyxin B | 14 (63.64) | 13 (56.52) | 0.63 | ||
| Ceftazidime-avibactam + aminoglycosides | 9 (40.91) | 6 (26.09) | 0.29 | ||
| Ceftazidime-avibactam + carbapenems | 1 (4.55) | 0 (0) | 0.49 | ||
| Ceftazidime-avibactam + polymyxin B | 2 (9.09) | 2 (8.70) | 1 | ||
OR, odds ratio; 95%CI, 95% confidence intervals; SD, standard deviation; IQR, inter-quartile range. Bolded p-values indicate P ≤ 0.05.
Variables showing P value ≤ 0.1 in the univariate analysis were further included in the multivariate model (binary logistic regression);
a Evaluated 48 hours before or 24 hours after the first positive blood culture, whichever is the highest;
b Including the biliary tract and enteric canal;
c Antibiotic treatment after BSI.
Mortality Predictor Analysis
Factors that may affect 28-day mortality in this CRPA BSI cohort were described in Table 2 . Multivariate analysis confirmed the following as independent factors: bla KPC carriage (odds ratio [OR], 4.9; 95% confidence interval [CI], 1.0-23.7; P = 0.05), Pitt bacteremia score (odds ratio [OR], 1.3; 95% confidence interval [CI], 1.0-1.6; P = 0.02), and underling hematological disease (odds ratio ([OR], 8.5; 95% confidence interval [CI], 1.6-46.4; P = 0.01).
Isolates’ Genetic Relatedness, Antimicrobial Susceptibility Testing, Antibiotic Resistance Genes, and Virulence Factors
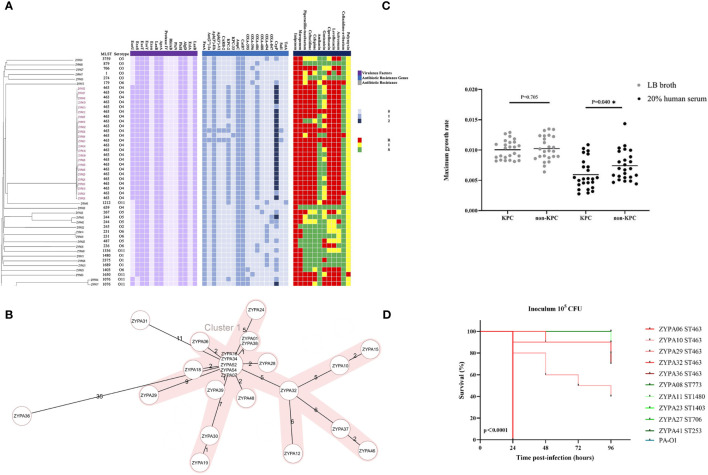
MLST and serotype analysis revealed that the 50 clinical strains belonged to 24 MLST types and eight serotypes, and P. aeruginosa ST463 strains were associated with serotype O4. The connections between serotypes and STs in other isolates are shown in Figure 3A . The same figure also depicts the antibiotic resistance genes identified in the CRPA genome. In our study, bla KPC was found in the genomes of 23/24 ST463 isolates, including 22 bla KPC-2 and 1 bla KPC-33; on the other hand, it was found in only one non-ST463 isolate (95.8% vs 3.8%, P < 0.001). The prevalence of the fluoroquinolone-resistance gene (crpP) was also significantly higher in the ST463 group than in the non-ST463 group, and most ST463 isolates had two copies of the crpP gene ( Figure 2A ). All ST463 strains carried bla OXA-486 β-lactamase gene, while the non-ST463 strains were genotyped with dispersed oxacillinases. Other resistance genes did not show differences between the two groups. We also found some significant differences in T3SS virulence factors between ST463 and non-ST463 CRPA strains ( Figure 2A ). All ST463 isolates coharbored exoU and exoS genes. However only 4/26 (15.4%) of the non-ST463 CRPA isolates carried exoU gene, and the other 22/26 non-ST463 strains carried the exoS gene without exoU.
Figure 3.
(A) Genetic relatedness, antibiotic resistance genes, virulence factor determinants, and antimicrobial susceptibility testing results of 50 CRPA BSI strains. These features are shown separately on the four blocks of the picture. The gray value indicates the copy number of the antibiotic resistance gene and virulence factors bla KPC-2 and ST463 are highly associated (22 out of 24; 91.7%). R, resistant; I, Intermediate; S, Sensitive; (B) Minimum spanning trees of 24 ST463 CRPA BSI strains from cgMLST analysis. Distance based on 4823 targets genes with pairwise ignore missing values. CgMLST profiles are represented by circles. The shaded parts represent the same cluster with a distance threshold of 10. The size of the circle is proportional to the number of isolates sharing the same cgMLST profile, with the biggest one including five isolates; (C) Neutrophil-killing resistance; at the maximum growth rate of KPC and non-KPC group; (D) Virulence potential in a G mellonella infection model. Five isolates of ST463 and five isolates of other STs representing various genetic background were randomly selected for the infection assay at 105 CFU. P values were calculated by log-rank (Mantel-Cox) test. Each line represents a single isolate. Isolate PA-O1 was used as the control. Abbreviation: CFU, colony-forming units.
Antimicrobial susceptibilities were summarized in Table 3 and Figure 2A . ST463 CRPA isolates were highly resistant to imipenem and meropenem (MIC50, 256 mg/L; 100% resistance). The MIC50s of carbapenems for ST463 CRPA isolates were 8- to 16-fold higher than for non-ST463 CRPA isolates: imipenem (MIC50, 32 mg/L; 100% resistance) and meropenem (MIC50, 16 mg/L; 84.6% resistance). Also, ST463 isolates show statistically significant difference in higher resistant rates to piperacillin-tazobactam (91.7% vs 26.9% resistance), ciprofloxacin (100% vs 38.5% resistance), levofloxacin (100% vs 42.3% resistance), ceftazidime (95.8% vs 19.2% resistance), cefepime (95.8% vs 30.8% resistance), and aztreonam (100% vs 42.3% resistance), to those of non-ST463 CRPA isolates (P < 0.001). Notably, all of the CRPA were less frequently resistant to ceftazidime-avibactam, amikacin and polymyxin.
Table 3.
Antimicrobial susceptibilities of 24 ST463 and 26 Non-ST463 Carbapenem-Resistant P. aeruginosa isolates.
| Antibiotic(s) | ST463 (n = 24) | Non-ST463 (n = 26) | P value (R%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Resistance rate (%) | MIC50 (μg/mL) | MIC90 (μg/mL) | MIC range (μg/mL) | Resistance rate (%) | MIC50 (μg/mL) | MIC90 (μg/mL) | MIC range (μg/mL) | ||
| Imipenem | 100 | 256 | 256 | 32 - 512 | 100 | 32 | 256 | 8 - 256 | 1 |
| Meropenem | 100 | 256 | 256 | 128 - 512 | 84.6 | 16 | 256 | 0.125 - 256 | 0.138 |
| Piperacillin-tazobactam | 91.7 | >256 | >256 | 16 - 512 | 26.9 | 16 | 256 | 4 - 512 | <0.001 |
| Ceftazidime | 95.8 | 256 | 256 | 8 - 256 | 19.2 | 8 | 256 | 2 - 128 | <0.001 |
| Cefepime | 95.8 | 128 | 256 | 16 - 512 | 30.8 | 8 | 128 | 2 - 256 | <0.001 |
| Amikacin | 12.5 | 8 | 256 | 2 - 256 | 0 | 8 | 16 | 1 - 32 | 0.206 |
| Gentamicin | 20.8 | 8 | 256 | 2 - 256 | 30.8 | 8 | 16 | 1 - 32 | 0.424 |
| Ciprofloxacin | 100 | 32 | 64 | 16 - 64 | 38.5 | 1 | 32 | 0.25 - 32 | <0.001 |
| Levofloxacin | 100 | 128 | 128 | 32 - 128 | 42.3 | 4 | 64 | 1 - 128 | <0.001 |
| Aztreonam | 100 | 256 | 256 | 32 - 512 | 42.3 | 16 | 256 | 1 - 256 | <0.001 |
| Ceftazidime-avibactam | 8.3 | 8 | 8 | 2 - 256 | 3.8 | 2 | 8 | 1 - 32 | 0.943 |
| Polymyxin | 0 | 1 | 1 | 1 - 2 | 3.8 | 1 | 2 | 1 - 4 | 1 |
Bolded p-values indicate P ≤ 0.05.
Based on sequence analysis of the whole genome, the phylogenetic tree showed that 22 of the 24 ST463 CRPA isolates belonged to the same cgMLST cluster 1, except for isolates of ZYPA36 and ZYPA31 ( Figure 3B ). For further core-genome SNPs, the average pairwise distance between two isolates is 99.95 SNPs and ranges from 1 to 2273 SNPs. Within the ST463 group, one strain genome (ZYPA01) was fully assembled for further analysis. pZYPA01, a unique plasmid, carried the sole resistance gene bla KPC-2, and no virulence genes were found ( Supplementary Figure S1A ). It could not be assigned to any known incompatibility group. Blast analysis of the pZYPA01 sequence revealed that it was highly similar to pPA1011 (GenBank accession number MH734334.1), a plasmid carrying carbapenem-resistant gene bla KPC-2 from an ST463 P. aeruginosa isolate in Hangzhou, China, with 99.1% identities and 99.0% query coverage ( Supplementary Figure S1A ). The plasmid pZYPA01 sequence was also highly similar to the plasmid sequence harboring the bla KPC-33 (pZYPA54), with 99.1% identities and 99.0% query coverage. The core bla KPC-2 platform Tn3-tnpA–ISKpn27–bla KPC-2–ISKpn6 had also been found in pPA1011 from P. aeruginosa, pKP048 from K. pneumoniae and pHS102707 from Escherichia coli ( Supplementary Figure S1B ). Unlike the mobile genetic element described in pPA1011, pKP048, and pHS102707 (Tn3–ISKpn8–bla KPC-2-ISKpn6), bla KPC-2 in pZYPA01 was carried on a 7-kb composite transposon-like element flanked by two IS26 elements (IS26–Tn3-tnpA–ISKpn27–bla KPC-2–ISKpn6–IS26), hence making it potentially transferable.
Virulence Assessment With Serum Killing and G. mellonella Infection Assays
In our study, all strains were tested in 20% human serum to investigate their capacity to resist the serum bactericidal activity. Subgroup analysis showed that the maximum growth rate of KPC-positive isolates was significantly lower compared to -negative isolates ( Figure 3C , P = 0.04). Although there was no statistical difference, the maximum growth rate was also lower in the ST463-CRPA group than in the non-ST463-CRPA (P = 0.24).
We randomly selected five representative ST463 isolates and five non-ST463 isolates to test the virulence properties of these strains. Continuous monitoring for 96 hours, even at the lowest concentration (105 cfu), the mortality rate of the ST463-infected larvae group was higher than in the non-ST463-infected. Of all isolates tested, the ST463 isolates PAZY06 were the most virulent strains in all inocula tested ( Figure 3D ).
Discussion
A recent increase in the detection rate of CRPA has been observed in Zhejiang Province, China, from 22% in 2015 to 32% in 2020; it is also significantly higher than in other provinces (Hu et al., 2019; Hu et al., 2021a; Hu et al., 2021b). In a recent report from a hospital in Zhejiang, ST463 CRPA emerged and has become the predominant CRPA clone among the population (Hu et al., 2021a). To date, there is still a lack of clinical studies on bloodstream infections of the new-onset KPC-producing CRPA belonging to ST463. Despite the retrospective and single-center nature of this study, it provides a uniquely detailed clinical and microbiological description of a KPC–producing ST463 CRPA BSI cohort from a Chinese hospital.
Conventionally, ST175, ST111, ST235, and ST395 are high-risk PA clones in clinics worldwide (Oliver et al., 2015). However, in our recent two-year study on the CRPA BSI cohort (2019-2020), none of the above international “high-risk” STs were isolated and ST463 has become the dominant CRPA clone in the population causing BSI (48.0%). Baseline characteristics showed no statistical differences in clinical variables across STs. However, 28-day mortality was significantly higher in ST463 cases than in non-ST463 cases. Of the 24 ST463 CRPA BSI cases in the cohort, 16 patients succumbed within 28 days. These BSIs were mainly from the respiratory tract. Indeed, several studies demonstrated that the initial site of infection affects prognosis, with surgery and pneumonia being associated with a particularly poor prognosis for P. aeruginosa bloodstream infections (Chatzinikolaou et al., 2000; Feng et al., 2017). In our study, bla KPC carriage, Pitt bacteremia score, and underlying hematological disease were three independent risk factors contributing to 28-day fatal outcomes. The survival curves also showed lower survival in KPC-positive ST463 CRPA-infected patients compared to those who are KPC-negative non-ST463 CRPA-infected ( Figures 2D, E ). The detection rate of bla KPC was also significantly higher in the ST463 group than in the non-ST463 group (95.8% vs 3.8%, P < 0.001). This suggests that the dominant clone of CRPA BSIs, ST463, has a high correlation with the bla KPC gene and the poor prognosis of BSIs.
Although metallo-β-lactamases (MBLs) were found to be the most frequent carbapenemases in CRPA, no MBL genes were found in either ST463 or non-ST463 isolates in this study. Moreover, the β-lactamase KPC was produced in almost all ST463 CRPA isolates. This is quite different from the results of our multicenter study conducted in China 10 years ago, when MBL genes including IMP and VIM were detected in 8.5% of CRPA isolates, while no KPC isolates were detected (Wang et al., 2010). Generally, KPC is detected from the Enterobacteriaceae family, including K. pneumoniae and E. coli; this leads to high levels of resistance to carbapenems and other β-lactam antibiotics. Therefore, we compared the surrounding sequences of bla KPC-containing plasmid sequences from different species including P. aeruginosa and Enterobacteriaceae. The results showed that each plasmid from various species presented core bla KPC-2 was franked by mobile genetic element ISKpn27 and ISKpn6 ( Supplementary Figure S1B ). This supports the hypothesis that bla KPC in P. aeruginosa might be acquired from Enterobacteriaceae (Queenan and Bush, 2007; Naas et al., 2013; Hu et al., 2021a).
All 24 ST463 (O4) CRPA isolates carried the exoU and exoS virulence genes, but not PldA, which was somewhat different from the reported strains from 2009 to 2018 (Hu et al., 2021a). Previously exoU and exoS were always reported to be mutually exclusive (Vareechon et al., 2017), while the coexistence of exoU and exoS may enhance the resistance of P. aeruginosa (Horna et al., 2019). In this study, all ST463 strains carried both exoS and exoU genes, while non-ST463 strains carried only one of them (15.4% exoU and 84.6% exoS), consistent with the results of higher resistance of ST463. On the other hand, the group of KPC-ST463-infected larvae had a higher mortality rate than other STs group. In 20% human serum, the maximum growth rate of KPC-positive CRPA was significantly lower compared to KPC-negative CRPA group. Thus, in vitro virulence phenotyping experiments showed that KPC was another independent risk factor associated with high lethality. We could further indicate that exoU and exoS virulence genes coexisting with bla KPC resistance gene in ST463 CRPA may be an important intrinsic cause of poor prognosis of clinical PA BSIs. This may lead to a new threat at the clinic.
In 2021, the Infectious Diseases Society of America (IDSA) recommended cefolozane-tazobactam, cefazidime-avibactam, and imipenem-cilastatin-relebactam as monotherapy first-line treatment options for difficult-to-treat P. aeruginosa (DTR-PA) infections (Tamma et al., 2021). In our study, the results of antimicrobial susceptibility testing showed that ST463 CRPA isolates were fairly susceptible to amikacin (87.5%) and ceftazidime-avibactam (91.7%). It is suggested that ceftazidime-avibactam or ceftazidime-avibactam combined with amikacin may be a good option for the treatment of infections caused by KPC-producing ST463 CRPA. For adult patients with severe infections, assuming normal renal and liver function, the recommended intravenous dose of ceftazidime-avibactam is 2.5 g every 8 hours, infused over 3 hours. However, more fluoroquinolone-resistant ST463 strains were detected compared to non-ST463 strains, as the prevalence of the crpP gene were also significantly higher in ST463 strains (mostly two copies of crpP). While no plasmid-mediated coding genes (e.g. qnr, aac (6’)-Ib-cr) were found, no other mechanisms of fluoroquinolone resistance such as chromosomal mutation in the DNA gyrase and topoisomerase II/IV-encoding genes (parC, parE, gyrA, and gyrB) were further investigated. Another noteworthy issuer, is that most MICs of polymyxin were 1 μg/mL and the effective therapeutic dose should be large and in combination (Nang et al., 2021). For patients on polymyxin B, the recommended loading dose is 2.0- 2.5 mg/kg based on total body weight (TBW) (equivalent to 20,000–25,000 IU/kg) over 1 hour. And the daily maintenance dose is 1.25- 1.5 mg/kg (equivalent to 12,500-15,000 IU/kg TBW) every 12 hours infused over 1 hour (Tsuji et al., 2019). High dose exposure to polymyxin has the potential to cause nephrotoxicity, hence limiting the therapeutic value (Lakota et al., 2018).
In conclusion, we described a BSI cohort by a novel high-risk KPC-PA ST463 clone that has been rapidly emerging and disseminating in East China in recent years. In the current study, we found a higher mortality rate in the ST463 CRPA BSI cohort than in the non-ST463 CRPA BSI. ST463 was the predominant epidemic type of KPC-producing CRPA. The independent risk factors for a 28-day outcome of CRPA BSI were bla KPC-carrying P. aeruginosa and hematological disease. Prompt and effective treatment of ST463 CRPA infection might reduce mortality, especially in patients with underlying hematological disease. These results also suggest that epidemiological surveillance is still needed to monitor the dynamic changes of high-risk clones, such as ST463 CRPA.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author Contributions
JW, YZ, MY, QYu, and HF collected clinical data. WS, HH, YC, YZ, HC, and PZ performed laboratory work. YJ, HH, and PZ performed whole-genome sequencing and bioinformatics analysis. TQ, HH, SZ, HF, and PZ analyzed clinical and microbiological data. TQ, HH, and QYa prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by research grants from the National Natural Science Foundation of China (No. NSFC81871689).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank all patients who participated in this study and all members of our institution for their help.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.756782/full#supplementary-material
References
- Bradbury R. S., Roddam L. F., Merritt A., Reid D. W., Champion A. C. (2010). Virulence Gene Distribution in Clinical, Nosocomial and Environmental Isolates of Pseudomonas Aeruginosa. J. Med. Microbiol. 59 (Pt 8), 881–890. doi: 10.1099/jmm.0.018283-0 [DOI] [PubMed] [Google Scholar]
- Chatzinikolaou I., Abi-Said D., Bodey G. P., Rolston K. V., Tarrand J. J., Samonis G. (2000). Recent Experience With Pseudomonas Aeruginosa Bacteremia in Patients With Cancer: Retrospective Analysis of 245 Episodes. Arch. Intern. Med. 160 (4), 501–509. doi: 10.1001/archinte.160.4.501 [DOI] [PubMed] [Google Scholar]
- Doi Y. (2019). Treatment Options for Carbapenem-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 69 (Suppl 7), S565–Ss75. doi: 10.1093/cid/ciz830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J., Balachandran P. (2009). Role of Pseudomonas Aeruginosa Type III Effectors in Disease. Curr. Opin. Microbiol. 12 (1), 61–66. doi: 10.1016/j.mib.2008.12.007 [DOI] [PubMed] [Google Scholar]
- Feltman H., Schulert G., Khan S., Jain M., Peterson L., Hauser A. R. (2001). Prevalence of Type III Secretion Genes in Clinical and Environmental Isolates of Pseudomonas Aeruginosa. Microbiol. (Reading) 147 (Pt 10), 2659–2669. doi: 10.1099/00221287-147-10-2659 [DOI] [PubMed] [Google Scholar]
- Feng J., Yin Z., Zhao Q., Zhao Y., Zhang D., Jiang X., et al. (2017). Genomic Characterization of Novel Incfii-Type Multidrug Resistant Plasmids P0716-KPC and P12181-KPC From Klebsiella Pneumoniae. Sci. Rep. 7 (1), 5830. doi: 10.1038/s41598-017-06283-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann J. B., Pfennigwerth N., Gatermann S. G., von Baum H., Essig A. (2018). KPC-2 Carbapenemase-Producing Pseudomonas Aeruginosa Reaching Germany. J. Antimicrob. Chemother. 73 (7), 1812–1814. doi: 10.1093/jac/dky105 [DOI] [PubMed] [Google Scholar]
- Halavaty A. S., Borek D., Tyson G. H., Veesenmeyer J. L., Shuvalova L., Minasov G., et al. (2012). Structure of the Type III Secretion Effector Protein Exou in Complex With its Chaperone Spcu. PloS One 7 (11), e49388. doi: 10.1371/journal.pone.0049388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A. R. (2009). The Type III Secretion System of Pseudomonas Aeruginosa: Infection by Injection. Nat. Rev. Microbiol. 7 (9), 654–665. doi: 10.1038/nrmicro2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horna G. A.-O., Amaro C., Palacios A., Guerra HA-OX, Ruiz J. A.-O. (2019). High Frequency of the Exou+/Exos+ Genotype Associated With Multidrug-Resistant “High-Risk Clones” of Pseudomonas Aeruginosa Clinical Isolates From Peruvian Hospitals. Sci. Rep. 9 (1), 10874. doi: 10.1038/s41598-019-47303-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. Y., Cao J. M., Yang Q., Chen S., Lv H. Y., Zhou H. W., et al. (2019). Risk Factors for Carbapenem-Resistant Pseudomonas Aeruginosa, Zhejiang Province, China. Emerg. Infect. Dis. 25 (10), 1861–1867. doi: 10.3201/eid2510.181699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.-y., Gu D.-x., Cai J.-c., Zhou H.-w., Zhang R. (2015). Emergence of KPC-2-Producing Pseudomonas Aeruginosa Sequence Type 463 Isolates in Hangzhou, China. Antimicrob. Agents Chemother. 59 (5), 2914–2917. doi: 10.1128/AAC.04903-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Liu C., Wang Q., Zeng Y., Sun Q., Shu L., et al. (2021. a). Emergence and Expansion of a Carbapenem-Resistant Pseudomonas Aeruginosa Clone are Associated With Plasmid-Borne Bla (KPC-2) and Virulence-Related Genes. mSystems 6 (3), e00154-21. doi: 10.1128/mSystems.00154-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F. P., Guo Y., Zhu D. M., Wang F., Jiang X. F., Xu Y. C., et al. (2021. b). CHINET 2020 Surveillance of Bacterial Resistance in China. Chin. J. Infect. Chemother. 21 (4), 377–387. doi: 10.16718/j.1009-7708.2021.04.001 [DOI] [Google Scholar]
- Lakota E. A., Landersdorfer C. B., Nation R. L., Li J., Kaye K. S., Rao G. G., et al. (2018). Personalizing Polymyxin B Dosing Using an Adaptive Feedback Control Algorithm. Antimicrob. Agents Chemother. 62 (7), e00483-18. doi: 10.1128/AAC.00483-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Luo Y. F., Williams B. J., Blackwell T. S., Xie C. M. (2012). Structure and Function of Oprd Protein in Pseudomonas Aeruginosa: From Antibiotic Resistance to Novel Therapies. Int. J. Med. Microbiol. 302 (2), 63–68. doi: 10.1016/j.ijmm.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zheng D., Jin Q., Chen L., Yang J. (2019). VFDB 2019: A Comparative Pathogenomic Platform With an Interactive Web Interface. Nucleic Acids Res. 47 (D1), D687–D692. doi: 10.1093/nar/gky1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Bonnin R. A., Cuzon G., Villegas M.-V., Nordmann P. (2013). Complete Sequence of Two KPC-Harbouring Plasmids From Pseudomonas Aeruginosa. J Antimicrob. Chemother. 68 (8), 1757–1762. doi: 10.1093/jac/dkt094 [DOI] [PubMed] [Google Scholar]
- Nang S. C., Azad M. A. K., Velkov T., Zhou Q. T., Li J. (2021). Rescuing the Last-Line Polymyxins: Achievements and Challenges. Pharmacol. Rev. 73 (2), 679–728. doi: 10.1124/pharmrev.120.000020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A., Mulet X., López-Causapé C., Juan C. (2015). The Increasing Threat of Pseudomonas Aeruginosa High-Risk Clones. Drug Resist. Update 21-22, 41–59. doi: 10.1016/j.drup.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Pang Z., Raudonis R., Glick B. R., Lin T. J., Cheng Z. (2019). Antibiotic Resistance in Pseudomonas Aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 37 (1), 177–192. doi: 10.1016/j.biotechadv.2018.11.013 [DOI] [PubMed] [Google Scholar]
- Papp-Wallace K. M., Bethel C. R., Distler A. M., Kasuboski C., Taracila M., Bonomo R. A. (2010). Inhibitor Resistance in the KPC-2 Beta-Lactamase, A Preeminent Property of This Class a Beta-Lactamase. Antimicrob. Agents Chemother. 54 (2), 890–897. doi: 10.1128/AAC.00693-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C., Cabot G., Gómez-Zorrilla S., Zamorano L., Ocampo-Sosa A., Murillas J., et al. (2015). Influence of Virulence Genotype and Resistance Profile in the Mortality of Pseudomonas Aeruginosa Bloodstream Infections. Clin. Infect. Dis. 60 (4), 539–548. doi: 10.1093/cid/ciu866 [DOI] [PubMed] [Google Scholar]
- Potron A., Poirel L., Nordmann P. (2015). Emerging Broad-Spectrum Resistance in Pseudomonas Aeruginosa and Acinetobacter Baumannii: Mechanisms and Epidemiology. Int. J. Antimicrob. Agents 45 (6), 568–585. doi: 10.1016/j.ijantimicag.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Queenan A. M., Bush K. (2007). Carbapenemases: The Versatile Beta-Lactamases. Clin. Microbiol. Rev. 20 (3), 440–458. table of contents. doi: 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa T., Shimizu M., Moriyama K., Wiener-Kronish J. P. (2014). Association Between Pseudomonas Aeruginosa Type III Secretion, Antibiotic Resistance, and Clinical Outcome: A Review. Crit. Care 18 (6), 668. doi: 10.1186/s13054-014-0668-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamma P. D., Aitken S. L., Bonomo R. A., Mathers A. J., van Duin D., Clancy C. J. (2021). Infectious Diseases Society of America Guidance on the Treatment of Extended-Spectrum Beta-Lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas Aeruginosa With Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 72 (7), 1109–1116. doi: 10.1093/cid/ciab295 [DOI] [PubMed] [Google Scholar]
- Thaden J. T., Park L. P., Maskarinec S. A., Ruffin F., Fowler V. G., Jr., van Duin D. (2017). Results From a 13-Year Prospective Cohort Study Show Increased Mortality Associated With Bloodstream Infections Caused by Pseudomonas Aeruginosa Compared to Other Bacteria. Antimicrob. Agents Chemother. 61 (6), e02671-16. doi: 10.1128/AAC.02671-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji B. T., Pogue J. M., Zavascki A. P., Paul M., Daikos G. L., Forrest A., et al. (2019). International Consensus Guidelines for the Optimal Use of the Polymyxins: Endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-Infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacotherapy 39 (1), 10–39. doi: 10.1002/phar.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vareechon C., Zmina S. E., Karmakar M., Pearlman E., Rietsch A. (2017). Pseudomonas Aeruginosa Effector Exos Inhibits ROS Production in Human Neutrophils. Cell Host Microbe 21 (5), 611–618.e5. doi: 10.1016/j.chom.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkty A., Alexander D. C., Karlowsky J. A., Nichol K., Embil J. (2019). Report of a KPC-Producing Pseudomonas Aeruginosa Isolate in Canada. J. Antimicrob. Chemother. 74 (6), 1748–1749. doi: 10.1093/jac/dkz064 [DOI] [PubMed] [Google Scholar]
- Wang J., Zhou J. Y., Qu T. T., Shen P., Wei Z. Q., Yu Y. S., et al. (2010). Molecular Epidemiology and Mechanisms of Carbapenem Resistance in Pseudomonas Aeruginosa Isolates From Chinese Hospitals. Int. J. Antimicrob. Agents 35 (5), 486–491. doi: 10.1016/j.ijantimicag.2009.12.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .