Abstract
We describe the synthesis and broad profiling of calcitroic acid (CTA) as vitamin D receptor (VDR) ligand. The x-ray co-crystal structure of the Danio Rerio VDR ligand binding domain in complex with CTA and peptide MED1 confirmed an agonistic conformation of the receptor. CTA adopted a similar conformation as 1,25(OH)2D3 in the binding pocket. A hydrogen bond with His333 and a water molecule was observed in the binding pocket, which was accommodated due to the shorter CTA side chain. In contrast, 1,25(OH)2D3 interacted with His423 and His333 due to its longer side chain. In vitro, the EC50 values of CTA and CTA-ME for VDR-mediated transcription were 2.89 μM and 0.66 μM, respectively, confirming both compounds as VDR agonists. CTA was further evaluated for interaction with fourteen nuclear receptors demonstrating selective activation of VDR. VDR mediated gene regulation by CTA in intestinal cells was observed for the VDR target gene CYP24A1. CTA at 10 μM upregulated CYP24A1 with similar efficacy as 1,25(OH)2D3 at 20 nM and 100-fold stronger compared to lithocholic acid at 10 μM. CTA reduced the transcription of iNOS and IL-1β in interferon γ and lipopolysaccharide stimulated mouse macrophages resulting in a reduction of nitric oxide production and secretion of IL-1β. These observed anti-inflammatory properties of 20 μM CTA were similar to 20 nM 1,25(OH)2D3.
Keywords: calcitroic acid, vitamin D receptor, CYP24A1, iNOS, IL-1β
1. Introduction
Several decades ago DeLuca identified calcitroic acid (CTA) [1] which together with calcioic acid are the two identified vitamin D metabolites bearing a carboxylic acid functionality [2]. CTA is constitutively formed in the urinary bladder and kidneys from 24,25,26,27-tetranor-1,23(OH)2D3 by mitochondrial P450 enzyme vitamin D3 24-hydroxylase (CYP24A1) (Scheme 1) [3, 4].
Scheme 1.
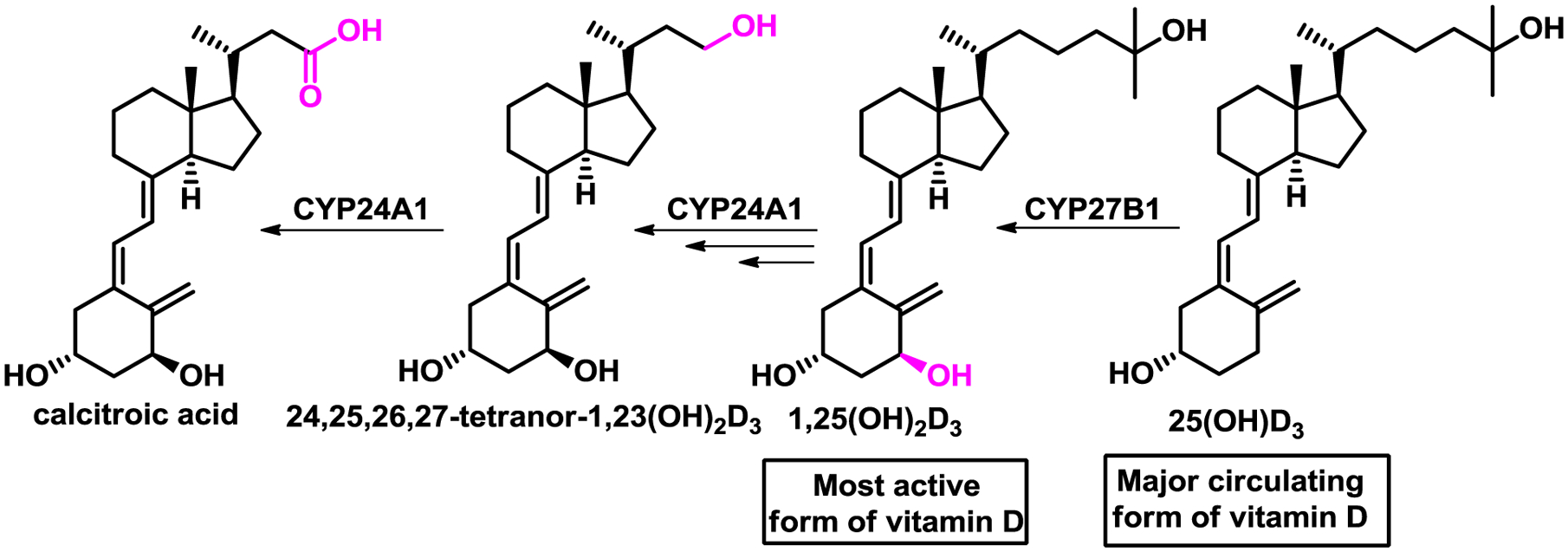
Metabolism of vitamin D analogs of calcitroic acid (CTA).
The enzymatic activity of CYP24A1 is not limited to the conversion of 24,25,26,27-tetranor-1,23(OH)2D3, as it oxidizes many other vitamin D metabolites including 1,25(OH)2D3, which has the highest affinity among all natural occurring vitamin D metabolites for the vitamin D receptor (VDR) [5]. VDR is a ligand-activated transcription factor that belongs to the superfamily of nuclear receptors [6]. VDR is expressed primarily in the epithelia of endocrine organs (e.g. parathyroid gland, mammary gland), digestive system, bronchi, kidneys, and thymus [7]. In addition, VDR can be found in leukocytes and bone [8, 9]. The downstream physiological effects mediated by liganded VDR have been partially elucidated using 1,25(OH)2D3 and include (among others) calcium homeostasis, cell proliferation, and differentiation [10]. In addition, VDR ligands play a pivotal role in reducing inflammation, which has been shown for psoriasis [11], inflammation-associated cancer [12], type 1 diabetes [13], multiple sclerosis [14], and inflammatory bowel disease [15]. It has been demonstrated that 1,25(OH)2D3 upregulated VDR target gene cathelicidin (including the human cationic antimicrobial protein), thus exhibiting antimicrobial effects that protect against pathogens such as Mycobacterium tuberculosis and Staphylococcus aureus [16]. Furthermore, 1,25(OH)2D3 inhibited the production of proinflammatory cytokines such as Il-17, IFNγ, and IL-21, in addition to upregulation of anti-inflammatory IL-10 in T cells [17]. Toll-like receptor expression was suppressed in monocytes in the presence of 1,25(OH)2D3 [18], and inhibition of the NF-κB pathway by 1,25(OH)2D3 included suppressed translocation and phosphorylation of IκB [19]. Anti-inflammatory pathways are activated in the presence of VDR agonists such as 1,25(OH)2D3, which is in turn produced from 25(OH)D3, the major circulatory form of vitamin D (Scheme 1) [20]. Importantly, the physiological concentration of 1,25(OH)2D3 in circulation is very low (35 pg/mL) and regulated by CYP24A1 via a negative feedback loop [21]. In addition, 1,25(OH)2D3 binds the vitamin D binding protein that enables circulation of many vitamin D metabolites in the blood [22]. As a result, the relationship between 1,25(OH)2D3 and CYP24A1 is mediated by VDR, which upon 1,25(OH)2D3 binding induces transcription of CYP24A1, the gene for CYP24A1 that catabolizes 1,25(OH)2D3 to CTA. Many studies have shown that CYP24A1, in addition to its constitutive expression in kidney and bladder, can be induced by vitamin D metabolites upon VDR binding in epithelia cells of other organs as well as immune cells. Thus, concentrations of 1,25(OH)2D3 are regulated locally and independently from kidney metabolism. CTA has been investigated in vitro and in vivo, especially relating to changes in calcium homeostasis [23, 24]. Recently, our group has shown that CTA among other compounds interacts with the ligand binding domain (LBD) of the VDR and induces recruitment of SRC-1 in a two-hybrid assay [25]. CTA also upregulated VDR target gene CYP24A1 in DU145 prostate cancer cells 9-fold at 7.5 μM, which was significantly lower than calcitriol at 20 nM giving a 110-fold induction. Here, we present the synthesis of CTA achieving a higher than reported yield for the last step. The interaction with VDR is demonstrated with an X-ray co-crystal structure of the CTA bound to zVDR LBD. Finally, we show that CTA selectively binds to VDR among other nuclear receptors and exhibits anti-inflammatory properties in vitro.
2. Material and Methods
2.1. Synthesis.
Ergocalciferol was purchased from AstaTech (44109). ((Z)-2-((3S,5R)-3,5-bis((tert-butyldimethylsilyl)oxy)-2-methylenecyclohexylidene)ethyl) diphenylphosphine oxide was obtained from ChemScene (CS-M1835). All moisture or oxygen-sensitive reactions were carried out under a dry nitrogen atmosphere. All reactions were monitored by thin-layer chromatography (TLC) using Merck 60 UV254 silica gel plates (Sigma-Aldrich). Products were purified by flash chromatography (SPI Biotage, silica gel 230–400 mesh). Compound masses were identified using a Shimadzu 2020 LC-MS (single quadrupole) instrument. NMR spectra were recorded on a Bruker Avance 500 MHz instrument. Optical rotations were recorded using Jasco DIP-370 Digital Polarimeter instrument in LCMS grade chloroform or methanol.
Methyl (R)-3-((1R,3aS,7aR,E)-4-((Z)-2-((3S,5R)-3,5-bis((tert-butyldimethylsilyl)oxy)-2-methylene cyclohexylidene)ethylidene)-7a-methyloctahydro-1H-inden-1-yl)butanoate (4).
A solution of n-butyllithium (1.6 M in hexanes, 1.56 mL, 2.50 mmol) was added dropwise to a THF solution of ((Z)-2-((3S,5R)-3,5-bis((tert-butyldimethylsilyl)oxy)-2-methylenecyclohexylidene) ethyl) diphenyl phosphine oxide (3) (1.84 g, 3.16 mmol, dried over 3Å molecular sieves in 5 ml THF overnight) at −78 °C. The solution turned deep red and was stirred for one hour at −78 °C. Raising the solution out of the bath briefly deepened the red color and ensured that the deprotonation was complete. After cooling back to −78 °C, methyl (R)-3-((1R,3aR,7aR)-7a-methyl-4-oxooctahydro-1H-inden-1-yl)butanoate (2) (0.42 g, 1.66 mmol, dried over 3Å molecular sieves in 5 ml THF overnight) was added dropwise. The solution was stirred for five hours at −78°C and then allowed to warm to room temperature. The reaction was treated with 10 mL water, diluted with tert-butylmethyl ether (TBME), and extracted with TBME twice (2 × 50 mL). The combined layers were washed with brine, dried over MgSO4, and concentrated. The crude product was purified by column chromatography (EtOAc: Hex 5:95) to yield 4 (0.66g, 65%) as a viscous clear oil; Rf 0.22 (EtOAc:Hex 3:97); 1H-NMR (500MHz, CDCl3) δ 0.04–0.10 (m, 12H), 0.58 (s, 3H), 0.89 (s, 18H), 1.00 (d, J = 6.3 Hz, 3H), 1.26–1.36 (m, 3H), 1.46–1.61 (m, 3H), 1.61–1.73 (m, 2H), 1.73–1.81 (m, 1H), 1.81–1.96 (m, 3H), 1.96–2.05 (m, 3H), 2.22 (dd, J = 12.6, 7.6 Hz, 1H), 2.40–2.48 (m, 2H), 2.80–2.86 (m, 1H), 3.66 (s, 3H), 4.17–4.23 (m, 1H), 4.35–4.40 (m, 1H), 4.84–4.89 (m, 1H), 5.16–5.20 (m, 1H), 6.03 (d, J = 11.2 Hz, 1H), 6.24 (d, J = 11.2 Hz, 1H); 13C-NMR: (125MHz, CDCl3) δ −5.1, −4.8, −4.68, −4.67, 12.0, 18.1, 18.2, 19.7, 22.1, 23.4, 25.8, 25.9, 27.7, 28.8, 34.1, 40.5, 41.3, 44.8, 45.8, 46.1, 51.3, 56.3 (2C), 67.5, 72.1, 111.3, 118.1, 123.1, 135.2, 140.5, 148.3, 173.84; m/z calculated for C36H65O4Si2 617.4416 [M + H],+ found 617.4401 =+ 21.0 (c 1.0, CHCl3)
Methyl (R)-3-((1R,3aS,7aR,E)-4-((Z)-2-((3S,5R)-3,5-dihydroxy-2-methylenecyclohexylidene) ethylidene)-7a-methyloctahydro-1H-inden-1-yl)butanoate or calcitroic acid methyl ester (5).
Tetrabutylammonium fluoride (1M in THF, 23 mL, 23.3 mmol) was added to a stirred solution of 4 (0.96 g, 1.55 mmol) in dry THF (20 mL) at room temperature. The solution was stirred overnight, treated with an aqueous NH4Cl solution (20 mL), and extracted with EtOAc (3×50 mL). The organic layers were combined, washed with brine (50 mL), and dried over MgSO4 and concentrated. The crude product was purified by column chromatography (EtOAc : Hex 7 : 3) to yield 5 as a slightly yellow oil (0.53 g, 88%); Rf 0.26 (EtOAc: Hex 7:3); 1H-NMR (500MHz, CDCl3) δ 0.57 (s, 3H), 0.98 (d, J = 6.4 Hz, 3H), 1.27–1.34 (m, 3H), 1.45–1.59 (m, 4H), 1.63–1.72 (m, 2H), 1.81–1.95 (m, 4H), 1.96–2.03 (m, 4H), 2.30 (dd, J = 14.0, 8.0 Hz, 1H), 2.43 (dd, J = 14.5, 3.5 Hz, 1H), 2.57 (dd, J = 13.7, 3.1 Hz, 1H), 2.82 (dd, J = 12.6, 4.3 Hz, 1H), 3.66 (s, 3H), 4.21 (m, 1H), 4.42 (dd, J = 7.6, 4.5 Hz, 1H), 4.98 (s, 1H), 5.32 (s, 1H), 6.02 (d, J = 11.0 Hz, 1H), 6.35 (d, J = 11.0 Hz, 1H); 13C-NMR: (125MHz, CDCl3) δ 12.0, 19.6, 22.2, 23.5, 27.6, 29.0, 34.1, 40.3, 41.3, 42.8, 45.2, 45.9, 51.4, 56.3 (2C), 66.7, 70.6, 111.8, 117.3, 124.6, 133.4, 142.5, 147.7, 174.0; m/z calculated for C24H36O4 389.2686 [M + H]+, found 389.2618; = −7.0 (c 1.0, CHCl3)
(R)-3-((1R,3aS,7aR,E)-4-((Z)-2-((3S,5R)-3,5-dihydroxy-2-methylenecyclohexylidene) ethylidene)-7a-methyloctahydro-1H-inden-1-yl)butanoic acid or calcitroic acid (6).
Calcitroic methyl ester (5) (0.52 g, 1.33 mmol) was dissolved in a solution of 10% NaOH in 9:1 MeOH:H2O (25 mL) and stirred at room temperature for two h. After completion, the reaction was neutralized with concentrated HCl to ~pH 7. Excess methanol was evaporated under reduced pressure. The reaction was diluted with H2O (25 mL), acidified with concentrated HCl to pH 1, and extracted with EtOAc (3 × 30 mL). The combined organic layers were washed with brine (25 mL), dried over MgSO4, concentrated, and purified by column chromatography (EtOAc) to yield 6 (0.40 g, 80%) as a yellow solid; Rf 0.14 (EtOAc); 1H-NMR (500MHz, CDCl3) δ 0.60 (s, 3H), 1.06 (d, J = 6.5 Hz, 3H), 1.30–1.41 (m, 3H), 1.48–1.61 (m, 3H), 1.67–1.76 (m, 3H), 1.86–1.98 (m, 4H), 1.98–2.12 (m, 4H), 2.34 (dd, J = 13.0, 6.5 Hz, 1H), 2.50 (dd, J = 15.0, 3.5 Hz, 1H), 2.62 (dd, J = 13.5, 3.2 Hz, 1H), 2.85 (dd, J = 13.6, 4.6 Hz, 1H), 4.26 (m, 1H), 4.46 (dd, J = 4.2, 7.6, 1H), 5.02 (s, 1H), 5.35 (s, 1H), 6.04 (d, J = 11.2 Hz, 1H), 6.39 (d, J = 11.2 Hz, 1H); 13C-NMR: (125MHz, CDCl3) δ 12.0, 19.7, 22.2, 23.5, 27.6, 29.0, 33.9, 40.3, 41.1, 42.8, 45.2, 45.9, 56.2, 56.3, 66.9, 70.8, 111.9, 117.2, 124.9, 133.1, 142.7, 147.6, 178.4; m/z calculated for C23H34O4 373.2373 [M + H]—, found 373.2335; = −6.0 (c 1.0, CHCl3)
2.2. Crystallization and Structure Determination.
cDNA encoding zVDR LBD (156–453 AA) was cloned into pET28b vector to generate N-terminal His-tag fusion proteins. Purification was carried out as previously described, including metal affinity chromatography and gel filtration [26]. The protein was concentrated using Amicon ultra-30 (Millipore) to 3–7 mg/mL and incubated with a 2-fold excess of ligand and a 3-fold excess of the coactivator MED1 NR2 peptide (640-NHPMLMNLLKDN-652). Crystals were obtained in 0.1 M Mes pH 6 and 1.8 M lithium sulfate. Protein crystals were mounted in a fiber loop and flash-cooled under a nitrogen flux after cryoprotection in 2.2 M lithium sulfate. Data collection from a single frozen crystal was performed at 100 K on the PX2 beamline at SOLEIL (France). The raw data were processed and scaled with XDS. The crystals belong to the space group P6522, with one LBD complex per asymmetric unit. The structure was solved and refined using BUSTER, Phenix [27], and iterative model building using COOT [28]. Crystallographic refinement statistics are presented in SI, Table S1. All structural figures were prepared using molecular operating environment (MOE).
2.3. Transcription Assay Protocol.
Human embryonic kidney (HEK293, ATCC CRL-3249) cells were cultured in DMEM/High Glucose (Hyclone, #SH3024301). Non-essential amino acids (Hyclone, #SH30238.01), 10 mM HEPES (Hyclone, #SH302237.01), penicillin and streptomycin (Hyclone, #SV30010), and 10% fetal bovine serum (Gibco, #10082147) were added to the media. Untreated media was combined with 0.78 μg of VDR-CMV plasmid, 7.9 μg of a CYP24A1-luciferase reporter gene, LipofectamineTM LTX (37.5 μL, Life Technologies #15338020), and PLUSTM reagent (12.5 μL) and incubated for 30 min. The solution was dispensed into wells of a 6-well plate (3 mL for each well). After 16 h at 37°C with 5% CO2, the cells were harvested and resuspended at 400,000 cell/mL. 40 μL of the suspension was added to each well of a sterile optical bottom 384-well plate, which was pre-treated with a 0.25% Matrigel for 5 min. The multi-well plate was centrifuged for 2 min at 600 xg, incubated for 4 h, and treated with 200 nL of different concentrations of CTA or CTA-ME in the presence or absence of 100 nM 1,25(OH)2D3 in DMSO using an EVO liquid handling system with a 100 nL pin tool (V&P Scientific). After 18 h at 37°C, 20 μL of Bright-Glo™ (Promega, Madison, WI) was added and luminescence measured with a Tecan Infinite M1000 reader. The assay was carried out with an n = 8 and analyzed by non-linear regression. The data are available in the supporting information.
2.4. Viability assay.
HEK293 cells were treated following the protocol for the transfection assay. After 18 h incubation at 37°C with 5% CO2, 20 μl of CellTiter-Glo™ (Promega, Madison, WI) was added and luminescence measured with a Tecan Infinite M1000 reader. The assay was carried out with an n = 8 and analyzed by non-linear regression. The data are available in the supporting information.
2.5. Transcription assay for LXR α and β, RARα.[29]
HEK293T cells were cultured as stated above. Transient transfection was carried out using Lipofectamine LTX reagent (Invitrogen) using the following plasmids: pFR-Luc plasmid (Stratagene), pRL-SV40 (Promega), and pFA-CMV-NR-LBD clones coding for Gal4 fusions of the respective human nuclear receptor ligand binding domains. Cells were then incubated with OptiMEM (Thermofisher) supplemented with penicillin (100 U/mL), streptomycin (100 μg/mL) and containing 1 μM CTA and 0.1% DMSO. After incubation for 16 h, Dual-Glo Luciferase Assay System (Promega) was used according to the manufacturer’s protocol to determine reporter activity. DMSO (0.1%) was used as negative control, reference agonists (1 μM TO901317 or 1 μM tretinoin) served as positive controls. All samples were set up in duplicates and the assay was performed in four biologically independent repeats.
2.6. Transcription assay for FXR.[30]
pcDNA3-hFXR, pSG5-hRXR, pGL3basic (Promega Corporation, Fitchburg, WI, USA) with a shortened construct of the promotor of the bile salt export protein (BSEP) cloned into the SacI/NheI cleavage site in front of the luciferase gene and pRL-SV40 (Promega) were used for the FXR assay. HeLa cells (German Collection of Microorganisms and Cell Culture GmbH, DSMZ) were grown in Dulbecco’s modified Eagle’s medium (DMEM) high glucose supplemented with 10% fetal calf serum (FCS), sodium pyruvate (1 mM), penicillin (100 U/mL) and streptomycin (100 μg/mL) at 37°C and 5% CO2. 24 h before transfection, HeLa cells were seeded in 96-well plates with a density of 8000 cells per well. 3.5 h before transfection, medium was changed to DMEM high glucose, supplemented with sodium pyruvate (1 mM), penicillin (100 U/mL), streptomycin (100 μg/mL) and 0.5 % charcoal-stripped FCS. Transient transfection of HeLa cells with BSEP-pGL3, pRL-SV40 and the expression plasmids pcDNA3-hFXR and pSG5-hRXR was carried out using calcium phosphate transfection method. 16 h after transfection, medium was changed to DMEM high glucose, supplemented with sodium pyruvate (1 mM), penicillin (100 U/mL), streptomycin (100 μg/mL) and 0.5% charcoal-stripped FCS. 24 h after transfection, medium was changed to DMEM without phenol red, supplemented with sodium pyruvate (1 mM), penicillin (100 U/mL), streptomycin (100 μg/mL), L-glutamine (2 mM) and 0.5% charcoal-stripped FCS, now additionally containing 0.1 % DMSO and the respective test compound or 0.1 % DMSO alone as untreated control. Following 24 h incubation with the test compounds, cells were assayed for luciferase activity using the Dual-Glo™ Luciferase Assay System (Promega) according to the manufacturer’s protocol. DMSO (0.1%) was used as negative control, reference agonist GW4064 (1 μM) served as positive control. All samples were set up in triplicates and the assay was performed in four biologically independent repeats.
2.7. Competition assay for PXR.[31]
Assay buffer containing 50 mM Tris, 50 mM KCl20 mM MgCl2, 1 mM CHAPS, 0.1 mg/mL BSA, 0.05 mM DTT, at pH 7.5 was used with 5 nM GST-hPXR-LBD, 5 nM Tb-anti-GST and 100 nM BODIPY FL vindoline at room temperature in 384-well black polypropylene plates. Calcitroic acid and PXR binding positive control TO901317 was added in DMSO with the final DMSO concentration at 0.4%. TR-FRET signals (10000 × 520 nm/490 nm) were recorded with an excitation at 340 nm after 30 minutes. The assay was carried out with an n = 8.
2.8. Coactivator binding assay for CAR.[32]
Assay buffer containing 50 mM Tris, 60 mM CaCl2, 0.1 mg/mL BSA, 5 mM DTT, at pH 7.5 was used with 5 nM GST-CAR-LBD, 5 nM Tb-anti-GST, and 100 nM fluorescein labeled PGC1α coactivator at room temperature in 384-well black polypropylene plates. Calcitroic acid and CAR binding positive control CITCO were added in DMSO with the final DMSO concentration at 0.4%. TR-FRET signals (10000 × 520 nm/495 nm) were recorded with excitation at 340 nm after 90 minutes. The assay was carried out with an n = 8.
2.9. Fluorescence Polarization Binding Assay for TRs, ERs, AR, RXRα and PPARγ [33].
Assay was conducted as described for VDR using the following changes: AR: AR-LBD (5 μM), Texas Red-labeled SRC2–3 (7 nM), and dihydrotestosterone (100 nM); ERα: ERα-LBD (1 μM), Texas Red-labeled SRC2–2 (7 nM), and estradiol (100 nM); ERβ: ERβ-LBD (1 μM), Texas Red-labeled SRC2–2 (7 nM), and estradiol (100 nM); TRα: TRα-LBD (1 μM), Texas Red-labeled SRC2–2 (7 nM), and triiodothyronine (100 nM); TRβ: TRβ-LBD (0.8 μM), Texas Red-labeled SRC2–2 (7 nM), and triiodothyronine (1 μM); PPARγ: PPARγ-LBD (5 μM), Texas Red-labeled DRIP2 (7 nM), and rosiglitazone (5 μM); RXRα: RXRα-LBD (1 μM), Texas Red-labeled SRC 2–3 (7 nM), and bexarotene (2 μM). After 3 h incubation at room temperature, analysis was conducted with a M1000 reader (Tecan) to detect fluorescence polarization at an Ex/Em wavelength of 595/615 nm. Two independent experiments were carried out in quadruplet.
2.10. Quantitative Real Time Polymerase Chain Reaction.
The human colon cancer cell line Caco2 (ATCC HTB-37) was cultured according to vendor’s protocol. The cells were plated in 3 mL using a 6-well plate (Corning 3516) at a density of 1 million cells per mL. Cells were incubated overnight at 37°C and dosed with compounds in DMSO at the following final concentrations: calcitriol (20 nM), calcitroic acid (CTA) (10 μM), and lithocholic acid (LCA) (10 μM). Cells were incubated for 18 h at 37°C followed by removal of the media and the addition of 300 μL of 0.25% trypsin. 700 μL of normal media was added to the trypsin cell suspension. The suspended cells were then transferred to Eppendorf 1.5 mL tubes and centrifuged at 300 xg for 3 min to form a cell pellet. The media was removed, resuspended in 350 μL of 1% β-mercaptoethanol in RLT buffer (Qiagen RNeasy Mini Kit 74104), and transferred directly onto QIAshredder spin columns (Qiagen 79654) and centrifuged for 2 min at 11,000 xg. To the homogenized lysate, 350 μL of 70% ethanol was added followed by transfer to the RNeasy spin column and centrifugation at 11,000 xg for 15 sec. The flowthroughs were discarded and the column washed with 0.7 mL Buffer RW1, 0.5 mL Buffer RPE x2, and the RNA eluted with 50 μL of RNA Storage Solution (Ambion, AM7001). The concentration and purity were determined by absorbance at 260 and 280 nm, respectively. Aliquots of RNA were used for the RT-PCR reaction with qScript One-Step SYBR Green (Quantabio #95087) using the following primers: GAPDH Forward primer 5’-GTC TCC TCT GAC TTC AAC AGC G-3’, Reverse primer 5’-ACC ACC CTG TTG CTG TAG CCA A-3’, product length = 131bp, CYP24A1 Forward primer 5’-CTT TGC TTC CTT TTC CCA GAA T-3’, Reverse primer 5’- CGC CGT AGA TGT CAC CAG TC-3’, product length = 207 bp. Reactions were carried out in 96-well PCR plates (Eppendorf 951022003) using a thermal cycling machine (Eppendorf AG22331 Hamburg Mastercycler) using the following time program: 10 min at 50 °C (reverse transcription, 5 min at 95 °C (activation step), and then cycling through 10 seconds at 95 °C and 30 seconds at 60 °C (annealing step) for 40 cycles.
2.11. Immunomodulation assays with RAW264.7 cells.
The murine macrophage cancer cell line RAW264.7 was cultured in DMEM (ATCC 30–2002) with 10% FBS. Cells were suspended at a cell density of one million cells per mL and a non-activated aliquot of 3 mL was dispensed into a 6-well plate (Corning 3516). The remaining cells were activated with interferon gamma (IFNγ, 150 units/mL) and lipopolysaccharide (LPS, 50 ng/mL) and dispensed into the remaining wells (3 mL/well). The following compounds were added at the final concentrations: calcitriol (20 nM), CTA (5 and 20 μM), dexamethasone (positive control, 1 μM), and DMSO (negative control). The cells were incubated for 18 h. 1) Griess Assay: Eight times 80 μL supernatant from each well were transferred to a 384-well clear-bottomed plate (CLS3700). 10 μL of sulfanilamide solution (Promega Griess Reagent System, TB229) was added and the plate shaken briefly and incubated in the dark for 10 min. 10 μL of NED solution (Promega Griess Reagent System, TB229) was added following by agitation and incubation for 10 min in the dark. Absorbance was read with a Tecan Infinite M1000 plate reader at 530 nm. The cytotoxicity of CTA on this cell line was conducted following the procedure for the “Viability assay”. RT-PCR experiments were carried out following the procedure for the “Quantitative Real Time Polymerase Chain Reaction Protocol”. The following primers were used: iNOS forward primer 5’- CAG CTG GGC TGT ACA AAC CTT −3’, reverse primer 5’- CAT TGG AAG TGA AGC GTT TCG −3’, product length = 95 bp; IL-1β forward primer 5’- AAG GGC TGC TTC CAA ACC TTT GAC −3’, reverse primer 5’-TGC CTG AAG CTC TTG TTG ATG TGC −3’, product length = 92 bp. IL1β production was detected with a HTRF IL1β cytokine assay (CisBio) in low volume 96 well plates (66PL96100) combining 16 μL of supernatant and 4 μL of HTRF detection reagents. After 2 h incubation, time resolved fluorescence was measured at 320 nm (Ex) and 620 (EM) lag time 150 μs and 320 nm (Ex) and 665 (EM) lag time 150 μs using a Tecan Infinite M1000 plate reader. Delta F% = ((mean ratiosample) – (mean rationegative))/(mean rationegative)*100. The ratio = rd650nm/rd620nm*10000 [34].
3. Results and Discussion
3.1. Improved synthesis of calcitroic acid (CTA) and its methyl ester (CTA-ME)
Calcitroic acid was synthesized using the procedure reported by Meyer et al. [35], with improvement reported recently by our group for the synthesis of calcioic acid [36]. Changes for the synthesis of calcitroic acid are outline in Scheme 2.
Scheme 2.
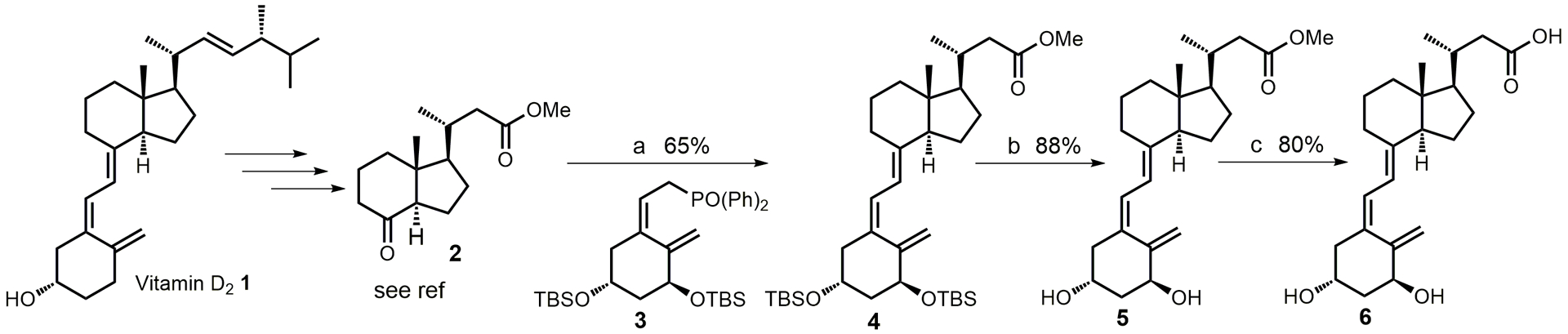
Synthesis of calcitroic acid. a) 1. n-BuLi (1.6 M in hexanes), 3, THF, −78°C, 1h; 2. 2, THF, −78°C to rt, 12h; b) TBAF (1 M in THF), THF, rt, 12h; l) NaOH (10% in water), methanol, rt, 1h.
Compound 2 was synthesized in 21% overall yield,[37] followed by Horner-Wadsworth-Emmons reaction that generated 4 in 65% yield. Deprotection in the presence of tetrabutylammonium fluoride yielded 5 (calcitroic acid methyl ester CTA-ME) in 88% yield. Similar yields were obtained using camphorsulfonic acid in methanol after 30 min [38]. The hydrolysis in the presence of sodium hydroxide instead of potassium hydroxide at room temperature generated CTA in a higher yield (80%).
3.2. VDR-mediated transcription induced by CTA and CTA-ME
Receptor binding of CTA and CTA-ME were investigated with HEK293 cells transfected with a plasmid to overexpress VDR and a luciferase reporter plasmid under control of a CYP24A1 promoter [33]. In the presence of CTA, we observed a moderate activation of gene expression that increased to a level of 30% in comparison to 100 nM 1,25(OH)2D3 (Figure 1, A).
Figure 1.
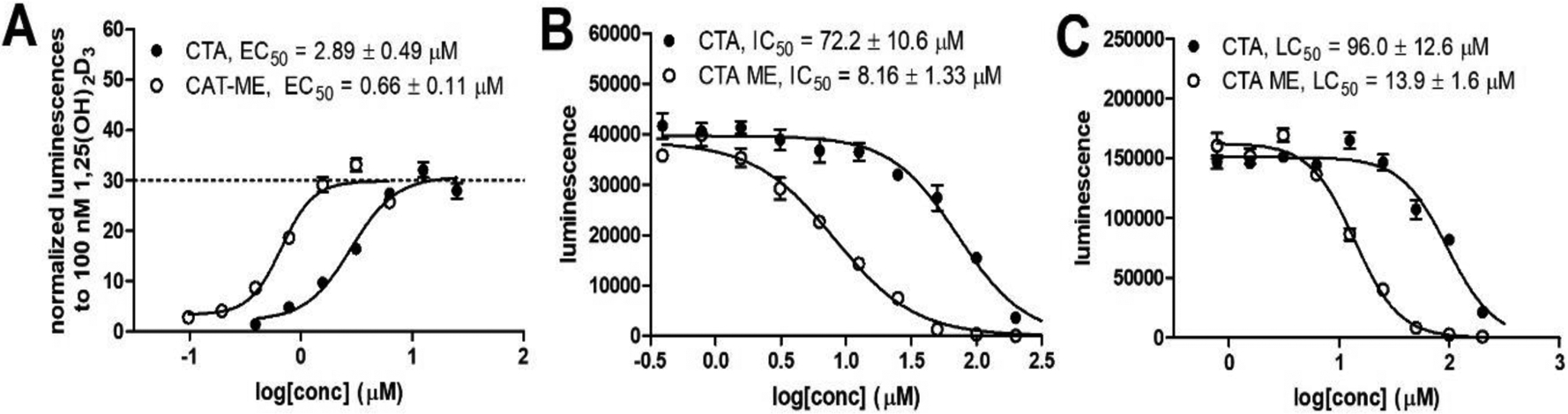
Transcriptional activity and toxicity of CTA and CTA-ME. A) Transcription assay with HEK293 cells that were transfected with a CMV-VDR plasmid and a luciferase reporter plasmid under control of a CYP24A1 promoter. Bright-Glo (Promega) was used for luminescence quantification after 18 h of treatment; B) Transcription assay was carried out in the presence of 100 nM 1,25(OH)2D3; C) Viability of HEK293 cell determined with Cell-Titer Glo (Promega) after 18-hour exposure to CTA and CTA-ME. Each concentration is depicted as the average of eight measurements with SEM followed by non-linear regression analysis.
The EC50 was 2.89 μM. The corresponding methyl ester (CTA-ME) exhibited a similar efficacy but a higher affinity with an EC50 of 660 nM. The same experiment was carried out in the presence of 100 nM 1,25(OH)2D3 to evaluate the antagonistic activity of CTA and CTA-ME. Both compounds reduced the luminescence signal with an IC50 of 72.2 μM and 8.16 μM, respectively. To verify if this reduction is due to toxicity, treated HEK293 cells were evaluated with Cell-titer Glo to measure intracellular APT. The determined LD50 values for CTA and CTA-ME of 96.0 μM and 13.9 μM were similar to IC50 values, confirming that the decrease of luminescence for CTA and CTA-ME in the presence of 1,25(OH)2D3 is likely due to toxicity and not antagonism. The cytotoxicity of both compounds also influenced the transcriptional activity in the absence of 1,25(OH)2D3, with increases to 30% followed by decreases. Thus, full VDR activation by CTA and CTA-ME potentially cannot be observed in this experiment due to toxicity.
3.3. CTA-VDR LBD crystal structure
The identification of the CTA binding site was accomplished by X-ray diffraction using crystals obtained with recombinantly expressed and purified Danio Rerio VDR (zVDR) ligand binding domain (LBD) (Figure 2). Similar to 1,25(OH)2D3, the hydroxy groups of CTA interacted with Tyr175, Ser265, Arg302 and Ser306 located in loop 1–2 and helices 3 & 5, respectively (Figure 2B).
Figure 2.
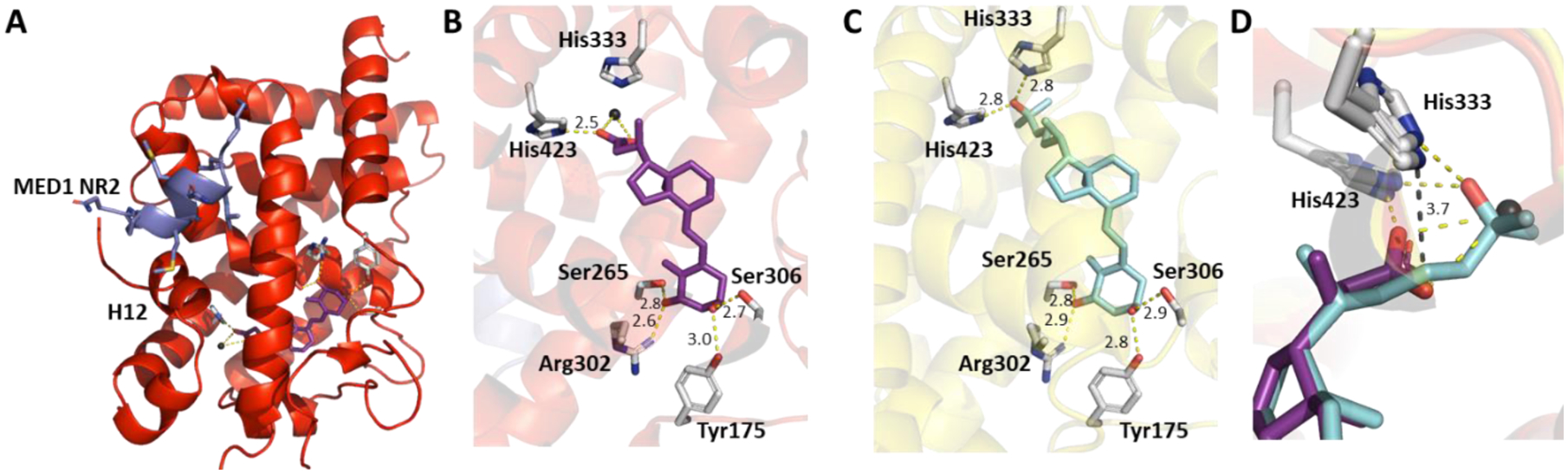
X-ray crystal structures of zVDR LBD and CTA. A) zVDR LBD, CTA and MED1-NR2 peptide (PDB 7OXU), B) zVDR LBD and CTA, C) zVDR LBD and 1,25(OH)2D3 (PDB 2HC4), D) D) Superposition of CTA and 1,25(OH)2D3. Black sphere corresponds to a water molecule Indicated distances are in Å. Hydrogen bonds formed by CTA and 1,25(OH)2D3 are shown as yellow dashed lines.
The distances of those four hydrogen bond interactions are almost identical to those found for 1,25(OH)2D3 (Figure 2C). From the crystal structure is appears that CTA interacts solely with His423 located on helix 11 due its location closer to the bicyclic ring system (Figure 2B) as well as to a water molecule. In contrast, 1,25(OH)2D3 interacts with His423 and His333, which is located in the 6–7 loop (Figure 2B). The dihedral angle of carbon 20 is similar for CTA and 1,25(OH)2D3 resulting in the very similar alignment of their methyl group (Figure 2D). The distance between the carboxylate and His333 is 3.7 Å, thus being too distant to form an additional hydrogen bond. However, the higher temperature factors of CTA carboxylate group suggest some flexibility. Nevertheless, only one hydrogen bond could be formed (the sole interaction with His423) that promotes a conformational change and realignment of helix 12 to enable coactivator binding. In the crystallization studies, a peptide with the sequence of the second nuclear receptor interaction domain (LxxLL motif) of MED1 (also termed DRIP205) adopts a helical conformation and interacts with CTA liganded VDR LBD (Figure 2A). Such recruitment of coactivator proteins is crucial for transcriptional activity of VDR [39], especially the second motif of MED1 or the second and third LxxLL motives of the SRC family having strong affinities for the liganded VDR [40, 41].
3.4. Selective CTA-VDR binding among other nuclear receptors.
Selective binding toward VDR LBD was evaluated with a panel of closely related nuclear receptors including VDR like receptors such as the pregnane X receptor (PXR) and the constitutive androstane receptor (CAR). At 10 or 50 μM, CTA did not bind to PXR or induce CAR-coactivator interactions (Figure 3). Other subfamily related receptors such as the thyroid receptors, retinoic acid receptor, peroxisome proliferator-activated receptor, liver X receptors, and farnesoid X receptor were also not activated by CTA. Lastly, other subfamily nuclear receptors such as the estrogen receptors, androgen receptor, and retinoid X receptor were likewise not activated by CTA. Thus, CTA binding to VDR LBD is selective among other nuclear receptors.
Figure 3.
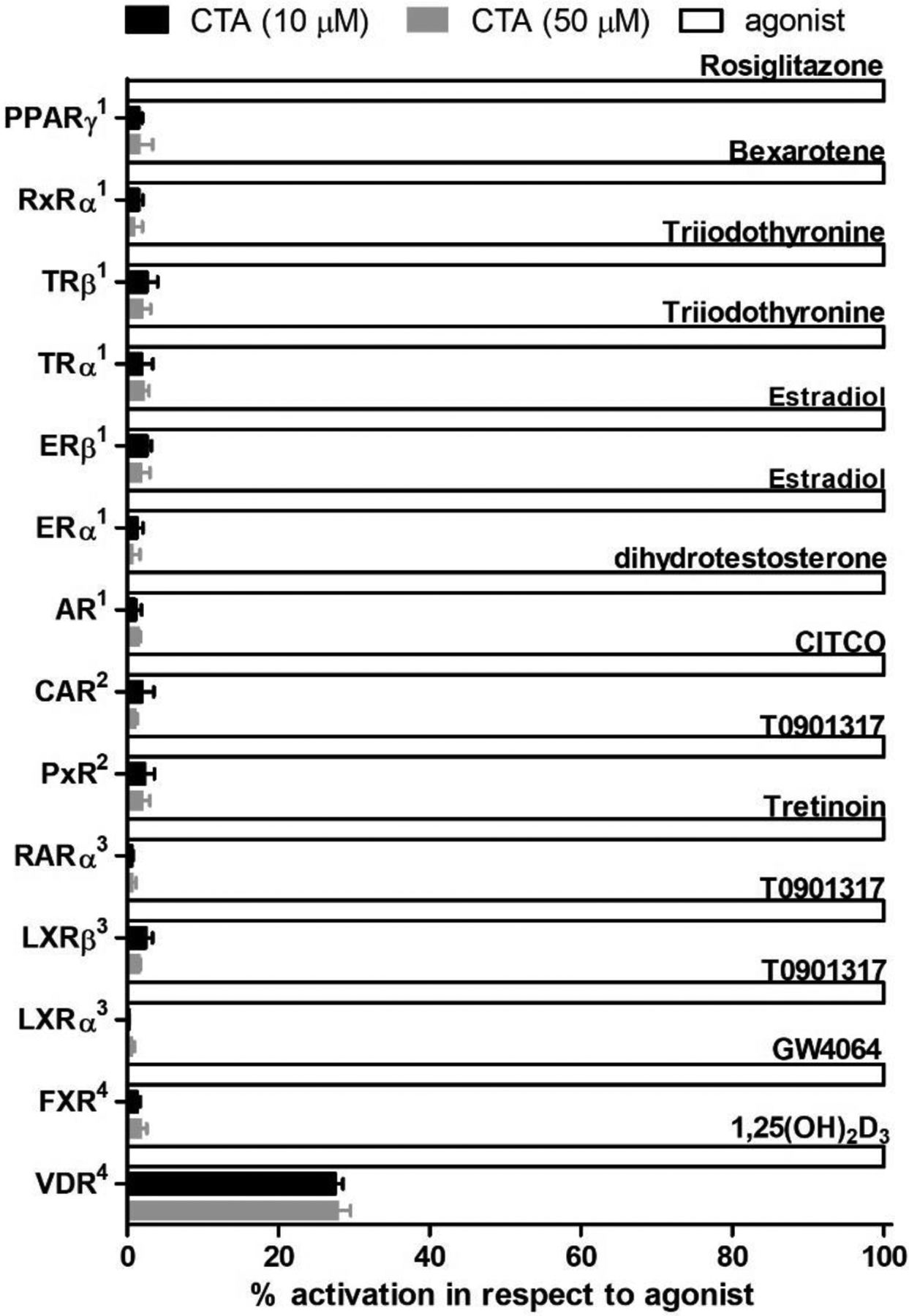
Nuclear receptor screen with CTA. 1Fluoresence polarization coactivator binding assay; 2TR-FRET binding assay; 3Cell-based 2-hybrid assay; 4Cell-based transcription assay.
3.5. Gene regulation by CTA
Transcriptional activity of CTA was investigated with Caco-2 cells. Here, cells were treated for 18 h with different VDR ligands, followed by mRNA isolation and detection by RT-PCR. Primer specificity was visualized by agarose gel electrophoresis. The results are summarized in Figure 4.
Figure 4.
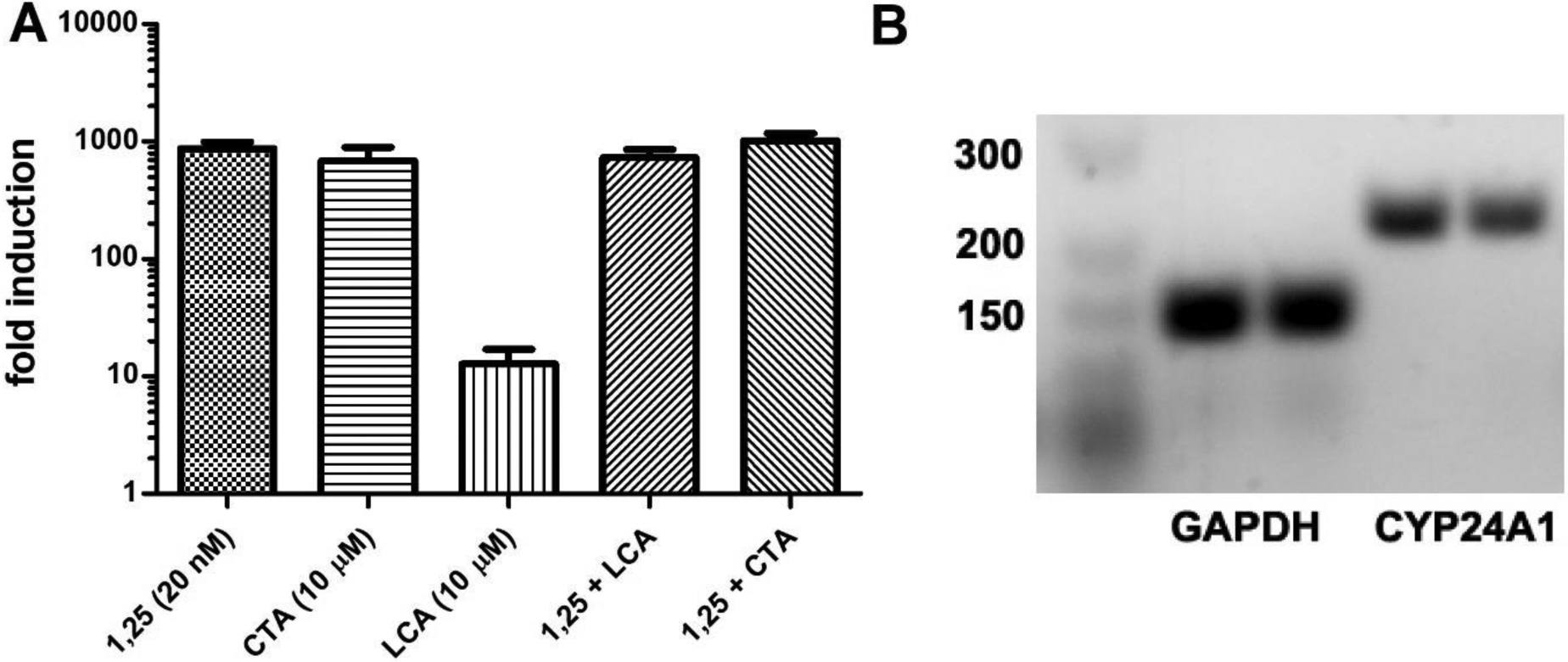
Transcriptional regulation of CYP24A1 in Caco2 cells induced a by VDR ligand treatment of 18 h. A) RT-PCR of isolated RNA for cells treated with 1,25(OH)2D3, CTA or lithocholic acid (LCA). CYP24A1 fold induction was calculated by 2−ΔΔCT using GAPDH as housekeeping gene and DMSO as vehicle. Data are depicted as the mean with SD from two independent experiments amplified in quadruplets. B) Agarose (2%) gel electrophoresis of PCR products after 50 cycles for CTA treated cells.
As reported, 1,25(OH)2D3 strongly induced CYP24A1 at 20 nM [42]. Similar induction of CYP24A1 was observed for CTA at 10 μM, confirming CTA as full VDR agonist. Endogenous VDR ligand lithocholic acid showed significantly lower induction of CYP24A1 [43]. In the presence of 1,25(OH)2D3, both CTA and LCA strongly induced the expression of CYP24A1, further excluding any antagonistic effects.
3.6. Anti-inflammatory properties of CTA
Macrophages are important immune cells that regulate inflammation by secreting pro- or anti-inflammatory cytokines. 1,25(OH)2D3 has been shown to reduce the secretion of proinflammatory cytokines such as tumor necrosis factor α (TNFα), interleukins 1β and 12 (IL-1β and IL-12), and inducible NO synthase (iNOS) in interferon γ (INFγ) and lipopolysaccharide (LPS) stimulated RAW264.7 mouse macrophages[44, 45]. To investigate possible anti-inflammatory effects of CTA, we conducted a similar study and quantified the levels of nitric oxide (NO), RNA expression of iNOS, and IL-1β and IL-1β protein. The results are summarized in Figure 5.
Figure 5.
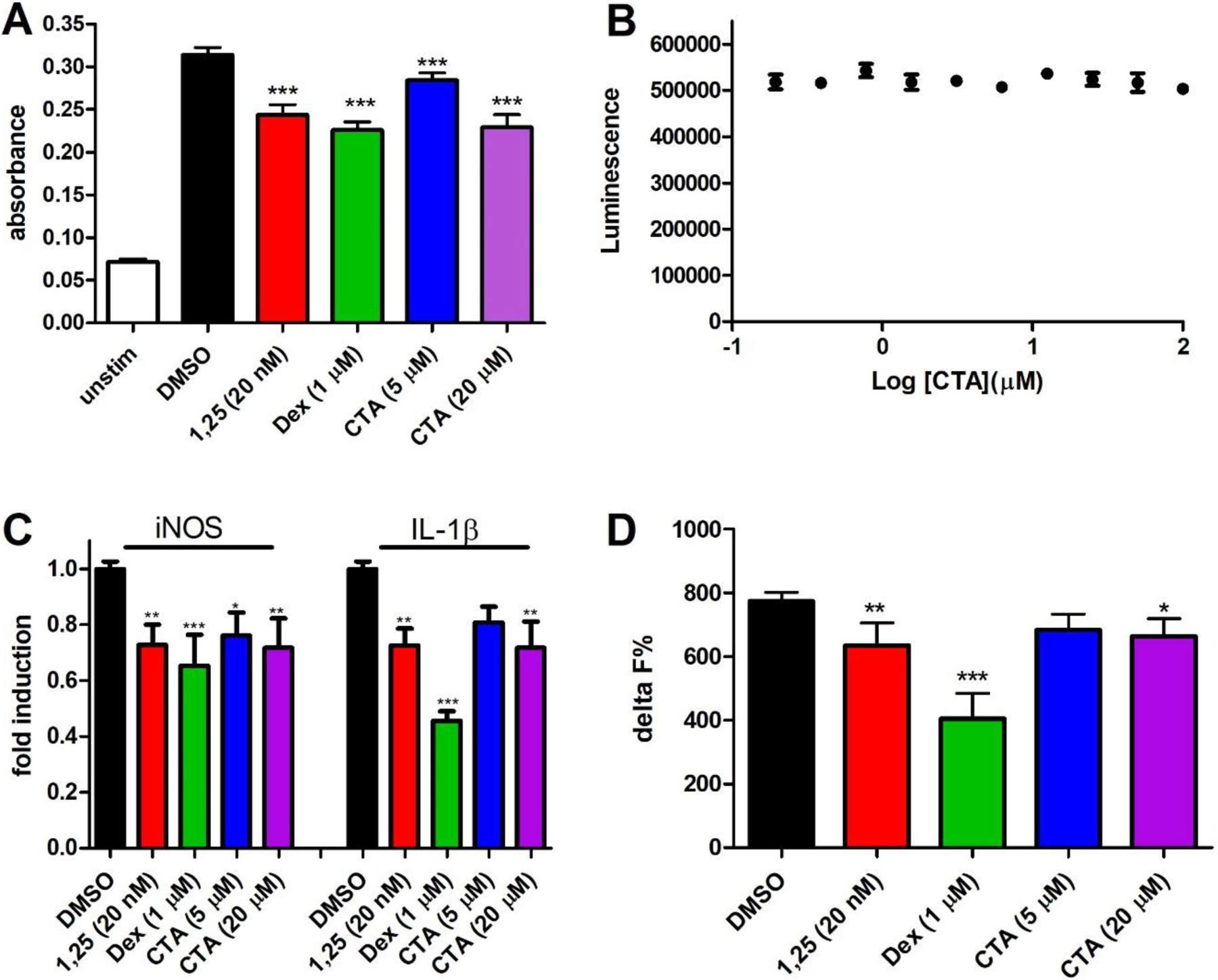
Reduction of inflammatory markers in INFγ/LPS-activated mouse macrophages (RAW264.7) by 1,25(OH)2D3 (1,25), dexamethasone (Dex), or calcitroic acid (CTA) after 24 h. A) Quantification of NO in media was conducted with a Griess Assay. The mean and SD reflect two independent assays with an n = 8. B) Cytotoxicity on RAW264.7 cells at different CTA concentrations were determined with CellTiter-Glo (luminescence). The data are given as mean and SD based on two independent assays with n = 4. C) RT-PCR was conducted with isolated RNA using a qScript One-Step SYBR Green assay. Fold induction was calculated by 2−ΔΔCT using GAPDH as housekeeping gene and DMSO as vehicle. Data are depicted as the mean with SD from two independent experiments amplified in quadruplets. D) IL-1β was quantified in media with a homogeneous time resolved fluorescence (HTRF) IL1β cytokine assay. Delta F% was calculated based on time resolved fluorescence and depicted as mean and SD of two independent assays with n = 8. Significance was determined by one way ANOVA using a Dunnett’s multiple comparison test with *, ** and ** indicating p < 0.05, p < 0.01 and p < 0.001 significance.
LPS is produced by Gram-negative bacteria and is recognized by macrophage Toll-like receptor 4 (TLR4) [46], which results in the activation of MAP kinase cascade and translocation of regulator nuclear factor-κB (NF-κB) into the nucleus to induce transcription of proinflammatory cytokines [47]. INFy signals through the Janus kinase (Jak)-signaling pathway and activates transcription via STAT1 to produce inflammatory factors such as iNOS [48]. 1,25(OH)2D3 has been shown to regulate the expression of many enzymes and receptors involved in macrophage-mediated inflammation through binding to nuclear transcription factor VDR. Some examples are the interaction with IKKβ to block NF-κB activation [49], attenuation of TLR signaling by enhancing negative feedback inhibition [50], and inhibition of COX-2 expression [51]. 24-hour treatment using 1,25(OH)2D3 (20 nM) in INFγ/LPS activated RAW264.7 macrophages reduced the production of NO in comparison to vehicle (Figure 5A). A significant reduction was also observed for CTA at 5 and 20 μM. Dexamethasone was used a positive control at 1 μM, which reduces inflammation by activating the glucocorticoid receptor signaling pathway. A luminescence viability assay demonstrated the absence of any cytotoxic effects of CTA at concentrations up to 100 μM in this cell line (Figure 5B). This demonstrated that CTA is less toxic towards RAW264.7 than HEK293 cells. Transcriptional regulation by 1,25(OH)2D3 and CTA were demonstrated with RT-PCR for iNOS and IL-1β (Figure 5C). The reduction of gene transcription was observed at 5 and 20 μM CTA. The reduction was comparable to 20 nM 1,25(OH)2D3. To demonstrate that reduction of transcription relates to reduced protein production for IL-1β, a homogeneous time resolved fluorescence assay was carried out (Figure 5D). Reduced IL-1β protein was observed for 1,25(OH)2D3 (20 nM) and CTA (20 μM) confirming the anti-inflammatory properties of CTA.
4. Conclusion
We confirmed that CTA is a VDR agonist and mediates biological effects similar to 1,25(OH)2D3, albeit at a 1,000-fold higher concentration. Efforts to detect CTA in blood of mice feed with a Tekland Rodent Diet (2.4 IU/g vitamin D3) were not successful using a tandem mass spectrometry method with a lower limit of detection of 6 nM [52]. Thus, it can be argued that typical CTA blood levels might not have any physiological effect. However, subcutaneous administration of CTA and CTA-ME have shown antirachitic activity [53], and we have demonstrated here the anti-inflammatory activity of CTA. Further studies will show if CTA might be therapeutically beneficial for inflammatory diseases of the intestines such as inflammatory bowel disease (IBD), for which 1,25(OH)2D3 has been shown to reduce the severity in vitamin D deficient IL-10 KO mice [54].
Supplementary Material
Acknowledgments
This work was supported by the University of Wisconsin Milwaukee, the Milwaukee Institute for Drug Discovery, the Shimadzu Laboratory of Applied and Analytical Chemistry, the UWM Research Growth Initiative, National Institutes of Health [R03DA031090], the UWM Research Foundation, the Lynde and Harry Bradley Foundation, and the Richard and Ethel Herzfeld Foundation. In addition, this work was supported by grant CHE-1625735 from the National Science Foundation, Division of Chemistry.
Abbreviation:
- CTA
calcitroic acid
- CTA-ME
calcitroic acid methyl ester
- 25OHD3
25-hydroxyvitamin D3
- CYP24A1
1α,25(OH)2D3-24-hydroxylase
- 1,25(OH)2D3
1α,25-dihydroxyvitamin D3
- VDR
vitamin D receptor
- n-BuLi
n-butyl lithium
- THF
tetrahydrofuran
- TBAF
tetrabutylammonium fluoride
- NaOH
sodium hydroxide
- SRC-2
steroid receptor coactivator 2
- TNFα
tumor necrosis factor α
- IL-1β and IL-12
interleukins 1β and 12
- NO
nitric oxide
- iNOS
inducible NO synthase
- LCA
lithocholic acid
- INFγ
interferon γ
- LPS
lipopolysaccharide
- HTRF
homogeneous time resolved fluorescence
- TLR4
Toll-like receptor 4
- TBME
tert-butylmethyl ether
- TLC
thin-layer chromatography
- Hex
hexanes
- EtOAc
ethyl acetate
- NMR
nuclear magnetic resonance
- CHCl3
chloroform
- NP-40
nonyl phenoxypolyethoxylethanol
- LBD
ligand binding domain
- ligand
binding domain
- DMSO
dimethyl sulfoxide
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- CMV
cytomegalovirus
References
- [1].Esvelt RP, Schnoes HK, DeLuca HF, Isolation and characterization of 1 alpha-hydroxy-23-carboxytetranorvitamin D: a major metabolite of 1,25-dihydroxyvitamin D3, Biochemistry 18(18) (1979) 3977–83. 10.1021/bi00585a021. [DOI] [PubMed] [Google Scholar]
- [2].Reddy GS, Omdahl JL, Robinson M, Wang G, Palmore GT, Vicchio D, Yergey AL, Tserng KY, Uskokovic MR, 23-carboxy-24,25,26,27-tetranorvitamin D3 (calcioic acid) and 24-carboxy-25,26,27-trinorvitamin D3 (cholacalcioic acid): end products of 25-hydroxyvitamin D3 metabolism in rat kidney through C-24 oxidation pathway, Arch Biochem Biophys 455(1) (2006) 18–30. 10.1016/j.abb.2006.08.021. [DOI] [PubMed] [Google Scholar]
- [3].Reddy GS, Tserng KY, Calcitroic acid, end product of renal metabolism of 1,25-dihydroxyvitamin D3 through C-24 oxidation pathway, Biochemistry 28(4) (1989) 1763–9. 10.1021/bi00430a051. [DOI] [PubMed] [Google Scholar]
- [4].Makin G, Lohnes D, Byford V, Ray R, Jones G, Target cell metabolism of 1,25-dihydroxyvitamin D3 to calcitroic acid. Evidence for a pathway in kidney and bone involving 24-oxidation, The Biochemical journal 262(1) (1989) 173–80. 10.1042/bj2620173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brumbaugh PF, Haussler MR, 1 Alpha,25-dihydroxycholecalciferol receptors in intestine. I. Association of 1 alpha,25-dihydroxycholecalciferol with intestinal mucosa chromatin, J Biol Chem 249(4) (1974) 1251–7. https://doi.org/. [PubMed] [Google Scholar]
- [6].Pike JW, Meyer MB, Lee SM, Onal M, Benkusky NA, Genone-Wide Perspectives on Vitamin D Receptor-Mediated Control of Gene Expression in Target Cells, in: Feldman D (Ed.), Vitamin D, Elsevier, London, 2018, pp. 142–167. [Google Scholar]
- [7].Wang YJ, Zhu JG, DeLuca HF, Where is the vitamin D receptor?, Archives of Biochemistry and Biophysics 523(1) (2012) 123–133. 10.1016/j.abb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- [8].Veldman CM, Cantorna MT, DeLuca HF, Expression of 1,25-dihydroxyvitamin D-3 receptor in the immune system, Archives of Biochemistry and Biophysics 374(2) (2000) 334–338. 10.1006/abbi.1999.1605. [DOI] [PubMed] [Google Scholar]
- [9].Wang Y, Zhu J, DeLuca HF, Identification of the vitamin D receptor in osteoblasts and chondrocytes but not osteoclasts in mouse bone, Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 29(3) (2014) 685–92. 10.1002/jbmr.2081. [DOI] [PubMed] [Google Scholar]
- [10].Brumbaugh PF, Haussler MR, 1Alpha,25-dihydroxyvitamin D3 receptor: competitive binding of vitamin D analogs, Life Sci 13(12) (1973) 1737–46. 10.1016/0024-3205(73)90120-3. [DOI] [PubMed] [Google Scholar]
- [11].Perez A, Raab R, Chen TC, Turner A, Holick MF, Safety and efficacy of oral calcitriol (1,25-dihydroxyvitamin D3) for the treatment of psoriasis, The British journal of dermatology 134(6) (1996) 1070–8. 10.1046/j.1365-2133.1996.d01-904.x. [DOI] [PubMed] [Google Scholar]
- [12].Kallay E, Buburuzan L, Vitamin D Inflammation and Cancer, in: Feldman D (Ed.), Vitamin D, Elsevier, London, 2018, pp. 892–904. [Google Scholar]
- [13].VanHerwegen A-S, Gysemans C, Mathieu C, Vitamin D and Diabetes, in: Feldman D (Ed.), Vitamin D, Elsevier, London, 2018, pp. 969–981. [Google Scholar]
- [14].H. CE, Nashold FE, Vitamin D and Multiple Sclerosis, in: Feldman D (Ed.), Vitamin D, Elsevier, London, 2019, pp. 990–1010. [Google Scholar]
- [15].Cantorna MT, Bruce D, Vitamin D and Inflammatory Bowel Disease, in: Feldman D (Ed.), Vitamin D, Elsevier, London, 2018, pp. 1025–1033. [Google Scholar]
- [16].White JH, Vitamin D as an inducer of cathelicidin antimicrobial peptide expression: Past, present and future, J Steroid Biochem 121(1–2) (2010) 234–238. 10.1016/j.jsbmb.2010.03.034. [DOI] [PubMed] [Google Scholar]
- [17].Jeffery LE, Burke F, Mura M, Zheng Y, Qureshi OS, Hewison M, Walker LSK, Lammas DA, Raza K, Sansom DM, 1,25-Dihydroxyvitamin D-3 and IL-2 Combine to Inhibit T Cell Production of Inflammatory Cytokines and Promote Development of Regulatory T Cells Expressing CTLA-4 and FoxP3, Journal of immunology 183(9) (2009) 5458–5467. 10.4049/jimmunol.0803217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dickie LJ, Church LD, Coulthard LR, Mathews RJ, Emery P, McDermott MF, Vitamin D-3 down-regulates intracellular Toll-like receptor 9 expression and Toll-like receptor 9-induced IL-6 production in human monocytes, Rheumatology 49(8) (2010) 1466–1471. 10.1093/rheumatology/keq124. [DOI] [PubMed] [Google Scholar]
- [19].Cohen-Lahav M, Shany S, Tobvin D, Chaimovitz C, Douvdevani A, Vitamin D decreases NFkappaB activity by increasing IkappaBalpha levels, Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association 21(4) (2006) 889–97. 10.1093/ndt/gfi254. [DOI] [PubMed] [Google Scholar]
- [20].Takeyama K.-i., Kitanaka S, Sato T, Kobori M, Yanagisawa J, Kato S, 25-hydroxyvitamin D3 1α-Hydroxylase and Vitamin D Synthesis, Science 277 (1997) 1827–1829. 10.1126/science.277.5333.1827. [DOI] [PubMed] [Google Scholar]
- [21].Tanaka Y, Castillo L, DeLuca HF, The 24-hydroxylation of 1,25-dihydroxyvitamin D3, J Biol Chem 252(4) (1977) 1421–4. https://doi.org/. [PubMed] [Google Scholar]
- [22].Bouillon R, Pauwels S, The vitamin D-binding protein, in: Feldman D (Ed.), Vitamin D, Elsevier, London, 2018, pp. 97–115. [Google Scholar]
- [23].Esvelt RP, Fivizzani MA, Paaren HE, Schnoes HK, DeLuca HF, Synthesis of calcitroic acid, a metabolite of 1.alpha.,25-dihydroxycholecalciferol, The Journal of Organic Chemistry 46(2) (1981) 456–458. 10.1021/jo00315a047. [DOI] [Google Scholar]
- [24].Yu OB, Arnold LA, Calcitroic Acid-A Review, ACS Chem Biol 11(10) (2016) 2665–2672. 10.1021/acschembio.6b00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Teske KA, Bogart JW, Sanchez LM, Yu OB, Preston JV, Cook JM, Silvaggi NR, Bikle DD, Arnold LA, Synthesis and evaluation of vitamin D receptor-mediated activities of cholesterol and vitamin D metabolites, Eur J Med Chem 109 (2016) 238–46. 10.1016/j.ejmech.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Belorusova AY, Eberhardt J, Potier N, Stote RH, Dejaegere A, Rochel N, Structural insights into the molecular mechanism of vitamin D receptor activation by lithocholic acid involving a new mode of ligand recognition, J Med Chem 57(11) (2014) 4710–9. 10.1021/jm5002524. [DOI] [PubMed] [Google Scholar]
- [27].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr D Biol Crystallogr 66(Pt 2) (2010) 213–21. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Emsley P, Cowtan K, Coot: model-building tools for molecular graphics, Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1) (2004) 2126–32. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- [29].Pollinger J, Gellrich L, Schierle S, Kilu W, Schmidt J, Kalinowsky L, Ohrndorf J, Kaiser A, Heering J, Proschak E, Merk D, Tuning Nuclear Receptor Selectivity of Wy14,643 towards Selective Retinoid X Receptor Modulation, J Med Chem 62(4) (2019) 2112–2126. 10.1021/acs.jmedchem.8b01848. [DOI] [PubMed] [Google Scholar]
- [30].Schmidt J, Klingler FM, Proschak E, Steinhilber D, Schubert-Zsilavecz M, Merk D, NSAIDs Ibuprofen, Indometacin, and Diclofenac do not interact with Farnesoid X Receptor, Sci Rep 5 (2015) 14782. 10.1038/srep14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lin W, Liu J, Jeffries C, Yang L, Lu Y, Lee RE, Chen T, Development of BODIPY FL vindoline as a novel and high-affinity pregnane X receptor fluorescent probe, Bioconjug Chem 25(9) (2014) 1664–77. 10.1021/bc5002856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lin W, Yang L, Chai SC, Lu Y, Chen T, Development of CINPA1 analogs as novel and potent inverse agonists of constitutive androstane receptor, Eur J Med Chem 108 (2016) 505–528. 10.1016/j.ejmech.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nandhikonda P, Lynt WZ, McCallum MM, Ara T, Baranowski AM, Yuan NY, Pearson D, Bikle DD, Guy RK, Arnold LA, Discovery of the first irreversible small molecule inhibitors of the interaction between the vitamin D receptor and coactivators, J Med Chem 55(10) (2012) 4640–51. 10.1021/jm300460c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Einhorn L, Krapfenbauer K, HTRF: a technology tailored for biomarker determination-novel analytical detection system suitable for detection of specific autoimmune antibodies as biomarkers in nanogram level in different body fluids, Epma J 6 (2015). 10.1186/s13167-015-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Meyer D, Rentsch L, Marti R, Efficient and scalable total synthesis of calcitroic acid and its C-13-labeled derivative, Rsc Adv 4(61) (2014) 32327–32334. 10.1039/c4ra04322g. [DOI] [Google Scholar]
- [36].Yu OB, Mutchie TR, Di Milo ES, Arnold LA, Synthesis and biological evaluation of calcioic acid, Steroids 154 (2020) 108536. 10.1016/j.steroids.2019.108536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yu OB, Part I: an Investigation of Calcitroic Acid and Its Phase II Conjugates. Part II: Toward the Development of a Novel Orally-available Asthma Treatment Targeting Gabaa Receptors in the Lungs, Department of Chemistry and Biochemistry, University of Wisconsin Milwaukee, 2021. [Google Scholar]
- [38].Filip B, Milczarek M, Wietrzyk J, Chodynski M, Kutner A, Antitumor properties of (5E,7E) analogs of vitamin D3, J Steroid Biochem Mol Biol 121(1–2) (2010) 399–402. 10.1016/j.jsbmb.2010.03.017. [DOI] [PubMed] [Google Scholar]
- [39].Pike JW, Meyer MB, Bishop KA, Regulation of target gene expression by the vitamin D receptor - an update on mechanisms, Rev Endocr Metab Disord 13(1) (2012) 45–55. 10.1007/s11154-011-9198-9. [DOI] [PubMed] [Google Scholar]
- [40].Teichert A, Arnold LA, Otieno S, Oda Y, Augustinaite I, Geistlinger TR, Kriwacki RW, Guy RK, Bikle DD, Quantification of the vitamin D receptor-coregulator interaction, Biochemistry 48(7) (2009) 1454–61. 10.1021/bi801874n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Belorusova AY, Bourguet M, Hessmann S, Chalhoub S, Kieffer B, Cianferani S, Rochel N, Molecular determinants of MED1 interaction with the DNA bound VDR-RXR heterodimer, Nucleic Acids Res 48(19) (2020) 11199–11213. 10.1093/nar/gkaa775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hobaus J, Fetahu I, Khorchide M, Manhardt T, Kallay E, Epigenetic regulation of the 1,25-dihydroxyvitamin D3 24-hydroxylase (CYP24A1) in colon cancer cells, J Steroid Biochem Mol Biol 136 (2013) 296–9. 10.1016/j.jsbmb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ, Vitamin D receptor as an intestinal bile acid sensor, Science 296(5571) (2002) 1313–1316. 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- [44].Chang JM, Kuo MC, Kuo HT, Hwang SJ, Tsai JC, Chen HC, Lai YH, 1-alpha,25-Dihydroxyvitamin D3 regulates inducible nitric oxide synthase messenger RNA expression and nitric oxide release in macrophage-like RAW 264.7 cells, J Lab Clin Med 143(1) (2004) 14–22. 10.1016/j.lab.2003.08.002. [DOI] [PubMed] [Google Scholar]
- [45].Wasnik S, Rundle CH, Baylink DJ, Yazdi MS, Carreon EE, Xu Y, Qin XZ, Lau KHW, Tang XL, 1,25-Dihydroxyvitamin D suppresses M1 macrophages and promotes M2 differentiation at bone injury sites, Jci Insight 3(17) (2018). 10.1172/jci.insight.98773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Takeda K, Akira S, Toll-like receptors in innate immunity, International Immunology 17(1) (2005) 1–14. 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- [47].Ci XX, Ren R, Xu K, Li HY, Yu QL, Song Y, Wang DC, Li RT, Deng XM, Schisantherin A Exhibits Anti-inflammatory Properties by Down-Regulating NF-kappa B and MAPK Signaling Pathways in Lipopolysaccharide-Treated RAW 264.7 Cells, Inflammation 33(2) (2010) 126–136. 10.1007/s10753-009-9166-7. [DOI] [PubMed] [Google Scholar]
- [48].Hu X, Chakravarty SD, Ivashkiv LB, Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms, Immunol Rev 226 (2008) 41–56. 10.1111/j.1600-065X.2008.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen Y, Zhang J, Ge X, Du J, Deb DK, Li YC, Vitamin D receptor inhibits nuclear factor kappaB activation by interacting with IkappaB kinase beta protein, J Biol Chem 288(27) (2013) 19450–8. 10.1074/jbc.M113.467670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chen Y, Liu W, Sun T, Huang Y, Wang Y, Deb DK, Yoon D, Kong J, Thadhani R, Li YC, 1,25-Dihydroxyvitamin D promotes negative feedback regulation of TLR signaling via targeting microRNA-155-SOCS1 in macrophages, Journal of immunology 190(7) (2013) 3687–95. 10.4049/jimmunol.1203273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wang Q, He Y, Shen Y, Zhang Q, Chen D, Zuo C, Qin J, Wang H, Wang J, Yu Y, Vitamin D inhibits COX-2 expression and inflammatory response by targeting thioesterase superfamily member 4, J Biol Chem 289(17) (2014) 11681–94. 10.1074/jbc.M113.517581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Di Milo ES, Determination of the Metabolism, Distribution, and Concentration of Calcitroic Acid, Department of Chemistry & Biochemistry, University of Wisconsin Milwaukee, 2021, pp. 1–137. [Google Scholar]
- [53].Esvelt RP, De Luca HF, Calcitroic acid: biological activity and tissue distribution studies, Arch Biochem Biophys 206(2) (1981) 403–13. 10.1016/0003-9861(81)90107-7. [DOI] [PubMed] [Google Scholar]
- [54].Cantorna MT, Munsick C, Bemiss C, Mahon BD, 1,25-Dihydroxycholecalciferol prevents and ameliorates symptoms of experimental murine inflammatory bowel disease, J Nutr 130(11) (2000) 2648–52. 10.1093/jn/130.11.2648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


