Abstract
The POU homeodomain protein UNC-86 and the LIM homeodomain protein MEC-3 are essential for the differentiation of the six mechanoreceptor neurons in the nematode Caenorhabditis elegans. Previous studies have indicated that UNC-86 and MEC-3 bind cooperatively to at least three sites in the mec-3 promoter and synergistically activate transcription. However, the molecular details of the interactions of UNC-86 with MEC-3 and DNA have not been investigated so far. Here we used a yeast system to identify the functional domains in UNC-86 required for transcriptional activation and to characterize the interaction of UNC-86 with MEC-3 in vivo. Our results suggest that transcriptional activation is mediated by the amino terminus of UNC-86, whereas amino acids in the POU domain mediate DNA binding and interaction with MEC-3. By random mutagenesis, we identified mutations that only affect the DNA binding properties of UNC-86, as well as mutations that prevent coactivation by MEC-3. We demonstrated that both the POU-specific domain and the homeodomain of UNC-86, as well as DNA bases adjacent to the proposed UNC-86 binding site, are involved in the formation of a transcriptionally active complex with MEC-3. These data suggest that some residues involved in the contact of UNC-86 with MEC-3 also contribute to the interaction of the functionally nonrelated POU protein Oct-1 with Oca-B, whereas other positions have different roles.
POU domain transcription factors are characterized by their bipartite DNA binding domain, consisting of a helix-turn-helix POU-specific domain (POUS) and an adjacent POU homeodomain (POUHD) (for reviews, see references 15 and 28). Both protein domains contact DNA and are necessary for high-affinity DNA binding (15). POU class IV is comprised of the mammalian Brn-3-encoding genes, the Drosophila I-POU/acj6 gene, and the Caenorhabditis elegans unc-86 gene (28). These are all expressed exclusively in the nervous system, but their expression is not limited to one specific neuronal cell type. In vitro studies have shown that POU class IV proteins bind very similar DNA sequences (12, 24). Therefore, it has been proposed that any differences in target gene activation are due to modulatory protein interactions (28). Most of the data for determination of the promoter specificity of POU proteins are derived from the nonrelated human POU class III protein Oct-1 (13, 41).
POU homeobox gene unc-86 is expressed in 57 neurons in adult C. elegans. These neurons comprise one-fifth of the animal's nervous system and represent 27 different functional classes (10). Consequently, null alleles of unc-86 result in several behavioral defects affecting mechanosensation, egg laying, chemosensation, and thermosensation and cause an uncoordinated phenotype (5, 10, 18, 26, 37). The diversity of neuron types that require UNC-86 raises the possibility that in different cell types, UNC-86 targets the promoters of different genes. This is supported by the fact that in at least two unc-86 mutants, only a subset of the unc-86-mediated behaviors is affected (S. Röhrig and R. Baumeister, unpublished observations). How UNC-86 exerts these selective effects on gene regulation in several distinct neural cell types remains unknown. Interactions with various other proteins are one way to achieve selectivity. For UNC-86, the only binding partner known so far is the LIM homeodomain protein MEC-3 (39).
During the development of the nervous system, UNC-86 functions in 10 neuroblasts to determine their correct cell lineage and subsequently in the development and differentiation of the six mechanoreceptor cells involved in body touch sensing (9). In these cells, UNC-86 binds to at least three regulatory sequences, CS1, CS2, and CS3, in the mec-3 promoter (35, 40, 42). A minimal mec-3 promoter that contains 311 bp including CS1, CS2, and CS3 is sufficient to direct reporter gene expression in the six mechanoreceptor cells. The LIM protein MEC-3 then cooperates with UNC-86 to maintain its own expression in a manner that is dependent on the presence of CS1-3 (38). Both proteins together regulate the expression of additional downstream genes (5, 8) that encode functionally crucial components of the mechanosensory neurons (39).
In order to investigate the DNA binding of UNC-86 and the protein interaction of UNC-86 with MEC-3, we developed an in vivo assay with the yeast Saccharomyces cerevisiae. For this purpose, we coexpressed unc-86 and mec-3 in the presence of a bona fide UNC-86 target site. We show here that upon binding to a mec-3 promoter site, UNC-86 activates transcription in yeast. We localized the activation domain (AD) and showed that upon MEC-3 interaction, transcriptional activation is strongly enhanced. By mutational mapping of the contact surface, we identified amino acid residues in UNC-86 that specifically interfere with the synergistic coactivation by MEC-3. These results are discussed with respect to data about other POU complexes.
MATERIALS AND METHODS
Yeast media and methods.
All S. cerevisiae strains were propagated by standard techniques. Yeast strains were grown at 30°C in liquid or on solid SC medium (synthetic medium) (30). For X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing plates, SC plates were supplemented with X-Gal at 400 μg/ml and buffered with 10× BU salt (0.26 M Na2HPO4, 0.25 M NaH2PO4). Transformants (11) were allowed to grow on SC media lacking the amino acids required for plasmid selection before further analysis. Plasmid isolation from S. cerevisiae was performed as previously described (36). Liquid β-galactosidase assays using chlorophenolred–β-d-galactopyranoside (Roche) as a substrate were performed as previously described (7).
AD mapping.
UNC-86–LexA fusions were generated by inserting the partial or full-length cDNA of unc-86 (3) into plasmid pBTM116 (2). Full-length unc-86 (nucleotides [nt] 1 to 1059) was inserted to generate plasmid pBY436, unc-86 (nt 425 to 1059) was inserted to generate plasmid pBY166 (UNC-86 amino acids [aa] 135 to 342), unc-86 (nt 130 to 1050) was inserted to generate plasmid pBY165 (UNC-86 aa 37 to 342), and unc-86 (nt 1 to 333) was inserted to create plasmid pBY135 (UNC-86 aa 1 to 104). All fusions were analyzed in yeast strain L40 (MATα his3Δ200 trp1-901 leu2-3,112 ade LYS::(lexAop)4-HIS3 URA::(lexAop)8-lacZ GAL4 gal80) (19).
Construction of S. cerevisiae strains RB101 and RB102.
Plasmid pBY180 was generated by inserting three tandem copies of 5′-GCATTCGAAATGCATTGCCCATAATG-3′ into plasmid pLacZi (Clontech), which contains a lacZ reporter gene under the control of the PCYC1 minimal promoter. Chromosomal integration of linearized pBY180 reconstitutes the ura3 locus of yeast strain RH1533 (Mat-α lys2-801 leu2-3,112 suc2-Δ9 ura3-52 MCL his3-Δ200 trp1-Δ901) (14) and was selected for by plating of the transformants on SC medium without uracil. The resulting strain, RH1533 URA::pBY180, was named RB101. Strain RB102 was constructed analogously using 5′-AATTGCATTCGAAATGAGCTGCCCATAATG-3′.
Plasmid constructions.
In order to generate unc-86 yeast expression vector pBY175, unc-86 cDNA was inserted into a modified pGBT9 vector (2) from which the GAL4 DNA binding domain was deleted. A partial unc-86 cDNA (nt 425 to 1059) was inserted into the modified pGBT9 vector described above in order to generate plasmid pBY4:40 encoding UNC-86ΔN (aa 135 to 342). unc-86 alanine substitution mutants were constructed by site-directed PCR mutagenesis (17). mec-3 AD yeast expression vector pBY117 was constructed by inserting the mec-3 cDNA derived from plasmid pTU47 (43) into plasmid pGAD10 (2). Plasmid pBY1107 was constructed by subcloning a partial mec-3 cDNA that lacks the regions coding for the LIM domains and the acidic domain into the pET-32a vector (Invitrogen).
PCR mutagenesis.
Mutations introduced into the unc-86 cDNA were named by the following scheme: the one-letter abbreviation of the amino acid, followed by its position in the protein, followed by the abbreviation of the amino acid substitution. POUS and POUHD of unc-86 were mutagenized using the following PCR conditions. A mixture of 2 mM each dATP, dTTP, and dCTP; 10 mM dGTP; 500 mM primers; 200 ng of pBY531 (identical to pBY175 but containing a BamHI restriction site introduced by silent mutagenesis at position 428) template DNA; 10 mM Tris-HCl; 50 mM KCl; 0.8% Nonidet P-40; and 2.5 mM MgCl2 was subjected to 2 min at 92°C, followed by 35 cycles of 30 s at 92°C, 30 s at 45°C, and 1 min at 72°C and a final elongation for 5 min at 72°C.
Introduction of unc-86 mutants into yeast via homologous recombination.
Gel-purified, mutagenized PCR products (500 ng) were transformed, together with 200 ng of linearized vector pBY531, into yeast strain RB101 expressing mec-3–GAL4 AD (GAD). Homologous recombination (27) between the PCR fragment and pBY531 was selected for on plates without leucine.
In vitro translation.
For in vitro translation of mutant, truncated, or wild-type unc-86, pCITE-4a (Novagen)-based constructs were used. In vitro transcription and translation were performed in rabbit reticulocyte lysate with the TNT T7 Expression System (Promega) and [35S]methionine in accordance with the manufacturer's instructions. The amount of protein was normalized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by autoradiography and PhosphorImager quantification.
Purification of recombinant MEC-3 protein.
E. coli was transformed with plasmid pBY1107 encoding a MEC-3 protein lacking the LIM domains and the acidic tail fused with six His tags to thioredoxin. After induction with 1 mM IPTG at 37°C for 4 h, recombinant MEC-3 was purified from the soluble fraction using Ni2+ nitrilotriacetic acid-agarose (Qiagen) in accordance with the instructions of the manufacturer.
Gel retardation assay.
One hundred nanograms of oligonucleotide RB135 (5′-GCATTCGAAATGCATTGCCCATAATG-3′) was labeled with 30 μCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham)–2 μl of 10× polynucleotide kinase buffer–1 μl (5 U) of polynucleotide kinase (MBI Fermentas) in 20 μl and incubated at 37°C for 60 min. The reaction was inactivated by heating at 65°C for 20 min, and excess [γ-32P]ATP was removed by passing the reaction over a G-25 column (Roche). The volume activity was determined in a scintillation counter; subsequent hybridization of complementary oligonucleotide RB136 was performed in a 10-fold excess by heating the reaction mixture to 95°C for 2 min and then cooling it to room temperature. Gel retardation experiments were performed for 30 min at room temperature as previously described (43). Equal amounts of mutant, truncated, and wild-type in vitro-translated UNC-86 protein were included in the reaction mixtures. After incubation, reaction mixtures were loaded onto a 6% polyarylamide acid gel in 0.5× TBE (45 mM Tris-borate, 0.5 mM EDTA) and subjected to electrophoresis at 150 V for 3 h at room temperature.
RESULTS
UNC-86 DNA and protein interactions can be analyzed in yeast in vivo.
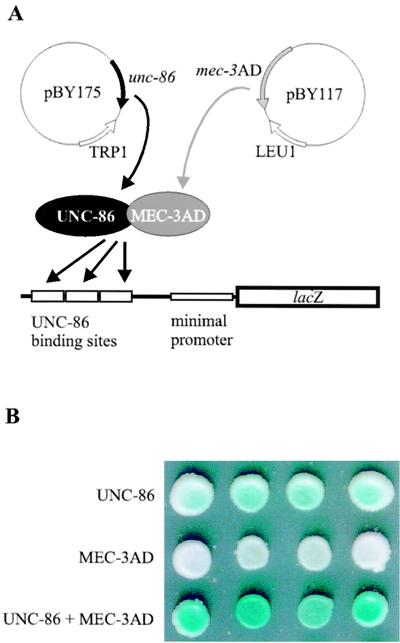
In order to facilitate the analysis of DNA binding and protein contacts of UNC-86, we developed an in vivo system to characterize UNC-86 mutants in S. cerevisiae. For this purpose, we constructed yeast strain RB101, which contains a lacZ reporter with a yeast minimal promoter under the control of three copies of the first 26 bp of mec-3 promoter element CS1 (5′-GCATTCGAAATGCATTGCCCATAATG-3′) (Fig. 1A; Table 1). It has been shown previously that this segment is sufficient for both UNC-86 binding and UNC-86-dependent binding of MEC-3 (25). The resulting S. cerevisiae strain, RB101, did not activate the lacZ reporter per se, as colonies on minimal plates did not turn blue even after 10 days of incubation in the presence of X-Gal (data not shown). In contrast, constitutive expression of unc-86 in RB101 resulted in light-blue colonies after a minimum of 3 days, indicating moderate lacZ reporter gene expression (Fig. 1B, row 1; Table 1). This expression was strongly enhanced (32-fold) by coexpression of the C. elegans mec-3 cDNA (Table 1). An even stronger level of activation was obtained (49-fold) when mec-3 was fused to a heterologous GAL4 AD (mec-3–GAD) (Fig. 1B, row 3; Table 1). In order to obtain maximum lacZ reporter gene expression, we performed all consecutive experiments with this mec-3–GAD variant. Activation is dependent on the presence of UNC-86 protein, since yeast colonies expressing only mec-3–GAD showed no lacZ expression (white colonies; Fig. 1B, row 2; Table 1). This indicates that UNC-86 can function as a transcriptional activator and that association with MEC-3 on DNA further increases transcriptional activation.
FIG. 1.
UNC-86 and MEC-3–GAD (here shown as MEC-3AD) interact in a yeast in vivo system. (A) Schematic representation of the yeast system. UNC-86 and MEC-3–GAD interact in the presence of appropriate DNA binding sites. (B) Four transformants of RB101, each expressing either UNC-86 (row 1) or MEC-3–GAD (row 2) alone or both proteins (row 3) were dotted on X-Gal plates, and color development was scored after 3 days at 30°C.
TABLE 1.
Interaction of UNC-86 and MEC-3 in an in vivo yeast system
| Yeast strain and UNC-86 variant | Relative activity (%)a | Relative activity (%)a with:
|
Fold increase
|
||
|---|---|---|---|---|---|
| MEC-3–GAD | MEC-3 | MEC-3–GADb | MEC-3c | ||
| RB101 | |||||
| None | 2.2 ± 1.4 | 2.1 ± 1 | |||
| Wild type | 100 ± 14 | 4,860 ± 230 | 3,250 ± 360 | 49 | 32 |
| UNC-86ΔN | 11.9 ± 0.7 | 557 ± 83 | 32.0 ± 2.1 | 47 | 3 |
| RB102 | |||||
| Wild type | 8.2 ± 1.6 | 8.5 ± 1.9 | 1 | ||
| UNC-86ΔN | 170 ± 13 | 430 ± 160 | 3 | ||
Activity is shown as the mean ± the standard error relative to the activity of wild-type UNC-86, which was set at 100%.
Values represent the ratio of the mean relative activity with MEC-3–GAD and the mean relative activity of the respective UNC-86 variant alone.
Values represent the ratio of the mean relative activity with MEC-3–GAD and the mean relative activity of the respective UNC-86 variant alone.
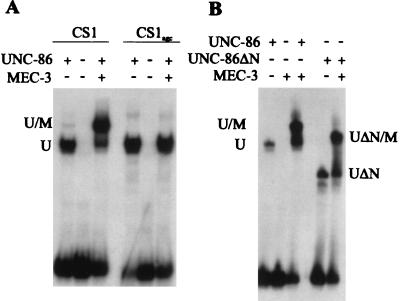
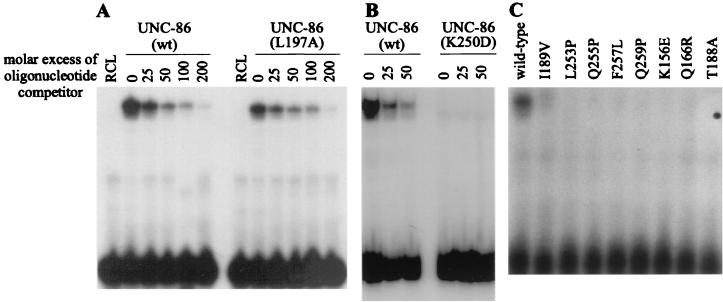
It has been proposed that UNC-86 binds to a spaced recognition site, CATtcgAAAT, in CS1 (35). In order to evaluate the contribution of bases flanking this motif to ternary complex formation, we constructed yeast strain RB102. This strain is genetically identical to RB101, except that we mutated the GCAT bases immediately 3′ of the proposed UNC-86 binding site to GAGC (CS1AGC) (35). These nucleotide exchanges did not affect lacZ reporter gene activation in the presence of UNC-86 alone. However, activation with UNC-86 and the MEC-3 AD together was only 3-fold, compared to 49-fold in RB101 (Table 1). By electrophoretic mobility shift assay (EMSA), we confirmed that the mutation does not affect the DNA binding of UNC-86 but severely impairs the interaction of MEC-3 with UNC-86 and/or CS1 DNA (Fig. 2A).
FIG. 2.
UNC-86 and MEC-3 interaction in vitro. U and UΔN designate the positions of the binary UNC-86 and UNC-86ΔN DNA complexes, whereas U/M and UΔN/M designate the ternary complex of the UNC-86 variant, MEC-3, and DNA. (A) Equal amounts of in vitro-translated UNC-86 wild-type protein were incubated with and without approximately 50 ng of recombinant MEC-3 protein and 25,000 cpm of the CS1 fragment and the CS1AGC fragment, respectively. (B) Equal amounts of in vitro-translated full-length UNC-86 and UNC-86ΔN proteins were incubated with and without approximately 50 ng of recombinant MEC-3 protein and the 26-bp fragment from CS1.
Next, we determined the domains of UNC-86 required for DNA binding and activation. For this purpose, we deleted the amino-terminal third of the UNC-86 protein (UNC-86ΔN) and performed EMSAs with CS1 DNA and MEC-3 protein. Neither binding to DNA nor the interaction of UNC-86 with MEC-3 was impaired by this amino-terminal deletion (Fig. 2B). In contrast, expression of UNC-86ΔN resulted in approximately eightfold lower activation of lacZ reporter gene expression in vivo (12% of wild-type activation; Table 1); however, the fold increase in activation obtained with MEC-3–GAD was similar to that obtained with wild-type UNC-86 (47-fold compared to 49-fold; Table 1). This strongly suggests that the amino-terminal deletion in UNC-86 eliminates a potential AD but does not compromise UNC-86 interaction with either MEC-3 or DNA. Interestingly, the synergistic activation by UNC-86ΔN and MEC-3 is strongly reduced (from 32-fold to 3-fold) when mec-3 is expressed without the Gal4p AD (Table 1).
UNC-86 contains an amino-terminal transactivation domain.
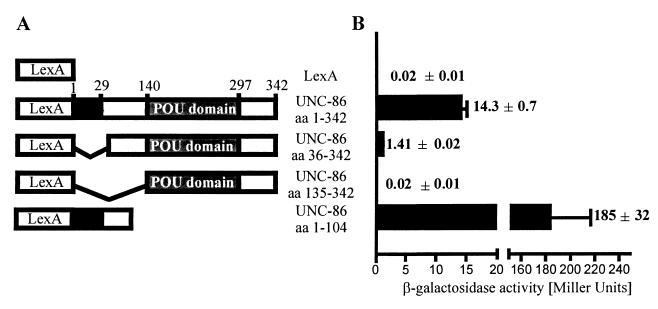
Our in vivo results obtained so far indicated that the ability of UNC-86 to activate transcription in yeast is dependent on the integrity of both the DNA binding domain and the amino terminus of the protein. In order to map the AD independently of DNA binding, we fused different fragments of the unc-86 cDNA to a portion of the lexA gene encoding the LexA DNA binding domain (aa 1 to 202) (Fig. 3A). lacZ reporter gene activation was determined quantitatively in S. cerevisiae strain L40, which contains a lacZ reporter gene controlled by a minimal promoter with multiple lexA operator sites (19). In this strain, the transcriptional activation of UNC-86 does not depend on its intrinsic DNA binding properties. The expression of a full-length unc-86–lexA fusion resulted in strong lacZ reporter gene activation, as determined by β-galactosidase assays (Fig. 3B). Removal of the 35 aa from the amino-terminal end of UNC-86 reduced lacZ reporter gene activation approximately 10-fold. Deletion of all of the amino acids amino terminal to the POU domain (UNC-86ΔN) resulted in complete loss of lacZ reporter gene expression. In contrast, a fusion containing only the first 104 aa of UNC-86 activated reporter expression 13-fold more strongly than did full-length UNC-86. From these data, we conclude that the amino terminus of UNC-86, but not the POU domain, is able to activate PolII-dependent transcription in yeast. In contrast, the POU domain is both necessary and sufficient for DNA binding and MEC-3–GAD interaction in our yeast model, as was suggested previously by in vitro studies (43).
FIG. 3.
Mapping of the UNC-86 AD. (A) Schematic representation of UNC-86 regions linked to the LexA DNA binding domain (aa 1 to 202). The amino acids from UNC-86 are as indicated (3). (B) Fusion constructs were transformed into yeast strain L40, and LacZ activity was quantified as described in Materials and Methods. The results of one representative experiment are shown along with the mean ± the standard error of triplicate determinations.
Both POUS and POUHD are involved in interactions with MEC-3.
The molecular nature of the UNC-86–MEC-3 interaction had not been analyzed previously in detail. Therefore, we conducted a screen to identify mutations in UNC-86 that interfere with the formation of the ternary complex. For this purpose, random PCR mutagenesis of POUS and POUHD of unc-86 was performed (see Materials and Methods).
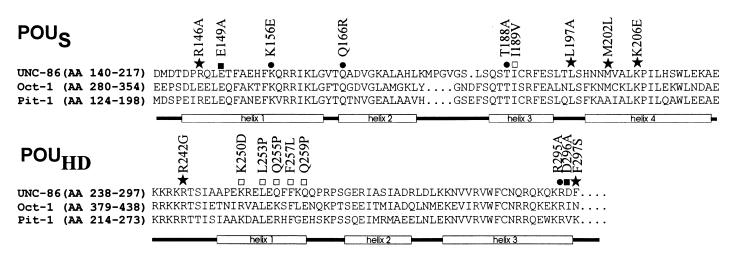
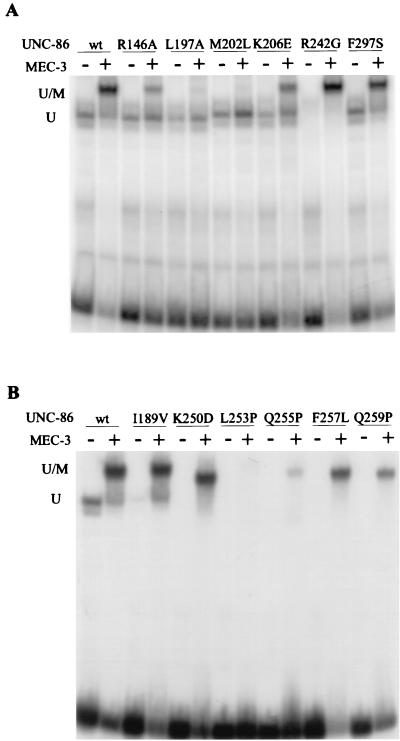
Transformants from a screen for POUS and POUHD mutants were randomly selected and streaked on plates containing X-Gal. The transformants were assigned to three different groups by distinguishing among white, light blue, and dark blue colonies. From a number of light blue and white colonies, we isolated the unc-86-encoding plasmids, retransformed them into yeast with and without the mec-3–GAD plasmid, and sequenced the respective unc-86 genes. The β-galactosidase activities obtained with unc-86 mutants containing single-site missense mutations in the presence or absence of the MEC-3–GAD were determined in a quantitative assay (Table 2). The differences in β-galactosidase activity were not due to protein levels, as determined by Western blot analyses (data not shown). The positions of the substituted amino acids in the respective mutants are depicted in Fig. 4. The UNC-86 mutants that result in light blue yeast colonies can be divided into two classes based on this analysis. The first (class I) is comprised of mutants L197A, M202L, and K206E, with amino acid substitutions in the fourth helix of POUS; mutant R242G, with a mutation at the amino terminus of POUHD; and F297S, with an amino acid substitution in the carboxy-terminal amino acid of POUHD (Fig. 4). All five mutants activated lacZ to an extent similar to that of wild-type UNC-86. Since these results suggested that these mutations do not influence the DNA binding of the respective mutants significantly, we next tested them in an EMSA with CS1 DNA (Fig. 5A and 6A). L197A, M202L, K206E, and F297S each formed a detectable DNA complex with CS1 with an intensity similar to that seen with wild-type UNC-86. No DNA complex was obtained with R242G.
TABLE 2.
Effects of amino acid substitutions in UNC-86 on lacZ reporter gene activation in yeast strain RB101
| UNC-86 variant | Relative activity (%)a | Relative activity with MEC-3–GAD (%)a | Fold increaseb |
|---|---|---|---|
| Wild type | 100 ± 14 | 4,860 ± 225 | 49 |
| E149Ac | 47.6 ± 9.5 | 1,950 ± 333 | 40 |
| D296Ac | 90.5 ± 42.9 | 4,670 ± 670 | 52 |
| Class I | |||
| R146Ac | 57.1 ± 9.5 | 885 ± 124 | 15 |
| L197A | 65.9 ± 5.5 | 247 ± 44 | 4 |
| M202L | 139 ± 15 | 474 ± 90 | 3 |
| K206E | 57.8 ± 6.8 | 517 ± 97 | 9 |
| R242G | 58.8 ± 4.1 | 983 ± 94 | 17 |
| F297S | 194 ± 14 | 603 ± 49 | 3 |
| Class II | |||
| I189V | 5.0 ± 0.6 | 538 ± 62 | 108 |
| K250D | 2.2 ± 0.3 | 92 ± 37 | 42 |
| L253P | 6.2 ± 0.6 | 233 ± 24 | 38 |
| Q255P | 14.6 ± 1.6 | 578 ± 43 | 40 |
| F257L | 8.2 ± 1.9 | 605 ± 63 | 74 |
| Q259P | 10.2 ± 0.4 | 279 ± 67 | 27 |
| Class III | |||
| K156E | 7.4 ± 1.3 | 7.7 ± 1.5 | 1 |
| Q166R | 4.3 ± 0.4 | 4.8 ± 0.6 | 1 |
| T188A | 8.4 ± 2.3 | 11.6 ± 2.2 | 1 |
| R295Ac | 18.1 ± 11.9 | 3.8 ± 1.0 | 0 |
| I189T R242G | 0.01 ± 0.01 | 0.01 ± 0.01 | 1 |
Activity is shown as the mean ± the standard error relative to the activity of wild-type UNC-86, which was set at 100%.
Values represent the ratio of the mean relative activity with MEC-3–GAD and the mean relative activity of the respective UNC-86 variant alone.
Analyzed in an alanine substitution screen.
FIG. 4.
Amino acid substitutions that affect UNC-86 ternary complex formation in yeast. The amino acid sequences of POUS and POUHD of UNC-86 (3), Oct-1 (32), and Pit-1 (20) are shown, with labeled boxes below the sequences depicting the α helices in the corresponding POU domain. Symbols: ★, mutation that interferes only with MEC-3–GAD coactivation (class I); □, mutation that influences transcriptional activation by UNC-86 (class II); ●, mutation that interferes with DNA binding and MEC-3–GAD coactivation (class III); ■, mutation that does not affect UNC-86 function.
FIG. 5.
Effects of amino acid substitutions in UNC-86 on DNA binding. Results of EMSAs of in vitro-translated wild-type UNC-86 (wt) and UNC-86 variants with the 26-bp fragment from CS1 are shown. (A and B) UNC-86 L197A mutant protein (class I) binds the 26-bp fragment to approximately the same extent as the wild-type protein, whereas UNC-86 K250D (class II) does not bind DNA. Titration experiments were performed using increasing amounts of unlabeled CS1 oligonucleotide as indicated. Lanes RCL contain labeled CS1 probe including reticulocyte lysate without translated UNC-86 protein. Equal amounts of protein were used in all reaction mixtures. (C) No or very weak binding of class II and III UNC-86 mutants to the CS1 oligonucleotide could be detected at the protein concentrations used.
FIG. 6.
Ternary complex formation by UNC-86 mutant proteins in vitro. (A) Equal amounts of in vitro-translated UNC-86 wild-type (wt) and UNC-86 class I mutant proteins were incubated with and without approximately 50 ng of recombinant MEC-3 protein and the 26-bp fragment from CS1. (B) Equal amounts of in vitro-translated UNC-86 wild-type (wt) and UNC-86 class II mutant proteins were incubated with and without approximately 50 ng of recombinant MEC-3 protein in the presence of 32P-labeled CS1 DNA. U represents the position of the binary complex of the UNC-86 variant bound to DNA, whereas U/M represents the ternary complex of the UNC-86 variant, MEC-3, and DNA.
All five mutations strongly affected the enhancement of lacZ reporter gene expression in the presence of the MEC-3–GAD (Table 2). The MEC-3–GAD-dependent lacZ activation of the respective mutants (4- to 17-fold activation) was greatly reduced compared to the 49-fold activation seen with wild-type UNC-86 (Table 2). In EMSAs, the L197A and M202L mutants did not form ternary complexes with MEC-3 and DNA (Fig. 6A) and the K206E mutant resulted in ternary complexes with MEC-3 and DNA represented by weaker bands than that observed with wild-type UNC-86. The complex formed with the mutant F297S was indistinguishable from the wild type. The mutant R242G behaved differently. Even though no DNA complex was obtained for this UNC-86 variant alone, a ternary complex with MEC-3 was formed. The data suggest that the residues at positions 197, 202, 206, and 297 are involved in the interaction and/or activation of UNC-86 with MEC-3 in the ternary complex but do not contribute to the DNA binding of UNC-86.
The second class of UNC-86 mutants (class II) that displayed reduced lacZ reporter gene expression includes I189V, with an amino acid substitution in the third α helix of POUS, and K250D, L253P, Q255P, F257L, and Q259P, with substitutions in the first α helix of POUHD (Fig. 4). When we expressed these variants without a mec-3–GAD gene in yeast, only very weak lacZ reporter gene expression was observed (Table 2). This suggests that these amino acid substitutions reduce the affinity of the respective UNC-86 proteins for the CS1 DNA element. In contrast, the increase in lacZ reporter activation after additional expression of mec-3–GAD was in the same range as that seen with wild-type UNC-86 (27- to 108-fold) (Table 2). In vitro DNA binding assays confirmed that these mutants have reduced DNA binding capacities (Fig. 5B and 6B). Although they were not able to bind CS1 alone, all but one of these mutants formed a detectable ternary complex with MEC-3 on CS1. We conclude that although the overall DNA binding affinity of these UNC-86 mutants is reduced, their ability to mediate MEC-3–GAD-dependent activation of transcription is not significantly diminished. We therefore suggest that these mutations do not interfere with binding of MEC-3–GAD to UNC-86.
From the white transformants, three members of a third class of mutants harboring single amino acid substitutions (class III) were identified: K156E, Q166R, and T188A with mutations in POUS (Fig. 4). These mutants showed a decreased ability to activate lacZ reporter gene expression compared to the wild-type UNC-86 protein (Table 2). In vitro DNA binding assays confirmed that these mutants display a decreased affinity for CS1 DNA (Fig. 5C). But in contrast to class II mutants, no increase in lacZ reporter gene expression upon addition of MEC-3–GAD was observed (Table 2). As these effects are not due to variations in protein levels (data not shown), these mutations may induce severe structural alterations in the protein, thereby rendering it nonfunctional. Alternatively, these mutations could also influence the ability of the protein to interact both with DNA and with MEC-3–GAD protein. The latter can be assumed for the I189R R242G double mutant that we isolated from this screen as a white transformant. As shown above, the single-site mutation R242G did not have a strong effect on transcriptional activation but eliminated MEC-3–GAD coactivation whereas the I189R mutation probably affects the reporter gene activation of UNC-86 itself (as does I189V) but not MEC-3–GAD interaction. We therefore conclude that the double mutant is defective for both functions.
UNC-86 and Oct-1 use similar residues to interact with their respective protein partners.
Our yeast screen has identified several residues in UNC-86 that may contact MEC-3 directly. In the POU protein Oct-1, two of these residues, L344 (corresponding to L197) and M349 (corresponding to M202), are part of the extended surface that interacts with the Oct-1 cofactor Oca-B. In order to further test the conservation of the contact interfaces, we substituted four additional residues in UNC-86 with alanine at positions that, in Oct-1, are involved in protein interactions (1, 21). We tested the ability of each mutant to activate transcription either alone or in conjunction with MEC-3–GAD in yeast strain RB101 (Table 2). The alanine substitution at position R295 resulted in a nonfunctional UNC-86 protein (class III) that did not activate the lacZ reporter gene, either alone or with MEC-3–GAD. Mutants E149A and D296A behaved similarly to wild-type UNC-86 both in the presence and in the absence of the MEC-3 AD. The R146A mutation, however, only reduced lacZ reporter gene activation of UNC-86 weakly but reduced coactivation by the MEC-3 AD from 49-fold to 15-fold. Consistent with a role of this position in the MEC-3 contact interface, the R146A mutant bound in an EMSA to CS1 element-like wild type UNC-86 but revealed reduced affinity for MEC-3 and DNA in the ternary complex, as indicated by a band weaker than that observed with wild-type UNC-86 (Fig. 6A).
DISCUSSION
The interaction of UNC-86 with MEC-3 on DNA can be monitored in yeast.
We wanted to understand how the transcriptional activation properties of UNC-86 are modulated by protein interactions. We therefore developed an in vivo system with S. cerevisiae. In contrast to mammalian cells, S. cerevisiae does not contain genes encoding POU proteins which would compete for artificially introduced DNA binding sites (33). Therefore, yeast provides an ideal system for studying POU protein interactions. It allows us to analyze ternary complex formation involving UNC-86, its interacting partner MEC-3, and single DNA binding motifs of the mec-3 promoter in vivo.
We have found that UNC-86, in contrast to the human POU protein Oct-1 (14), activates transcription in yeast. The data from the LexA–UNC-86 fusion constructs (Fig. 3) suggest that activation is mediated by the amino terminus of UNC-86. This region contains a 29-aa region termed the POU IV box, which is the only sequence element outside the POU domain that is conserved between UNC-86 and the homologous Brn-3 proteins (34). The POU IV box has already been implicated in transcriptional activation by the Brn-3 proteins (4, 31). We found no evidence of the presence of a strong activator region in the UNC-86 POU domain, as has been suggested for Brn-3a (4).
MEC-3 incorporation into the ternary complex is dependent on the DNA context.
In order to contribute to lacZ reporter gene activation, MEC-3 has to be recruited to DNA by the UNC-86 POU domain. UNC-86 binding to CS1 is a prerequisite of MEC-3 activation, since MEC-3–GAD could not activate lacZ expression on its own.
The first 26 bp of the CS1 mec-3 promoter element used in our study contain both a CATnnnAAAT motif and an overlapping octameric consensus motif (AAATGCAT) for UNC-86 binding (35, 40, 42). Both motifs are repeated in promoter element CS2, which, together with CS1, has been shown to be required both for UNC-86-dependent establishment and UNC-86- and MEC-3-dependent maintenance of mec-3 expression in C. elegans (35, 40, 42). Here we show that UNC-86 can still activate transcription in vivo on the CS1agc element in which the nucleotides CAT in the octamer motif are changed to AGC. We suggest that the nucleotides CAT are not a prerequisite for UNC-86 binding to CS1, either in vivo or in vitro (our results and reference 35). Therefore, UNC-86 most likely interacts with the CATtcgAAAT motif, as suggested previously (39, 46). We show here that the CAT bases 3′ adjacent to this motif are crucial for MEC-3-dependent activation in yeast and for the incorporation of MEC-3 into the UNC-86–DNA complex. A similar requirement for DNA target site-dependent recruitment has also been suggested for the formation of the Oct-1–Oca-B complex (6, 23). Whether MEC-3 interacts directly with DNA or induces a conformational change in UNC-86 that results in an extension or shift of its DNA binding surface has to be the subject of future studies. The importance of the octamer motif in CS1 and CS2 is further supported by the evolutionary conservation of this sequence among C. elegans, C. briggsae, and C. remanei (40, 42).
MEC-3 enhances the DNA binding of UNC-86.
We identified several mutations that strongly reduced the affinity of UNC-86 for DNA in vitro (Fig. 6B), resulting in very weak transcriptional activation in vivo (Table 2). However, coexpression of mec-3–GAD along with these unc-86 variants in the yeast system was still found to cooperatively increase lacZ reporter gene activation by a factor similar to that obtained with the wild type (Table 2). All but one of the mutant proteins were able to form a ternary complex with the MEC-3 protein in vitro (Fig. 6B).
None of the positions affected by these mutations have previously been reported to directly contact DNA in other POU proteins. The K250D mutation alters the charge of a solvent-exposed residue, while three of the other mutations that cluster in the first α helix of POUHD introduce prolines (Fig. 4), which may result in a kink in this helix. A slightly altered structure of POUHD could impair the DNA binding of these mutants. Interestingly, mutations that affect DNA binding but not ternary complex formation have not been identified in previous mutational analyses of POU proteins (1, 29). From these data, we conclude that MEC-3 interaction may enhance the binding affinity of UNC-86, either by inducing a conformational change in the protein or because MEC-3 itself contributes to DNA binding.
Amino acids in both POU subdomains of UNC-86 influence the interaction with MEC-3.
Five amino acid substitutions in POUS and POUHD of UNC-86 (Fig. 4) result in mutants that bind DNA in vitro like wild-type UNC-86 and also activate transcription in vivo to a similar extent. However, when coexpressed with mec-3–GAD, these mutants did not display an increase in lacZ reporter gene activation as seen with wild-type UNC-86 (Table 2). This suggests that we have identified amino acids in both POUS and POUHD of UNC-86 that are required for the interaction with MEC-3 or for the synergistic activation of the UNC-86–MEC-3 heterodimer.
The structures of the POU domains of Oct-1 and Pit-1 are strikingly similar, even though they display only 52% amino acid identity (16). No structural data are available for UNC-86, but from the 45 and 44% sequence identity of the UNC-86 POU domain with those of Oct-1 and Pit-1, respectively, one could assume that it folds very similarly to the POU domains of these proteins (16). Based on these similarities, we compared the locations of mutations in UNC-86 with their respective positions in Oct-1 and Pit-1 (21, 22).
The residues corresponding to R146, L197, and M202 in Oct-1 and F297 in Pit-1 (compare Fig. 4) have been implicated in direct protein contacts (1, 13, 21, 29). Alanine substitutions at E286, L344, and M349 in Oct-1 result in loss of interaction with Oca-B (1, 13, 21), and K60 (F297) is involved in protein contacts in the Pit-1 crystal structure (21). Consistent with a similar function, L197A and M202L did not form ternary complexes in an EMSA and the ternary complex formation of R146A appeared weaker than that of wild-type UNC-86. As both the leucine (position 197) and methionine (position 202) are conserved in the functionally nonrelated UNC-86, Pit-1, and Oct-1 proteins (Fig. 4) (16), one might hypothesize that they are part of a general protein docking surface conserved among POU proteins. K206E may also be part of this docking surface, since this residue is located close to positions 202 and 197 in the structure of the POU domain, and ternary complex formation of the K206E mutant was weaker than with the wild type. There are no data available from Oct-1 or Pit-1 POU complexes that link this position to protein interactions.
F297S, on the other hand, did not affect DNA and MEC-3 binding of UNC-86 in an EMSA, but in vivo coactivation with MEC-3–GAD was reduced from 49-fold to 3-fold. This suggests that F297S is an important discriminator of protein interactions and coactivation. It is consistent with such a role that this position is distinct in the different POU proteins and is located in the vicinity of other residues that, in both Oct-1 and Pit-1, affect DNA or protein interactions. In contrast, E289A and I437A in Oct-1 prevented the interaction with Oca-B (1), whereas the equivalent substitutions in UNC-86 (E149A and D296A) affected neither DNA binding nor MEC-3 interactions. We conclude that Oct-1 and UNC-86 share some of the residues of their surface for protein interactions, but other positions are obviously functionally distinct.
An unexpected result was obtained for position 242. In the cocrystal structures of Pit-1 and Oct-1, this conserved arginine is involved in minor-groove base contacts (21, 22). Consistent with a similar role in UNC-86, R242G did not bind to DNA in vitro, although in vivo, this mutant only showed twofold reduced transcriptional activation. Even more strikingly, R242G was capable of ternary complex formation like the wild type whereas our in vivo data suggest fivefold reduced coactivation with MEC-3–GAD. We have no explanation for this discrepancy. We cannot exclude the possibility that the DNA binding of UNC-86 R242G is enhanced by intrinsic yeast proteins. Alternatively, the R242G mutation could have a negative effect on the DNA binding of UNC-86 and at the same time lead to strong enhancement of the amino-terminal transactivation domain of UNC-86. The latter case would suggest a novel role for the POU domain in transcriptional regulation. While the POU domain does not possess activating properties, it may participate in the regulation of the amino-terminal AD of UNC-86.
Synergistic activation between UNC-86 and MEC-3 may require an extensive interaction surface.
The cooperative binding of MEC-3 and UNC-86 strongly enhanced transcriptional activation in yeast, corroborating the previously reported results of in vitro transcription assays (25). However, a comparison of our in vivo and in vitro data clearly demonstrates that the affinity of UNC-86 and MEC-3 in the ternary complex with DNA in vitro does not always correlate with the transcriptional activity of the complex in vivo. For example, binding of UNC-86 to MEC-3 is not significantly affected by deletion of the amino-terminal half of UNC-86. This led us (this study) and others (43) to propose that UNC-86 binding to MEC-3 is mediated by the POU domain only. However, we found that the UNC-86ΔN protein, which lacks all residues N terminal to the POU domain, has almost lost its capability for synergistic transcriptional activation with MEC-3 in vivo whereas, strikingly, this synergism with the MEC-3 AD is not affected. This suggests that under in vivo conditions, UNC-86 and MEC-3 are involved in more complex interactions than previously thought. These require binding of MEC-3 to the UNC-86 POU domain but obviously also involve additional interactions between the UNC-86 amino terminus and an as yet unidentified domain of MEC-3. The latter interaction can be replaced in our yeast system by providing a heterologous AD (Gal4p) with the MEC-3–GAD fusion. The nature of the synergistic activation of UNC-86–MEC-3 is unknown and has to be the subject of future studies.
ACKNOWLEDGMENTS
We thank W. Hammerschmidt for the modified pGBT9 vector and the yeast strain RH1533, M. Chalfie for pTU47, and R. Grosschedl and the members of the Kolanus, Meisterernst, and Baumeister labs for helpful suggestions and comments on the manuscript.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to R.B.
REFERENCES
- 1.Babb R, Cleary M A, Herr W. Oca-B is a functional analog of VP16 but targets a separate surface of the Oct-1 POU domain. Mol Cell Biol. 1997;17:7295–7305. doi: 10.1128/mcb.17.12.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel P L, Chien C T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Vol. 14. Oxford, England: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 3.Baumeister R, Liu Y, Ruvkun G. Lineage-specific regulators couple cell lineage asymmetry to the transcription of the C. elegans POU gene unc-86 during neurogenesis. Genes Dev. 1996;10:1395–1410. doi: 10.1101/gad.10.11.1395. [DOI] [PubMed] [Google Scholar]
- 4.Budhram-Mahadeo V, Morris P J, Lakin N D, Theil T, Ching G Y, Lillycrop K A, Möröy T, Liem R K H, Latchman D S. Activation of the α-internexin promoter by the Brn-3a transcription factor is dependent on the N-terminal region of the protein. J Biol Chem. 1995;270:2853–2858. doi: 10.1074/jbc.270.6.2853. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie M, Au M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 1989;243:1027–1033. doi: 10.1126/science.2646709. [DOI] [PubMed] [Google Scholar]
- 6.Chang J F, Philipps K, Lundback T, Gstaiger M, Ladbury J E, Luisi B. Oct-1 POU and octamer DNA co-operate to recognize the Bob-1 transcription co-activator via induced folding. J Mol Biol. 1999;288:941–952. doi: 10.1006/jmbi.1999.2711. [DOI] [PubMed] [Google Scholar]
- 7.Clontech Laboratories, Inc. Clontech manual: MATCHMAKER One-Hybrid System Protocol. Palo Alto, Calif: Clontech Laboratories, Inc.; 1997. [Google Scholar]
- 8.Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- 9.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 10.Finney M, Ruvkun G, Horvitz H R. The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell. 1988;55:757–769. doi: 10.1016/0092-8674(88)90132-8. [DOI] [PubMed] [Google Scholar]
- 11.Gietz R D, Woods R A. High efficiency transformation with lithium acetate. 1994. pp. 121–134. . Guide to yeast genetics and molecular biology. Oxford Unirversity Press, Oxford, England. [Google Scholar]
- 12.Gruber C A, Rhee J M, Gleiberman A, Turner E E. POU domain factors of the Brn-3 class recognize functional DNA elements which are distinctive, symmetrical, and highly conserved in evolution. Mol Cell Biol. 1997;17:2391–2400. doi: 10.1128/mcb.17.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gstaiger M, Georgiev O, van Leeuwen H, van de Vliet P, Schaffner W. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 1996;15:2781–2790. [PMC free article] [PubMed] [Google Scholar]
- 14.Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens C M. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- 15.Herr W, Cleary M A. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 16.Herr W, Sturm R A, Clerc R G, Corcoran L M, Baltimore D, Sharp P A, Ingraham H A, Rosenfeld M G, Finney M, Ruvkun G, Horvitz H R. The POU domain: a large conserved region in the mammalian Pit-1, Oct-1, Oct-2 and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 17.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Hodgkin J A, Horvitz H R, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingraham H A, Chen R, Mangalam H J, Elsholtz H P, Flynn S E, Lin C R, Simmons D M, Swanson L, Rosenfeld M G. A tissue specific transcription factor containing a homeo domain specifies a pituitary phenotype. Cell. 1988;55:519–529. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson E M, Li P, Leon-del-Rio A, Rosenfeld M G, Aggarwal A K. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 22.Klemm J D, Rould M A, Aurora R, Herr W, Pabo C O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 23.Krapp A, Strubin M. B-cell coactivator OBF-1 exhibits unusual transcriptional properties and functions in a DNA-bound Oct-1 dependent fashion. Mol Cell Biol. 1999;19:4247–4254. doi: 10.1128/mcb.19.6.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P, He X, Gerrero M R, Mok M, Aggarwal A, Rosenfeld M G. Spacing and orientation of bipartite DNA-binding motifs as potential functional determinants for POU domain factors. Genes Dev. 1993;7:2483–2496. doi: 10.1101/gad.7.12b.2483. [DOI] [PubMed] [Google Scholar]
- 25.Lichtsteiner S, Tjian R. Synergistic activation of transcription by UNC-86 and MEC-3 in Caenorhabditis elegans embryo extracts. EMBO J. 1995;14:3937–3945. doi: 10.1002/j.1460-2075.1995.tb00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature. 1995;376:344–348. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- 27.Petermann R, Mossier B M, Aryee D N T, Kovar H. A recombination based method to rapidly assess specificity of two-hybrid clones in yeast. Nucleic Acids Res. 1998;26:2252–2253. doi: 10.1093/nar/26.9.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan A K, Rosenfeld M G. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- 29.Sauter P, Matthias P. Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol Cell Biol. 1998;18:7397–7409. doi: 10.1128/mcb.18.12.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 31.Smith M D, Morris P J, Latchman D S. The Brn-3c transcription factor contains a neuronal-specific activation domain. Neuroreport. 1998;9:851–856. doi: 10.1097/00001756-199803300-00016. [DOI] [PubMed] [Google Scholar]
- 32.Sturm R A, Das G, Herr W. The ubiquitous octamer binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 33.Svetlov V V, Cooper T G. Compilation and characteristics of dedicated transcription factors in Saccharomyces cerevisiae. Yeast. 1995;11:1439–1484. doi: 10.1002/yea.320111502. [DOI] [PubMed] [Google Scholar]
- 34.Theil T, McLean-Hunter S, Zornig M, Möröy T. Mouse Brn-3 family of POU transcription factors: a new aminoterminal domain is crucial for the oncogenic activity of Brn-3a. Nucleic Acids Res. 1993;21:5921–5929. doi: 10.1093/nar/21.25.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Way J C. Promoter sequences for the establishment of mec-3 expression in the nematode Caenorhabditis elegans. Mech Dev. 1996;56:183–196. doi: 10.1016/0925-4773(96)00523-0. [DOI] [PubMed] [Google Scholar]
- 36.Ward A C. Single-step purification of shuttle vectors from yeast for high frequency back-transformation into E. coli. Nucleic Acids Res. 1990;18:5319. doi: 10.1093/nar/18.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward S, Thomson N, White J G, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- 38.Way J C, Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 1989;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- 39.Way J C, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- 40.Way J C, Wang L, Run J A, Wang A. The mec-3 gene contains cis-acting elements mediating positive and negative regulation in cells produced by asymmetric cell division in C. elegans. Genes Dev. 1991;5:2199–2211. doi: 10.1101/gad.5.12a.2199. [DOI] [PubMed] [Google Scholar]
- 41.Wong M W, Henry R W, Ma B, Kobayashi R, Klages N, Matthias P, Strubin M, Hernandez N. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol Cell Biol. 1998;18:368–377. doi: 10.1128/mcb.18.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue D, Finney M, Ruvkun G, Chalfie M. Regulation of the mec-3 gene by the C. elegans homeoproteins UNC-86 and MEC-3. EMBO J. 1992;11:4969–4979. doi: 10.1002/j.1460-2075.1992.tb05604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue D, Tu Y, Chalfie M. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science. 1993;261:1324–1328. doi: 10.1126/science.8103239. [DOI] [PubMed] [Google Scholar]