ABSTRACT.
Multiplex bead assays (MBAs) for serologic testing have become more prevalent in public health surveys, but few studies have assessed their test performance. As part of a trachoma study conducted in a rural part of Ethiopia in 2016, dried blood spots (DBS) were collected from a random sample of 393 children aged 0 to 9 years, with at least two separate 6-mm DBS collected on a filter card. Samples eluted from DBS were processed using an MBA on the Luminex platform for antibodies against 13 antigens of nine infectious organisms: Chlamydia trachomatis, Vibrio cholera, enterotoxigenic Escherichia coli, Cryptosporidium parvum, Entamoeba histolytica, Camplyobacter jejuni, Salmonella typhimurium Group B, Salmonella enteritidis Group D, and Giardia lamblia. Two separate DBS from each child were processed. The first DBS was run a single time, with the MBA set to read 100 beads per well. The second DBS was run twice, first at 100 beads per well and then at 50 beads per well. Results were expressed as the median fluorescence intensity minus background (MFI–BG), and classified as seropositive or seronegative according to external standards. Agreement between the three runs was high, with intraclass correlation coefficients of > 0.85 for the two Salmonella antibody responses and > 0.95 for the other 11 antibody responses. Agreement was also high for the dichotomous seropositivity indicators, with Cohen’s kappa statistics exceeding 0.87 for each antibody assay. These results suggest that serologic testing on the Luminex platform had strong test performance characteristics for analyzing antibodies using DBS.
INTRODUCTION
Serologic testing of dried blood spots (DBS) is increasingly used in resource-limited settings for surveillance of neglected tropical diseases.1 DBS are relatively easy and inexpensive to collect, store, and transport, making them an attractive biospecimen. Seroprevalence studies can provide information about ongoing transmission of infection through estimation of seroconversion rates among young children. Testing is most commonly done with either a multiplex bead assay (MBA) or an enzyme-linked immunosorbent assay (ELISA).2 The MBA is particularly efficient because it allows simultaneous serological testing of exposure to a variety of pathogens from a single specimen. It also provides robust data by measuring the signal for bound antibody antigen on multiple beads for each antigen—typically 100 beads are read per antigen per specimen, and the median fluorescence intensity (MFI) among all readings is reported for each antigen. Yet despite the promise of MBA platforms, little has been published regarding the precision of these testing methods when measuring seroprevalence of antibodies.3 In the present study, we assess the precision of an MBA platform, testing the variability of results when performing the test on two DBS from the same child, and when altering the number of beads used to determine the MFI of the response.
METHODS
Ethics.
Ethical approval was obtained from the University of California, San Francisco; Emory University; the Ethiopian Ministry of Science and Technology; and the Food, Medicine, and Health Care Administration and Control Authority of Ethiopia. Given the high level of illiteracy in the study area, guardians of all participants provided verbal informed consent. CDC staff were determined not to be engaged.
Study design.
The present study was embedded in a randomized trial in the WagHemra Zone of Ethiopia, a rural setting with hyperendemic trachoma.4,5 DBS collected at the baseline study visit in January and February 2016 were assessed for antibodies against a panel of antigens at the Centers for Disease Control and Prevention (Atlanta, GA) in February and March 2017. Subsequently, a simple random sample of blood spots was selected for a repeatability study and processed at the same laboratory in June 2017.
Sample collection.
A random sample of approximately 60 children aged 0 to 9 years per community from each of 40 study communities provided DBS as part of the parent trial in January and February 2016 in the Amhara region of Ethiopia.4 Blood spots were collected by finger-prick by one of eight individuals drawn from local health facilities who had prior experience collecting blood samples. The filter paper card (TropBio Pty Ltd, Sydney, Australia) used for blood collection had six 6-mm extensions, each designed to collect 10 µL of blood (Figure 1). Each card was allowed to dry at room temperature, placed into its own individual plastic bag, and packaged with desiccant. Cards were stored for 11 months in a –20°C freezer at the Amhara Public Health Institute in Ethiopia, kept at ambient temperature for 11 days while being shipped to Atlanta in January 2017, and finally stored in a –80°C freezer until processed.
Figure 1.

Filter paper used for dried blood spot collection. For each child, a trained worker attempted to fill five consecutive extensions out of the six total extensions of the filter paper card (TropBio Pty Ltd, Sydney, Australia), with extension 1 filled first, then extension 2, and so on. This figure appears in color at www.ajtmh.org.
Multiplex assay.
The Luminex platform was used for multiplex antibody testing (Bio-Rad, Hercules, CA). The assay has been described in detail previously.6 For the present study, a bead set was developed at the CDC consisting of beads coupled to 13 antigens (Table 1). Antigens were selected based on the aims of the parent study.4 Most antigens were tagged with glutathione S-transferase (GST) fusion proteins and purified through a glutathione column; beads coated only in GST were included as a negative control. Beads were added to each well of a 96-well plate. A single 6-mm spot per card was eluted in 1,600 µL of phosphate-buffered saline containing 0.5% casein, 0.3% Tween 20, 0.02% sodium azide, 0.5% polyvinyl alcohol, 0.8% polyvinylpyrrolidone, and 3 µg/mL Escherichia coli extract (to block binding of anti–E. coli antibody to beads and lower background levels). DBS eluate from a single 6-mm spot per card was added to the beads and incubated. The bound antibody was detected by adding anti-human immumoglobulin (Ig)G (Southern Biotech, Birmingham, AL) and anti-human IgG4 (Southern Biotech) biotinylated detection antibodies, followed by R-phycoerythrin-labeled streptavidin (Invitrogen, Carlsbad, CA). Plates were read on a BioPlex 200 instrument equipped with Bio-Plex Manager 6.0 software (BioRad, Hercules, CA), with the software collecting data on a prespecified, programmable number of beads per antigen before moving to the next well. The test output was the MFI minus background (MFI-BG), with background values derived from beads run with buffer alone. As per the protocol, any well with a high GST response (i.e., the negative control) would have been excluded, although no specimens in this study had a high GST response. The MFI-BG values were classified as seropositive or seronegative according to a threshold based on one of three methods depending on the antigen: 1) receiver operating characteristics (ROC) curves, 2) a standard curve using previously established cutoff values, or 3) the third standard deviation above the mean of samples taken from individuals at very low risk of infection.7,8 Methodologies to determine seropositivity thresholds are summarized in Table 1.
Table 1.
Antigens included in multiplex bead assay
| Antigen | Organism | Cutoff method* | Cutoff |
|---|---|---|---|
| pgp36 | Chlamydia trachomatis | ROC† | 1,113 |
| CT6946 | Chlamydia trachomatis | ROC† | 337 |
| CTBc | Vibrio cholera | ||
| ETEC LTB‡ | Heat labile toxin-producing enterotoxigenic Escherichia coli | ||
| Cp1711 | Cryptosporidium parvum | Standard curve§ | 182 |
| Cp2311 | Cryptosporidium parvum | Standard curve§ | 323 |
| LecA12 | Entamoeba histolytica | Mean +3 SD‖ | 28 |
| p1813 | Camplyobacter jejuni | ||
| p3913 | Camplyobacter jejuni | ||
| Sal Bc | Salmonella typhimurium Group B | ||
| Sal Dc | Salmonella enteritidis Group D | ||
| VSP314 | Giardia lamblia | Mean +3 SD‖ | 167 |
| VSP514 | Giardia lamblia | Mean +3 SD‖ | 295 |
Blank cells indicate that the antibody response was not classified as seropositive or seronegative.
Receiver operating characteristics (ROC) curve derived from a panel of external standards that were previously classified as positive or negative by MBA, using Youden’s index to determine threshold.7
Sigma Chemical Co., St. Louis, MO.
Threshold defined using a 2-fold serial dilution curve with previously established unit cutoff values.8
The third standard deviation above the mean from a sample of 86 volunteers from the CDC who did not travel internationally.
Parameters altered between runs.
Three separate runs of the assay were performed for each child’s filter paper. The laboratory, operator, and apparatus were identical between the three runs, and the assay methodology was identical except for those parameters listed in Table 2 (i.e., which DBS extension was tested, the number of beads added and read, and the calendar month the assay was performed).
Table 2.
Interrun assay parameters
| Sample source* | Run A DBS extension 1 |
Run B DBS extension 2 |
Run C DBS extension 2 |
|---|---|---|---|
| Beads added per well | 2,500 | 2,500 | 1,250 |
| Beads per antigen read per well† | 100 | 100 | 50 |
| Dates of processing | Feb–March 2017 | June 2017 | June 2017 |
DBS = dried blood spot
The extension number indicates the order in which blood was applied to the extensions in the field.
The software can be programmed to collect data on a prespecified number of beads before moving to the next well on the 96-well plate.
Statistical analysis.
Precision was assessed by comparing pairs of measurements differing in a single parameter, with two main pairwise comparisons: A) different blood spot extensions from the same child (i.e., run A versus run B in Table 2) and 2) altering the number of beads used to make a result determination for the same DBS eluent (i.e., run B versus run C in Table B). Bland-Altman plots were constructed depicting the mean difference and 95% limits of agreement (LOA) for each pairwise comparison. One-way, single-measurement intraclass correlation coefficients (ICCs) were calculated from linear mixed models to estimate the variability in pairs of continuous MFI-BG measurements.9 Agreement between seropositivity values was assessed with Cohen’s kappa statistic. We based sample size considerations on calculation of a two-sided confidence interval for a correlation coefficient; 393 samples would provide a 99% confidence interval of 0.05 assuming a correlation coefficient of 0.90.
RESULTS
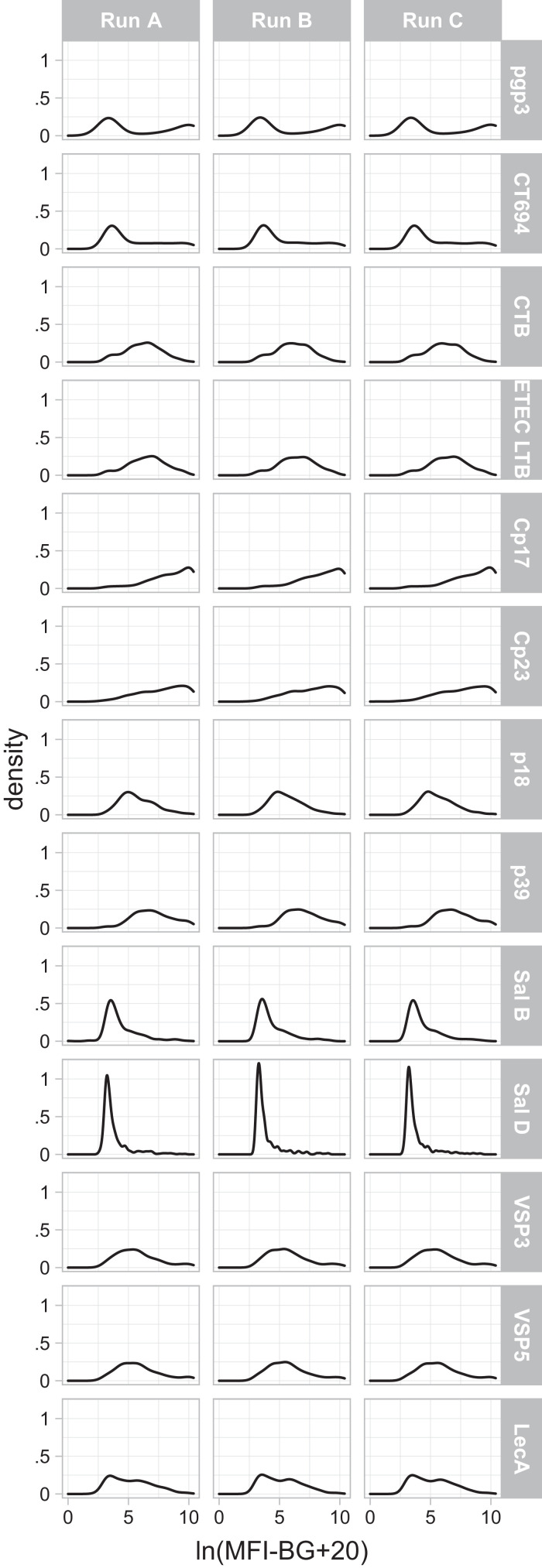
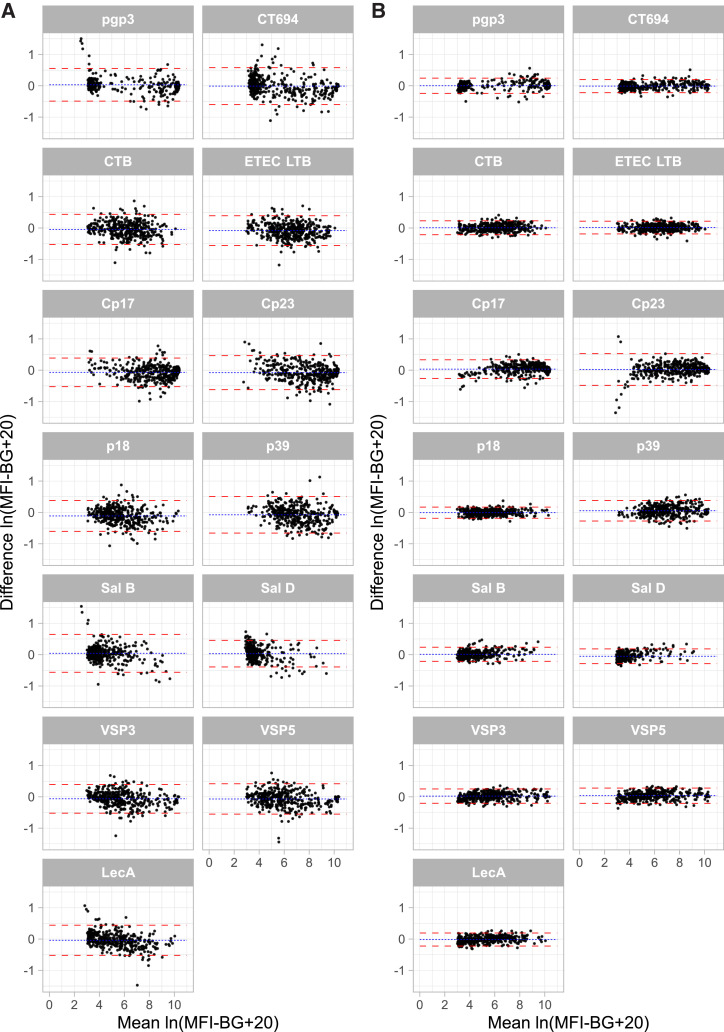
DBS from 393 children were run, 392 of which contributed data for all 13 antigens for all three test runs and are included in the main analysis. Data from one child was excluded due to a labeling error; analyses including this child provided nearly identical results. The distribution of MFI-BG values obtained during the first run is shown in Figure 2 for each of the 13 antigens included. Bland Altman plots did not reveal marked evidence of heteroskedasticity (i.e., differences in intertest variability dependent on the magnitude of the MFI-BG) for either of the pairwise comparisons of interest (Figure 3). The mean differences, 95% LOAs, and ICCs are shown in Table 3; ICCs exceeded 0.95 for each pairwise comparison except for the two Salmonella antigens, which had slightly lower estimates of repeatability when comparing different blood samples from the same child (i.e., different extensions of the filter paper card). ICCs for the 50- versus 100-bead comparison were slightly higher than ICCs comparing measurements from two blood samples.
Figure 2.
Distributions of median fluorescence intensity minus background (MFI-BG) values across runs. MFI-BG values are natural log-transformed for visual comparability across antigens and are increased by a constant value of 20 to account for negative MFI-BG values before natural log-transformation.
Figure 3.
Bland-Altman plots. Two pairwise comparisons are depicted: (A) eluate 1 vs. eluate 2 (i.e., two blood spots from the same child) and (B) 50 vs. 100 beads per well (i.e., the number of beads used by the assay’s software to calculate the raw median fluorescence intensity minus background (MFI-BG) result. Dashed lines depict the 95% limits of agreement. For visual comparability across antigens the mean and difference values are depicted as natural log-transformed MFI-BG values, after adding a constant value of 20 to account for negative MFI-BG values before natural log-transformation. The eluate comparisons exclude four extreme values (1 pgp3, 1 p39, and 2 Sal B); the bead comparisons exclude 1 extreme value (Cp23). This figure appears in color at www.ajtmh.org.
Table 3.
Precision of dried blood spot serology results from a multiplex bead assay
| Eluate 1 vs. Eluate 2 (Run A vs. Run B) |
100 beads vs. 50 beads (Run B vs. Run C) |
|||||
|---|---|---|---|---|---|---|
| Antigen | Mean difference | 95% LOA | ICC | Mean difference | 95% LOA | ICC |
| pgp3 | −204.3 | (–3,405.3, 2,996.7) | 0.990 | 167.5 | (–2,009.9, 2,344.9) | 0.995 |
| CT694 | −339.8 | (–2,896.3, 2,216.6) | 0.979 | 44.0 | (–1,043.3, 1,131.2) | 0.996 |
| CTB | −124.3 | (–1,322.7, 1,074.0) | 0.968 | 20.2 | (–445.8, 486.3) | 0.995 |
| ETEC LTB | −225.2 | (–1,613.2, 1,162.8) | 0.972 | 34.1 | (–513.8, 582.0) | 0.995 |
| Cp17 | −643.6 | (–4,521.6, 3,234.5) | 0.981 | 320.2 | (–2,246.6, 2,886.9) | 0.992 |
| Cp23 | −690.6 | (–4,691.2, 3,309.9) | 0.970 | 324.1 | (–1,845.4, 2,493.6) | 0.991 |
| LecA | −134.1 | (–1,124.0, 855.8) | 0.977 | 9.9 | (–393.0, 412.8) | 0.996 |
| p18 | −177.3 | (–1,460.2, 1,105.7) | 0.978 | 9.9 | (–526.3, 546.0) | 0.996 |
| p39 | −434.0 | (–3,840.9, 2,973.0) | 0.958 | 220.4 | (–1,612.0, 2,052.9) | 0.987 |
| Sal B | −101.0 | (–1,700.4, 1,498.3) | 0.850 | 40.8 | (–584.6, 666.2) | 0.967 |
| Sal D | −36.8 | (–850.0, 776.3) | 0.907 | 9.0 | (–242.2, 260.3) | 0.988 |
| VSP3 | −252.5 | (–2,153.2, 1,648.2) | 0.981 | 82.2 | (–1,002.4, 1,166.7) | 0.994 |
| VSP5 | −274.7 | (–2,241.5, 1,692.1) | 0.983 | 101.5 | (–1,083.1, 1,286.1) | 0.994 |
ICC = intraclass correlation coefficient; LOA = limits of agreement. Values represent pairwise comparisons of raw median fluorescence intensity minus background values from 392 children. Eluate 1 was taken from the first blood spot extension and eluate 2 from the second blood spot extension. The bead comparison (e.g., 50 vs. 100) refers to the number of bead reads per well used by the software algorithm.
Exploratory analyses of the measurement pairs from run A and run B (i.e., two blood spots from the same child) showed that the absolute differences in measurement disagreement correlated well between most antigens (i.e., if a child’s response to a given antigen was higher in run A than run B then the response to a different antigen was also likely to be higher in run A than run B; Supplemental Figure 1). Although it is not clear why agreement for Sal B and Sal D was lower than that of the other antigens, the vast majority of disagreements were seen in older children (Supplemental Figure 2).
Test results were classified as seropositive or seronegative for several antigens based on external standards (Table 1). Seroprevalence estimates were similar for each of the antigens regardless of testing run (Table 4). Estimates of Cohen’s kappa were greater than 0.87 for each pairwise comparison. Similar to the ICCs for the raw MFI-BG results, agreement was slightly higher for the 50- versus 100-bead comparison relative to the comparison of two blood samples.
Table 4.
Agreement in seroprevalence results
| Seroprevalence | Cohen’s kappa (95% CI) | ||||||
|---|---|---|---|---|---|---|---|
| Antigen | Run A N = 392 |
Run B N = 392 |
Run C N = 392 |
Eluate 1 vs. Eluate 2 (Run A vs. Run B) |
100 beads vs. 50 beads (Run B vs. Run C) |
||
| pgp3 | 42.6% | 42.3% | 42.3% | 0.995 | (0.985 − 1.000) | 0.990 | (0.975–1.000) |
| CT694 | 36.5% | 37.0% | 37.0% | 0.967 | (0.941–0.993) | 0.989 | (0.974–1.000) |
| Cp17 | 92.3% | 91.8% | 92.1% | 0.947 | (0.887–1.000) | 0.983 | (0.949–1.000) |
| Cp23 | 81.6% | 81.9% | 81.6% | 0.915 | (0.863–0.967) | 0.957 | (0.920–0.994) |
| LecA | 71.7% | 71.7% | 72.2% | 0.899 | (0.850–0.947) | 0.975 | (0.950–0.999) |
| VSP3 | 57.4% | 57.7% | 56.9% | 0.917 | (0.877–0.957) | 0.943 | (0.909–0.976) |
| VSP5 | 45.4% | 45.2% | 42.6% | 0.871 | (0.822–0.920) | 0.928 | (0.890–0.965) |
CI = confidence interval. Cohen’s kappa shown for pairwise comparisons of 392 children. Eluate 1 was taken from the first blood spot extension and eluate 2 from the second blood spot extension. The bead comparison (e.g., 50 vs. 100) refers to the number of bead reads per well used by the software algorithm.
DISCUSSION
Repeatability refers to the similarity of results from multiple runs of the same test, when the laboratory, operator, and testing apparatus are the same and the interval between tests is short. In contrast, reproducibility refers to the situation when laboratory, operator, and apparatus are all different and the testing interval is long.10 Each is important because repeatability indicates the minimum variability of the test when performed under the same conditions, whereas reproducibility provides information about the generalizability of the test under different testing conditions. Each assumes the tests are performed on replicate specimens. However, sample collection technique may also affect the variability of antibody testing results, and thus it is also important to assess the accuracy of an assay when analyzing different specimens from the same subject. In the present study, we assessed an estimate of intermediate precision closer to repeatability because we kept most laboratory parameters constant. We intentionally altered two parameters: which blood sample was used for testing and how many beads were read by the software before providing a result. We found that the variability associated with each of these testing parameters was very low, providing confidence in the accuracy of the Luminex testing platform on dried blood spots.
Few reproducibility studies of MBAs have been done for the antigens of interest, although the results of the present study are consistent with a previous study that found an ICC of 0.997 on a set of 45 DBS from a trachoma-endemic area.3 The finding of high agreement between two samples from the same child is an important finding to support the validity of serologic testing for trachoma and gastrointestinal pathogens. Although field collections of DBS are done in a standardized fashion according to a protocol, in practice there is variability between subjects and even between separate samples from the same subject. It is likely that individual samples differ in the amount of serum (i.e., the liquid component of blood devoid of coagulation factors), which could increase variability in testing. For example, it is possible that samples collected at a longer time interval following the finger prick may have more clotting factors than samples collected immediately after the prick. Or children may become uncooperative, leading to variability in the overall volume of blood collected on the blood spot. The finding here that two blood spots from the same child have high agreement for this multiplex serologic assay provides reassurance that serologic antibody testing is a repeatable technique in the low- and middle-income countries where it may have the greatest application.
The 50- versus 100-bead comparison, which had runs performed on the same eluate, had higher agreement than the comparison of eluates from two blood samples, although it should be noted that more time had elapsed between the two runs for this latter comparison. The 50- versus 100-bead comparison was performed to determine whether a less data-intensive assay could perform similarly to the routine assay, with the rationale that the 50-bead run would offer efficiencies to a laboratory in terms of time and reagent requirements. Indeed, the beads are the most expensive part of the MBA, so the finding of high agreement between the 50-bead and 100-bead assays could mean large cost-savings for laboratories. This might be especially important because a likely scenario for such a multiplex antibody assay would be in a large public health survey, after which samples would be processed in large numbers. As this application is expanded to more populations, lower limits of bead counts for various antigens should be evaluated for further cost savings.
This study had some limitations. All repeat testing was done in one laboratory with experienced technicians who helped develop these specific assays. This makes it difficult to assess the reproducibility of the assay in other laboratories, especially in laboratories that may have less experience with the Luminex platform. Similarly, all samples were collected by an experienced group of field workers in Ethiopia who had specialized training in collecting blood samples. It is possible that more novice field workers would have less standardization of technique (i.e., more variability in the amount of blood obtained on each of the different DBS extensions), which could lead to more variability. A limited number of parameters were altered during the repeated runs of the assay; it is possible that altering other parameters would lead to more substantial interassay variability. It is unclear how generalizable this will be to antigens from a wider variety of pathogens, and thus similar validation studies should be done for expanded antigen panels. Finally, the data only provide information about the quality of the test performance, not the “correctness” of the results or whether the test is fit-for-purpose.
Multiplex serologic testing on the Luminex platform promises to be an efficient method for screening populations for a large number of diseases with a single blood sample. Although ongoing studies are important to determine the antibody kinetics surrounding seroconversion and seroreversion, such studies would be less interpretable if the assay itself were not repeatable. The high repeatability demonstrated in the present study lends confidence to the use of multiplex serologic assays for public health surveys, at least for the antigens included in the present study.
Supplemental figures
ACKNOWLEDGMENTS
Purified Cp17, Cp23, VSP-3, VSP-5, p18, and p39 antigens were kindly provided by Jeffrey Priest (US CDC), and LecA antigen was kindly provided by William Petri (University of Virginia) and Joel Herbein (TechLab).
Note: Supplemental figures appear at www.ajtmh.org.
References
- 1. Arnold BF, Scobie HM, Priest JW, Lammie PJ, 2018. Integrated serologic surveillance of population immunity and disease transmission. Emerg Infect Dis 24: 1188–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gwyn S, Cooley G, Goodhew B, Kohlhoff S, Banniettis N, Wiegand R, Martin DL, 2017. Comparison of platforms for testing antibody responses against the Chlamydia trachomatis antigen Pgp3. Am J Trop Med Hyg 97: 1662–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaur H, Dize L, Munoz B, Gaydos C, West SK, 2018. Evaluation of the reproducibility of a serological test for antibodies to Chlamydia trachomatis pgp3: a potential surveillance tool for trachoma programs. J Microbiol Methods 147: 56–58. [DOI] [PubMed] [Google Scholar]
- 4. Wittberg DM. et al. , 2021. WASH Upgrades for Health in Amhara (WUHA): study protocol for a cluster-randomised trial in Ethiopia. BMJ Open 11: e039529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aiemjoy K, Aragie S, Wittberg DM, Tadesse Z, Callahan EK, Gwyn S, Martin D, Keenan JD, Arnold BF, 2020. Seroprevalence of antibodies against Chlamydia trachomatis and enteropathogens and distance to the nearest water source among young children in the Amhara region of Ethiopia. PLoS Negl Trop Dis 14: e0008647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goodhew EB. et al. , 2012. CT694 and pgp3 as serological tools for monitoring trachoma programs. PLoS Negl Trop Dis 6: e1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Migchelsen SJ. et al. , 2017. Defining seropositivity thresholds for use in trachoma elimination studies. PLoS Negl Trop Dis 11: e0005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Priest JW, Moss DM, 2020. Measuring cryptosporidium serologic responses by multiplex bead assay. Methods Mol Biol 2052: 61–85. [DOI] [PubMed] [Google Scholar]
- 9. Shrout PE, Fleiss JL, 1979. Intraclass correlations: uses in assessing rater reliability. Psychol Bull 86: 420–428. [DOI] [PubMed] [Google Scholar]
- 10. Menditto A, Patriarca M, Magnusson B, 2007. Understanding the meaning of accuracy, trueness and precision. Accredit Qual Assur 12: 45–47. [Google Scholar]
- 11. Moss DM, Montgomery JM, Newland SV, Priest JW, Lammie PJ, 2004. Detection of cryptosporidium antibodies in sera and oral fluids using multiplex bead assay. J Parasitol 90: 397–404. [DOI] [PubMed] [Google Scholar]
- 12. Houpt E, Barroso L, Lockhart L, Wright R, Cramer C, Lyerly D, Petri WA, 2004. Prevention of intestinal amebiasis by vaccination with the Entamoeba histolytica Gal/GalNac lectin. Vaccine 22: 611–617. [DOI] [PubMed] [Google Scholar]
- 13. Schmidt-Ott R, Brass F, Scholz C, Werner C, Gross U, 2005. Improved serodiagnosis of Campylobacter jejuni infections using recombinant antigens. J Med Microbiol 54: 761–767. [DOI] [PubMed] [Google Scholar]
- 14. Priest JW, Moss DM, Visvesvara GS, Jones CC, Li A, Isaac-Renton JL, 2010. Multiplex assay detection of immunoglobulin G antibodies that recognize Giardia intestinalis and Cryptosporidium parvum antigens. Clin Vaccine Immunol 17: 1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.