ABSTRACT.
Japanese encephalitis virus (JEV) is transmitted between swine, migratory birds, and Culex mosquitoes, and has circulated indigenously in Asia for almost a century. Despite being the country with the highest JEV diversity, surveillance targeting of Indonesia’s vectors is scarce. This study collected mosquitoes from several locations in Tabanan Regency, Bali Island, Indonesia. We captured and classified 3,032 adult Culex mosquitoes into seven species, with Culex vishnui subgroup mosquitoes making up approximately 90% of the total. Japanese encephalitis virus was identified by next-generation sequencing (NGS) analysis of a Cx. vishnui mosquito pool. Genetic and phylogenetic analysis revealed the JEV as genotype (G) IV. The nucleotide identity was 99% with other JEV GIV isolates obtained from swine sera in 2017 on Bali Island and from a human patient in Australia with a travel history to Bali in 2019. This finding indicated that JEV GIV persists in restricted areas and is circulating between swine-mosquito vectors.
Japanese encephalitis virus (JEV) has been reported to be mainly distributed in Asia and Western Pacific regions and maintained between swine, birds, and Culex mosquitoes.1 This virus is conventionally classified into five different genotypes (GI–GV) based on the envelope gene sequence but when the information is available, its entire genome sequence is the preferred choice for classification. Four out of five JEV genotypes have been recorded in Indonesia,2 most of which were partially sequenced in 1980s,3 thus indicating the country has the highest genotype diversity globally.4 Notwithstanding, disease surveillance targeting of Indonesia’s vectors is scarce.
Virological surveillance targeting the vectors has been confirmed to facilitate early detection before human case incidence or to act as indicator during low-incidence scenarios,5,6 and it may be applicable in areas with past evidence of JEV presence in Indonesia, such as Bali Island.7 In this study, we conducted cross-sectional JEV surveillance by collecting mosquitoes in several pig sheds in Tabanan Regency, Bali Island, Indonesia, and subjected them to an RNA virus isolation protocol.
Sweep nets and mouth aspirators were used to collect mosquitoes. Sampling was done in the early hours of dusk between 6 pm and 8 pm, coinciding with peak biting activity, on the July 2 and 3, 2019. Mosquitoes were collected from pig sheds close to rice fields and residential areas in Tabanan Regency, Bali Island (Table 1 and Figure 1), and kept for at least 3 days under insectary conditions (temperature and humidity controlled, with 10% sugar ad libitum) to allow for complete blood digestion. On the fourth day, live mosquitoes were killed by low temperatures using dry ice packs and stored at −80°C until use. Mosquitoes were identified morphologically according to established identification keys,8 sorted by species and sex, and stored in 2-mL microtubes (Eppendorf, Hamburg, Germany) in a −80°C freezer.
Table 1.
Collection sites and GPS coordinates
| Collection site group | Name of village and subdistrict in Tabanan Regency | GPS coordinates | Collection date |
|---|---|---|---|
| 1 | Sudimara, Tabanan | 8.5774S 115.0867E | July 2, 2019 |
| Gubug, Tabanan | 8.5592S 115.0929E | ||
| 2 | Gubug, Tabanan | 8.5586S 115.0928E | July 2, 2019 |
| Gubug, Tabanan | 8.5594S 115.0937E | ||
| 3 | Nyitdah, Kediri | 8.5811S 115.1223E | July 3, 2019 |
| Nyitdah, Kediri | 8.5819S 115.1216E | ||
| 4 | Nyitdah, Kediri | 8.5803S 115.1222E | July 3, 2019 |
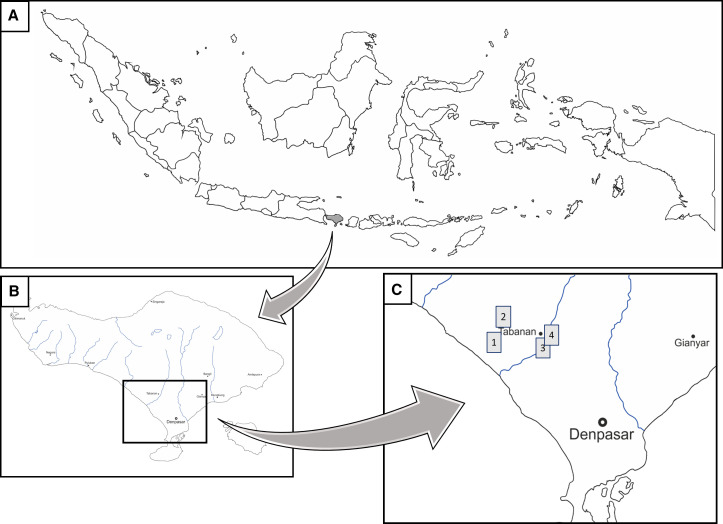
Figure 1.
Collection site map in Tabanan Regency, Bali Island, Indonesia. (A) Map of the Indonesian archipelago, (B) Bali Island, and (C) magnification of four collection sites in Tabanan Regency. The gray boxes depict the four groups of collection site in Tabanan Regency. The map templates were modified with permission from d-maps.com.
The Aedes albopictus C6/36 cell line (European Collection of Authenticated Cell Cultures) was used for virus isolation of 2,191 mosquitoes (Table 2) and were subjected to virus isolation protocol, as described previously.9 In brief, selected mosquito pools were homogenized, filtered, inoculated onto monolayers of C6/36 cells in 24-well culture plates, and incubated for 7 days. After two rounds of blind passages, the culture supernatant was collected and stored at −80°C. Phase-contrast microscopy was used to observe any cytopathic effects (CPEs) before harvesting the third supernatant.
Table 2.
The list of mosquitoes processed in this study
| Genus | Species | No. of pools tested (1-25♀/pool) | No. of mosquitoes tested | Japanese encephalitis virus (JEV) isolated |
|---|---|---|---|---|
| Culex | pseudovishnui | 63 | 1,566 | |
| vishnui | 24 | 562 | 1 | |
| tritaenioryhnchus | 2 | 15 | ||
| vishnui subgroup | 3 | 43 | ||
| quinquefasciatus | 1 | 1 | ||
| bitaeniorhynchus | 1 | 1 | ||
| gelidus | 1 | 3 | ||
| Total | 95 | 2,191 | 1 |
Harvested supernatants were combined (12 pools per sample group) and nuclease-treated before RNA extraction by ISOGEN II (Nippon Gene, Toyama, Japan). The library preparation, next-generation sequencing (NGS) procedure, and de novo assembly were conducted using the Neb next ultra RNA first-strand synthesis module (New England BioLabs, Ipswich, MA), MiniSeq (Illumina, San Diego, CA) and CLC Genomics Workbench version 11 (QIAGEN, Venlo, Limburg, Netherlands), respectively, as previously described.9
To confirm the JEV pool source, viral RNA was extracted from the final supernatant of the virus isolation process using the rapid viral RNA screening method described in Faizah et al.10 Detergent mixture was used for extraction and reverse transcription-polymerase chain reaction (RT-PCR) using a PrimeScript One-Step RT-PCR Kit version 2 (Takara Bio, Shiga, Japan) was used for sequence amplification. A primer set targeting the JEV envelope gene (JEV955f: 5′-TGYTGGTCGCTCCGGCTTA-3′; JEV1606R: 3'-GACYTYGAMCCACGGTCAT-5′) was used.
GENETYX version 13 (Genetyx Corp., Osaka, Japan) and MEGA version 6.06 were used for phylogenetic tree construction. Complete envelope gene sequences were used for alignment in the E-MAFFT online service with slow progressive method, G-INS-1. Phylogenetic analysis was conducted using the Tamura-Nei model (TN93) with Gamma distribution and Invariant sites (G + I) by maximum likelihood statistical method with 1,000 replications of the bootstrap test. GenBank accession number of the obtained JEV sequence is LC579814.
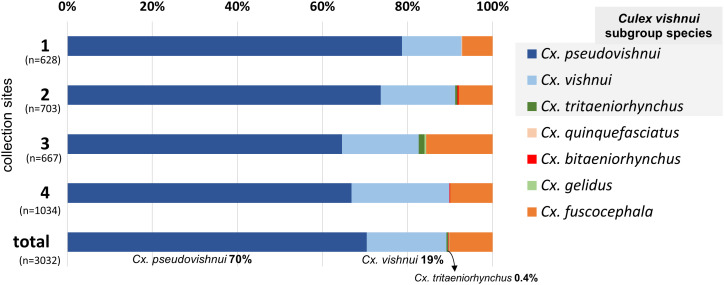
Results showed that out of 3,613 identified adult mosquitoes that were captured alive, 3,032 were Culex species (Figure 2). Among Culex, the three species of Culex. vishnui subgroup mosquitoes made up to 90%, with Culex pseudovishnui being dominant (70%), followed by Cx. vishnui (20%), and Culex tritaeniorhynchus (< 1%) (Figure 2).
Figure 2.
Species composition of Culex mosquitoes captured in four collection site groups in Tabanan Regency. Culex vishnui subgroup mosquitoes were dominant in all locations, accounting for up to 90% of the captured Culex mosquitoes. This figure appears in color at www.ajtmh.org.
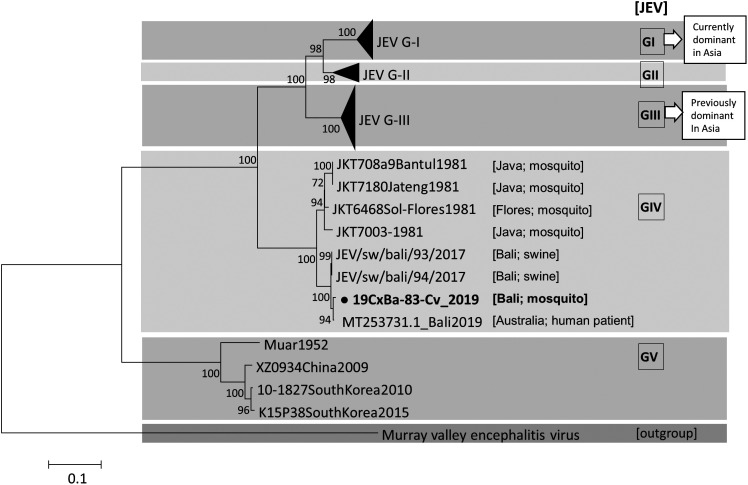
Results from virus isolation showed that after three subcultures in C6/36 cells, one out of 24 Cx. vishnui pools were confirmed to contain propagated JEV (Table 2). The origin of the JEV-positive pool was collection site 4 in Nyitdah, Kediri (Figure 1C, Table 1). The obtained genome length of the JEV in this study was 10,857 nucleotides, covering a single large polyprotein, and showed 99% BLASTn identity with two JEV GIV strains (JEV/sw/Bali/93/2017 and Bali 2019) and 95% with another GIV strain collected in 1981 (JKT6468). Phylogenetic analysis using the complete E gene substantiated the current isolate as GIV (Figure 3). The minimum infection rate (MIR) is defined as follows: (number of pools with JEV-confirmed/total number of mosquitoes tested) × 1,000. The MIR in this study was 1.8 (one positive pool/562 Cx. vishnui mosquitoes processed) for JEV in Cx. vishnui only.
Figure 3.
Maximum likelihood phylogeny of Japanese encephalitis virus (JEV) using nucleotide sequences in E gene (1500 nt). Tamura-Nei (TN93) model with gamma distribution and invariant sites was applied, along with 1,000 bootstrap replications. The Murray Valley encephalitis virus sequence was used as an outgroup. JEV GIV strains are shown, along with their collection source information inside the brackets, that is, [island; host]. λ signifies the sequence from this study.
Surveillance targeting the primary vector is critical in determining possible virus circulation and the disease risk because of the positive correlation between vector density and JEV occurrence in humans.11 Sample collection period was coincided with the start of the dry season in Tabanan Regency, with precipitation between 21 and 50 mm.12 The abundance of Cx. pseudovishnui and Cx. vishnui, coupled with low numbers of Cx. tritaeniorhynchus, was consistent with previous reports of a low density of Cx. tritaeniorhynchus mosquitoes during dry seasons in Tamil Nadu, India,13 and Jambi Province, Indonesia.2
Regarding the JEV isolation from Cx. vishnui, studies have documented the JEV isolations from the dominant species at each location for each season.2,13,14 JEV isolates were obtained from Cx. vishnui in July in Vietnam14 and from Cx. gelidus in June in Indonesia,2 in parallel with Cx. Tritaeniorhynchus’ scant presence. Here, the JEV was isolated from Cx. vishnui, which was the second most common Culex species captured in July. Future entomological studies to understand the seasonal dynamics of Culex mosquitoes in Indonesia along with Cx. pseudovishnui’s JEV GIV vector competence will be essential.
In this study, the JEV was isolated using C6/36 cells, following a successful JEV isolation method from mosquitoes collected in Japan.9 However, here we found that using C6/36 cells may not be advisable for JEV isolation from mosquitoes collected in Indonesia. Most of the pools had severe CPEs, making it challenging to distinguish JEV solely by CPE observation under phase-contrast microscopy. The NGS was thus performed by combining pool supernatants to confirm JEV presence, followed by RT-PCR confirmation to identify the pool source. Supriyono et al.15 reported similar results with distinct CPEs observed in C6/36 cells that were mainly caused by mosquito viruses circulating in Indonesia. Furthermore, mosquito-specific flaviviruses presence may also disrupt the RT-PCR analysis targeting the flaviviruses’ RdRP region. Although conventional PCR targeting the NS3 gene and subsequent sequencing have proven effective in detecting JEV genotypes2 in the past, the apparent lack of information on the complete genome sequences of known or detected genotype strains has necessitated the current approach of using virus isolation protocols and NGS analysis for in-depth classification and characterization. Virus isolation and NGS analysis for virus surveillance have therefore proven effective, particularly in areas where the circulating genotype is underreported.
JEV GIV detection in this study gives credence to the previous notion that this genotype is restricted to limited areas.4,16 In contrast, the Garjito et al2 report on the first detection of JEV GI in Indonesia (Sumatra Island) was an indication of possible JEV GI dissemination in Indonesia from other Asian countries through commercial routes. In the 1980s, JEV GIV strains were detected in Java, Bali, and Flores Island.3 After a long period of no detection, presumably caused by the lack of surveillance, the JEV GIV strains were recently detected in swine sera collected in Denpasar in 2017,16 in a human patient in 2019 in Queensland, Australia, with recent travel history to Bali Island,17 and Cx. vishnui in 2019 (this study). These detections all pointed to Bali Island as the source of infection, and the phylogenetic tree showed a robust correlation between the three studies. There seems to be active circulation between the amplifier host (swine), the vector (mosquitoes), and humans as the dead-end host in this area. As our current literature is lacking about JEV GIV information, which is the most restricted genotype available in the world, further studies may be required as there may be differences in the species of the maintenance hosts or mosquito vectors.
Although findings in this study reaffirms the notion of active local circulation and risk of JEV GIV on Bali Island, it may not reflect the strain of JEV circulating on other Indonesian islands. Java and Flores Island, in particular, where JEV GIV has previously been reported, will be an interesting study area to deepen our understanding of global JEV genotype distribution and the possible risk of dissemination. Finally, this study highlights the importance of mosquito surveillance in determining the Culex species in areas around pig sheds and supports the evidence of active JEV GIV circulation. Robust and periodic entomological surveillance on Bali Island, and in Indonesia in general, is therefore strongly suggested.
ACKNOWLEDGMENTS
We would like to sincerely thank the staffs of local health authorities and also residents of Tabanan Regency, Bali Island, for valuable assistance and guidance during the mosquito collections.
References
- 1. Campbell G. et al. , 2011. Estimated global incidence of Japanese encephalitis. Bull World Health Organ 89: 766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garjito TA, Prihatin MT, Susanti L, Prastowo D, Sa’adah SR, Taviv Y, Satoto TBT, Waluyo J, Manguin S, Frutos R, 2019. First evidence of the presence of genotype-1 of Japanese encephalitis virus in Culex gelidus in Indonesia. Parasit Vectors 12: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schuh AJ, Guzman H, Tesh RB, Barrett ADT, 2013. Genetic diversity of Japanese encephalitis virus isolates obtained from the Indonesian archipelago between 1974 and 1987. Vector Borne Zoonotic Dis 13: 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Solomon T, Ni H, Beasley DWC, Ekkelenkamp M, Cardosa MJ, Barrett ADT, 2003. Origin and evolution of Japanese encephalitis virus in Southeast Asia. JVI 77: 3091–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calzolari M. et al. , 2015. West Nile virus surveillance in 2013 via mosquito screening in northern Italy and the influence of weather on virus circulation. Coffey LL, ed. PLoS ONE 10: e0140915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Madewell ZJ, Hemme RR, Adams L, Barrera R, Waterman SH, Johansson MA, 2019. Comparing vector and human surveillance strategies to detect arbovirus transmission: a simulation study for Zika virus detection in Puerto Rico. Sunil S, ed. PLoS Negl Trop Dis 13: e0007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kari K. et al. , 2006. A hospital-based surveillance for Japanese encephalitis in Bali, Indonesia. BMC Med 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reuben R, Tewari SC, Hiriyan J, Akiyama J, 1994. Illustrated keys to species of Culex (Culex) associated with Japanese encephalitis in Southeast Asia (Diptera: Culicidae). Mosq Systematics 26: 75. [Google Scholar]
- 9. Faizah AN. et al. , 2020. Deciphering the virome of Culex vishnui subgroup mosquitoes, the major vectors of Japanese encephalitis, in Japan. Viruses 12: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faizah AN. et al. , 2020. Evaluating the competence of the primary vector, Culex tritaeniorhynchus, and the invasive mosquito species, Aedes japonicus japonicus, in transmitting three Japanese encephalitis virus genotypes. PLoS Negl Trop Dis 14: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murty US. et al. , 2002. Seasonal prevalence of Culex vishnui subgroup, the major vectors of Japanese encephalitis virus in an endemic district of Andhra Pradesh, India. J Am Mosq Control Assoc 18: 290–293. [PubMed] [Google Scholar]
- 12. Meteorological, Climatological, and Geophysical Agency , 2019. Badan Meteorologi, Klimatologi, dan Geofisika—Bali. Available at: http://iklim.bali.bmkg.go.id/2020/01/10/informasi-iklim-agustus-2019/ (in Indonesian). Accessed August 23, 2020.
- 13. Kuwata R. et al. , 2013. Surveillance of Japanese encephalitis virus infection in mosquitoes in Vietnam from 2006 to 2008. Am J Trop Med Hyg 88: 681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramesh D. et al. , 2015. Seasonal abundance and role of predominant Japanese encephalitis vectors Culex tritaeniorhynchus and Cx. gelidus Theobald in Cuddalore district, Tamil Nadu. Indian J Med Res 142: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Supriyono et al. , 2020. Mosquito-borne viruses, insect-specific flaviviruses (family flaviviridae, genus flavivirus), Banna virus (family reoviridae, genus seadornavirus), Bogor virus (unassigned member of family permutotetraviridae), and alphamesoniviruses 2 and 3 (family mesoniviridae, genus alphamesonivirus) isolated from Indonesian mosquitoes. J Vet Med Sci 82: 1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuwata R. et al. , 2020. Distribution of Japanese encephalitis virus, Japan and Southeast Asia, 2016–2018. Emerg Infect Dis 26: 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pyke AT. et al. , 2020. A case of Japanese encephalitis with a fatal outcome in an Australian who traveled from Bali in 2019. Trop Med Infect Dis 5: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]