Abstract
Ribonuclease 7 (RNase 7) is an antimicrobial peptide that prevents urinary tract infections (UTI); however, it is yet unknown how RNASE7 genetic variations affect its antimicrobial activity and its mitigation of UTI risk. This study determined whether the RNASE7 SNP rs1263872 is more prevalent in children with UTI and defined how rs1263872 affects RNase 7’s antimicrobial activity against uropathogenic E. coli (UPEC). We performed genotyping for rs1263872 in 2 national UTI cohorts, including children enrolled in the Randomized Intervention for Children with Vesicoureteral Reflux trial or the Careful Urinary Tract Infection Evaluation study. Genotypes from these cohorts were compared with those of female controls with no UTI. To assess whether rs1263872 affects RNase 7’s antimicrobial activity, we generated RNase 7 peptides and genetically modified urothelial cultures encoding wild-type RNase 7 and its variant. Compared with controls, girls in both UTI cohorts had an increased prevalence of the RNASE7 variant. Compared with the missense variant, wild-type RNase 7 peptide showed greater bactericidal activity against UPEC. Wild-type RNase 7 overexpression in human urothelial cultures reduced UPEC invasive infection compared with mutant overexpression. These results show that children with UTI have an increased prevalence of RNASE7 rs1263872, which may increase UTI susceptibility by suppressing RNase 7’s antibacterial activity.
Keywords: Infectious disease
Keywords: Innate immunity, UTI/pyelonephritis, Urology
Introduction
Urinary tract infections (UTI) are one of the most common bacterial infections. In children, 8% of girls will develop a UTI by 7 years of age and up to 50% of affected children will suffer from at least 1 UTI recurrence. In acute cases, UTI can lead to kidney injury, bacteremia, and urosepsis. Long-term UTI complications include hypertension, proteinuria, renal fibrosis, and chronic kidney disease. Although all children are susceptible to UTI, evidence suggests that different variables influence UTI risk. Depending on age, vesicoureteral reflux (VUR), bowel and bladder dysfunction, and sexual activity augment UTI susceptibility (1, 2). However, these conditions do not completely explain why some people develop UTI, more severe UTI, or recurrent UTI. Identifying additional UTI risk factors may improve the health of children and reduce healthcare costs.
Recently, sizable advancements have been made in identifying host responses that protect the kidneys and bladder from uropathogens. Evidence suggests the innate immune system provides the front-line defense against UTI. Among these innate defenses, antimicrobial peptides (AMPs) play important roles in shielding the urothelium from uropathogenic E. coli (UPEC) — the most common pathogen causing UTI (3). AMPs are small cationic peptides produced and secreted by cells involved in host defense. In the kidney and urinary tract, AMPs are produced by the bladder urothelium and kidney tubules. Some AMPs are produced at high basal concentrations to prevent bacterial attachment while others are induced to facilitate UPEC clearance. When AMP production is disrupted, UTI risk increases. In humans, cathelicidins, defensins, and ribonucleases are the major AMP families (3, 4).
Previously, we identified ribonuclease 7 (RNase 7) as an essential and abundant human AMP that shields the urinary tract from invasive infection (5–7). RNase 7 has antimicrobial activity against Gram-positive and Gram-negative uropathogens, including UPEC and multidrug resistant (MDR) UPEC (5, 7). RNase 7’s catalytic activity is not required for its bactericidal activity. Instead, RNase 7’s antimicrobial functions are dependent on its binding affinity for LPSs on the bacterial cell wall as well as its ability to permeate and disrupt bacterial membranes. These mechanisms of bacterial membrane disruption differ from those of antibiotics, which kill or prevent bacterial replication by inhibiting cell wall synthesis, DNA replication, RNA transcription, or protein synthesis (8–10).
While genetic variations in AMP genes have been associated with infection propensity, no published data to our knowledge interrogate how RNASE7 genetic variations effect UTI risk. Thus, the objectives of this study were to (a) evaluate the frequency of the RNASE7 SNP rs1263872 in clinical cohorts comprising female participants with UTI and controls without UTI and (b) to define the effect of SNP rs1263872 on the antimicrobial activity of the encoded RNase 7 peptide against UPEC. SNP rs1263872 has a C-to-G nucleotide substitution and confers a proline-to-alanine amino acid substitution at amino acid position 103 (P103A) of the RNase 7 peptide. This substitution predicts a change in RNase 7’s secondary structure and possibly its antimicrobial activity (11).
Results and Discussion
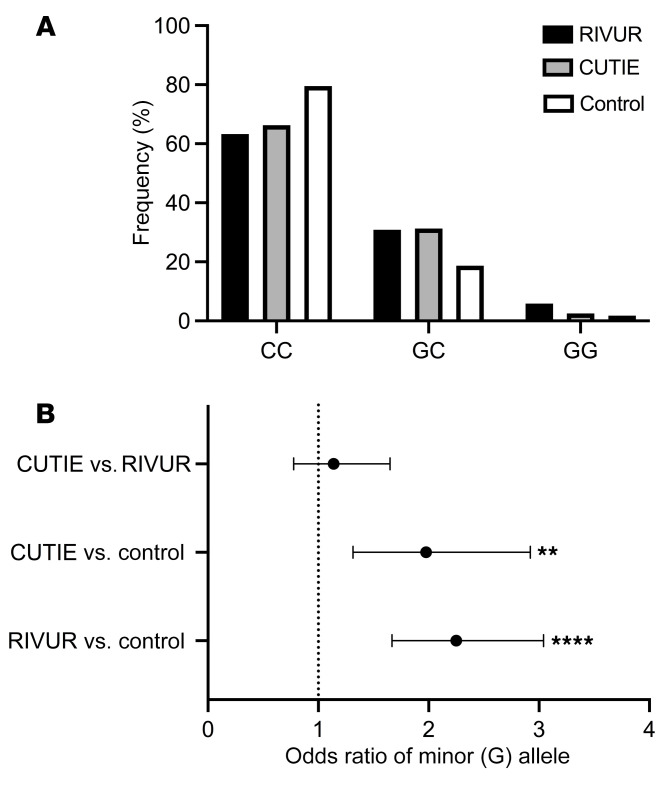
We queried the dbSNP (https://www.ncbi.nlm.nih.gov/snp/) and identified 5 nonsynonymous variants in the coding region of the RNASE7 gene. At the time of our search, rs1263872 was the most prevalent SNP with a minor allele frequency of 0.07–0.19 (11). To our knowledge, there are no published data that link this SNP to clinical outcomes. Next, we performed genotyping for rs1263872 on controls without UTI and children with UTI enrolled in the Randomized Intervention for Children with Vesicoureteral Reflux (RIVUR) trial or the Careful Urinary Tract Infection Evaluation (CUTIE) study, as outlined in the Supplemental Methods (12, 13). Allele frequencies of rs1263872 in the control cohort were found to be in Hardy-Weinberg equilibrium and did not differ from those reported in dbSNP (11). Genotype frequencies are shown (Figure 1). Compared with controls, we identified an increased prevalence of the minor G allele in both UTI cohorts. Thus, we postulated that this missense polymorphism leads to an amino acid substitution that negatively affects RNase 7’s antimicrobial activity. In the RIVUR cohort we did not detect a statistical difference in the prevalence of the minor G allele between the low-grade, nondilating VUR and higher-grade, dilating VUR groups. Additionally, we did not identify a correlation with the minor G allele and kidney scarring following UTI (data not shown).
Figure 1. Distribution and odds ratio of minor RNASE7 allele in clinical cohorts.
(A) Relative percentage of each genotype across study cohorts. Female participants enrolled in the RIVUR (black bars, n = 424) and CUTIE (gray bars, n = 160) studies have a higher prevalence of GC and GG genotypes compared with controls (white bars, n = 482). 63.3% and 66.3% of participants in the RIVUR and CUTIE studies had the CC genotype, respectively. In contrast, 79.5% of controls had the CC genotype. (B) Odds ratios comparing the presence of the minor allele (influence on UTI risk) in different cohorts. Comparing the presence of the minor allele in the RIVUR cohort with that controls resulted in an odds ratio of 2.25 (95% CI 1.67–3.04, P < 0.0001). Comparing the presence of the minor allele in the CUTIE cohort and that in controls resulted in an odds ratio of 1.98 (95% CI 1.32–2.92, P = 0.0011). No difference was seen between the RIVUR and CUTIE cohorts (odds ratio 1.14, 95% CI 0.78–1.65), indicating that VUR status does not affect UTI risk in relation to genotype. All comparisons were made using Fisher’s exact test. **P < 0.01, ****P < 0.0001.
Next, we generated RNase 7 peptides to reflect RNASE7 SNP rs1263872 wild-type (RNASE7103Pro) and its missense variant (RNASE7103Ala). To test these peptides’ antimicrobial activity against E. coli, bactericidal activity assays were performed by coincubating each RNase 7 peptide with E. coli commensal strain MG-1655 or UPEC clinical strains from phylogenetic group B2 — the most predominant E. coli clade associated with UTI (14–16). We determined the O serogroup in all tested UPEC strains and assessed the expression of 10 virulence factors contributing to UTI pathogenicity (17). Published sequencing data identify added genomic differences between MG1655 and UPEC strains CFT073, 536, and UTI89 (18–20). In addition, the functional activity of type 1 pili, which promote UPEC adherence to urothelial cells, was detected by hemagglutination in the presence and absence of mannose (21). Genotyping these clinical strains supports data showing that UPEC is a heterogeneous pathotype (Table 1). When these E. coli strains were incubated with RNase 7 peptides, our results show that the minor allele RNASE7103Ala had diminished bactericidal activity compared with wild-type RNASE7103Pro (Figure 2A). We did not observe a correlation between UPEC genotype and RNase 7 susceptibility, as RNase 7 killed each strain similarly. Future studies are needed to identify UPEC virulence factors that may confer a fitness advantage.
Table 1. Characteristics of E. coli strains used in this study.
Figure 2. Compared with wild-type RNase 7, RNASE7103Ala has reduced antimicrobial activity.
(A) E. coli strains were incubated with 2.5 μM RNASE7103Ala (diamonds) or wild-type RNASE7103Pro (triangles). An irrelevant peptide (circles) served as control. After incubation, UPEC strains were plated on LB agar and colonies were enumerated. Results are from 4 to 5 experiments performed in duplicate (n = 4–5). Data shown represent percentages of remaining colony-forming units (mean ± SEM). (B) Displacement of LPS-bound Bodipy TR Cadaverine with 0.5 μM RNase A (circles, negative control), RNASE7103Ala (diamonds), and wild-type RNASE7103Pro (triangles). Results show the mean and SEM from 4 experiments performed in duplicate (n = 4). (C and D) UROtsa cells were infected with retroviral constructs expressing wild-type RNASE7103Pro, RNASE7103Ala, or empty vector. Mean ± SEM. (C) Representative Western blot, probed for RNase 7 and GAPDH. (D) ELISA quantified RNase 7 in culture media. Results are from 3 experiments performed in triplicate (n = 3). Mean ± SEM. (E) Extracellular bactericidal activity assays were performed using culture media from empty vector (circles), RNASE7103Ala (diamonds), and RNASE7103Pro (triangles) retrovirus-infected cells, as described in the Supplemental Methods. Graphs show the mean UPEC survival and SEM. Results are from 4 to 5 experiments performed in triplicate (n = 4–5)(mean ± SEM). (F and G) UPEC attachment (F) and invasion (G) assays were performed as outlined in the Supplemental Methods. Shown are the percentage of UPEC adhering to the cellular surface and invading empty vector– (circles), RNASE7103Ala- (diamonds), and RNASE7103Pro-transduced (triangles) UROtsa cells. Results are from 5 to 6 experiments performed in quadruplicate (n = 5–6) (mean ± SEM). ND, not detected. (H) To account for the attenuated attachment of MDR12 and UTI89ΔfimH, the ratio of UPEC attachment in RNASE7103Ala- (diamonds) and RNASE7103Pro-overexpressing (triangles) cells compared with empty vector–expressing cells is shown. (A, B, and D–G) Pairwise comparisons were made by 1-way ANOVA with Tukey’s modification or (H) Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001.
Because rs1263872 is predicted to cause a change in RNase 7’s secondary structure, we determined if this amino acid substitution (P103A) affects the ability of RNase 7 to bind the bacterial cell wall. We assessed the ability of the RNASE7103Ala and RNASE7103Pro to bind LPS using the Bodipy TR Cadaverine probe (BC). BC binds the lipid A portion of LPS and measures the competitive displacement of BC by LPS-binding molecules such as RNase 7. When compared with RNASE7103Pro peptide, RNASE7103Ala showed reduced affinity binding toward LPS (Figure 2B). This finding may explain the reduced antibacterial capacity of RNASE7103Ala. Prior reports suggest that LPS binding is required for RNase 7’s antibacterial activity (8).
To confirm this phenotype in vitro, human UROtsa cells were stably infected with retroviral constructs expressing these RNASE7 alleles or empty vector. UROtsa cells have minimal endogenous RNase 7 expression (7). Western blot confirmed cellular RNase 7 overexpression and ELISA quantified secretion into the media (Figure 2, C and D). To evaluate extracellular UPEC bactericidal activity, we collected 3-day-old culture media from confluent RNase 7–expressing cells and control cells. Isolated media was inoculated with UPEC pyelonephritis strain 536, cystitis strains MDR12 and UTI89, and the avirulent mutant UTI89ΔfimH. After a 90-minute incubation, survival of all tested UPEC strains decreased in media isolated from wild-type RNASE7103Pro-expressing cells compared with that in mutant RNASE7103Ala-expressing cells (Figure 2E). To assess if cellular RNase 7 shields the urothelium from bacterial challenge, confluent UROtsa cells were challenged with these same UPEC strains. The percentage of UPEC adhering to and invading wild-type RNASE7103Pro-overexpressing cells was significantly reduced compared with mutant RNASE7103Ala (Figure 2, F and G). UPEC MDR12, which demonstrated little functional type 1 pili activity by hemagglutination, and the attenuated UTI89ΔfimH showed reduced urothelial binding and minimal urothelial invasion. However, UPEC attachment ratios were comparable to those of UTI89 — suggesting that RNase 7 does not interfere with type 1 pilus-meditated bacterial adhesion (Figure 2H). These results suggest that extracellular and cellular bacterial killing is the major upstream antimicrobial mechanism of RNase 7. Additionally, they further demonstrate that wild-type RNASE7103Pro has enhanced antimicrobial activity against multiple UPEC strains compared with its variant, RNASE7103Ala — suggesting that the RNASE7 rs1263872 minor allele can effect UPEC susceptibility.
The role of host defense peptides, such as RNase 7, in UTI prevention and UPEC clearance is a relatively new concept. Our research group and others have shown that bacteriostatic iron-regulatory peptides, such as hepcidin and lipocalin 2, or bactericidal AMPs, such as cathelicidin, defensins, and ribonucleases, prevent UTI (3, 4). To our knowledge, this is the first report describing a SNP in an AMP gene that may affect UTI susceptibility. Previously, we demonstrated that α-defensin copy number variations in the DEFA1A3 gene may affect UTI risk. In a subset of children in the RIVUR study, we found low DEFA1A3 copy numbers associated with recurrent UTI — indicating that AMP copy number polymorphisms may increase UTI severity (22).
Data in this study suggest that a SNP altering RNase 7’s primary structure can increase UPEC susceptibility. The data generated here, in conjunction with our previously published findings (6, 7), suggest that RNase 7 has a key role in preventing UTI. In addition to its antimicrobial activity against UPEC and MDR UPEC, our data show that overexpressing RNase 7 in vitro reduces the ability of UPEC to attach to or invade urothelial cells. When we subjected transgenic, humanized RNase 7–expressing mice to experimental UTI, RNase 7’s in vivo antibacterial activity promoted UPEC clearance (7). Conversely, silencing urothelial RNase 7 expression in vitro increased invasive UPEC infection and blocking urinary RNase 7 antibacterial activity increased UPEC growth in human urine (6). In addition, our published data demonstrate that female participants with recurrent UTI have lower urinary RNase 7 concentrations compared with controls, and urinary RNase 7 concentrations negatively correlate with the number of UTI (7). Together, these data provide evidence that altering RNase 7’s structure, expression, or antimicrobial activity affects UTI susceptibility.
We acknowledge that this study has limitations. The RIVUR and CUTIE studies primarily comprised non-Hispanic, White female participants of European ancestry (12, 13). Thus, our results must be cautiously interpreted in children of different sex, race, or ethnicity. While the use of recombinant RNase 7 peptides and in vitro tissue culture experiments suggest the RNase 7 proline-to-alanine substitution negatively affects RNase 7’s bactericidal activity, these results may not translate in vivo. To fully define the use of RNase 7 as a UTI prognostic, a large, prospective analysis in children with different UTI risk phenotypes and racial/sex profiles is needed to determine if children with RNASE7 genetic variations have an increased propensity to develop recurrent UTI, more severe UTI, or atypical infections. These prospective studies may identify at-risk children with abnormal RNase 7 profiles, who benefit from close UTI surveillance, antibiotic prophylaxis, or new approaches to boost endogenous UTI defenses (7).
To conclude, a growing body of evidence suggests that RNase 7 has an important role in UTI prevention. Findings in this study show that the RNASE7 SNP rs1263872, which encodes an RNase 7 peptide with reduced antibacterial activity, is more common in females participants with UTI. These data provide a foundation that may ultimately help clinicians make more informed decisions regarding UTI treatment or prevention in children at risk for UTI complications.
Methods
See Supplemental Methods for details on study cohorts and study methods.
Study approval.
Research in human participants was performed in accordance with the principles of the World Medical Association’s Declaration of Helsinki and approved by the University of Tennessee Health Science Center IRB (protocol 14-03325-XP) and Nationwide Children’s IRB (protocol 07-00383).
Author contributions
DSH and JDS supervised the project and contributed to the design and interpretation of all experiments. KP and SC performed the human genotyping experiments and analysis. AS completed the UPEC genotyping. TE performed the PINCO and hemagglutination experiments. JDS and CMV generated the RNase 7 recombinant peptides and performed the UPEC bactericidal assays, respectively. KP, TE, ALS, DSH, and JDS wrote the manuscript with input from all coauthors.
Supplementary Material
Acknowledgments
JDS, ALS, and DSH are supported by the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases grants R01DK115737, R01DK106286, and R01DK117934.
Version 1. 11/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(22):e149807.https://doi.org/10.1172/JCI149807.
Contributor Information
Tad Eichler, Email: tad.eichler@nationwidechildrens.org.
Aaron Simoni, Email: aaron.simoni@nationwidechildrens.org.
Steven Creacy, Email: screacy@transnetyx.com.
David S. Hains, Email: dhains@iu.edu.
References
- 1.Becknell B, et al. The diagnosis, evaluation and treatment of acute and recurrent pediatric urinary tract infections. Expert Rev Anti Infect Ther. 2015;13(1):81–90. doi: 10.1586/14787210.2015.986097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korbel L, et al. The clinical diagnosis and management of urinary tract infections in children and adolescents. Paediatr Int Child Health. 2017;37(4):273–279. doi: 10.1080/20469047.2017.1382046. [DOI] [PubMed] [Google Scholar]
- 3.Becknell B, et al. Amplifying renal immunity: the role of antimicrobial peptides in pyelonephritis. Nat Rev Nephrol. 2015;11(11):642–655. doi: 10.1038/nrneph.2015.105. [DOI] [PubMed] [Google Scholar]
- 4.Zasloff M. Antimicrobial peptides, innate immunity, and the normally sterile urinary tract. J Am Soc Nephrol. 2007;18(11):2810–2816. doi: 10.1681/ASN.2007050611. [DOI] [PubMed] [Google Scholar]
- 5.Spencer JD, et al. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int. 2013;83(4):615–625. doi: 10.1038/ki.2012.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spencer JD, et al. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. 2011;80(2):174–180. doi: 10.1038/ki.2011.109. [DOI] [PubMed] [Google Scholar]
- 7.Eichler T, et al. Ribonuclease 7 shields the kidney and bladder from invasive uropathogenic Escherichia coli infection. J Am Soc Nephrol. 2019;30(8):1385–1397. doi: 10.1681/ASN.2018090929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torrent M, et al. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 2010;277(7):1713–1725. doi: 10.1111/j.1742-4658.2010.07595.x. [DOI] [PubMed] [Google Scholar]
- 9.Koten B, et al. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS One. 2009;4(7):e6424. doi: 10.1371/journal.pone.0006424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becknell B, Spencer JD. A review of ribonuclease 7’s structure, regulation, and contributions to host defense. Int J Mol Sci. 2016;17(3):E423. doi: 10.3390/ijms17030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoberman A, et al. Antimicrobial prophylaxis for children with vesicoureteral reflux. N Engl J Med. 2014;370(25):2367–2376. doi: 10.1056/NEJMoa1401811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keren R, et al. Risk factors for recurrent urinary tract infection and renal scarring. Pediatrics. 2015;136(1):e13–e21. doi: 10.1542/peds.2015-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181(1):261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 15.Moreno E, et al. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol. 2008;46(8):2529–2534. doi: 10.1128/JCM.00813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, et al. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J Clin Microbiol. 2002;40(11):3951–3955. doi: 10.1128/JCM.40.11.3951-3955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiles TJ, et al. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85(1):11–19. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brzuszkiewicz E, et al. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci U S A. 2006;103(34):12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd AL, et al. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol. 2007;189(9):3532–3546. doi: 10.1128/JB.01744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99(26):17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hultgren SJ, et al. Role of type 1 pili and effects of phase variation on lower urinary tract infections produced by Escherichia coli. Infect Immun. 1985;50(2):370–377. doi: 10.1128/iai.50.2.370-377.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwaderer AL, et al. Polymorphisms in α-defensin-encoding DEFA1A3 associate with urinary tract infection risk in children with vesicoureteral reflux. J Am Soc Nephrol. 2016;27(10):3175–3186. doi: 10.1681/ASN.2015060700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.