Abstract
Metabolic pathways regulate immune responses and disrupted metabolism leads to immune dysfunction and disease. Coronavirus disease 2019 (COVID-19) is driven by imbalanced immune responses, yet the role of immunometabolism in COVID-19 pathogenesis remains unclear. By investigating 87 patients with confirmed SARS-CoV-2 infection, 6 critically ill non–COVID-19 patients, and 47 uninfected controls, we found an immunometabolic dysregulation in patients with progressed COVID-19. Specifically, T cells, monocytes, and granulocytes exhibited increased mitochondrial mass, yet only T cells accumulated intracellular reactive oxygen species (ROS), were metabolically quiescent, and showed a disrupted mitochondrial architecture. During recovery, T cell ROS decreased to match the uninfected controls. Transcriptionally, T cells from severe/critical COVID-19 patients showed an induction of ROS-responsive genes as well as genes related to mitochondrial function and the basigin network. Basigin (CD147) ligands cyclophilin A and the SARS-CoV-2 spike protein triggered ROS production in T cells in vitro. In line with this, only PCR-positive patients showed increased ROS levels. Dexamethasone treatment resulted in a downregulation of ROS in vitro and T cells from dexamethasone-treated patients exhibited low ROS and basigin levels. This was reflected by changes in the transcriptional landscape. Our findings provide evidence of an immunometabolic dysregulation in COVID-19 that can be mitigated by dexamethasone treatment.
Keywords: Inflammation, Metabolism
Keywords: Mitochondria, Monocytes, T cells
Introduction
In February 2020, Zhu and colleagues described a novel coronavirus named 2019-nCoV detected in patients with pneumonia of unknown cause in Wuhan, China (1, 2). Infection with the severe acute respiratory syndrome coronavirus (SARS-CoV-2) causes COVID-19, an illness of varying degrees. The vast majority of COVID-19 cases present with a mild or moderately symptomatic infection. However, a subset of individuals progress to develop severe disease and there is increasing evidence that critical cases are driven by dysregulated host immune responses to SARS-CoV-2 infection, allowing virus persistence and causing lung damage and systemic inflammation (3–6). Infectious diseases change metabolic processes, e.g., in infected cells to support viral replication or in immune cells fighting the infection. Codo and colleagues reported that SARS-CoV-2–infected monocytes upregulate glycolysis and elevated glucose levels support viral replication and proinflammatory cytokine expression (7). These data suggest a link between cellular metabolism and systemic host metabolism for disease progression. In line with this, patients with type 2 diabetes have a greater risk of developing severe disease (8). Obesity, together with age, is the major risk factor for diabetes, hyperglycemia, and dyslipidemia that can result in an imbalance in T cell subpopulations (9). Interestingly, systemic metabolic changes were observed in COVID-19 patients (10) and metabolic treatment with cholesterol-lowering drugs such as statins can reduce COVID-19 mortality (11).
T cells are central players in adaptive immunity and are essential for controlling viral infections, including SARS-CoV-2 (12). To initiate an immune response, pathogen-specific T cells need to activate and to expand. This is accompanied by an extensive metabolic reprogramming, and activated T cells increase expression of nutrient transporters such as glucose transporter GLUT1 and enzymes involved in glycolysis (13). Furthermore, effector T cells contain higher numbers of mitochondria than naive cells (14). Mitochondria produce ATP via oxidative phosphorylation (OXPHOS) and are important for cellular processes such as apoptosis (14) and antiviral defenses (15). Besides nutrients, OXPHOS requires oxygen, which is often limited in patients with COVID-19 (16). Extraordinarily low blood-oxygen levels are often found in infected patients and thus immune cells experience hypoxia and may induce hypoxia-inducible factors (HIFs) such as HIF-1α, a transcriptional effector of the hypoxic response. Some studies showed that HIF-1α functions as a negative regulator of T cell responses and that O2 is required for T cell effector functions (17). To preserve energy production and effector functions, T cells experiencing low O2 and glucose levels can enhance fatty acid catabolism as a metabolic escape mechanism (18, 19).
Reduced oxygen saturation and hypoxia lead to generation and accumulation of reactive oxygen species (ROS) by mitochondria (20). Disrupted mitochondrial metabolism and oxidative stress can drive heart disease (21) and immune dysfunction in HIV patients (22). Furthermore, ROS play an important role in the pathogenesis of viral infections and sepsis, and it has been suggested that COVID-19 pathogenesis is also related to hypoxia and oxidative stress (2, 23). As mitochondrial superoxide leads to aberrant T cell development, Case et al. suggested that manipulation of mitochondrial superoxide levels may significantly alter clinical outcomes of patients with viral infections (24). Recent data suggested that immunometabolic dysregulation in COVID-19 might affect mitochondria and might lead to apoptosis (7, 25, 26). However, it remains unclear how these changes are mediated, whether they are pharmacologically targetable, and which surrogate parameters can be used to detect immunometabolic dysfunction in COVID-19.
Nevertheless, treatment of COVID-19 patients with antioxidants has already been proposed (27). An alternative therapeutic target is CD147, also called basigin or extracellular matrix metalloproteinase inducer (EMMPRIN). Basigin is a transmembrane glycoprotein that interacts with several extracellular and intracellular partners. Basigin is also shed from the membrane (28, 29) and soluble basigin is a marker of inflammation (30). Moreover, basigin can mediate the entry of various viruses (31–33), including SARS-CoV and SARS-CoV-2 (34, 35). Intriguingly, viral proteins often do not directly interact with basigin but bind to cyclophilins (CyPs), which are basigin ligands (31, 34). Cyclophilin A (CyPA) plays a critical role in viral replication and CyP inhibitors such as cyclosporine A (CsA) or alisporivir can block viral replication (31, 36, 37). Given the roles of basigin and CyPs in viral diseases, clinical trials blocking basigin or CyPs are underway for treating hospitalized COVID-19 patients (NCT04275245, NCT04586153 [a phase II/III trial], and CYCLOVID trial NCT04608214). Interestingly, the basigin ligand CyPA has been described as the most relevant marker for COVID-19 (38). Apart from its role in virus entry, basigin tightly associates with GLUT1, the amino acid transporter CD98, the lactate transporters (monocarboxylate transporters; MCT1, -3, and -4), and CD44, and is recognized by lectins such as galectin-3 and E-selectin (39). These molecular interactions explain the central role of basigin in energy metabolism, cell motility, and activation.
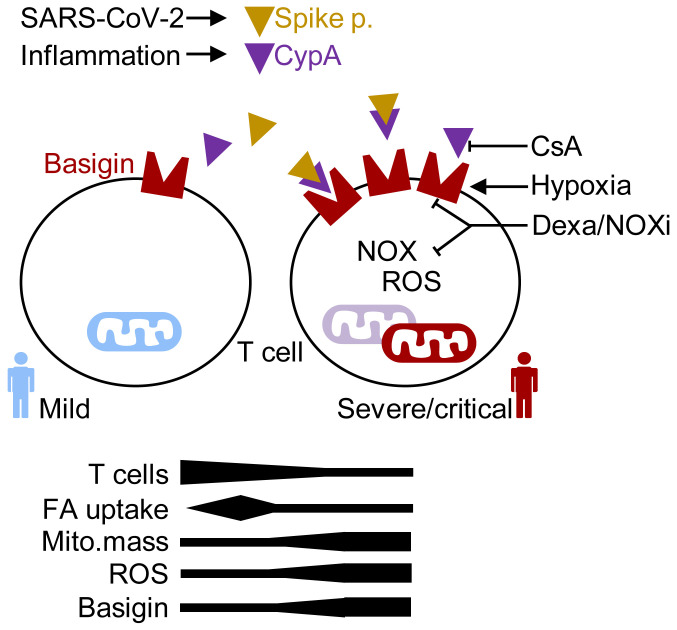
Metabolic features of immune cells strongly determine the outcomes of an immune response and a variety of publications reported dysregulated innate and adaptive immune pathways in COVID-19 patients. Up to now, immune cell metabolism was mainly analyzed in cryopreserved samples, which might blunt the metabolic phenotype. Therefore, we studied immune cell metabolism in freshly isolated blood samples of COVID-19 patient subgroups with different degrees of disease severity with or without preexisting metabolic comorbidities. Our data show T cell metabolic changes in mitochondrial mass, architecture, and ROS production as well as in fatty acid uptake in the course of disease that are (a) not related to classical activation markers, (b) stage specific, (c) basigin related, and (d) reversed in recovering patients to reach levels of healthy controls and non–COVID-19 patients. Finally, basigin ligation induced ROS, and dexamethasone treatment mitigated ROS accumulation and basigin expression. Our data show that immunometabolic dysregulation contributes to the dysfunctional T cell response in COVID-19 patients.
Results
ROS accumulation in T cells from progressed COVID-19 patients is reversible and correlates with changes in mitochondrial mass and architecture.
COVID-19 patients with severe disease experience immune dysregulation (40) and hypoxia (2). Hypoxia and other factors might induce ROS generation to drive the immunopathology of COVID-19. We therefore analyzed intracellular ROS levels in peripheral blood immune cell populations (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI148225DS1) from age-matched, noninfected controls (see Methods, Study participants), asymptomatic and mildly symptomatic SARS-CoV-2–positive subjects (no symptoms/mild group; Supplemental Tables 1 and 2), moderately ill (moderate; Supplemental Tables 1 and 2), severely to critically ill patients (severe/critical group; Supplemental Table 1), patients recovering from a severe/critical COVID-19 (recovering group; Supplemental Table 1), and non–COVID-19 patients requiring intensive care (ICU non-COVID; Supplemental Table 3). To accurately depict the in vivo metabolic state, samples were processed freshly without cryopreservation or density gradient centrifugation.
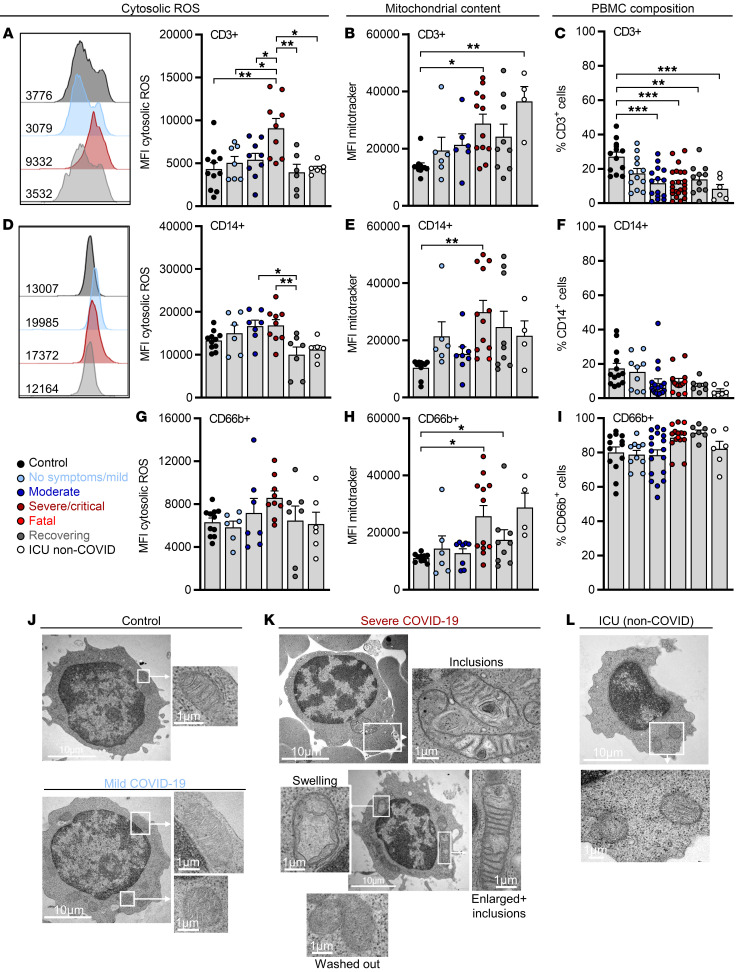
Intracellular ROS accumulation was observed in T cells from COVID-19 patients, specifically in those with severe/critical disease. In contrast, T cell ROS levels were low in patients recovering from a critical COVID-19 and matched the uninfected controls and critically ill non–COVID-19 patients (Figure 1A). Changes in redox balance can accompany mitochondrial metabolic reprogramming (20) and we observed an increase in mitochondrial mass in T cells from critically ill patients (Figure 1B). Increased mitochondrial mass correlated with low T cell frequencies, which remained low in recovering patients (Figure 1C and Supplemental Figure 1B). Moreover, in studying the frequencies of CD4+ and CD8+ T cells, we observed no changes specific to the severe/critical COVID-19 group, suggesting that both subsets are affected. Nevertheless, increased CD8+ T cell frequencies were observed in patients with moderate disease (Supplemental Figure 1C). Metabolic defects might have been induced selectively in SARS-CoV-2–specific T cells. The frequencies of SARS-CoV-2–specific T cells are reported to be approximately 0.1% of all T cells (41). In line with this, we used SARS-CoV-2–derived M, N, and S peptides and detected an ELISpot response translating to approximately 0.01% cells (thus approximately 0.1% T cells) being SARS-CoV-2 specific within peripheral blood mononuclear cells (PBMCs) (Supplemental Figure 1D). A general shift in mitochondrial content and ROS levels, affecting all analyzed cells in the studied samples, argues against a limitation to SARS-CoV-2–specific T cells.
Figure 1. ROS accumulation in T cells from progressed COVID-19 patients is reversible and correlates with changes in mitochondrial mass and architecture.
Blood was drawn and processed the same day. Erythrocytes were removed and immune cells were stained for population-specific surface markers. The results show comparisons between specified COVID-19 patient subgroups, critically ill non–COVID-19 patients (ICU non-COVID), and healthy controls. (A) ROS levels, (B) mitochondrial content in CD3+ T cells, and (C) percentage of CD3+ cells among viable cells. MFI, median fluorescence intensity. (D) Cytosolic ROS levels, (E) mitochondrial content in CD14+ monocytes, and (F) proportion of CD14+ monocytes among the CD11b+ myeloid population. (G) Cytosolic ROS levels, (H) mitochondrial content in CD66b+ granulocytes, and (I) proportion of CD66b+ granulocytes among CD11b+ myeloid cells. Shown are representative histograms of ROS staining in CD3+ and CD14+ cells of each group. Shown is the median + SEM, and each symbol represents 1 donor. *P < 0.05; **P < 0.01; ***P < 0.001 by 1-way ANOVA with Bonferroni’s multiple comparisons test. Analysis of mitochondrial structure by electron microscopy in lymphocytes from healthy control cells, a COVID-19 patient with mild symptoms (J), of 2 COVID-19 patients with severe symptoms (K), and a critically ill non–COVID-19 patient (L). Scale bars: 10 μm. Shown are representative examples.
While high basal ROS levels were observed in CD14+ monocytes, no significant increase in the severe/critical group was observed. Nevertheless, low monocyte ROS levels were present in recovering patients and critically ill non–COVID-19 patients (Figure 1D). Mitochondrial mass was increased in CD14+ monocytes from critically ill COVID-19 patients, similar to T cells (Figure 1E). However, consistent with the high ROS tolerance of monocytes (42), monocyte and monocyte subpopulation frequencies were not significantly impaired in blood of COVID-19 patients (Figure 1F and Supplemental Figure 1E). Similarly, increased mitochondrial mass, but no ROS accumulation or changes in blood frequencies of CD66b+ granulocytes, was observed in COVID-19 patients in the severe/critical group (Figure 1, G–I).
To correlate the observed metabolic phenomena with mitochondrial morphology, electron microscopy of PBMCs from critically ill COVID-19 patients was performed. In contrast to controls and cells from COVID-19 patients with mild disease (Figure 1J), the majority of lymphocytes from severe COVID-19 patients displayed alterations in the mitochondrial structure. We detected mitochondria with membrane inclusions, mitochondrial swelling (dilated cristae), or mitochondria with almost invisible (washed out) cristae, indicating the accumulation of damaged mitochondria. In addition, enlarged mitochondria were observed (Figure 1K). No mitochondrial defects were detected in cells from critically ill non–COVID-19 patients (Figure 1L). Moreover, the majority of monocytes showed no signs of structural aberrations (Supplemental Figure 1F).
Lastly, while patients with preexisting diabetes and/or obesity showed a further decrease in T cell counts, neither ROS accumulation nor mitochondrial content of T cells was related to preexisting comorbidities (Supplemental Figure 1, G and H).
Collectively, T cells from patients with severe COVID-19 accumulated ROS and showed an increased mitochondrial mass but low peripheral blood counts. These immunometabolic features were reversible and correlated with a disrupted mitochondrial morphology.
ROS-inducible and basigin-related gene expression in T cells from patients with severe COVID-19 suggests basigin as a marker of T cell metabolic dysfunction.
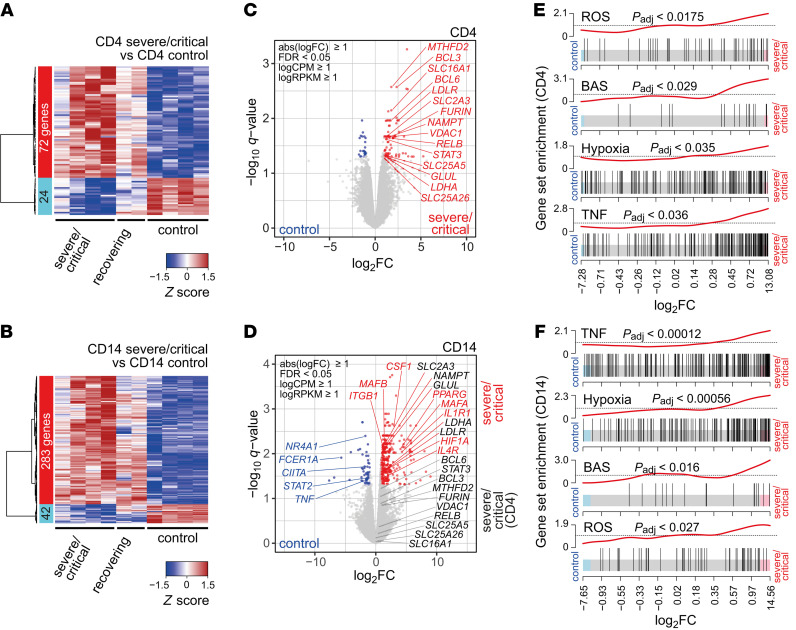
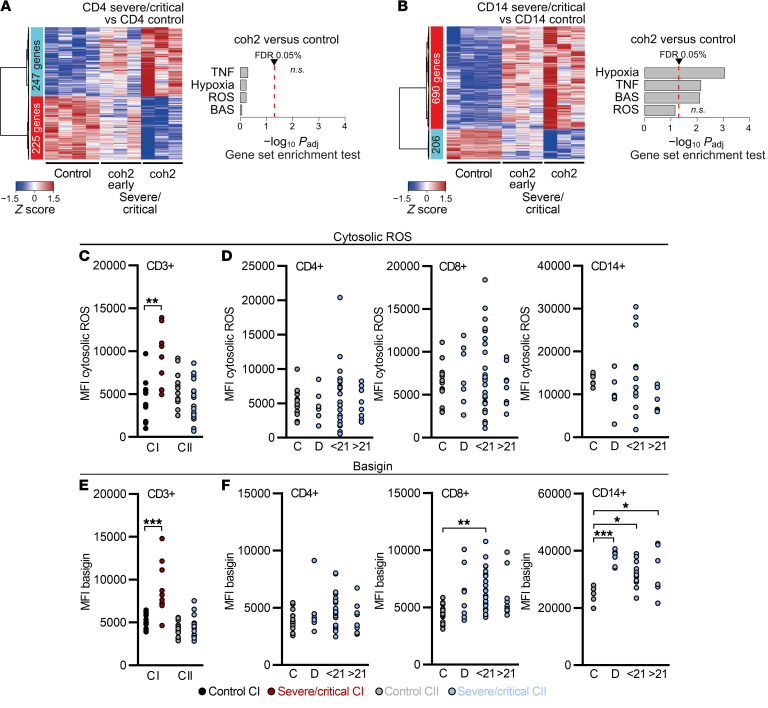
To determine whether the observed dysregulation in COVID-19 associates with transcriptional changes, we performed RNA sequencing (RNA-seq) of T cells and monocytes from COVID-19 patients (4 severe/critical patients; 2 recovering patients) and healthy controls (4 individuals). CD4+ T cells from COVID-19 patients from the severe/critical group showed significantly increased expression of 72 genes, while 24 genes were downregulated (Figure 2A). CD14+ monocytes showed a COVID-19–specific upregulation of 283 genes and downregulation of 42 genes (Figure 2B). In both immune cell populations, metabolism-related genes were upregulated; however, the top hits specific to severe COVID-19 (later referred to as hallmarks) varied between monocytes and T cells. Whereas CD4+ T cells and monocytes shared a COVID-19–specific upregulation of nicotinamide phosphoribosyltransferase (NAMPT), which protects against cell death induced by mitochondrial damage (43), other mitochondria-related proteins, such as voltage-dependent anion-selective channel protein 1 (VDAC1), a key regulator of mitochondrial metabolism and cellular apoptosis (26, 44), or methylenetetrahydrofolate dehydrogenase/cyclohydrolase (MTHFD2), involved in redox homeostasis, were strongly increased in T cells only (Figure 2, C and D). The lactate-transporting MCT1 (also known as SLC16A1) was upregulated in CD4+ T cells, whereas lactate dehydrogenase A (LDHA) expression was elevated in monocytes. Interestingly, CD4+ T cells from COVID-19 patients also showed an increased expression of furin (FURIN), which was reported to activate SARS-CoV-2 (ref. 45 and Figure 2C).
Figure 2. ROS-inducible and basigin-related gene expression in immune cells from patients with severe COVID-19.
Blood samples from COVID-19 patients (4 ICU, 2 recovering) and 4 healthy controls were enriched for PBMCs and cryopreserved. Cells were sorted according to specific surface markers, and RNA was extracted and subjected to stranded total RNA sequencing. Heatmaps presenting hierarchically clustered and scaled expression data of CD4+ T cells (A) or CD14+ monocytes (B). Shown are differentially expressed genes in severe/critical patients versus controls filtered for absolute logFC > 1, logCPM and logRPKM > 1, and FDR < 0.05. Each column corresponds to an individual. The list and expression values of all regulated genes are in the supplemental material as well as a list of all read counts. Volcano plots of significantly up- or downregulated genes (red or blue) in CD4+ T cells (C) or CD14+ monocytes (D) of severe/critical patients with additional genes upregulated in CD4+ T cells but not in CD14+ monocytes (black). Genes with known function in transcriptional or metabolic processes are highlighted. Gene set enrichment analysis of candidate HALLMARK pathways in CD4+ T cells (E) or CD14+ monocytes (F). HALLMARK pathways include REACTIVE_OXYGEN_SPECIES_PATHWAY (ROS), TNFA_SIGNALING_VIA_NFKB (TNF), HYPOXIA, and basigin-interacting proteins (BAS), defined by STRING analysis (see Supplemental Figure 2). Enrichment P values were adjusted for multiple testing using Benjamini-Hochberg correction.
Although the overlap on the single-gene level was limited, a gene set enrichment approach (46) identified pathways mutually regulated by both immune cell populations. CD4+ T cells and monocytes showed a significant enrichment of genes related to TNF signaling and, importantly, of genes associated with cellular responses to ROS-induced stress and hypoxia (Figure 2, E and F). In addition, we observed a signature of genes encoding proteins from the basigin network (Figure 2, E and F, and Supplemental Figure 2).
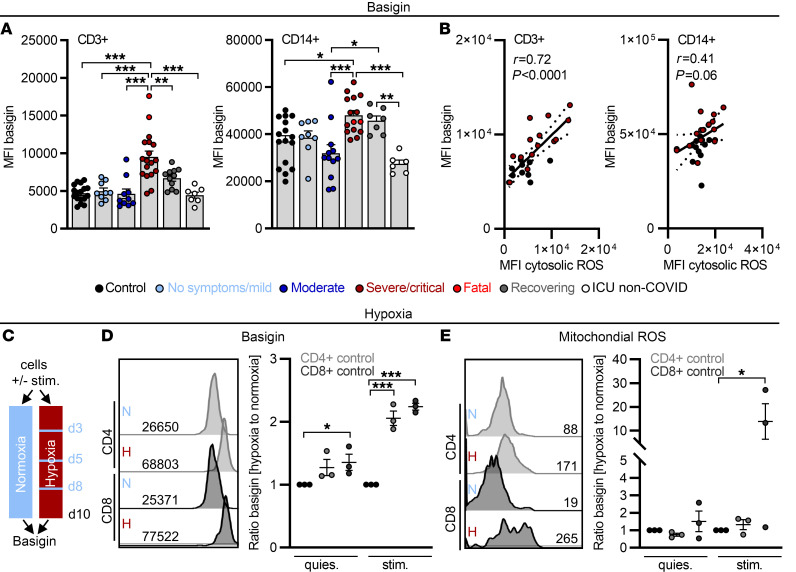
Confirming the data obtained on the transcriptional level, T cell and monocyte basigin surface expression increased with disease severity (Figure 3A). In line with the observed T cell metabolic recovery in convalescent patients, basigin expression decreased during convalescence and was low in critically ill non–COVID-19 patients (Figure 3A and Supplemental Figure 3A). Elevated basigin expression was detected in both T cell subpopulations, but not in granulocytes (Supplemental Figure 3B). Transcriptome analyses suggested basigin as a marker of T cell immunometabolic dysregulation. In line with this, accumulation of cytosolic ROS positively correlated with T cell basigin levels, indicating a biological interdependence (Figure 3B). Monocytes showed high basal basigin expression but its correlation with ROS levels was weak (Figure 3B).
Figure 3. Elevated basigin expression correlates with cytosolic ROS in severe/critical COVID-19 patients and can be triggered by hypoxia.
(A–E) Blood was drawn and processed the same day. (A) Basigin expression determined on CD3+ cells and CD14+ cells. MFI, median fluorescence intensity. (B) Correlation of ROS and basigin in CD3+ and CD14+ cells, calculated by Spearman’s r, with single data points for controls in black and severe/critical patients in red. (C) Control samples were cultured with (stim.) or without (quies.) anti-CD3/anti-CD28 Dynabeads in normoxia or hypoxia for 10 days interrupted by 1-hour periods of normoxia on days 3, 5, and 8. On day 10, basigin expression (D) and mitochondrial ROS (E) were analyzed. Values normalized to corresponding treatment under normoxia are displayed. Each symbol represents 1 donor. In A, D, and E, summarized data are displayed as mean + SEM. *P < 0.05; **P < 0.01; ***P < 0.001 by 1-way ANOVA with Bonferroni’s multiple comparisons test. Representative histograms are shown in D and E.
Severely ill COVID-19 patients experience recurrent hypoxic phases (2) and hypoxia has been previously shown to induce basigin expression on tumor cells (47). We therefore cultured unstimulated and stimulated control donor T cells in hypoxia, interrupted by short normoxic phases (Figure 3C). Hypoxia induced T cell basigin expression and this effect was boosted by T cell receptor stimulation (Figure 3D). Lastly, basigin induction was accompanied by production of mitochondrial ROS in T cells cultured in recurrent hypoxia, suggestive of mitochondrial defect induction (Figure 3E).
Taken together, our results show that T cells and monocytes from severely ill COVID-19 patients increase transcription of ROS-inducible and hypoxia-related genes as well as genes of the basigin network. Moreover, both immune cell populations show high expression of basigin. Basigin is induced by recurrent hypoxia and might serve as a cell-surface marker of T cell immunometabolic dysregulation in COVID-19.
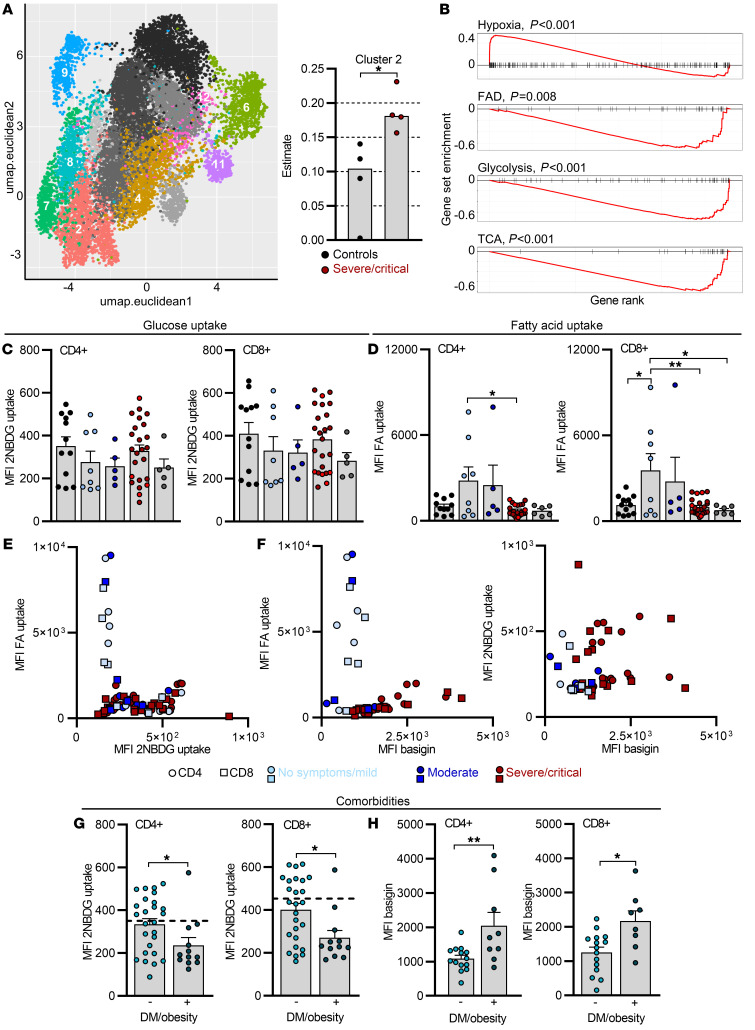
Increased fatty acid uptake is limited to T cells from mild COVID-19 and correlates with low basigin expression.
Increasing evidence points to basigin as a regulator of cellular metabolism, as it tightly associates with transporters of amino acids and glucose, likely coordinating glucose and amino acid metabolism (39). In addition, basigin signaling can inhibit fatty acid metabolism via modulation of PPARα in tumor cells (48). We performed digital tissue deconvolution (DTD) to dissect the heterogeneity and metabolic pathways of T cells from COVID-19 patients using publicly available single-cell transcriptomic data (49). Leiden clustering was used to identify 14 unique clusters, 8 of which could be found using DTD in the CD4+ T cell bulk sequencing data. Cluster 2, but no other clusters, was enriched in T cells from severely ill COVID-19 patients (Figure 4A and Supplemental Figure 4A) and was characterized by expression of hypoxia-related genes. Interestingly, this cluster was also characterized by low expression of genes involved in fatty acid metabolism, glycolysis, and the TCA cycle (Figure 4B).
Figure 4. Increased fatty acid uptake is limited to T cells from mild COVID-19 and correlates with low basigin expression.
(A) Leiden clustering was used to identify 14 unique clusters in publicly available single-cell RNA-seq data for CD4+ COVID-19 PBMCs, and 8 were detected in CD4+ T cell bulk sequencing data (Figure 2, A, C, and E) using digital tissue deconvolution. These are shown in colors in the UMAP embedding, while clusters estimated as zero are in gray scale. For better clarity, only 10% of the used data are shown. Cluster 2 estimates in CD4+ T cells from controls and severe/critical COVID-19 patients are shown on the right. (B) Gene set enrichment analysis for selected gene sets. The horizontal axis shows the rank of each gene in cluster 2 compared to all others. Vertical marks show the position of a gene from the gene set in the ranking, and the red lines show either over (positive values) or under representation (negative values). FAD, fatty acid degradation. (C–H) Blood was drawn and processed the same day. Determination of 2NBDG uptake (C) and BODIPY 500/510 C1, C12 uptake (D) in CD4+ and CD8+ T cells, and correlation of both from corresponding measurements (E). FA, fatty acid; MFI, median fluorescence intensity. (F) Fatty acid uptake and 2NBDG uptake in CD4+ and CD8+ T cells plotted against basigin expression from the same sample. Comparison of 2NBDG uptake (G) and basigin expression (H) in CD4+ and CD8+ T cells in diabetic (DM)/obese (+) versus nondiabetic/obese (–) COVID-19 patients. Each symbol represents 1 donor. In C, D, G, and H, summarized data are displayed as mean + SEM. *P < 0.05; **P < 0.01 by 1-way ANOVA with Bonferroni’s multiple comparisons test (C and D) or Mann-Whitney U test (G and H).
Functional metabolic studies were performed next and glucose, fatty acid uptake, and the expression of the amino acid transporter CD98 of T cells from COVID-19 patients was assessed in fresh patient samples. T cell expression of CD98 was similar among the patient groups (Supplemental Figure 4B). DTD suggested low glucose metabolism of COVID-19 T cells and in line with this, glucose uptake of CD4+ and CD8+ T cells was low and comparable to quiescent T cells from uninfected controls (Figure 4C). Consistent with a metabolically quiescent phenotype, the expression of markers of activation and differentiation such as CD25 or PD-1 was similar in T cells from COVID-19 patients and controls (Supplemental Figure 4, C and D). Furthermore, no differences were detected in the expression of the chemokine receptor CCR7 on bulk CD4+ and CD8+ T cells (Supplemental Figure 4E) and no significant accumulation of CD4+FOXP3+ Treg cells was observed (Supplemental Figure 4F).
In line with the transcriptional data, we measured low fatty acid uptake in T cells from severely ill COVID-19 patients, but interestingly, an increased fatty acid uptake was observed in CD4+ and CD8+ T cells from patients with no or mild symptoms (Figure 4D). Suggestive of a metabolic switch, high fatty acid uptake was specific to T cells with low glucose uptake and vice versa (Figure 4E). We next assessed the dynamics of this metabolic phenotype after complete disease resolution. Fatty acid uptake of CD4+ and CD8+ T cells as well as of their respective subsets was analyzed in 3 COVID-19 patients from 1 family with mild symptoms over the course of 7 weeks after the onset of symptoms (Supplemental Figure 4G). Interestingly, the capacity of T cells to take up fatty acids decreased over time. Notably, this phenomenon occurred 4 weeks after the onset of symptoms and was observed in bulk CD8+ and CD4+ T cells as well as in major T cell subpopulations (Supplemental Figure 4H). Focusing on the time point with high fatty acid uptake at week 2 (w2), we did not observe any changes in cytosolic ROS accumulation (Supplemental Figure 4I). However, w2 T cells from mild COVID-19 patients showed an increased mitochondrial mass, most prominently in the effector subtypes (Supplemental Figure 4J) and pooled analyses showed a positive correlation between mitochondrial mass and fatty acid uptake (Supplemental Figure 4K). Finally, T cells with high fatty acid uptake were low in basigin expression and vice versa, and no increased glucose uptake was observed in T cells with high basigin expression (Figure 4F).
As T cell glucose uptake was variable, we asked whether preexisting patient comorbidities contribute. Interestingly, diabetic and obese patients showed decreased glucose uptake by T cells compared with controls, but increased basigin expression (Figure 4, G and H). In contrast, separation of patients according to cardiovascular disease or age did not show significant differences in immune cell glucose or fatty acid uptake (Supplemental Figure 4, L and M).
Taken together, in contrast to basigin upregulation, our results show that the nutrient uptake capacity of T cells from severely ill COVID-19 patients was low, especially in patients with metabolic comorbidities. In addition, increased T cell fatty acid uptake was a hallmark of COVID-19 patients with no or mild symptoms and decreased during convalescence.
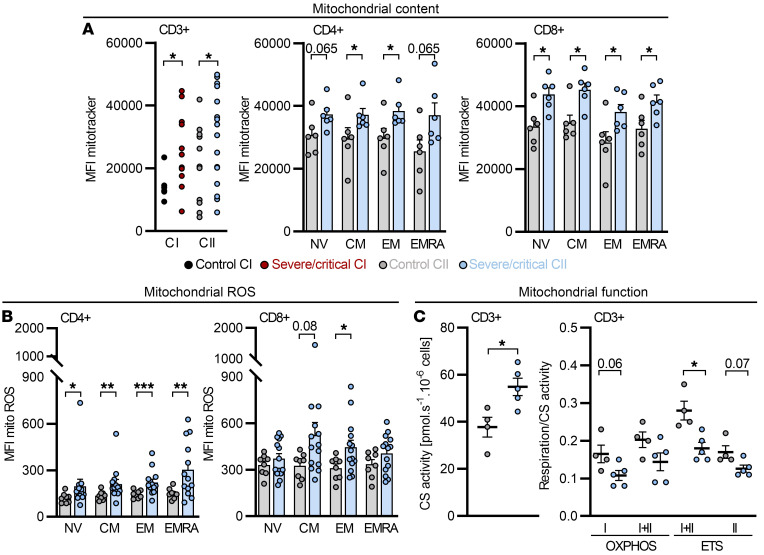
Dexamethasone-treated critically ill COVID-19 patients show altered mitochondrial function, but normalized ROS and basigin levels in CD4+ T cells.
Severely/critically ill COVID-19 patients showed disturbed T cell mitochondrial morphology, which might have been related to an impaired fatty acid metabolism. Thus, we investigated T cell mitochondrial respiratory function in a second cohort of COVID-19 patients (CII; Supplemental Table 2). All patients in the severe/critical group of cohort II received an early dexamethasone treatment, analogous to the RECOVERY trial (50). Consistent with previous observations, severe COVID-19 associated with a notable T cell lymphopenia (Supplemental Figure 5A), affecting all T cell subpopulations (Supplemental Figure 5B). Comparative transcriptome analyses of cohort I and II samples from the severe/critical group and healthy controls by t-distributed stochastic neighbor embedding (tSNE) and principal component analysis (PCA) showed clustering of CD14+ monocytes and CD4+ T cells of both cohorts (Supplemental Figure 5C). Similar to cohort I, an increased mitochondrial mass but also elevated mitochondrial ROS production of both bulk T cells and naive and memory T cell subsets of severe/critically ill patients was detectable in cohort II (Figure 5, A and B). Importantly, mitochondrial ROS levels were most likely not a consequence of increased mitochondrial mass, as these 2 parameters did not correlate (Supplemental Figure 5D).
Figure 5. Increased mitochondrial content, mitochondrial ROS, and disturbed mitochondrial function in T cells from COVID-19 patients cannot be ameliorated by dexamethasone.
A second cohort (CII) of severe/critical COVID-19 patients receiving dexamethasone was analyzed. (A–C) Blood was drawn and processed the same day. (A) Mitochondrial content in T cells from controls and severe/critical patients of CII was analyzed beyond 19 days after intensive care unit (ICU) hospitalization and compared to cohort I (CI) and in CD4+ and CD8+ T cell subpopulations of CII. MFI, median fluorescence intensity. (B) Subset-specific expression of mitochondrial ROS in CII. (C) Citrate synthase (CS) activity, determined in T cells in which mitochondrial function was analyzed. Mitochondrial oxidative phosphorylation (OXPHOS) capacity of complex I (I) and complex I+II (I+II), capacity of the electron transfer system (I+II, ETS; after uncoupling), and ETS capacity driven by complex II (II, ETS; after rotenone administration) were determined by high-resolution respirometry. (A–C) Each symbol represents 1 donor, and summarized data are displayed as mean + SEM. *P < 0.05; **P < 0.01; ***P < 0.001 by Mann-Whitney U test (A and B subset-specific comparison).
Consistent with an increased mitochondrial mass, COVID-19 T cells showed elevated levels of the mitochondrial enzyme citrate synthase. Importantly, these changes did not result in increased mitochondrial respiration and instead, the respiratory activity normalized to citrate synthase activity was lower in COVID-19 T cells. Specifically, we observed a decreased capacity of the electron transport system (ETS) and a trend toward a reduced capacity for OXPHOS (CI+II ETS and CI+II OXPHOS, respectively; Figure 5C). Mitochondrial structure was studied next using electron microscopy. Lymphocytes from 9 critically ill COVID-19 patients showed, similar to cohort I, the presence of mitochondrial aberrations. However, analyzing paired samples from patients revealed a reduction in the occurrence of mitochondria with inclusions after dexamethasone treatment (Supplemental Figure 5E).
Besides increased mitochondrial content, high basigin and cytosolic ROS levels were markers of cohort I on the transcriptional and protein levels. Thus, we investigated whether dexamethasone might have affected these parameters. Transcriptomic analyses comparing controls and patients shortly after and beyond 3 weeks of dexamethasone treatment, a time point comparable to cohort I samples (Figure 2, A and B), were performed next. We observed that signatures of TNF, hypoxia, ROS, and basigin pathways, previously detectable in T cells, disappear in samples after dexamethasone treatment (Figure 6A). CD14+ monocytes maintained the transcriptomic profile of hypoxia and TNF and basigin pathways; however, an enrichment of ROS-related genes was lost (Figure 6B). The transcriptomic data were reflected on the metabolic level, as T cells did not accumulate ROS, in contrast to cohort I, after analyzing samples taken at comparable time points after hospitalization (Figure 6C). Next, we investigated these parameters in a time-dependent manner during dexamethasone treatment in CD4+ and CD8+ T cells and monocytes (D, day 1 to day 10), in the first 21 days after therapy (<21) and beyond (>21). ROS levels did not increase, suggesting durable dexamethasone effects (Figure 6D). In contrast to cohort I, basigin expression was not elevated in CD3+ T cells of COVID-19 patients (Figure 6E). In CD4+ T cells, basigin expression remained low over time, whereas CD8+ T cells reexpressed basigin in bulk and in all memory subpopulations after dexamethasone therapy cessation (Figure 6F and Supplemental Figure 5F). In monocytes, basigin levels remained high at all time points analyzed (Figure 6F). Thus, T cells from severely ill COVID-19 patients showed subpopulation-independent metabolic changes with impaired mitochondrial function and production of mitochondrial ROS. However, treatment with dexamethasone might have mitigated T cell cytosolic ROS accumulation in vivo.
Figure 6. Dexamethasone mitigates ROS accumulation and basigin expression and related gene signatures in COVID-19 patient T cells.
(A and B) Cohort 2 (coh2) blood samples were enriched for PBMCs, cryopreserved, and processed (see Figure 2). Heatmaps presenting hierarchically clustered and scaled expression data (filtered for absolute logFC > 1, logCPM and logRPKM > 1, and FDR < 0.05) and gene set enrichment analysis of the HALLMARK pathways, as analyzed in CI (see Figure 2, E and F) of CD4+ T cells (A) or CD14+ monocytes (B). Shown are differentially expressed genes in severe/critical patients taken in the first 14 days after dexamethasone cessation (coh2 early) or beyond (coh2 late, time point comparable to CI; 3 samples each time point, 2 paired early and late patient samples). Displayed is the comparison with CI controls (see Figure 2, A–F). Each column corresponds to an individual sample. A list of all read counts can be found in the supplemental material. (C–F) Blood samples were drawn and processed the same day. (C) Cytosolic ROS levels of CD3+ T cells from controls and severe/critical patients of CI and CII analyzed in the same period of time after ICU hospitalization (see Figure 5A). MFI, median fluorescence intensity. (D) Cytosolic ROS levels in CD4+ and CD8+ T cells and CD14+ monocytes of CII grouped according to time point analyzed. (E) Basigin expression of CD3+ T cells from controls and severe/critical patients of CI and CII analyzed in the same period of time after ICU hospitalization (see Figure 5A). Basigin expression of CD4+ and CD8+ T cells and CD14+ monocytes of CII grouped according to time point analyzed (F). Each symbol in C–F represents 1 donor. *P < 0.05; **P < 0.01; ***P < 0.001 by Mann-Whitney U test (C and E) or 1-way ANOVA with Bonferroni’s multiple comparisons test (D and F).
T cell cytosolic ROS accumulation is induced upon basigin ligation and can be mitigated by dexamethasone treatment.
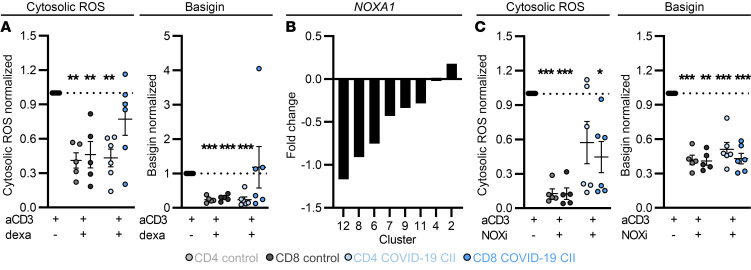
Basigin and cytosolic ROS were upregulated in T cells from COVID-19 patients and this phenotype was partially blunted in cohort II. To assess whether dexamethasone can modulate ROS and basigin expression, we cultured PBMCs from COVID-19 patients and healthy controls in vitro. In CD4+ and CD8+ T cells from controls, dexamethasone significantly reduced ROS and basigin expression. In patients, a significant downregulation was only observed in CD4+ T cells, reflecting the data from ex vivo patient analyses (Figure 7A). Basigin can be shed and soluble basigin levels were significantly higher in supernatants of cultured cells from COVID-19 patients (Supplemental Figure 6A). Following dexamethasone therapy, soluble basigin was reduced in supernatants from COVID-19 samples (Supplemental Figure 6A). A similar trend could be observed in plasma of patients after dexamethasone treatment (Supplemental Figure 6B).
Figure 7. Dexamethasone and NOX inhibition mitigate T cell ROS accumulation and basigin expression in vitro.
(A and C) Blood of donors was drawn and processed the same day. PBMCs were isolated and treated as indicated (for concentrations see Methods). (A) After 72 hours, PBMCs were stained for CD4 and CD8 and cytosolic ROS and basigin levels were analyzed. Data were normalized to anti-CD3 (aCD3) stimulation of the respective donor. (B) Log(fold change) of expression estimates for NOXA1 were determined for each cluster (as in Figure 4A and Supplemental Figure 4A). (C) PBMCs were processed as in A. Data normalized to anti-CD3 stimulation of the respective donor are shown. Summarized data are displayed as mean + SEM, with each symbol representing 1 donor. *P < 0.05; **P < 0.01; ***P < 0.001 by 1-way ANOVA with Bonferroni’s multiple comparisons test (A and C; T cell population specific).
Transcriptomic data identified cluster 2 to be enriched in severe COVID-19 patient T cells. We observed increased expression of NOXA1, which activates several NADPH oxidases (NOXs), leading to ROS production (51, 52) in this cluster (Figure 7B). NOXs can be targeted by dexamethasone (53, 54) and the effects of the NOX inhibitor diphenyleneiodonium on T cell ROS levels were assessed next. NOX inhibition decreased cytosolic ROS levels in T cells from healthy controls and CD8+ T cells from patients even better than dexamethasone; however, it had only a limited effect in patient CD4+ T cells (Figure 7C). Nevertheless, basigin expression was reduced in control and patient T cells (Figure 7C).
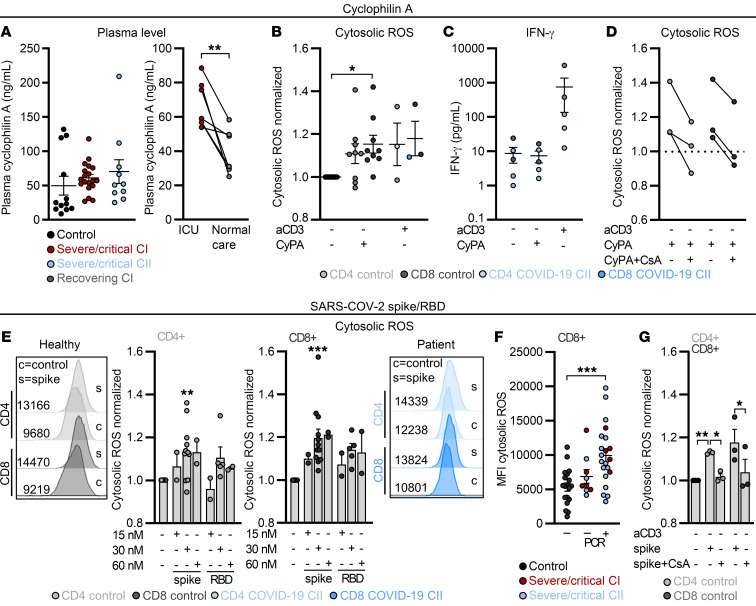
Lastly, we tested whether basigin ligation can trigger cytosolic ROS accumulation. Basigin is a receptor for CyPs (31) and we found CyPA to be present in the plasma of healthy donors and COVID-19 patients (Figure 8A). CyPA is involved in systemic inflammation and we observed a decrease in CyP plasma levels in recovering COVID-19 patients (Figure 8A). Importantly, addition of CyPA increased T cell cytosolic ROS levels such that they were comparable to those produced by T cell receptor stimulation using soluble anti-CD3 (aCD3; Figure 8B), whereas IFN-γ levels were only increased through aCD3 stimulation but not CyPA, indicating a nonclassical stimulation (Figure 8C). Intracellular ROS elevation could be reverted by cyclosporine A (CsA), which blocks CyP activity (Figure 8D). CsA and CyPA were preincubated together before administration to allow a direct interaction. Moreover, dexamethasone reduced the CyPA-induced ROS increase in CD4+ T cells of controls and one patient, but failed in CD8+ T cells from the same patient (Supplemental Figure 6C).
Figure 8. ROS can be triggered by cyclophilin A and SARS-CoV-2 spike protein and is mitigated by cyclosporine A in vitro.
(A) Cyclophilin A (CyPA) levels were determined in plasma of controls and severe/critical patients of cohort I and II and in paired patient samples taken at the intensive care unit (ICU) and after returning back to normal care unit. **P < 0.01 by Wilcoxon’s test. (B–G) Blood was drawn and processed the same day. PBMCs were treated for 24 hours as indicated. (B) The impact of CyPA on cytosolic ROS was determined. Data normalized to unstimulated cells of the respective donor are shown. (C) IFN-γ in culture supernatants was determined by ELISA. (D) The effect of CsA on CyPA-induced cytosolic ROS was determined. Data normalized to unstimulated T cells of the respective donor are shown. (E) Cytosolic ROS in CD4+ and CD8+ T cells in the presence of increasing concentrations of the SARS-CoV-2 spike protein (spike) or respective receptor binding domain (RBD). Data normalized to unstimulated T cells of the respective donor are displayed. Representative histograms for controls and 1 patient are displayed. (F) Cytosolic ROS levels in controls and severe/critical COVID-19 patients PCR negative or positive for SARS-CoV-2 are displayed. MFI, median fluorescence intensity. (G) Impact of CsA on spike-induced cytosolic ROS levels was determined in CD4+ and CD8+ T cells. Data normalized to unstimulated T cells of the respective donor are displayed. Each symbol represents 1 donor, and summarized data are displayed as mean ± SEM (A–C and F) or mean + SEM (E and G). *P < 0.05; **P < 0.01; ***P < 0.001 by 1-way ANOVA with Bonferroni’s multiple comparisons test (B, F, and G; T cell population–specific comparison in B and G) or Mann-Whitney U test (E).
As basigin interacts with various viruses, including SARS-CoV and SARS-CoV-2 (34, 35), we next incubated T cells with the SARS-CoV-2 spike protein. SARS-CoV-2 spike protein induced ROS in T cells, similarly to CyPA, in a concentration-dependent manner (Figure 8E), whereas the receptor binding domain of the spike protein (RBD) had only limited impact on ROS levels. No significant effect on basigin expression was observed (Supplemental Figure 6D). SARS-CoV-2 spike had no effect on CD14+ monocyte ROS production (Supplemental Figure 6E). Interestingly, CD8+ T cells from severely/critically ill COVID-19 patients with detectable (by PCR) SARS-CoV-2 infection showed high cytosolic ROS levels, while SARS-CoV-2–negative patients did not differ from healthy controls (Figure 8F). These data support our finding that the spike protein itself induces ROS production. As CyPA binds to a variety of viral proteins, we investigated whether it might serve as an adaptor for spike protein binding to basigin by applying CsA. As for CyPA, the spike protein and CsA were preincubated together before administration. CsA alone had no impact on ROS (data not shown); however, it did block the spike-induced ROS increase (Figure 8G).
Collectively, the data show that severe COVID-19 associates with T cell mitochondrial defects and is hallmarked by high T cell basigin expression and ROS accumulation. Basigin is induced by hypoxia, coordinates with cytosolic ROS pathways to induce mitochondrial damage, and can be targeted using dexamethasone and NOX inhibition. Lastly, basigin ligation with CyPA and SARS-CoV-2 spike protein induces T cell cytosolic ROS accumulation.
Discussion
It is now clear that the role of cellular metabolic pathways goes beyond energy production. Intriguingly, immune cell metabolism can steer immune cell function and thereby affect the outcome of inflammation (55). Here, we describe an immune cell metabolic phenotype with high accumulation of ROS and elevated mitochondrial content and basigin expression in T cells and monocytes of critically ill COVID-19 patients. Transcriptionally, T cells and monocytes displayed a higher expression of genes of the basigin network and genes involved in the cellular ROS response. T cells showed a disturbed mitochondrial morphology and function and these changes were accompanied by T cell lymphopenia. Analysis of T cells from patients treated analogously to the RECOVERY trial (50), receiving a 10-day dexamethasone treatment, revealed lower ROS and basigin levels. Consistently, those pathways were not enriched on the transcriptional level. However, CD8+ T cell basigin was reexpressed and ROS levels increased in some patients upon treatment termination. Interestingly, elevated ROS levels were observed mainly in patients positive for SARS-CoV-2. Finally, we can show that ROS is induced in vitro in T cells by SARS-CoV-2 spike protein or CyPA, both described as basigin ligands. A direct interaction of both proteins is suggested by our finding that CsA administration, which blocks CyPA activity, inhibits ROS induction in the presence of the spike protein.
Yet, what is the cause of the oxidative stress in T cells? COVID-19 patients can suffer from extensive hypoxia (2) and COVID-19 pathogenesis is most likely related to hypoxia, which can lead to superoxide radical and peroxide generation by mitochondria. In line with this, we observed an increase in basigin and mitochondrial ROS levels in T cells upon a 10-day period of hypoxia interrupted by phases of normoxia. Nevertheless, cytosolic ROS may not only result from hypoxia or mitochondrial dysfunction. NOXs represent a possible source for ROS and have been implicated in altered host survival and T cell phenotype after influenza infection (51). Consistent with this, we found NOXA1, a NOX activator, to be upregulated in a subpopulation of CD4+ T cells, as identified by single-cell and bulk sequencing approaches and specific to severely ill COVID-19 patients. Moreover, NOX inhibition reduced ROS levels in T cells from healthy controls and critically ill COVID-19 patients.
Confocal images of endothelial cells treated with recombinant SARS-CoV-2 S1 protein revealed mitochondrial fragmentation and attenuated mitochondrial function (56). These results suggest that not only hypoxia but also exposure to viral proteins may contribute to the mitochondrial dysregulation in T cells. A profound impact of viral proteins on cellular metabolism is further supported by our data, which show that spike protein can induce cytosolic ROS in T cells. As immune cells exhibit almost no ACE2 expression, we conclude that basigin, which is described as a SARS-CoV-2 receptor, is involved either directly or via interaction with CyPA, a basigin ligand that has been shown to interact with different types of viral proteins (31, 34). Interestingly a meta-analysis from 32 data sets searching for host genes implicated in human betacoronavirus infection found that the top-ranked gene was PPIA, which encodes CyPA (38). As treatment of vascular smooth muscle cells with CyPA can augment ROS production (57), we exposed T cells to CyPA, which resulted in ROS production and could be prevented by applying CsA. Moreover, the effects of spike protein on ROS production by T cells were mitigated by CsA, indicating a direct interaction of both proteins.
Generation of ROS has been shown to stabilize CD147 expression in tumor cells under glucose-restricted conditions (47). This has not yet been reported for immune cells, but might explain the strong correlation between cytosolic ROS levels and basigin expression, accompanied by transcriptional changes reflecting ROS stress and hypoxia. Interestingly, the upregulation of basigin membrane levels in T cells was not reflected by the RNA-seq analyses, which is in line with data from by De Saedeleer et al. showing that in tumor cells, ROS stabilizes basigin posttranslationally (47). A link between ROS and basigin expression is supported by our data showing that upon treatment with a NOX inhibitor both basigin and ROS levels are reduced.
High basigin levels may lead to suppressed T cell function and proliferation, as splenocytes from basigin-deficient mice are more active in mixed lymphocyte reactions (58). Furthermore, basigin-related high ROS levels can induce cell death (59), possibly explaining the reduced T cell numbers in COVID-19 patients that were described here and by others (40, 60). COVID-19 T cells possibly try to counteract cell death, as indicated by the elevated expression of BCL3 and its regulator STAT3 that promote T cell survival (61) and NAMPT, which protects against cell death induced by mitochondrial damage (43). In contrast to T cell lymphopenia, the immunometabolic dysregulation with high ROS and basigin levels was reversed in the early recovery phase, when severely and critically ill COVID-19 patients improve and are no longer ventilated, indicating that hypoxia may be one important underlying factor for disease outcome. In line with this, our data show that hypoxia induces basigin expression in T cells, which may set the stage for basigin ligands such as CyP or SARS-CoV-2 spike protein, which in turn induce a metabolic shift in COVID-19 T cells.
Interestingly, T cell frequencies were significantly lower in diabetic and/or obese patients, and T cells and myeloid cells of these patients exhibited decreased glucose uptake. Acquired insulin resistance of T cells was reported in diabetic patients previously (62). In contrast, cardiovascular disease or age had no impact on T cell numbers and glucose uptake, suggesting an interplay between systemic and T cell metabolism in patients with metabolic disorders. Given the high levels of systemic inflammation, the findings of low glucose and fatty acid uptake by T cells and myeloid cells were unexpected, especially as hypoxia can promote fatty acid uptake (18). Increased fatty acid uptake was only found in T cells from SARS-CoV-2–infected patients with none or mild symptoms compared with critically ill patients, suggesting that in this disease stage, a T cell metabolic switch is still possible. Interestingly, we detected a certain T cell metabolic exclusivity, as high fatty acid uptake was specific to T cells with low glucose uptake and vice versa. Moreover, T cell fatty acid uptake capacity did not decrease until 4 weeks after the onset of symptoms of mild COVID-19, thus reaching beyond the time point of infection resolution. We speculate that disturbed mitochondrial function in severely ill patients may prevent fatty acid degradation, thereby reducing fatty acid uptake. Recently, in a metabolomic analysis of sera from patients with COVID-19, Thomas et al. described increased circulating levels of fatty acids along with increased markers of oxidant stress (e.g., methionine sulfoxide, cystine) which could reflect impaired mitochondrial fatty acid oxidation (19). They also suggested potential metabolic therapeutic targets, e.g., through dietary supplementation with taurine/N-acetylcysteine.
Our data support the hypothesis that COVID-19 pathogenesis is related to hypoxia and oxidative stress in T cells, as it has been proposed for monocytes and neutrophils (23). Laforge et al. suggested that ROS-induced tissue damage, thrombosis, and red blood cell dysfunction could be targeted with free-radical scavengers. Such an approach could also ameliorate T cell function and is currently being tested in an ongoing study treating COVID-19 patients with antioxidant drugs such as vitamin C (27).
Importantly, clinical use of the glucocorticoid dexamethasone has become a standard of care and is clearly beneficial for COVID-19 patients (50, 63) but its mechanisms of action are not yet fully understood and its effects on immune cells are broad. Besides its widely established role as immunosuppressant, dexamethasone can support T cell viability (64). In macrophages, dexamethasone induced mitochondrial ROS and apoptosis in vitro (65) and in vivo dexamethasone treatment increased ROS production in pancreatic islets (66). Surprisingly, in our experiments, treatment with dexamethasone was effective in preventing intracellular ROS accumulation and basigin elevation in T cells of COVID-19 patients in vitro. Notably, similar results were obtained when studying COVID-19 patients under dexamethasone treatment. During the 10 days of application, ROS levels and basigin levels were not different from controls. Dexamethasone exerts broad effects on immune cells, but previous results (53, 54) and our data suggest that NOX inhibition might contribute. After the end of treatment ROS remained stable; however, basigin increased again in CD8+ T cells and in SARS-CoV-2–positive patients ROS levels were elevated. As the spike protein itself triggers ROS production in T cells, we propose that patients with a detectable viral load might benefit from a prolonged dexamethasone treatment.
Taken together, our results show that the effects of dexamethasone in COVID-19 patients are at least partially due to an immunometabolic modulation. Future studies should address whether other agents specifically targeting T cell metabolism and ROS production could improve the outcomes of critically ill COVID-19 patients. Based on our findings that spike protein and CyPA induce ROS production in T cells, which could be counteracted by dexamethasone and CsA, we propose that drugs such as alisporivir, a nonimmunosuppressive CyP inhibitor and cyclosporine analogue, could support the treatment of COVID-19 patients. Alisporivir has been shown to prevent hepatitis C virus protein–mediated decreases in cell respiration, collapse of mitochondrial membrane potential, and overproduction of ROS (67). A phase II proof-of-concept clinical trial of antiviral alisporivir (CYCLOVID trial NCT04608214) for treating hospitalized COVID-19 patients has been initiated. Based on in vitro data showing that alisporivir reduced SARS-CoV-2 RNA production in a dose-dependent manner in Vero E6 cells (37), the reduction in COVID-19 viral load in alisporivir-treated patients will be the trial’s primary objective. In light of our results it would be interesting to also focus on immune cell metabolism in these patients, as besides viral entry, stimulation of immune cells by circulating viral proteins may also be involved in the pathogenesis of COVID-19.
We propose here that hypoxia-induced basigin triggers cytosolic ROS production, contributing to metabolic dysfunction and a dysregulation of inflammation in COVID-19 patients. As cell-surface basigin expression is specific to T cells with severe immunometabolic dysregulation, it might serve as a prognostic parameter and a therapeutic target in COVID-19. Furthermore, our study strengthens the rationale for the use of agents that reduce oxidative stress in COVID-19 therapy.
Methods
Further information can be found in Supplemental Methods.
Study participants.
This study involved 140 participants consisting of 87 patients with confirmed SARS-CoV-2 infection, detected in a nasopharyngeal swab or a respiratory sample using routinely established RT-PCR and 47 uninfected controls. All patients with positive SARS-CoV-2 testing were included, and one patient with first diagnosis of leukemia was excluded.
Cohort I COVID-19 patients are described in Supplemental Table 1 and did not receive dexamethasone treatment. Cohort I controls were asymptomatic healthy individuals with a median age of 50 years (IQR 37.25–54.5) and male sex in 42%.
Cohort II COVID-19 patients are described in Supplemental Table 2 and severely ill patients were all treated with dexamethasone. Cohort II controls were asymptomatic healthy individuals with a median age of 51 years (IQR 39–55) and male sex in 54%.
Critically ill non–COVID-19 patients were treated at an intensive care unit for severe stroke or brain injury requiring mechanical ventilation (Supplemental Table 3) and did not receive dexamethasone.
COVID-19 severity was assessed as described previously (4, 10). Details can be found in Supplemental Methods.
Sample collection and processing.
All whole-blood samples analyzed for immune cell frequencies and single-cell metabolic assays were analyzed freshly without cryopreservation. Blood was collected and erythrocytes were lysed using ACK buffer. Details can be found in Supplemental Methods.
Determination of immune cell populations.
For analysis of surface marker expression, 0.2 × 106 to 1 × 106 cells/tube were stained with respective surface markers (staining procedures, list of antibodies, and lineage-defining markers listed in the Supplemental Methods). The gating strategy is displayed in Supplemental Figures 1A and 5B.
ELISpot assay for T cell detection.
Human venous blood was collected in lithium-heparin tubes and processed according to the instructions of the commercial T-SPOT.TB test (Oxford Immunotec). The ELISpot plates and chemistry, except the specific SARS-CoV-2 antigens, were derived from the T-SPOT.TB test. Medium was used as negative control and phytohemagglutinin (PHA) as positive control according to the manufacturer’s protocol. Peptides from SARS-CoV-2 were used for antigen stimulation.
Peptides of SARS-CoV-2 membrane glycoprotein (Pep. M), nucleocapsid phosphoprotein (Pep. N), and surface glycoprotein (Pep. S) are mainly 15-mers with 11 amino acid overlap. In brief, 250,000 PBMCs were stimulated with the peptide antigens and antigen-specific T cell–released IFN-γ was determined after contact with antigen-presenting cells. Details are in Supplemental Methods.
Isolation and in vitro culture of PBMCs.
PBMCs were isolated using Ficoll density gradient centrifugation and cultured in RPMI 1640 supplemented with 5 % autologous plasma, 2 mM stable glutamine, 50 U/mL penicillin, and 50 mg/mL streptomycin. Cells were stimulated with 5 μg/mL anti-CD3 antibody (clone OKT3) and treated with dexamethasone (500 nM), human CyPA (100 nM), diphenyleneiodonium chloride (DPI; 500 nM, dissolved in DMSO), CsA (1 μM), recombinant SARS-CoV-2 spike protein (15–60 nM), or SARS-CoV-2 RBD B1.351 (15–120 nM, dissolved in PBS). If CsA was administered together with CyPA or the spike protein, CsA and the respective protein were preincubated for 5 to 10 minutes and then added to PBMC cultures. At indicated time points, PBMCs were stained for surface marker expression and cytosolic ROS. To mimic phases of normoxia and reduced oxygen availability as observed in critically ill COVID-19 patients, PBMCs were cultured in 0.5% O2 in a hypoxic workstation in the presence or absence of anti-CD3/anti-CD28–coated beads at a cell to bead ratio of 1:1 for 10 days interrupted by 1-hour periods of normoxia on days 3, 5 and 8; medium was changed on day 5. After 10 days, PBMCs were stained for mitochondrial ROS and with antibodies against CD3, CD4, CD8, and basigin and analyzed by flow cytometry. For selected assays, cells were cryopreserved until analysis. After thawing, cells were cultured in standard medium with 10% human AB serum, with or without the addition of anti-CD3/anti-CD28–coated beads at a cell to bead ratio of 1:1.
Single-cell metabolic assays.
Basigin expression was determined by anti-CD147 surface staining simultaneously with the staining for immune cell populations. Cytosolic ROS were determined using 2′,7′-dichlorofluorescein diacetate; mitochondrial ROS using MitoSOX Red; mitochondrial content using MitoTracker Green FM; fatty acid uptake using BODIPY 500/510 C1, C12 dye; and glucose uptake using 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-D-glucose (2NBDG). Details are in Supplemental Methods.
Mitochondrial respiratory activity (high-resolution respirometry).
For analysis of mitochondrial function, T cells were isolated from PBMCs by untouched magnetic bead cell separation using the Pan T cell isolation kit according to the manufacturer’s protocol (Miltenyi Biotec). OXPHOS capacity and capacity of the electron transfer system were analyzed in a 2-channel titration injection respirometer at 37°C in mitochondrial respiration medium (MiR05) (68). Details are in Supplemental Methods.
Determination of citrate synthase activity.
Citrate synthase activity was determined in CD3+ T cells taken directly from the oxygraph chamber at the end of the experiment, snap-frozen, and stored. Citrate synthase activity was measured spectrophotometrically. Details are in Supplemental Methods.
ELISA.
IFN-γ, soluble basigin, and plasma CyPA were determined by ELISA. Details are in Supplemental Methods.
Electron microscopic analyses.
For transmission electron microscopic analyses, cell pellets were fixed in Karnovsky’s fixative and embedded in 4% low-melting-temperature agarose. For details regarding embedding, dehydration, sectioning, and analysis, refer to the Supplemental Methods.
Cell sorting and RNA-seq library preparation.
For RNA-seq experiments, cryopreserved PBMCs were thawed and stained with anti-CD3, anti-CD4, anti-CD8, and anti-CD14 antibodies. Live, single cells with leukocyte FSC/SSC characteristics were sorted into CD14+CD3– monocytes and CD14–CD3+CD8– CD4+ T cells by 4-way sorting on a BD FACSAria IIu. For details regarding reagents, RNA isolation, quantification, quality control, and dsDNA library preparation, refer to Supplemental Methods.
RNA-seq analysis.
Sequencing reads were mapped to human (GRCh38) using STAR v2.5.3a (69). The human GRCh38 genome index incorporated gene annotation from GENCODE 44 (release 27) to aid in spliced alignment. Details about respective counts, read count generation, differential expression analyses, heatmap and volcano plot generation, and gene enrichment can be found in the Supplemental Methods.
Deconvolution analysis.
The deconvolution of our bulk RNA-seq was carried out with DTD (70) using machine learning techniques and relying on single-cell data for training and constructing a reference matrix that defines the cell types that the bulks are deconvolved into. We used published single-cell RNA-seq data from Ren et al. (49), obtained from the already quality-controlled cells available at the repository (71). Refer to the Supplemental Methods for more information about the data set details, data processing, clustering, and comparison of cluster estimates and metabolic gene data set description.
Statistics.
GraphPad Prism was used for statistical analyses, using ANOVA with Bonferroni’s post hoc test and Mann-Whitney U test. A P value of less than 0.05 was considered significant: *P < 0.05; **P < 0.01; ***P < 0.001. Correlations were computed using Spearman’s correlation test. The following statistical tests were used for clinical parameters: Kruskal-Wallis (age), χ2 test according to Pearson (all categorical variables), Student’s t test (ferritin), or 1-way ANOVA (leukocytes and all other laboratory values).
Study approval.
The study was approved by the Institutional Review Board of University Hospital Regensburg (no. 20-1785-101) and conformed to the principles outlined in the Declaration of Helsinki.
Author contributions
PJS and KR conceived the project, performed experiments, analyzed data, and wrote the manuscript. MK conceived the project and wrote the manuscript. KS analyzed data and supervised experiments. J Klitzke, NB, SMD, KK, J Köstler, MP, and AD performed experiments and analyzed data. MS, JS, and RS analyzed single-cell and bulk sequencing data. CB, CM, AP, GS, JR, IU, and KF performed experiments. LS, DL, BG, FG, ML, MM, P Hau, C Bohr, RB, AG, BS, F Hanses, F Hitzenbichler, and WH provided critical resources. RW provided SARS-CoV-2 RBD B1.351 protein. JJ, DH, FL, TP, DW, and HP provided critical resources and supervised experiments. HS and C Brochhausen performed EM experiments. P Hoffmann performed cell sorting and MR performed transcriptome analyses.
Supplementary Material
Acknowledgments
We thank Marcus Kielmanowicz, Monika Wehrstein, Barbara Stemmer, and Rüdiger Eder for technical assistance. We thank the members of the Regensburg Center for Interventional Immunology (RCI) FACS Core Facility for expert technical assistance. RNA sequencing was conducted at the NGS Core of the RCI. This work was supported by the Bavarian Ministry of Science and Arts. PJS is supported by the Else Kröner Fresenius Foundation. KR, MK, PJS, C Brochhausen, MH, and PH are supported by the Bavarian Ministry of Science and Arts. MK and DW were funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, project number 324392634-TRR 221). JJ is supported by the Bavarian Ministry of Science and the Arts in the framework of the Bavarian Research Network “New Strategies Against Multi-Resistant Pathogens by Means of Digital Networking–bayresq.net.” AD is supported by the Marga and Walter Boll Foundation (220-05.01-15). JS, MS, and RS are supported by BMBF (eMed grant no. 031A428A).
Version 1. 11/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(22):e148225.https://doi.org/10.1172/JCI148225.
Contributor Information
Peter J. Siska, Email: peter.siska@gmx.at.
Sonja-Maria Decking, Email: Sonja-Maria.Decking@ukr.de.
Nathalie Babl, Email: nathalie.kauer@ukr.de.
Carina Matos, Email: carina.matos@ukr.de.
Christina Bruss, Email: christina.bruss@ukr.de.
Katrin Singer, Email: singer.katrin@freenet.de.
Jana Klitzke, Email: Jana.Klitzke@stud.uni-regensburg.de.
Marian Schön, Email: marian.schoen@ukr.de.
Jakob Simeth, Email: jakob.simeth@ukr.de.
Heiko Siegmund, Email: heiko.siegmund@ukr.de.
Ines Ugele, Email: ines.ugele@ukr.de.
Alexander Dietl, Email: alexander.dietl@ukr.de.
Kristina Kolodova, Email: kristina.kolodova@ukr.de.
Louisa Steines, Email: louisa.steines@ukr.de.
Katharina Freitag, Email: Katharina.Freitag@stud.uni-regensburg.de.
Alice Peuker, Email: alice.peuker@ukr.de.
Gabriele Schönhammer, Email: gabriele.schoenhammer@ukr.de.
Johanna Raithel, Email: johanna.raithel@ukr.de.
Bernhard Graf, Email: Bernhard.Graf@klinik.uni-regensburg.de.
Florian Geismann, Email: florian.geismann@ukr.de.
Matthias Lubnow, Email: matthias.lubnow@ukr.de.
Matthias Mack, Email: matthias.mack@klinik.uni-regensburg.de.
Peter Hau, Email: peter.hau@klinik.uni-regensburg.de.
Christopher Bohr, Email: christopher.bohr@ukr.de.
Ralph Burkhardt, Email: ralph.burkhardt@ukr.de.
Andre Gessner, Email: andre.gessner@klinik.uni-regensburg.de.
Bernd Salzberger, Email: bernd.salzberger@ukr.de.
Ralf Wagner, Email: ralf.wagner@ukr.de.
Frank Hanses, Email: frank.hanses@ukr.de.
Florian Hitzenbichler, Email: florian.hitzenbichler@ukr.de.
Daniel Heudobler, Email: daniel.heudobler@ukr.de.
Florian Lüke, Email: florian.lueke@ukr.de.
Tobias Pukrop, Email: Tobias.Pukrop@klinik.uni-regensburg.de.
Wolfgang Herr, Email: Wolfgang.Herr@klinik.uni-regensburg.de.
Daniel Wolff, Email: Daniel.Wolff@ukr.de.
Rainer Spang, Email: rainer.spang@ukr.de.
Hendrik Poeck, Email: hendrik.poeck@ukr.de.
Petra Hoffmann, Email: petra.hoffmann@ukr.de.
Jonathan Jantsch, Email: jonathan.jantsch@ukr.de.
Christoph Brochhausen, Email: christoph.brochhausen@ukr.de.
Dirk Lunz, Email: dirk.lunz@ukr.de.
Michael Rehli, Email: Michael.Rehli@klinik.uni-regensburg.de.
Marina Kreutz, Email: marina.kreutz@ukr.de.
Kathrin Renner, Email: kathrin.renner-sattler@ukr.de.
References
- 1.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Biasi S, et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11(1):3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadjadj J, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JS, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5(49):eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebillard RM, et al. Identification of SARS-CoV-2-specific immune alterations in acutely ill patients. J Clin Invest. 2021;131(8):e145853. doi: 10.1172/JCI145853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Codo AC, et al. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1alpha/glycolysis-dependent axis. Cell Metab. 2020;32(3):437–446. doi: 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayres JS. A metabolic handbook for the COVID-19 pandemic. Nat Metab. 2020;2(7):572–585. doi: 10.1038/s42255-020-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Candia P, et al. Type 2 diabetes: how much of an autoimmune disease? Front Endocrinol. 2019;10:451. doi: 10.3389/fendo.2019.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song JW, et al. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020;32(2):188–202. doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XJ, et al. In-hospital use of statins is associated with a reduced risk of mortality among individuals with COVID-19. Cell Metab. 2020;32(2):176–187. doi: 10.1016/j.cmet.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Bert N, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 13.Geltink RIK, et al. Unraveling the complex interplay between T cell metabolism and function. Annu Rev Immunol. 2018;36:461–488. doi: 10.1146/annurev-immunol-042617-053019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ron-Harel N, et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 2016;24(1):104–117. doi: 10.1016/j.cmet.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seth RB, et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Couzin-Frankel J. The mystery of the pandemic’s ‘happy hypoxia’. Science. 2020;368(6490):455–456. doi: 10.1126/science.368.6490.455. [DOI] [PubMed] [Google Scholar]
- 17.McNamee EN, et al. Hypoxia and hypoxia-inducible factors as regulators of T cell development, differentiation, and function. Immunol Res. 2013;55(1–3):58–70. doi: 10.1007/s12026-012-8349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. Enhancing CD8+ T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell. 2017;32(3):377–391. doi: 10.1016/j.ccell.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas T, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5(14):e140327. doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jezek J, et al. Reactive oxygen species and mitochondrial dynamics: the yin and yang of mitochondrial dysfunction and cancer progression. Antioxidants (Basel) 2018;7(1):13. doi: 10.3390/antiox7010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou B, Tian R. Mitochondrial dysfunction in pathophysiology of heart failure. J Clin Invest. 2018;128(9):3716–3726. doi: 10.1172/JCI120849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Younes SA, et al. Cycling CD4+ T cells in HIV-infected immune nonresponders have mitochondrial dysfunction. J Clin Invest. 2018;128(11):5083–5094. doi: 10.1172/JCI120245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laforge M, et al. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat Rev Immunol. 2020;20(9):515–516. doi: 10.1038/s41577-020-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Case AJ, et al. Elevated mitochondrial superoxide disrupts normal T cell development, impairing adaptive immune responses to an influenza challenge. Free Radic Biol Med. 2011;50(3):448–458. doi: 10.1016/j.freeradbiomed.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, et al. Metabolic defects of peripheral T cells in COVID-19 patients. J Immunol. doi: 10.4049/jimmunol.2100068. [published online May 28, 2021]. [DOI] [PubMed] [Google Scholar]
- 26.Thompson EA, et al. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021;34(11):108863. doi: 10.1016/j.celrep.2021.108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavezzi A, et al. COVID-19: hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin Pract. 2020;10(2):1271. doi: 10.4081/cp.2020.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egawa N, et al. Membrane type 1 matrix metalloproteinase (MT1-MMP/MMP-14) cleaves and releases a 22-kDa extracellular matrix metalloproteinase inducer (EMMPRIN) fragment from tumor cells. J Biol Chem. 2006;281(49):37576–37585. doi: 10.1074/jbc.M606993200. [DOI] [PubMed] [Google Scholar]
- 29.Tang Y, et al. Tumor-stroma interaction: positive feedback regulation of extracellular matrix metalloproteinase inducer (EMMPRIN) expression and matrix metalloproteinase-dependent generation of soluble EMMPRIN. Mol Cancer Res. 2004;2(2):73–80. [PubMed] [Google Scholar]
- 30.Kaushik DK, et al. Evaluating soluble EMMPRIN as a marker of disease activity in multiple sclerosis: studies of serum and cerebrospinal fluid. PLoS One. 2016;11(10):e0163802. doi: 10.1371/journal.pone.0163802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pushkarsky T, et al. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc Natl Acad Sci U S A. 2001;98(11):6360–6365. doi: 10.1073/pnas.111583198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanarsdall AL, et al. CD147 promotes entry of pentamer-expressing human cytomegalovirus into epithelial and endothelial cells. mBio. 2018;9(3):e00781-18. doi: 10.1128/mBio.00781-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe A, et al. CD147/EMMPRIN acts as a functional entry receptor for measles virus on epithelial cells. J Virol. 2010;84(9):4183–4193. doi: 10.1128/JVI.02168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J Infect Dis. 2005;191(5):755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K, et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;5(1):283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka Y, et al. Suppression of coronavirus replication by cyclophilin inhibitors. Viruses. 2013;5(5):1250–1260. doi: 10.3390/v5051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Softic L, et al. Inhibition of SARS-CoV-2 infection by the cyclophilin inhibitor alisporivir (Debio 025) Antimicrob Agents Chemother. 2020;64(7):e00876-20. doi: 10.1128/AAC.00876-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkinson N, et al. Dynamic data-driven meta-analysis for prioritisation of host genes implicated in COVID-19. Sci Rep. 2020;10(1):22303. doi: 10.1038/s41598-020-79033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muramatsu T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J Biochem. 2016;159(5):481–490. doi: 10.1093/jb/mvv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucas C, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sattler A, et al. SARS-CoV-2-specific T cell responses and correlations with COVID-19 patient predisposition. J Clin Invest. 2020;130(12):6477–6489. doi: 10.1172/JCI140965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cristofanon S, et al. Oxidation-dependent maturation and survival of explanted blood monocytes via Bcl-2 up-regulation. Biochem Pharmacol. 2008;76(11):1533–1543. doi: 10.1016/j.bcp.2008.07.042. [DOI] [PubMed] [Google Scholar]
- 43.Garten A, et al. Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol Metab. 2009;20(3):130–138. doi: 10.1016/j.tem.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsujimoto Y, Shimizu S. VDAC regulation by the Bcl-2 family of proteins. Cell Death Differ. 2000;7(12):1174–1181. doi: 10.1038/sj.cdd.4400780. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann M, et al. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liberzon A, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Saedeleer CJ, et al. Glucose deprivation increases monocarboxylate transporter 1 (MCT1) expression and MCT1-dependent tumor cell migration. Oncogene. 2014;33(31):4060–4068. doi: 10.1038/onc.2013.454. [DOI] [PubMed] [Google Scholar]
- 48.Li J, et al. CD147 reprograms fatty acid metabolism in hepatocellular carcinoma cells through Akt/mTOR/SREBP1c and P38/PPARα pathways. J Hepatol. 2015;63(6):1378–1389. doi: 10.1016/j.jhep.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 49.Ren X, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184(7):1895–1913. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.RECOVERY Collaborative Group. et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hofstetter AR, et al. NADPH oxidase 1 is associated with altered host survival and T cell phenotypes after influenza a virus infection in mice. PLoS One. 2016;11(2):e0149864. doi: 10.1371/journal.pone.0149864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panday A, et al. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol Immunol. 2015;12(1):5–23. doi: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu FF, et al. Dexamethasone and tofacitinib suppress NADPH oxidase expression and alleviate very-early-onset ileocolitis in mice deficient in GSH peroxidase 1 and 2. Life Sci. 2019;239:116884. doi: 10.1016/j.lfs.2019.116884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huo Y, et al. Dexamethasone inhibits the Nox-dependent ROS production via suppression of MKP-1-dependent MAPK pathways in activated microglia. BMC Neurosci. 2011;12:49. doi: 10.1186/1471-2202-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Neill LA, et al. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16(9):553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lei Y, et al. SARS-CoV-2 spike protein impairs endothelial function via downregulation of ACE 2. Circ Res. 2021;128(9):1323–1326. doi: 10.1161/CIRCRESAHA.121.318902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Satoh K, et al. Cyclophilin A enhances vascular oxidative stress and the development of angiotensin II-induced aortic aneurysms. Nat Med. 2009;15(6):649–656. doi: 10.1038/nm.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hahn JN, et al. The role of EMMPRIN in T cell biology and immunological diseases. J Leukoc Biol. 2015;98(1):33–48. doi: 10.1189/jlb.3RU0215-045R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hildeman DA, et al. T cell apoptosis and reactive oxygen species. J Clin Invest. 2003;111(5):575–581. doi: 10.1172/JCI200318007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin C, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bauer A, et al. The NF-kappaB regulator Bcl-3 and the BH3-only proteins Bim and Puma control the death of activated T cells. Proc Natl Acad Sci U S A. 2006;103(29):10979–10984. doi: 10.1073/pnas.0603625103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lau EYM, et al. Type 2 diabetes is associated with the accumulation of senescent T cells. Clin Exp Immunol. 2019;197(2):205–213. doi: 10.1111/cei.13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aguas R, et al. Potential health and economic impacts of dexamethasone treatment for patients with COVID-19. Nat Commun. 2021;12(1):915. doi: 10.1038/s41467-021-21134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franchimont D, et al. Positive effects of glucocorticoids on T cell function by up-regulation of IL-7 receptor alpha. J Immunol. 2002;168(5):2212–2218. doi: 10.4049/jimmunol.168.5.2212. [DOI] [PubMed] [Google Scholar]
- 65.Ai F, et al. Dexamethasone induces aberrant macrophage immune function and apoptosis. Oncol Rep. 2020;43(2):427–436. doi: 10.3892/or.2019.7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roma LP, et al. Pancreatic islets from dexamethasone-treated rats show alterations in global gene expression and mitochondrial pathways. Gen Physiol Biophys. 2012;31(1):65–76. doi: 10.4149/gpb_2012_011. [DOI] [PubMed] [Google Scholar]
- 67.Quarato G, et al. The cyclophilin inhibitor alisporivir prevents hepatitis C virus-mediated mitochondrial dysfunction. Hepatology. 2012;55(5):1333–1343. doi: 10.1002/hep.25514. [DOI] [PubMed] [Google Scholar]
- 68.Pesta D, Gnaiger E. High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol Biol. 2012;810:25–58. doi: 10.1007/978-1-61779-382-0_3. [DOI] [PubMed] [Google Scholar]
- 69.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gortler F, et al. Loss-function learning for digital tissue deconvolution. J Comput Biol. 2020;27(3):342–355. doi: 10.1089/cmb.2019.0462. [DOI] [PubMed] [Google Scholar]
- 71. Peking University. Summary — COVID19. http://covid19.cancer-pku.cn/ Accessed February 25, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.