Abstract
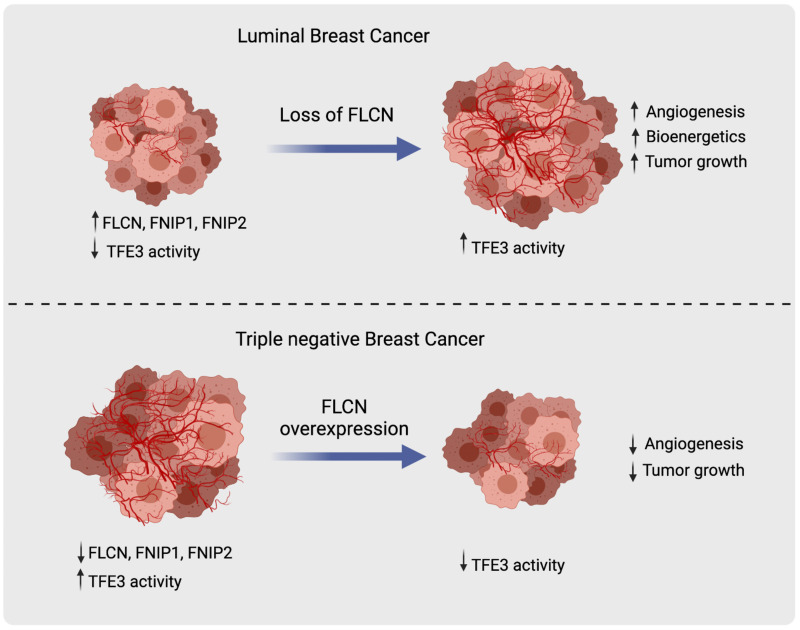
Growing tumors exist in metabolically compromised environments that require activation of multiple pathways to scavenge nutrients to support accelerated rates of growth. The folliculin (FLCN) tumor suppressor complex (FLCN, FNIP1, FNIP2) is implicated in the regulation of energy homeostasis via 2 metabolic master kinases: AMPK and mTORC1. Loss-of-function mutations of the FLCN tumor suppressor complex have only been reported in renal tumors in patients with the rare Birt-Hogg-Dube syndrome. Here, we revealed that FLCN, FNIP1, and FNIP2 are downregulated in many human cancers, including poor-prognosis invasive basal-like breast carcinomas where AMPK and TFE3 targets are activated compared with the luminal, less aggressive subtypes. FLCN loss in luminal breast cancer promoted tumor growth through TFE3 activation and subsequent induction of several pathways, including autophagy, lysosomal biogenesis, aerobic glycolysis, and angiogenesis. Strikingly, induction of aerobic glycolysis and angiogenesis in FLCN-deficient cells was dictated by the activation of the PGC-1α/HIF-1α pathway, which we showed to be TFE3 dependent, directly linking TFE3 to Warburg metabolic reprogramming and angiogenesis. Conversely, FLCN overexpression in invasive basal-like breast cancer models attenuated TFE3 nuclear localization, TFE3-dependent transcriptional activity, and tumor growth. These findings support a general role of a deregulated FLCN/TFE3 tumor suppressor pathway in human cancers.
Keywords: Angiogenesis, Metabolism
Keywords: Breast cancer, Cancer
Introduction
Breast cancer is the first leading cause of cancer deaths in women worldwide, with thousands dying from the disease each year (1). Gene expression profiling classifies human breast cancers into different subtypes, including luminal, HER2+, and basal-like (2–4). Around 10% to 20% of breast cancer patients are diagnosed with basal-like tumors, a high percentage of which are triple-negative breast cancers (TNBCs), an extremely heterogeneous disease lacking estrogen and progesterone receptors, as well as human epidermal growth factor receptor 2 (5). The TNBC subtype lacks effective targeted therapy options, and hence is associated with the worst prognostic outcomes for breast cancer patients (6).
A general characteristic of cancer cells is the capability to obtain nutrients from a nutrient-deprived environment and to use these nutrients to sustain their transformed state and increase cell proliferation (7). We have previously shown that folliculin (FLCN) plays an important role in mediating an AMPK-dependent resistance to several energy-depleting stresses, including nutrient deprivation, oxidative stress, anoxia, and hyperosmotic stresses (8–12). FLCN is an AMPK-binding partner (13, 14) identified as a tumor suppressor protein responsible for the Birt-Hogg-Dubé (BHD) syndrome in humans (15). The interaction of FLCN with AMPK is mediated by 2 homologous FLCN-binding proteins, called FNIP1 and FNIP2, where at least FLCN and one of the FNIPs is required for the full function of the complex (13, 15). We have previously shown that FLCN loss results in constitutive AMPK activation, which enhances survival to several metabolic stresses (8–11). Metabolic changes following FLCN loss include enhanced ATP production and an increase in metabolic intermediates derived from induction of mitochondrial oxidative phosphorylation (OXPHOS) and aerobic glycolysis. Metabolic rewiring following FLCN loss is dependent on the AMPK-dependent activation of peroxisome proliferator–activated receptor γ–coactivator 1α (PGC-1α) and subsequently hypoxia-inducible factor 1α (HIF-1α) (12).
HIF-1α, a well-defined hypoxia-responsive factor, activates diverse pathways that regulate cellular metabolism, angiogenesis, proliferation, and drug resistance (16, 17). HIF pathway activation in tumor cells is an important stimulus for blood vessel growth, where it regulates the expression of multiple proangiogenic genes, primarily that encoding vascular endothelial growth factor (VEGF) (18). Interestingly, studies have shown that TNBC tumors express high levels of intratumoral VEGF (19), possess high microvessel density (20), and display VEGF gene amplification compared with non-TNBC tumors (21), suggesting a marked angiogenic dependency in TNBC tumorigenesis.
In addition to HIF-1α activation, several studies support a role for FLCN in TFE3 regulation, where FLCN loss induces TFE3 nuclear translocation and subsequent transcriptional activation (22–25). TFE3, a transcription factor belonging to the microphthalmia/transcription factor E (MiT/TFE) family of transcription factors, has recently emerged as a global regulator of cell survival and metabolic reprogramming (26, 27). TFE3 regulates the expression of target genes involved in cellular processes ranging from cell-specific differentiation to basic cellular energy homeostasis, including autophagy, lysosomal biogenesis, and lipid homeostasis (28). Indeed, TFE3 activation and nuclear accumulation have been shown to be tightly regulated by cellular nutrient/energy status (28). Under nutrient-rich conditions, TFE3 is hyperphosphorylated, predominantly by mammalian target of rapamycin (mTOR), and remains inactive within the cytoplasm. Conversely, upon nutrient depletion, TFE3 becomes dephosphorylated and translocates to the nucleus where it induces the expression of genes in the coordinated lysosomal expression and regulation (CLEAR) network (29). Interestingly, the link between FLCN and mTOR has been previously proposed, where FLCN was identified as a guanosine triphosphate–activating (GTP-activating) protein (GAP) for Ras-related GTPase (Rag) C/D, which ultimately activates mTOR (30–32), implicating FLCN as a positive regulator of the mTOR signaling pathway. TFE3 and other conserved family members (TFEB and MITF) can act as oncogenes in renal cancer, melanoma, and pancreatic cancer (26).
Given that loss of FLCN mediates resistance to energy-depleting stresses encountered during tumor growth through several pathways, we investigated the previously unexplored role of FLCN in breast cancer. In this study, we report that FLCN and its binding partners FNIP1 and FNIP2 are downregulated, while AMPK and TFE3 transcriptional targets are elevated in TNBCs when compared with less aggressive luminal breast cancer subtypes. We show that FLCN loss in luminal breast cancer cells enhances tumor growth in a TFE3-dependent manner. Conversely, FLCN overexpression in basal-like TNBC models attenuates the nuclear localization and transcriptional activity of TFE3 and leads to impaired tumor growth. We further show that, beyond its impact on autophagy and lysosomal biogenesis, FLCN loss or downregulation activates TFE3, which subsequently engages a PGC1-α/HIF-1α–dependent induction of OXPHOS, glycolysis, and angiogenesis pathways that promote aggressive tumor growth.
Results
FLCN, FNIP1, and FNIP2 downregulation is recurrent in TNBC when compared with luminal subtypes.
FLCN was identified as a classic tumor suppressor gene when second-hit mutations were identified in BHD-associated renal tumors (33). Thus, we first investigated FLCN expression levels across different tumor types compared to their corresponding normal tissue. Using The Cancer Genome Atlas (TCGA) data set, we show that FLCN levels are substantially lower across several important human tumor types compared with their normal tissue counterparts (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI144871DS1). Given that patients affected with BHD syndrome are at risk of developing renal, skin, and colon tumors (34), we were particularly interested in studying the unexplored role of FLCN in invasive breast carcinoma.
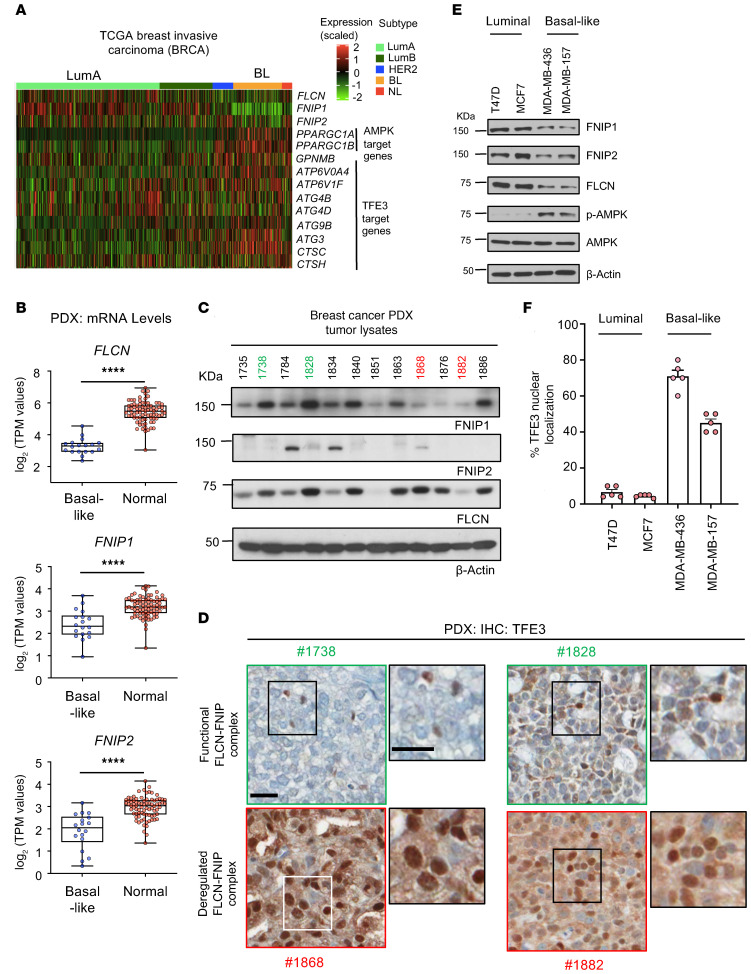
Using the invasive breast carcinoma TCGA data set, we show that FLCN, FNIP1, and FNIP2 levels are all significantly downregulated in TNBC compared with the less aggressive luminal subtypes (Figure 1A and Supplemental Figure 1, B–D). Additionally, we show that the downstream targets of AMPK (PPARGC1A and PPARGC1B) and TFE3 (ATP6V0A4, ATP6V1F, ATG4B, ATG4D, ATG9B, ATG3, CTSC, and CTSH) are upregulated in TNBC (Figure 1A). Analysis of RNA-sequencing data from a panel of 37 breast cancer patient–derived xenografts (PDXs) (35) revealed that the expression levels of FLCN, FNIP1, and FNIP2 are all significantly downregulated in basal-like samples when compared with normal breast tissue (Figure 1B). Moreover, immunoblot analysis of 12 representative basal-like PDX tumor lysates show reduced expression of at least one of the components of the FLCN-FNIP1-FNIP2 complex across the tumor samples (Figure 1C). To determine the relevance of a functional versus deregulated FLCN-FNIP1-FNIP2 complex, we assessed the localization and activity of TFE3 in the tumor samples. In a functional FLCN complex setting, such as Goodman Cancer Research Centre PDX samples 1738 and 1828 (GCRC1738 and GCRC1828), where at least FLCN and one of the FNIPs are highly expressed, we show by immunohistochemistry (IHC) that TFE3 is mainly localized in the cytoplasm, where it remains inactive (Figure 1D). In contrast, in tumors where the FLCN complex is deregulated (GCRC1868 and GCRC1882: loss or reduced expression of FLCN, FNIP1, and FNIP2) we show that TFE3 is mainly localized to the nucleus (Figure 1D). Notably, higher-magnification images demonstrate some nuclear TFE3 staining in the functional FLCN complex setting, which we attribute to the tumor-infiltrating immune cells. Nuclear TFE3 is transcriptionally active, as evident by the upregulation of GPNMB, which is a well-known downstream TFE3 target (ref. 36 and Supplemental Figure 1, E and F). Finally, we examined FLCN expression across different breast cancer cell lines using a published data set (37) representing luminal and basal-like subtypes, and show that FLCN levels are significantly downregulated in basal-like compared with luminal subtypes (Supplemental Figure 1G). To identify breast cancer models for further functional studies, we selected a panel of cell lines representing luminal A (MCF7 and T47D) and basal-like TNBC subtypes (MDA-MB-436 and MDA-MB-157). In agreement with results from TGCA data set and PDX models, we show that FLCN, FNIP1, and FNIP2 are more highly expressed in luminal A versus TNBC cell lines (Figure 1E). AMPK activation (p-Thr172-AMPK) is associated with reduced FLCN, FNIP1, and FNIP2 levels (Figure 1F). Immunofluorescence staining revealed that TFE3 is localized to the nucleus to a greater extent in TNBC subtypes compared with luminal subtypes (Figure 1F and Supplemental Figure 1H).
Figure 1. FLCN and its binding partners FNIP1 and FNIP2 are downregulated in basal-like breast cancer compared with luminal subtypes.
(A) TCGA analysis of invasive breast carcinoma comparing the expression of FLCN, FNIP1, FNIP2, and the downstream targets of AMPK and TFE3 in basal-like breast cancer (TNBC) compared with luminal subtypes. The different subtypes are color coded, where light green is luminal A, dark green is luminal B, dark blue is HER2+, orange is basal-like (BL), and red is normal-like (NL) subtype. (B) RNA expression of FLCN, FNIP1, and FNIP2 in basal patient–derived xenografts (PDXs) compared with normal GTEx breast tissue. Significance was determined using Student’s t test. ****P < 0.0001. (C) Immunoblot analysis showing the expression levels of FNIP1, FNIP2, and FLCN in PDX tumor lysates from patients with TNBC. Each number represents a PDX model derived from a different breast cancer patient. The numbers highlighted in green (1738 and 1828) represent PDXs with a functional FLCN-FNIP1-FNIP2 complex, while those in red (1868 and 1882) represent PDXs with a deregulated complex. Actin was used as a loading control. (D) Immunohistochemistry analysis of TFE3 for the selected PDX models representing the deregulated FLCN-FNIP1-FNIP2 complex in red (1868 and 1868) and the functional complex in green (1738 and 1828). Scale bars: 20 μm. (E) Immunoblot analysis indicating expression levels of FNIP1, FNIP2, FLCN, p-Thr172-AMPK (representing the activated form of AMPK), and total AMPK in breast cancer cell lines representing luminal (T47D and MCF7) and TNBC (MDA-MB-436 and MDA-MB-157) cells. Actin was used as a loading control. Blots are representative of at least 3 independent experiments. (F) Immunofluorescence analysis showing the percentage nuclear localization of TFE3 in luminal (T47D and MCF7) compared with TNBC (MDA-MB-436 and MDA-MB-157) cells. Results represent the mean ± SEM from at least 3 independent experiments performed in triplicate.
Loss of FLCN in luminal breast cancer cell lines activates AMPK and induces TFE3 nuclear localization and activation.
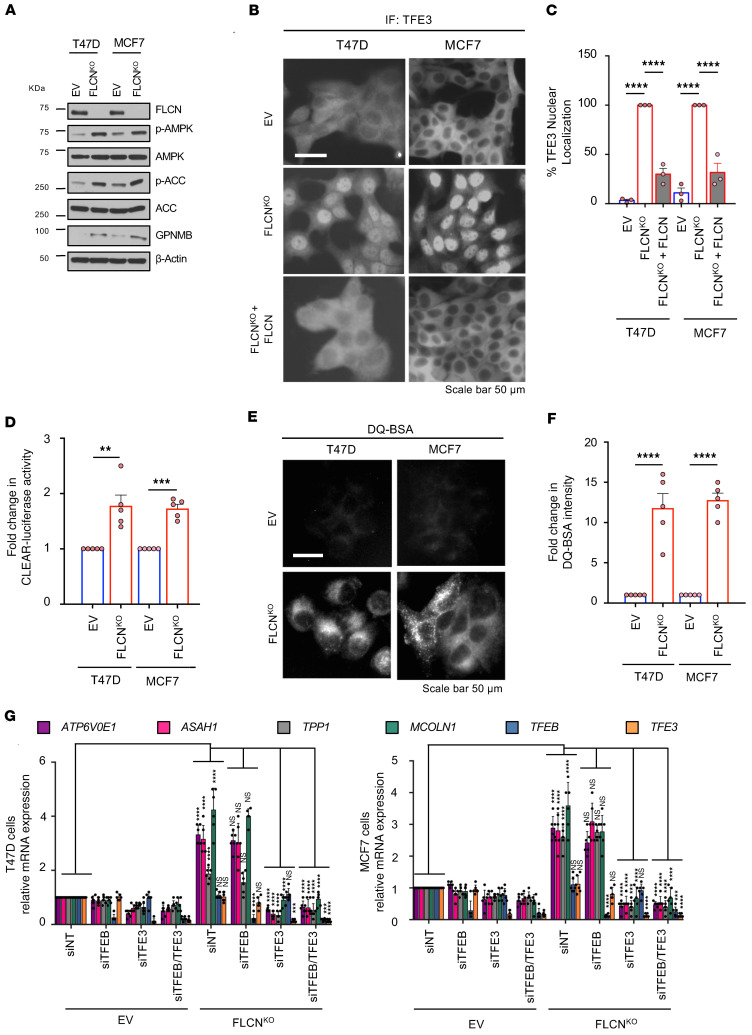
We next asked whether FLCN loss in luminal breast cancer cells (MCF7 and T47D) impacts tumor growth. To investigate this, we knocked out FLCN using CRISPR/Cas9 genome editing approaches. For each cell line, we employed 2 different guide RNAs targeting FLCN. Individual clones were selected, FLCN loss was verified by immunoblot, and a reconstituted pool (n = 4 clones) was generated to minimize the possibility of clonal effects (Figure 2A). Consistent with our previous studies (9–12), we show that loss of FLCN in both cell lines activates AMPK, as shown by phosphorylation of AMPK (p-Thr172-AMPK) and its substrate ACC (p-Ser79-ACC). Additionally, we show that GPNMB is induced upon FLCN loss, indicating transcriptional activation of TFE3 (Figure 2A). Notably, GPNMB is highly abundant in many tumors, including TNBC, and was shown to be implicated in tumor growth, angiogenesis, and poor prognosis of TNBC (38). Immunofluorescence staining revealed that TFE3 is 100% localized within the nucleus of both MCF7 and T47D FLCN-KO cells and reexpression of FLCN restored TFE3 cytoplasmic localization (Figure 2, B and C). To further investigate TFE3 transcriptional activity, we used a luciferase reporter containing the TFE3 consensus promoter motif (CLEAR), where we report an approximately 2-fold enhanced transcriptional activity in T47D and MCF7 FLCN-KO luminal cell lines (Figure 2D). Given that one of the important readouts of TFE3 activation is enhanced lysosomal activity (29), cells were assayed for their ability to process dye-quenched bovine serum albumin (DQ-BSA). DQ-BSA is a self-quenched fluorescent substrate that enters the cell through endocytosis and fluoresces upon lysosomal degradation (39). We show a significant, approximately 10-fold increase in DQ-BSA fluorescence intensity, indicating enhanced lysosomal activity in FLCN-KO cells (Figure 2, E and F). Furthermore, we assessed the expression of known TFE3 target genes involved in autophagy and lysosomal biogenesis (ATP6V0E1, ASAH1, TPP1, and MCOLN1), all of which contain the CLEAR motif consensus sequence in their promoter regions and show a significant increase upon FLCN loss in both T47D and MCF7 cells (Figure 2G). To investigate whether this observed increase is TFE3 dependent, we transiently reduced TFE3 expression using small interfering RNA (siRNA) and show a significant decrease in the expression of all the assessed genes (Figure 2G).
Figure 2. Loss of FLCN in luminal breast cancer cell lines activates AMPK and induces TFE3 nuclear localization and transcriptional activation.
(A) Immunoblot analysis of FLCN and downstream signaling molecules in empty vector (EV) control and CRISPR/Cas9-mediated FLCN-knockout (FLCNKO) T47D and MCF7 cells. β-Actin was used as a loading control. (B) Representative immunofluorescence images showing the localization of TFE3 in EV, FLCNKO, and reexpression of FLCN in T47D and MCF7 cells. Scale bar: 20 μm. (C) Quantitative analysis of the immunofluorescence results in D showing the percentage of TFE3 nuclear localization in EV, FLCNKO, and reexpression of FLCN inT47D and MCF7 cells. Results represent the mean ± SEM of at least 3 independent experiments, each performed in triplicate. Significance was determined using Student’s t test. ****P < 0.0001. (D) Fold change in TFE3 transcriptional activity, as determined by CLEAR-luciferase promoter activity normalized against CMV-Renilla, in EV and FLCNKO T47D and MCF7 cells. Data represent the average ± SEM of 3 independent experiments, each performed in triplicate. Significance was determined using Student’s t test. **P < 0.01, ***P < 0.001. (E) Representative images of EV and FLCNKO T47D and MCF7 cells after 1 hour of incubation with DQ-BSA-Red followed by a 2-hour chase in complete cellular media prior to fixation. Scale bar: 20 μm. Images are representative of at least 3 independent experiments. (F) Relative lysosomal activity, as determined by DQ-BSA assay, in EV and FLCNKO T47D and MCF7 cells upon treatment as indicated in E. Results represent the mean ± SEM of at least 3 independent experiments, each performed in triplicate. Significance was determined using Student’s t test. ****P < 0.0001. (G) Relative TFE3 and TFEB mRNA levels and their lysosomal and autophagy target gene mRNA levels measured by RT-qPCR in EV and FLCNKO T47D (left) and MCF7 (right) cells transfected with nontargeting (NT) siRNA control, or siRNA targeting TFEB or TFE3, or both. Data represent the average ± SEM of 3 independent experiments, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. ***P < 0.001; ****P < 0.0001. NS, not significant.
Since many TFE3 target genes are also regulated by TFEB, we assessed the contribution of TFEB to the enhanced autophagy/lysosomal biogenesis responses observed upon FLCN loss using siRNAs targeting TFEB alone, TFE3 alone, or both simultaneously. Interestingly, downregulation of TFEB alone had no effect on the expression level of the assessed genes in FLCN-KO cells, and TFEB/TFE3 double knockdown produced similar effects to those produced by downregulation of TFE3 alone (Figure 2G). It is worth noting that expression levels of TFEB are much lower than TFE3 in human breast cancer cell lines (http://www.proteinatlas.org). Altogether, these results suggest that TFE3 may be more dynamically involved in the FLCN/AMPK pathway than TFEB in this context. In light of these observations, we have focused specifically on the role of TFE3 in breast tumor phenotypes associated with loss of FLCN.
Loss of FLCN in MCF7 cells enhances cellular metabolism in a TFE3-dependent manner.
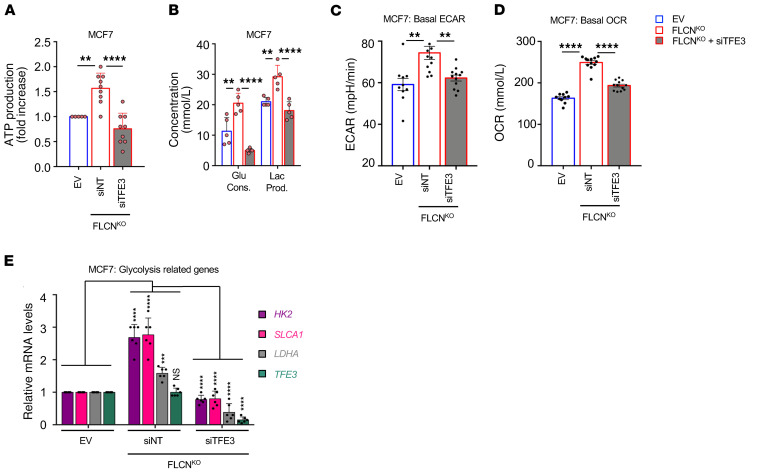
A fundamental characteristic of cancer cells is to couple nutrient consumption to macromolecular biosynthesis and energy production to facilitate tumor growth and survival (40). We have previously shown that FLCN loss induces an AMPK-dependent increase in resistance to several energy-depleting stresses, and this is likely to contribute to cellular metabolic adaptation (8–10, 12). However, the link between metabolic reprograming following FLCN loss and TFE3 activation has not been studied to date to our knowledge. We show a significant 1.7-fold increase in ATP levels in FLCN-KO MCF7 cell lines, which was abolished following transient knockdown of TFE3 (Figure 3A). In parallel, we show an increase in glucose consumption and lactate production (Figure 3B), and an enhanced extracellular acidification rate (ECAR) (Figure 3C) and oxygen consumption rate (OCR) (Figure 3D) under basal culture conditions in FLCN-KO MCF7 cell lines compared with empty vector (EV) controls. Notably, all of these metabolic changes were dependent on TFE3 activity, as TFE3 downregulation in FLCN-KO cells mitigated these effects (Figure 3, B–D). Finally, we assessed the expression of common glycolytic genes (HK2, SLCA1, and LDHA) in MCF7 cells and show a significant increase in FLCN-KO cells compared with EV, which was abolished following TFE3 downregulation (Figure 3E). Taken together, our results show that loss of FLCN in luminal breast cancer cells engages metabolic reprogramming toward increased cellular bioenergetics, which can provide a metabolic advantage to cells to facilitate growth and survival.
Figure 3. Loss of FLCN in MCF7 cells enhances cellular metabolism in a TFE3-dependent manner.
(A) Fold change in ATP levels in empty vector (EV) and FLCN-knockout (FLCNKO) MCF7 cells transfected with nontargeting (NT) siRNA control or siRNA targeting TFE3, after 48 hours of transfection as measured by CellTiter-Glo Luminescent Cell Viability Assay. Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. **P < 0.01; ****P < 0.0001. (B) Glucose consumption and lactate production levels in the cellular media were measured using a NOVA Bioanalysis flux analyzer in EV and FLCNKO MCF7 cells transfected with nontargeting (NT) control siRNA or siRNA targeting TFE3 after 48 hours of transfection. Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. **P < 0.01; ****P < 0.0001. (C and D) Basal extracellular acidification rate (ECAR) (C) and oxygen consumption rate (OCR) (D) in EV and FLCNKO MCF7 cells transfected with NT control siRNA or siRNAs targeting TFE3, after 48 hours of transfection, measured by Seahorse Bioscience XF96 extracellular flux analyzer. Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. **P < 0.01; ****P < 0.0001. (E) Relative mRNA levels of TFE3 and glycolysis-related genes measured by RT-qPCR in EV and FLCNKO MCF7 cells transfected with nontargeting (NT) control siRNA or siRNA targeting TFE3. Data represent the average ± SEM of n = 6 independent experiments, each performed in triplicate, where each point represents the average of the triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. ***P < 0.001; ****P < 0.0001. NS, not significant.
Loss of FLCN in luminal breast cancer cells enhances tumor growth.
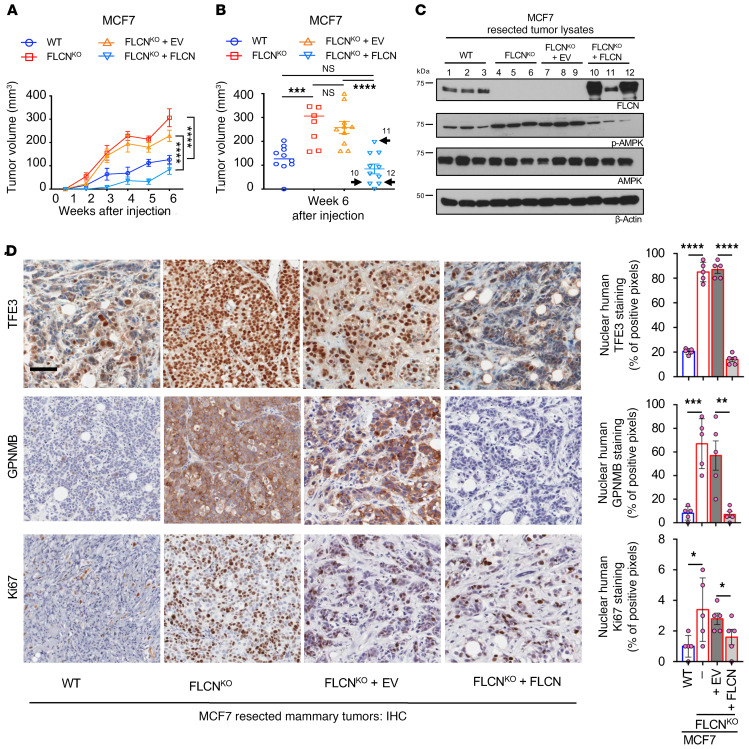
Next, we investigated the impact of FLCN loss on breast tumor growth. FLCN-KO MCF7 and T47D luminal breast cancer cells exhibited significantly increased tumor growth as compared with wild-type (WT) control cells when injected into the mammary fat pads of NOD SCID gamma (NSG) mice (Figure 4, A and B, and Supplemental Figure 2A). Importantly, reexpression of FLCN in MCF7 FLCN-KO cells restored tumor growth kinetics to those observed with parental MCF7 breast cancer cells (Figure 4, A and B). In line with the in vitro data, characterization of these tumors following resection showed enhanced AMPK activity (p-Thr172-AMPK) upon FLCN loss, which occurred in both MCF7 and T47D cells (Figure 4C and Supplemental Figure 2B). Again, elevated AMPK activity was abrogated upon reexpression of FLCN in MCF7 FLCN-KO cells (Figure 4C). Notably, we show that tumors 10 and 12 had higher expression of FLCN, as evident by immunoblot analysis (Figure 4C), which was associated with lower tumor volumes (Figure 4B). Conversely, the weak FLCN expression achieved in tumor 11 (Figure 4C) was associated with a much greater tumor volume (Figure 4B).
Figure 4. Loss of FLCN in luminal breast cancer cells enhances tumor growth.
(A) Growth curves of tumors of WT (blue), FLCN-knockout (FLCNKO) (red), FLCNKO plus EV (orange), and FLCNKO FLCN-reexpressing (turquoise) MCF7 cells injected in mammary fat pads (MFP) of NSG mice over the course of 6 weeks. Data represent the mean volumes ± SEM of each cohort measured each week (n = 10 mice in each cohort). Significance was determined using repeated-measures 1-way ANOVA. ****P < 0.0001. (B) Individual volumes of tumors as recorded 6 weeks after injection in cells indicated in A. Data represent the average volume ± SEM of each cohort recorded 6 weeks after injection (n = 10 mice in each cohort). Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. ***P < 0.001; ****P < 0.0001. NS, not significant. Samples 10 and 12 represent tumors with higher FLCN expression compared with sample 11, where FLCN expression was lower (refer to panel C). (C) Immunoblot analysis of WT, FLCNKO, FLCNKO plus EV, and FLCNKO FLCN-reexpressing MCF7 tumors resected 6 weeks after injection. Three representative samples were run from each cohort. β-Actin was used as a loading control. (D) Representative images of immunohistochemistry (IHC) staining for human TFE3, GPNMB, and Ki67 in WT, FLCNKO, FLCNKO plus EV, and FLCNKO FLCN-reexpressing MCF7 tumors resected 6 weeks after injection (left). Scale bar: 50 μm. Quantification of IHC results showing the percentage of TFE3 nuclear localization, positive GPNMB staining, and positive Ki67 staining (right). Data represent mean quantifications ± SEM of IHC images from at least 5 different mice. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
IHC analysis demonstrates that TFE3 is localized to the nucleus in FLCN-KO MCF7 and T47D tumors and is transcriptionally active, as shown by an enhanced GPNMB expression (Figure 4D and Supplemental Figure 2C). TFE3 localization reverted to the cytoplasm and GPNMB levels were reduced upon FLCN reexpression in MCF7 FLCN-KO cells (Figure 4D). In keeping with increased tumor growth, we report a significant increase in nuclear Ki67 staining in FLCN-KO luminal breast tumors (Figure 4D and Supplemental Figure 2C), which was suppressed following FLCN expression in the MCF7 model (Figure 4D). IHC staining for cleaved caspase-3, a marker of apoptosis, failed to detect any significant differences in MCF7 or T47D cells as a result of FLCN loss (Supplemental Figure 2D). These results indicate that loss of FLCN in luminal breast cancer tumors potentially enhances proliferation without altering apoptosis.
An angiogenic profile emerges following FLCN loss in luminal breast cancer cells.
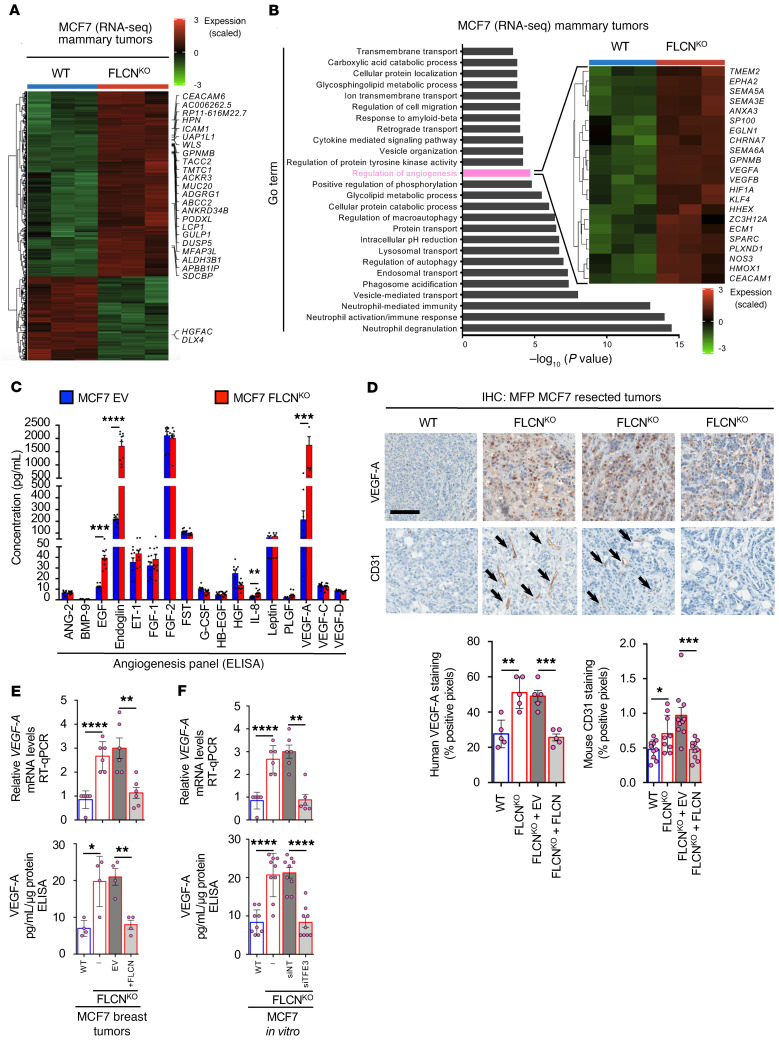
We have shown that FLCN loss induces the nuclear localization and activation of TFE3, which is shown to be involved in several cellular stress pathways (29), all of which could potentially contribute to the enhanced tumor growth observed in FLCN-deficient luminal breast cancer cells. Hence, we wanted to elucidate the genes and pathways involved in the growth of FLCN-deficient tumors. We performed RNA-sequencing analysis on MCF7 WT and FLCN-KO mammary tumors and report significant differences in the gene expression profile in both cohorts (Figure 5A). Specifically, Gene Ontology (GO) analysis following RNA sequencing revealed enrichment in several pathways, many of which we have recently reported in FLCN-deficient cells (8), including autophagy, lysosomal biogenesis, and innate immune responses (Figure 5B and Supplemental Table 2). Importantly, regulation of the angiogenesis pathway was one of the top hits in FLCN-KO tumors compared with their controls (Figure 5B and Supplemental Table 3). This was of interest since, to our knowledge, a direct link between FLCN, TFE3, and angiogenesis has yet to be reported. We show a significant increase in many angiogenesis-associated genes in FLCN-KO tumors compared with their WT controls, including HIF1A, EGLN1, SEMA, TMEM2, EPHA2, NOS3, VEGFA, and VEGFB (Figure 5B) (18). To further evaluate this angiogenic profile, we assessed a panel of secreted angiogenic factors by multiplex ELISA in FLCN-proficient and -deficient tumor lysates. We observed a multiple-fold increase in several of these factors, including EGF, endoglin, IL-6, and VEGF-A, in FLCN-KO compared with FLCN-expressing MCF7 cells (Figure 5C). IHC analyses of the resected MCF7 and T47D tumors confirmed a significant increase in VEGF-A staining in FLCN-KO cells compared with their controls (Figure 5D and Supplemental Figure 3, A and B). FLCN reexpression in MCF7 FLCN-KO cells caused a reduction in VEGF-A levels (Figure 5D). We also show that the number of intratumoral vessels, as determined by mouse CD31 staining, was significantly increased in FLCN-KO cells compared with their controls (Figure 5D and Supplemental Figure 3, A and C), which was also lost following FLCN reexpression in MCF7 FLCN-KO cells (Figure 5D).
Figure 5. Loss of FLCN in luminal breast cancer cells promotes an angiogenic profile.
(A) Heatmap representing differential gene expression in WT and FLCN-knockout (FLCNKO) MCF7 tumors following RNA-sequencing analysis. Each column represents a different mouse from each cohort, where blue is WT and red is FLCNKO. Fold increase was normalized against EV and color coded (dark red indicates 3-fold or more increase, light green indicates 3-fold or more decrease, black indicates no change). (B) Gene enrichment scores for significantly upregulated pathways in FLCNKO compared with WT MCF7 tumors highlighting regulation of angiogenesis as a differentially induced pathway, with the heatmap specifically showing the upregulation of angiogenesis-related genes. Each column represents a different mouse from each cohort, where blue is WT and red is FLCNKO. Fold increase was normalized against EV and color coded (dark red indicates 3-fold or more increase, light green indicates 3-fold or more decrease, black indicates no change). (C) Fold increase in 17 angiogenic/growth factors detected in WT and FLCNKO MCF7 tumor lysates using human angiogenesis array. Data represent the average values of 10 mice in each of the indicated cohorts. Significance was determined using Student’s t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. (D) Representative images of immunohistochemistry (IHC) staining for human VEGF-A, mouse CD31, and mouse F4/80 of WT, FLCNKO, FLCNKO plus EV, and FLCNKO FLCN-reexpressing MCF7 tumors resected 6 weeks after injection (top). Scale bar: 50 μm. Quantification of IHC results showing percentage positive VEGF-A staining, positive CD31 staining, and positive F4/80 staining (bottom). Data represent mean quantifications ± SEM of IHC images from at least 5 different mice. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. *P < 0.05; **P < 0.01; ***P < 0.001. (E) Relative human VEGFA mRNA levels measured by RT-qPCR in WT, FLCNKO, FLCNKO plus EV, and FLCNKO FLCN-reexpressing MCF7 tumors (top). Concentration of human VEGF-A in EV and FLCNKO cells transfected with siRNA targeting TFE3 in MCF7 cells, and FLCNKO FLCN-reexpressing MCF7 tumor lysates, as measured by ELISA (bottom). Data represent the average ± SEM of at least 5 different mice from each cohort, performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. *P < 0.05; **P < 0.01; ****P < 0.0001. (F) Relative human VEGFA mRNA levels measured by RT-qPCR in EV, FLCNKO, and FLCNKO cells transfected with nontargeting (NT) control siRNA or siRNA targeting TFE3 in MCF7 cells (top). Concentration of human VEGF-A in EV and FLCNKO cells transfected with siRNA targeting TFE3 in MCF7 cells, and FLCNKO FLCN-reexpressing MCF7 tumor lysates, as measured by ELISA (bottom). Data represent the average ± SEM of at least n = 3, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. **P < 0.01; ****P < 0.0001.
Given that early initiation of tumor angiogenesis is required to support rapidly growing tumors (41), we performed a short-term in vivo experiment to assess the degree of angiogenesis at earlier time points during tumor growth. MCF7 EV and FLCN-KO tumors were resected 2 weeks after injection into NSG mice. In agreement with our previous observations (Figure 4A), we show that tumor volumes were larger in FLCN-KO MCF7 cells (Supplemental Figure 3D). Interestingly, FLCN-KO MCF7 tumors also exhibited a prominent red coloration (Supplemental Figure 3D), which may reflect the increased vascularization we see in end-stage tumors. Indeed, IHC staining of these early tumors revealed a significant increase in VEGF-A and CD31 staining in FLCN-KO cells compared with their control (Supplemental Figure 3, E–G).
Given that VEGF-A is a key player in tumor-associated angiogenesis, we validated VEGF-A expression and its dependence on FLCN and TFE3 in MCF7 breast cancer cells. Using real-time quantitative PCR (RT-qPCR) and ELISA, we show that VEGF-A was expressed at higher levels in MCF7 FLCN-KO tumors compared with FLCN-expressing controls, and that VEGF-A levels were reduced following reexpression of FLCN (Figure 5E). Interestingly, we showed via transient TFE3 knockdown that this increase in VEGF-A levels in MCF7 FLCN-KO cells is TFE3 dependent (Figure 5F). Thus, tumor-derived VEGF-A is likely to contribute to the enhanced angiogenic response observed in FLCN-null breast cancer cells.
Loss of FLCN in luminal breast cancer cells enhances tumor growth in a TFE3-dependent manner.
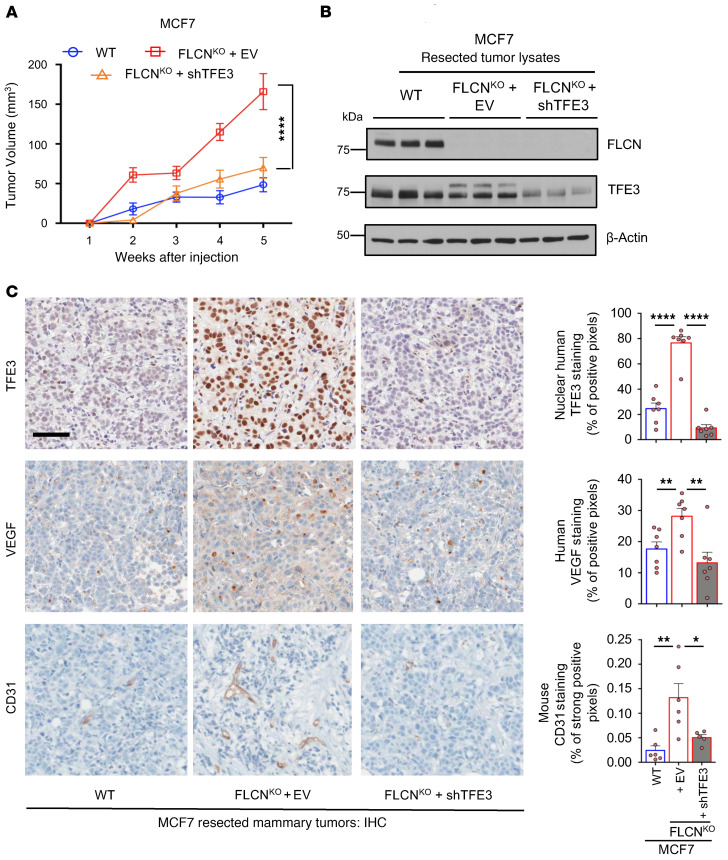
We next investigated whether the enhanced tumor growth in FLCN-KO cells was dependent on TFE3 transcriptional activity. Accordingly, we stably reduced TFE3 expression levels in MCF7 FLCN-KO cells by shRNA and showed that the increase in tumor growth observed following FLCN loss is abolished by the concomitant reduction of TFE3 expression (Figure 6A). Characterization of these tumors following resection confirmed loss of FLCN and reduced TFE3 expression (Figure 6B). The residual signal for TFE3 observed in whole-tumor lysates likely represents stromal contamination, as no TFE3 expression was detected in the MCF7 FLCN-KO cells expressing TFE3-targeting shRNAs in vitro (Supplemental Figure 4). Additionally, our IHC analysis of the resected tumors shows that TFE3 is localized to the nucleus in FLCN-KO MCF7 cells compared with WT controls, which was ablated upon downregulation of TFE3 (Figure 6C). Moreover, we report an increase in VEGF-A and CD31 staining in FLCN-KO cells compared with EV, both of which were significantly reduced upon downregulation of TFE3 (Figure 6C). These results indicate that loss of FLCN in luminal breast cancer tumors enhances tumor growth and angiogenesis in a TFE3-dependent manner.
Figure 6. Loss of FLCN in luminal breast cancer cells enhances tumor growth and promotes angiogenesis in a TFE3-dependent manner.
(A) Growth of mammary tumors in mice injected with WT (blue), FLCN-knockout (FLCNKO) (red), and FLCNKO plus shTFE3 (orange) MCF7 cells over the course of 5 weeks. Data represent the mean tumor volumes ± SEM of each cohort measured each week (n = 10 mice in each cohort). Significance was determined using repeated-measures 1-way ANOVA. ****P < 0.0001. (B) Immunoblot analysis of WT, FLCNKO, and FLCNKO plus shTFE3 MCF7 tumors resected 5 weeks after injection. Three representative samples were run from each cohort. β-Actin was used as a loading control. (C) Left: Representative images of the immunohistochemistry (IHC) staining for human TFE3, human VEGF-A, and mouse CD31 in WT, FLCNKO, and FLCNKO plus shTFE3 MCF7 tumors resected 6 weeks after injection. Scale bar: 50 μm. Right: Quantification of IHC results showing the percentage TFE3 nuclear localization, VEGF-A staining, and CD31 staining in the indicated cohorts. Data represent the mean quantifications ± SEM of IHC images from at least 5 different mice. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. *P < 0.05; **P < 0.01; ****P < 0.0001.
Loss of FLCN in luminal breast cancer cells activates a HIF-1α–dependent angiogenesis program in a TFE3-dependent manner.
One of the main transcription factors involved in glycolytic and angiogenic responses is HIF-1α (42). HIF-1α regulates several hundred genes, including numerous glycolytic genes and VEGFs. Interestingly, we have previously shown that loss of FLCN in cells provides an energetic advantage due to metabolic reprograming that engages aerobic glycolysis, a process that we have shown to be dependent on the activation of PGC-1α, a potent metabolic sensor upstream of HIF-1α (12).
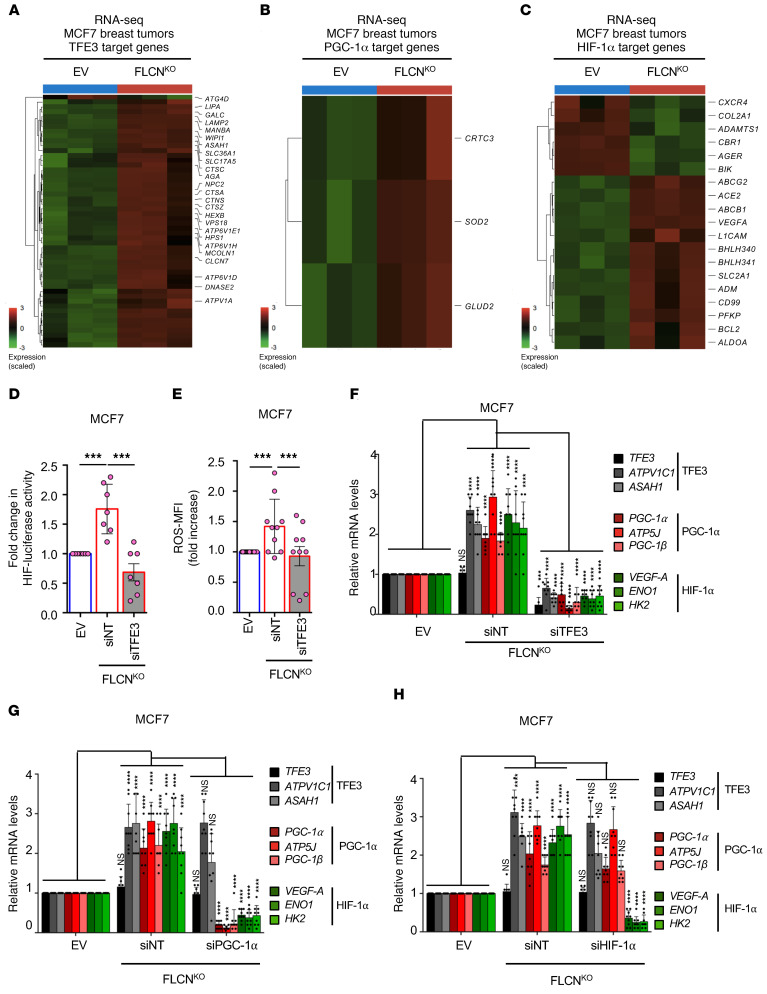
Our data suggest that FLCN loss induces a TFE3-dependent angiogenic program, including upregulation of VEGF-A. Given that HIF-1α is one of the main transcription factors involved in VEGF-A expression, we next investigated the dependency of VEGF-A expression on the PGC-1α/HIF-1α axis, and whether it is linked to TFE3 activity. Our RNA-sequencing analyses revealed that TFE3, PGC-1α, and HIF-1α downstream targets are all upregulated in FLCN-KO MCF7 tumors compared with FLCN-expressing controls (Figure 7, A–C). To further corroborate these results and assess the role of TFE3 in PGC-1α/HIF-1α pathway induction, we show that the HIF-1α transcriptional activity is elevated in FLCN-KO MCF7 cells and this is abrogated upon the downregulation of TFE3 (Figure 7D). We have previously shown that enhanced cellular reactive oxygen species (ROS) induced HIF transcriptional activity and drove Warburg metabolic reprogramming in an AMPK- and PGC-1α–dependent manner (12). We quantified the relative levels of cellular ROS using the general oxidative stress indicator CM-H2DCFDA. We observed that loss of FLCN is associated with a significant 1.7-fold increase in intracellular ROS levels, which we show to be TFE3 dependent (Figure 7E). We next verified the RNA-sequencing results by RT-qPCR, showing that loss of FLCN in MCF7 cells induces the upregulation of several TFE3, PGC-1α, and HIF-1α target genes (Figure 7F). Importantly, downregulation of TFE3 in FLCN-KO cells significantly abrogated the observed induction of TFE3 (ATPV1C1 and ASAH1), PGC-1α (ATP5J and PGC-1β), and HIF-1α (ENO1 and HK2) target genes (Figure 7F), revealing an important role of TFE3 in controlling the PGC-1α/HIF-1α pathway. To investigate this pathway further, we downregulated PGC-1α in FLCN-KO cells and observed a significant decrease in both PGC-1α and HIF-1α target genes, but the levels of TFE3 target genes were unaffected (Figure 7G). Similarly, downregulation of HIF-1α in FLCN-KO cells abrogated only the HIF-1α target genes, but the levels of both PGC-1α and TFE3 target genes were unaffected (Figure 7H). Together, these results indicate that loss of FLCN in luminal MCF7 cells induces TFE3 activity, which acts upstream of the PGC-1α/HIF-1α pathway. Importantly, we have identified an oncogenic pathway where TFE3 acts as a master regulator of autophagy and lysosomal biogenesis on one hand, while controlling PGC1-α/HIF-1α pathways involving OXPHOS, glycolysis, and angiogenesis, all of which could be contributing to tumor growth.
Figure 7. Loss of FLCN in luminal breast cancer cells activates a HIF-1α–dependent angiogenic program in a TFE3-dependent manner.
(A–C) Heatmaps representing the differential TFE3 (A), PGC-1α (B), and HIF-1α (C) target gene expression in WT and FLCN-knockout (FLCNKO) MCF7 tumors following RNA-sequencing analysis. Each column represents gene expression from a different mouse from each cohort, where blue indicates WT and red indicates FLCNKO tumors. Fold increase was normalized against EV and color coded (dark red indicates 3-fold or more increase, light green indicates 3-fold or more decrease, black indicates no change). (D) Fold change in HIF-1α transcriptional activity, as determined by HIF-1α luciferase promoter activity normalized to CMV-Renilla, in EV and FLCNKO MCF7 cells transfected with nontargeting (NT) control siRNA or siRNA targeting TFE3. Data represent the average ± SEM of n = 7 independent experiments. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. ***P < 0.001. (E) Relative mean fluorescence intensity of the total cellular reactive oxygen species (ROS) in EV and FLCNKO MCF7 cells transfected with NT control siRNA or siRNA targeting TFE3, as measured by flow cytometry. Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Significance was determined using Student’s t test. ***P < 0.001. (F–H) Relative TFE3, PGC-1α, and HIF-1α downstream target gene mRNA levels measured by RT-qPCR in EV and FLCNKO MCF7 cells transfected with NT control siRNA or siRNA targeting TFE3 (F), PGC-1α (G), or HIF-1α (H). Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. ***P < 0.001; ****P < 0.0001. NS, not significant.
FLCN overexpression in basal-like breast cancer cells restores TFE3 cytoplasmic localization and attenuates tumor growth.
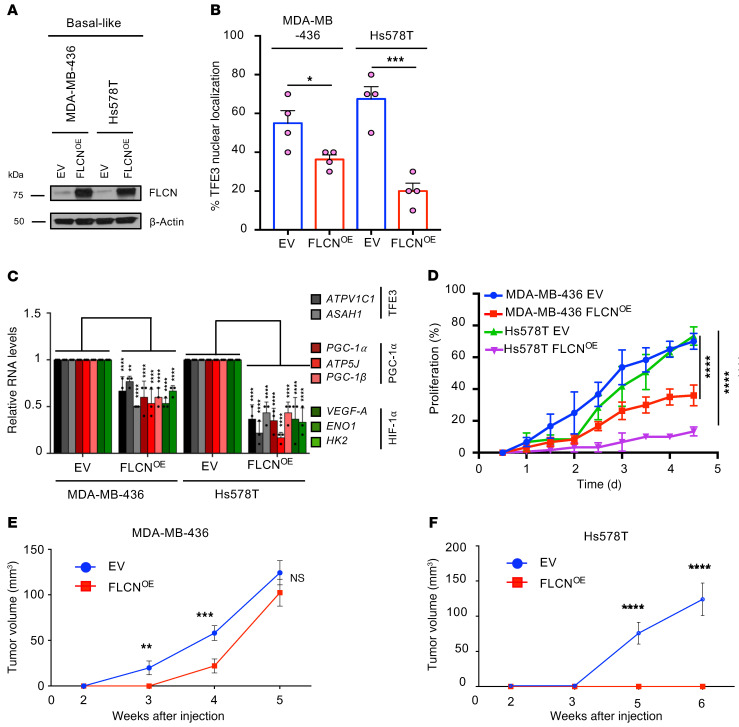
We have shown that loss of FLCN in luminal breast cancer cells, which typically express high levels of FLCN, enhances tumor growth in a TFE3-dependent manner. We next investigated the effect of increasing FLCN expression in basal-like TNBC cell lines, which typically express lower FLCN levels when compared with luminal cell lines (Figure 1). Accordingly, we stably overexpressed FLCN in 2 basal-like TNBC cell lines, MDA-MB-436 and Hs578T, both of which exhibit low FLCN levels and predominant TFE3 nuclear localization (Figure 8, A and B). Interestingly, immunofluorescence staining revealed that TFE3 nuclear localization is significantly impaired in both MDA-MB-436 and Hs578T following FLCN overexpression (Figure 8B). To investigate the impact of FLCN overexpression on the angiogenic pathway, we assessed the relative expression of TFE3, PGC-1α, and HIF-1α target genes by RT-qPCR and report a significant decrease in the levels of these genes in both MDA-MB-436 and Hs578T cell lines (Figure 8C). Reduced nuclear TFE3 localization and target gene activation were associated with a significant decrease in cellular proliferation in vitro, which was more profound in FLCN-overexpressing Hs578T cells (Figure 8D).
Figure 8. FLCN overexpression in basal-like breast cancer cells restores TFE3 cytoplasmic localization and attenuates tumor growth.
(A) Immunoblot analysis of empty vector (EV) and FLCN-overexpressing (FLCNOE) MDA-MB-436 and Hs578T basal-like breast cancer cells. β-Actin was used as a loading control. (B) Quantitative analysis of the immunofluorescence data showing the percentage of TFE3 nuclear localization in EV and FLCNOE MDA-MB-436 and Hs578T cells. Data represent the average ± SEM of n = 4 independent experiments. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. *P < 0.05; ***P < 0.001. (C) Relative TFE3, PGC-1α, and HIF-1α downstream target gene mRNA levels measured by RT-qPCR in EV and FLCNOE MDA-MB-436 and Hs578T cells. Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. **P < 0.001; ****P < 0.0001. (D) The percentage proliferation of EV and FLCNOE MDA-MB-436 and Hs578T cells over 5 days, as monitored and analyzed by an IncuCyte Live Cell Analysis System. Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Significance was determined using repeated-measures 1-way ANOVA. ****P < 0.0001. (E and F) Growth of mammary tumors in mice injected with WT (EV) (blue) or FLCNOE cells (red) in MDA-MB-436 (E) and Hs578T (F) cell models over the course of 5 to 6 weeks. Data represent the mean tumor volumes ± SEM of each cohort measured each week (n = 10 mice in each cohort). Significance was determined using repeated-measures 1-way ANOVA. **P < 0.01, ***P < 0.001, ****P < 0.0001. NS, not significant.
Importantly, FLCN overexpression also impaired mammary tumor growth in vivo, inducing a more profound effect in the Hs578T cells when compared with the MDA-MB-436 model (Figure 8, E and F). Interestingly, Hs578T breast cancer cells overexpressing FLCN can only be detected as residual cancer cells at the site of injection as compared with control cells (Supplemental Figure 5, A and B). In contrast, MDA-MB-436 cells that overexpress FLCN exhibit an early growth defect that is rapidly overcome, resulting in growth rates and tumor volumes that were similar to those of their EV controls at the endpoint (Figure 8E). This likely reflects the fact that FLCN overexpression has a more modest effect in reducing the nuclear localization of TFE3, TFE3 downstream gene activation, and in vitro proliferation in MDA-MB-436 cells when compared with Hs578T cells (Figure 8, B–D). Indeed, while expression was still detectable in end-stage MDA-MB-436 mammary tumors (Supplemental Figure 5C), FLCN levels were significantly reduced in end-stage tumors when compared with the levels detected in MDA-MB-436 cells just prior to injection (Supplemental Figure 5D). Notably, both MDA-MB-436 control and FLCN-overexpressing tumors displayed a similar degree of nuclear TFE3 staining in end-stage mammary tumors (Supplemental Figure 5, E and F). These data suggest that there is strong selective pressure to maintain TFE3 nuclear localization in rapidly growing cancer cells.
Collectively, these results demonstrate that expression of exogenous FLCN in basal-like TNBC cells impairs TFE3 nuclear localization and TFE3-dependent transcription, which results in impaired tumor growth.
Discussion
TNBC remains the most challenging breast cancer subtype to treat, potentially due to a lack of targeted therapies that underscores the need to better understand the molecular pathways that contribute to the growth and metastatic progression of this aggressive disease. In this study, we report that the FLCN/TFE3 pathway is dysregulated in the TNBC subtype, where FLCN and its binding partners FNIP1 and FNIP2 are downregulated and AMPK and TFE3 downstream targets are elevated compared with the less aggressive luminal subtype. We show that loss of FLCN in luminal subtypes promotes tumor growth through the activation of several pathways that enhance tumor growth, including metabolic reprograming, autophagy, lysosomal biogenesis, and engagement of an angiogenic program, all of which we report to be regulated by the MiT/TFE family member TFE3. Importantly, we show that FLCN overexpression in TNBC models attenuates TFE3 nuclear localization and transcriptional activity, leading to impaired tumor cell growth in vitro. Hs578T breast cancer cells overexpressing FLCN failed to grow into palpable tumors and could only be detected as residual deposits of cancer cells at endpoint. In contrast, FLCN overexpression in MDA-MB-436 cells only transiently exhibited impaired tumor growth. Notably, exogenous FLCN levels were reduced in end-stage MDA-MB-436 tumors, which correlated with increased nuclear TFE3 localization. Together, these data argue for a strong selective pressure to maintain TFE3 nuclear localization in rapidly growing TNBC cells.
Growing tumors exist in metabolically compromised environments and require activation of multiple pathways to scavenge nutrients to support accelerated rates of growth. We have previously shown that loss of FLCN increases the AMPK-dependent resistance to several energy-depleting stresses, including nutrient deprivation, oxidative stress, heat, anoxia, hyperosmotic stresses, obesity, and pathogen infection (8–12). In line with our previous studies, we report here that loss of FLCN in luminal breast cancer cells prompts metabolic reprogramming that increases cellular bioenergetics. These metabolic changes encompass an increase in ATP production, enhanced glycolysis, mitochondrial respiration, elevated autophagic flux, and increased lysosomal biogenesis. Importantly, we show that all such changes, which improve the metabolic fitness of FLCN-deficient cells, is driven through TFE3 transcriptional activation.
In this work, we report several pathways affected by FLCN loss in luminal breast cancers, all of which have been independently linked to tumorigenesis. Indeed, TFE3 nuclear localization and activation (43), PGC1-α induction and mitochondrial biogenesis (44), ROS production (45), and HIF1-α activation (46) have all been described as steps leading to tumor development in different cellular systems.
The role of MiT/TFE family members in cancer has also been previously reported. For example, oncogenic TFEB and TFE3 gene fusions have been described in renal cell carcinoma (47). TFEB and TFE3 have also been shown to play a crucial role in regulating autophagy/lysosome function, which promotes the growth of pancreatic ductal adenocarcinoma (26). More recently, TFE3 was shown to exert protumorigenic roles in a Kras-dependent, non–small-cell lung cancer model through activation of autophagy/lysosomes (48).
In addition to these well-documented cellular phenotypes for TFE3 in cancer, we reveal a consequence of FLCN loss, which is the induction of multiple bioenergetic programs including OXPHOS, glycolysis, and angiogenesis in breast cancers, which are controlled by PGC1-α/HIF-1α downstream of TFE3. Moreover, knockdown of TFE3 in the context of FLCN loss impairs tumor growth and mitigates these responses.
HIF1-α is a key factor in the regulation of VEGF and other angiogenic factors. IHC analysis of human tumor biopsies reveals that HIF-1α overexpression is a common feature of many solid cancers (42) and associates with increased tumor VEGF expression and vascularization (49, 50). Interestingly, a role for MiT/TFE factors in the regulation of angiogenesis was first hypothesized following the observation that TFEB-KO mice die prenatally due to a defect in placental vascularization (51). Thus, our results demonstrate that FLCN loss causes the nuclear localization of TFE3, which in turn engages the PGC-1α/HIF1-α axis to increase the expression of angiogenic factors such as VEGF-A.
The relative contributions of metabolic reprogramming, autophagy/lysosomal biogenesis, or increased angiogenesis to the progrowth phenotype observed with FLCN-deficient luminal breast cancer cells remains to be elucidated. It is entirely possible that all 3 processes are required. One could speculate that during initial stages of tumor development, all solid tissues require a proper vasculature that grants oxygen, nutrients, and waste disposal. Hence, early activation of angiogenic processes is mandatory to sustain the deregulated proliferation of tumor cells. When nutrients become limiting, cancer cells may benefit from the enhanced metabolic flexibility (increased glycolysis and OXPHOS-dependent metabolism) observed in FLCN-null cancer cells. Moreover, metabolic stresses may be overcome through the utilization of nonconventional energy sources that are mobilized through activation of nutrient scavenging pathways involving autophagy and the lysosome, both of which are induced by TFE3. Thus, aggressive/metastatic cancers are highly reliant on constitutive activation of these pathways to degrade and recycle cellular materials. Strikingly, we show that cellular metabolism, autophagy/lysosomal biogenesis, increased glycolysis, OXPHOS-dependent metabolism, and angiogenic pathways can be regulated by TFE3. It is likely that each of these processes will be differentially activated, depending on the environmental cues and energetic demands of the growing tumor. FLCN-deficient breast cancer cells permanently activate each of these processes, and hence are provided with an advantage over FLCN-proficient control cells, which contributes to increased tumor growth.
In the work presented here, we uncover a signaling pathway that links TFE3 activity to the HIF-1α/PGC-1α pathway via FLCN loss or downregulation. Interestingly, the components of the FLCN tumor suppressor complex are rarely mutated in sporadic human cancers, with the exception of somatic mutations in rare cases of renal cell carcinoma, colorectal cancer, and thyroid oncocytoma (52–55). In this study, we show that downregulation of FLCN is not only limited to breast cancer but is recurrent in many human cancers. The mechanism of FLCN/FNIP downregulation will be the subject of further research. Collectively, these findings have wider implications for a general role of a deregulated FLCN tumor suppressor pathway in human cancers in which TFE3 or HIF activity is known to be induced.
Methods
Cell lines and cell culture.
MCF7, T47D, MDA-MB-436, MDA-MB-157, and Hs578T breast cancer cells were obtained from the American Type Culture Collection (ATCC). Cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Wisent, 319-005CL) supplemented with 10% fetal bovine serum (FBS) (Wisent, 080-150), 100 U/mL penicillin plus 100 μg/mL streptomycin (Wisent, 450-201-EL), and 50 μg/mL gentamycin (Wisent, 450-135) in 5% CO2 at 37°C. For gene-silencing experiments, breast cancer cells were seeded in 6-cm dishes and transfected with 10 nM siRNA duplexes using Lipofectamine RNAiMax (Invitrogen, 13778030) according to the manufacturer’s instructions. The following siRNA SMARTpools were used: human TFEB (locus ID, 7942) (Dharmacon, L-009798-00-0005), human TFE3 (locus ID, 7030) (Dharmacon, L-00933-00-0005), human PPARGC1A (QIAGEN, GeneSolution, GS10891), human HIF-1α (locus ID, 3091) (Dharmacon, L-004018-00-0005) and siControl (Dharmacon, D-001810-10-05). Experiments were performed 48 to 72 hours after transfection. Stable knockdown of TFE3 in MCF7 FLCN-KO breast cancer cells was achieved using the Mission lentivirus shRNA empty vector (shEV), shTFE3 (Sigma-Aldrich, TRCN0000232151). For FLCN rescue or overexpression experiments, the human FLCN cDNA was cloned into pLenti CW57-MCS1-P2A-MCS2-BLAST (a gift from Adam Karpf, Addgene plasmid 80921).
Generation of knockout lines.
CRISPR/Cas9 guide RNA targeting sequences for human FLCN were identified bioinformatically using the CRISPR Design Tool available at http://chopchop.cbu.uib.no/, Zhang Lab). Two different guide RNA sequences were used to target the first expressed exon in the gene. Targeting sequences used were TCGCACATGTCCGACTTTTT and GCGGGCTGCTGGACTCGACGC. Targeting sequences were cloned into the lentiCRISPR plasmid (http://www.addgene.org/49535/) as previously described (56). Lentivirus was produced for the FLCN targeting sequences as well as an empty lentiCRISPR vector for control lines. Lentiviral transfer plasmids were cotransfected along with VSV-G envelope (https://www.addgene.org/12259/) and packaging plasmids (https://www.addgene.org/12260/), into HEK293T cells using Lipofectamine LTX (Invitrogen, 15338-500). Media were changed after 24 hours and virus-containing media were collected and centrifuged 72 hours after transfection. For the MCF7 and T47D cell lines, we performed single-cell cloning for each FLCN guide RNA, and once FLCN-KO cells were verified by immunoblotting, a pool of 4 clones was generated in an effort to eliminate clonal effects.
Reagents, chemicals, and antibodies.
N-acetyl-L-cysteine (NAC) (Sigma-Aldrich, A7250) was dissolved in 1× PBS to a stock concentration of 1 M and pH adjusted to 7.4. NAC was used at a final concentration of 5 mM. β-Estradiol (Sigma-Aldrich, E8875) was dissolved in 100% ethanol to a final concentration of 3 mg/mL. This solution was then diluted in canola oil to 20 μg/mL, which was then used to subcutaneously inject mice during the tumor growth experiments, once per week at 1 μg per mouse.
Antibodies used for immunoblotting were against β-actin (Santa Cruz Biotechnology, sc-47778), AMPKα (Cell Signaling Technology, 2532), human FLCN (Cell Signaling Technology, 3967), human FNIP1 (Abcam, ab61395), human FNIP2 (Sigma-Aldrich, SAB3500010), p-AMPKα (Thr172) (Cell Signaling Technology, 2531), ACC (Cell Signaling Technology, 3676), p-ACC (S79) (Cell Signaling Technology, 3661), GPNMB (Cell Signaling Technology, 38313), and TFE3 (Cell Signaling Technology, 14779S; and Sigma-Aldrich, HPA023881). Antibodies used for IHC were against the following proteins: human GPNMB (Cell Signaling Technology, 38313), human TFE3 (Sigma-Aldrich, HPA023881), mouse F4/80 (Cell Signaling Technology, 70076), human cleaved caspase-3 (Cell Signaling Technology, 9661), mouse CD31 (Dianova, AF5149-01; and Cell Signaling Technology, 77699), human Ki67 (Cell Signaling Technology, 9449), and human VEGF-A (Dako Technology, M7273).
Luciferase reporter assays.
Cells were seeded in 6-well plates and transfected for 6–8 hours with 1 μg of the 4XCLEAR-luciferase reporter plasmid (Addgene, 66800) or firefly luciferase HIF activity reporter pGL2-TK-HRE plasmid (gift from G. Melillo, NCI, Frederick, Maryland, USA) and 0.1 μg of CMV-Renilla Luciferase plasmid (Promega, E2261) using 5 μL of polyethylenimine (PEI) (Polysciences, 23966-1) at 1 mg/mL stock concentration. Proteins were extracted using 100 μL of Passive Lysis Buffer from the Dual-Luciferase reporter assay system (Promega, E1980) according to the manufacturer’s instructions and assayed using FLUOstar Omega (BMG Labtech). Samples were normalized against nontransfected controls and CMV-Renilla values.
DQ-BSA assay.
T47D and MCF7 cells were seeded at 500,000 cells per well. The next day, the cells were incubated with 5 μg/mL DQ Red BSA (Thermo Fisher Scientific, D12051) for 1 hour and washed twice with 37°C PBS. Cells were then fixed and stained with DAPI (0.1 μg/mL) in PBS for 15 minutes at room temperature. PBS-washed dishes were covered with coverslips and imaged with an Axioskop microscope (Zeiss). Gray pixels from pictures acquired were then quantified using ImageJ (NIH).
RT-qPCR in mammalian cells.
MCF7 and T47D cells were seeded in triplicate in 6-well plates at 5 × 105 cells per well in DMEM supplemented with 10% FBS. After incubation for 24 hours at 37°C and 5% CO2, cells were collected and total RNA was isolated and purified using a Total RNA Mini Kit (Geneaid) according to the manufacturer’s instructions. For RT-qPCR analysis, 1 μg of total RNA was reverse transcribed using the SuperScript III kit (Invitrogen). SYBR Green reactions using the SYBR Green qPCR supermix (Invitrogen) and specific primers (available upon request) were performed using an AriaMAX Real-time PCR system (Agilent Technologies). Relative expression of mRNAs was determined after normalization against housekeeping gene RPLP0 or B2M. Oligonucleotide sequences of the primers used for RT-qPCR can be found in Supplemental Table 1.
Protein extraction and immunoblotting.
For AMPK immunoblotting, cells were washed twice with cold PBS, lysed in AMPK lysis buffer (10 mM Tris-HCl [pH 8.0], 0.5 mM CHAPS, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, 5 mM NaF, 0.1 mM Na3VO4, 1 mM benzamidine, 5 mM NaPPi) supplemented with complete protease inhibitor (Roche) and DTT (1 mM), and cell lysates were cleared by centrifugation at 13,000g. For all other immunoblotting, cells were washed twice with cold PBS and lysed directly in RIPA light buffer (50 mM Tris-HCl [pH 8.8], 150 mM NaCl, 1% NP40, 0.1% SDS, 0.1% Triton X-100, 5 mM EDTA). Proteins were resolved in SDS-PAGE gels and revealed by Western blotting using the antibodies listed above.
Immunofluorescence.
Cells were washed with PBS and fixed in Petri dishes with 3.7% formaldehyde at room temperature for 30 minutes. After fixation, cells were washed twice with PBS and then permeabilized with 0.3% Triton X-100 in PBS at room temperature for 15 minutes. Cells were incubated in 5% BSA in PBS for 1 hour and then with anti-TFE3 primary antibody in 1.5% BSA in PBS for 2 hours at 37°C. Cells were washed 3 times with PBS and incubated with the corresponding secondary antibodies conjugated to Alexa Fluor 488 in 1.5% BSA in PBS for 30 minutes at 37°C. Cells were washed 3 times with PBS and incubated with DAPI (0.1 μg/mL) in PBS for 15 minutes at room temperature. PBS-washed dishes were coverslipped and observed with a Zeiss Axioskop microscope.
Metabolic assays.
Glucose production and lactate consumption were measured using a NOVA Bioanalysis flux analyzer or the Eton Bioscience kit (Eton Bioscience). Briefly, cells were plated at 500,000 cells/well in triplicate in 6-well plates in DMEM for 24 hours. Conditioned media were collected, spun down at 13,000g for 5 minutes, and transferred to new tubes in which the media were analyzed using the NOVA Bioanalysis flux analyzer. OCR and ECAR measurements were obtained using an XF96 Extracellular Flux Analyzer (Seahorse Bioscience). In brief, MCF7 EV, FLCN-KO, and FLCN-KO cells treated with TFE3 siRNA were plated at 10,000 cells/well in growth medium for 24 hours. After 24 hours, cells were incubated in nonbuffered DMEM containing 25 mM glucose and 2 mM glutamine in a CO2-free incubator at 37°C for 2 hours to allow for temperature and pH equilibration before loading into the XF96 apparatus. XF assays consisted of sequential mix (3 minutes), pause (3 minutes), and measurement (5 minutes) cycles, allowing for determination of OCR/ECAR every 10 minutes.
ATP quantification.
Cells were plated in triplicate at 10,000 cells/well in 96-well plates. After 24 hours, cells were lysed and mixed for 10 minutes (CellTiter-Glo luminescent cell viability assay, Promega). Luminescence was measured using Fluostar Omage (BMG Labtech) directly in the plates.
IHC.
Mammary tumors were fixed overnight in 4% paraformaldehyde at 4°C. After washing with 1× PBS, tumors were embedded in paraffin and sectioned by the GCRC Histology Core. Sections were stained using routine IHC protocols provided by the GCRC Histology Core using a Ventana BenchMark ULTRA system (Roche). Briefly, for Ventana: Sections were stained using routine IHC protocols provided by the GCRC Histology Core using a Ventana BenchMark ULTRA system (Roche). Slides were deparaffinized in EZ prep buffer for 8 minutes at 75°C. Antigen retrieval was performed by incubating slides in cell conditioning buffer 1 (CC1) at 95°C for 44 minutes. Slides were then blocked with the included Inhibitor CM at 37°C for 8 minutes. Incubation with primary antibody was conducted at 37°C for 32 minutes. Incubation with secondary antibody was performed by applying 1 drop of OmniMap anti-Rb HRP on slides for 16 minutes. Staining was revealed by adding one drop of DAB CM for 8 minutes. Slides were then incubated for 5 minutes with 1 drop of Copper CM followed by counterstain with hematoxylin for 8 minutes. Post counterstaining was performed for 8 minutes with Bluing Reagent. Slides were then dehydrated using increasing concentrations of ethanol, cleaned in xylene, and mounted using Acrytol mounting media. Staining was quantified with the Imagescope software (Aperio) using the positive pixel count algorithm (GPNMB, F4/80, CD31, and cleaved caspase-3) and nuclear algorithm (TFE3 and Ki67).
Quantification of soluble VEGF-A.
For in vitro studies, MCF7 cells were seeded in triplicate in 6-well plates at 5 × 105 cells per well in DMEM supplemented with 10% FBS. After incubation for 24 hours at 37°C and 5% CO2, conditioned media were collected, spun down at 13,000g for 5 minutes. VEGF-A levels in conditioned media were then assessed by employing a human VEGF-A quantikine ELISA kit (R&D Systems, DVE00) according to the manufacturer’s instructions. For in vivo studies, 1 μg of tumor lysates from each condition was used to assess levels of VEGF-A by employing the same human VEGF-A quantikine ELISA kit according to the manufacturer’s instructions.
Multiplex assay human angiogenesis array.
The Discovery Assay simultaneously measures 17 angiogenic/growth factors in a single microwell. The multiplex assay was performed by using the Bio-Plex 200 system (Bio-Rad Laboratories, Inc.) and a Milliplex Mouse Cytokine/Chemokine kit (Millipore). The 32-plex consisted of angiopoietin-2, BMP-9, EGF, endoglin, endothelin-1, FGF-1, FGF-2, FLCN, G-CSF, HB-EGF, HGF, IL-8, leptin, PLGF, VEGF-A, VEGF-C, and VEGF-D. The change in the cytokine levels in FLCN-KO medium was normalized against their respective WT medium.
Mammary fat pad injections.
For in vivo studies, 5 × 106 MCF7 cells were suspended in a 50:50 mixture of 1× PBS/Matrigel (Corning, 354248) and injected into the fourth mammary fat pads of 6- to 8-week-old NSG female mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) (Jackson Laboratory, 005557). Forty-eight hours before tumor inoculation, mice were injected subcutaneously with 1 μg of β-estradiol (Sigma-Aldrich, E8875) dissolved in corn oil and β-estradiol injections were repeated once per week until the experimental endpoint. Mammary tumors were monitored by palpation every few days and tumor volumes were calculated from weekly caliper measurements. Tumors were resected and harvested when tumor volumes reached between 150 and 300 mm3. Mice were housed in the McGill Animal Care Facility in standard cages with food and water ad libitum. Mice were maintained at 22°C to 24°C on a 12-hour light/12-hour dark cycle.
ROS potential.
Cellular ROS levels were determined using the general oxidative stress indicator CM-H2DCFDA (Invitrogen). Briefly, subconfluent adherent cells were incubated for 30 minutes at 37°C with CM-H2DCFDA dye. Cells were collected and analyzed using a BD FACSDiva analyzer. Cells not incubated with the dye or pretreated with 100 μM H2O2 were used as negative and ROS-positive controls, respectively.
IncuCyte cell proliferation assay.
Cells were seeded at 1 × 104 per well in a 6-well plate, which was then incubated at 37°C with 5% CO2 and monitored on the IncuCyte Live Cell Analysis System (Sartorius). After incubation for the indicated times, live-cell images were obtained using a 10× objective lens (4 images per well) within the instrument and cell density was analyzed using the IncuCyte software.
RNA extraction for RNA sequencing.
MCF7 EV and FLCN-KO mammary fat pad tumors were resected 6 weeks after injection, flash frozen, and stored in liquid nitrogen. RNA was extracted from 3 mammary tumors in each cohort using TRIzol and purified using QIAGEN RNeasy columns. RNA samples were processed for RNA-sequencing analysis at Genome Québec.
RNA-sequencing analysis.
Adaptor sequences and low-quality-score bases (Phred score < 30) were first trimmed using Trimmomatic (57). The resulting reads were aligned to the human genome reference sequence (GRCh38) using STAR (58). Read counts were obtained using HTSeq (59) and are represented as a table that reports, for each sample (columns), the number of reads mapped to a given gene (rows). For all downstream analyses, genes exhibiting low expression levels with an average read count lower than 10 across all samples were excluded, resulting in 19,133 genes in total. The R package limma (60) was used to identify differences in gene expression levels between WT and FLCN-KO samples. Nominal P values were corrected for multiple testing using the Benjamini-Hochberg method. The complete list of differentially expressed genes is presented in Supplemental Table 2.
Gene set enrichment analysis.
We used Enrichr (61) (https://amp.pharm.mssm.edu/Enrichr/) to test for enrichment of functionally annotated gene sets among the differentially expressed genes. The complete GO enrichment results are reported in Supplemental Table 3.
Data and software availability.
The RNA-sequencing and microarray data are deposited in NCBI’s Gene Expression Omnibus database (GEO GSE163791). PDX breast cancer RNA-sequencing data were obtained from Savage et al. (32): GEO GSE14276. Normal breast gene expression levels were obtained from the Genotype-Tissue Expression (GTEx) Portal (https://gtexportal.org/home/). TCGA breast cancer data were obtained from Firehose Broad GDAC (illuminahiseq_rnaseqv2-RSEM_genes_normalized - http://gdac.broadinstitute.org/ Accessed March 17, 2019). The intrinsic molecular breast cancer subtyping was obtained according to Paquet and Hallett (62).
Statistics.
Data are expressed as mean ± SEM. Statistical analyses for all data were performed using 2-tailed Student’s t test for comparisons between 2 groups, 1-way or 2-way ANOVA with Bonferroni’s correction for comparisons between 3 or more groups, and log-rank Mantel-Cox test for survival plots, using GraphPad Prism 7 software. The data were assumed normal as tested by the Shapiro-Wilk normality test. Statistical significance is indicated in figures (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001) or included in the supplemental tables, with a P value of less than 0.05 considered significant. In vitro studies were biologically repeated at least 3 times in triplicate. The numbers of animals in each experiment are indicated in the figure legends.
Study approval.
All mouse studies were approved by the Animal Resource Centre at McGill University and comply with guidelines set by the Canadian Council of Animal Care.
Author contributions
LEH, MB, PMS, and A Pause conceived and designed the experiments. LEH, MB, M Paquette, A Pacis, and HK performed, collected, and assembled the experiments. LEH, MB, PMS, and A Pause wrote the manuscript. LEH, MB, M Paquette, HK, M Park, PMS, and A Pause revised the manuscript critically for important intellectual content.
Supplementary Material
Acknowledgments
LEH and MB were supported by Fonds de la Recherche du Québec- Santé (FRQS) PhD and postdoctoral fellowships, respectively. M Paquette was supported by a Canadian Institutes of Health Research (CIHR) PhD fellowship. A Pause acknowledges that funding for this work was supported by grants from CIHR (PJT-165829) and the Cancer Research Society (CRS 79664). PMS and A Pause acknowledge funding from a Terry Fox New Frontiers Program Project Grant (TFRI-251427). PMS acknowledges funding from a CIHR grant (PJT-247494). PMS is a McGill University William Dawson Scholar. We acknowledge Matthew Annis and Sebastien Tabaries for help with animal work. Microscopy image processing and analysis for this manuscript was performed in the McGill University Life Sciences Complex Advanced BioImaging Facility (ABIF). We also thank the McGill Core Flow Cytometry Facility (McGill University) for their technical support and expertise. We would like to thank the Canadian Centre for Computational Genomics Genome Canada–funded bioinformatics platform and the GCRC Histology Core Facility (McGill University) for their services. PDX banking was performed by the Breast Cancer Functional Genomics Group breast tissue and data bank at the GCRC (Research Institute of the McGill University Health Centre) in collaboration with the Réseau de Recherche sur le cancer (FRQS), which is affiliated with the Canadian Tumor Repository Network (CTRNet). We further acknowledge support from the GCRC Metabolomics Core Facility (McGill University), which is supported by the Canada Foundation for Innovation, the Terry Fox Research Institute, and the Québec Breast Cancer Foundation.
Version 1. 11/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(22):e144871.https://doi.org/10.1172/JCI144871.
Contributor Information
Leeanna El-Houjeiri, Email: leeanna.el-houjeiri@mail.mcgill.ca.
Marco Biondini, Email: marco.biondini@mail.mcgill.ca.
Mathieu Paquette, Email: mathieu.paquette2@mail.mcgill.ca.
Helen Kuasne, Email: hellen.kuasne@mcgill.ca.
Alain Pacis, Email: alain.pacis@computationalgenomics.ca.
Morag Park, Email: morag.park@mcgill.ca.
Arnim Pause, Email: arnim.pause@mcgill.ca.
References
- 1.Siegl Rl, et al. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foulkes WD, et al. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.El-Houjeiri L, et al. The transcription factors TFEB and TFE3 link the FLCN-AMPK signaling axis to innate immune response and pathogen resistance. Cell Rep. 2019;26(13):3613–3628. doi: 10.1016/j.celrep.2019.02.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Possik E, et al. FLCN and AMPK confer resistance to hyperosmotic stress via remodeling of glycogen stores. PLoS Genet. 2015;11(10):e1005520. doi: 10.1371/journal.pgen.1005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Possik E, et al. Folliculin regulates ampk-dependent autophagy and metabolic stress survival. PLoS Genet. 2014;10(4):e1004273. doi: 10.1371/journal.pgen.1004273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan M, et al Chronic AMPK activation via loss of FLCN induces functional beige adipose tissue through PGC-1alpha/ERRalpha. Genes Dev. 2016;30(9):1034–1046. doi: 10.1101/gad.281410.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan M, et al. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J Clin Invest. 2014;124(6):2640–2650. doi: 10.1172/JCI71749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baba M, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103(42):15552–15527. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takagi Y, et al. Interaction of folliculin (Birt-Hogg-Dube gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 2008;27(40):5339–5347. doi: 10.1038/onc.2008.261. [DOI] [PubMed] [Google Scholar]
- 15.Tee AR, Pause A. Birt-Hogg-Dube: tumour suppressor function and signalling dynamics central to folliculin. Fam Cancer. 2013;12(3):367–372. doi: 10.1007/s10689-012-9576-9. [DOI] [PubMed] [Google Scholar]
- 16.Wilson GK, et al. Hypoxia inducible factors in liver disease and hepatocellular carcinoma: current understanding and future directions. J Hepatol. 2014;61(6):1397–1406. doi: 10.1016/j.jhep.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 17.Keith B, et al. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krock BL, et al. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. 2011;2(12):1117–1133. doi: 10.1177/1947601911423654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linderholm B, et al. Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer. Ann Oncol. 2009;20(10):1639–1646. doi: 10.1093/annonc/mdp062. [DOI] [PubMed] [Google Scholar]
- 20.Mohammed RA, et al. Lymphatic and blood vessels in basal and triple-negative breast cancers: characteristics and prognostic significance. Mod Pathol. 2011;24(6):774–785. doi: 10.1038/modpathol.2011.4. [DOI] [PubMed] [Google Scholar]
- 21.Andre F, Zielinski C. Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents. Ann Oncol. 2012;23(suppl_6):vi46–vi51. doi: 10.1093/annonc/mds195. [DOI] [PubMed] [Google Scholar]
- 22.Betschinger J, et al. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153(2):335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200(4):475–491. doi: 10.1083/jcb.201209135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petit CS, et al. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202(7):1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada S, et al. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev. 2016;30(22):2551–2564. doi: 10.1101/gad.287953.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perera RM, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastore N, et al. Nutrient-sensitive transcription factors TFEB and TFE 3 couple autophagy and metabolism to the peripheral clock. EMBO J. 2019;38(12):e101347. doi: 10.15252/embj.2018101347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martina JA, et al. Novel roles for the MiTF/TFE family of transcription factors in organelle biogenesis, nutrient sensing, and energy homeostasis. Cell Mol Life Sci. 2014;71(13):2483–2497. doi: 10.1007/s00018-014-1565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raben N, Puertollano R TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu Rev Cel Dev Biol. 2016;32:255–278. doi: 10.1146/annurev-cellbio-111315-125407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen K, et al. Cryo-EM structure of the human FLCN-FNIP2-Rag-ragulator complex. Cell. 2019;179(6):1319–1329. doi: 10.1016/j.cell.2019.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence RE, et al. Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science. 2019;366(6468):971–977. doi: 10.1126/science.aax0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petit CS, et al. Recruitment of folliculin to lysosomes supports the amino acid–dependent activation of Rag GTPases. J Cell Biol. 2013;202(7):1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vocke CD, et al. High frequency of somatic frameshift BHD gene mutations in Birt-Hogg-Dube–associated renal tumors. J Natl Cancer Inst. 2005;97(12):931–935. doi: 10.1093/jnci/dji154. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt LS, Linehan WM. Molecular genetics and clinical features of Birt-Hogg-Dube syndrome. Nat Rev Urol. 2015;12(10):558–569. doi: 10.1038/nrurol.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savage P, et al. Chemogenomic profiling of breast cancer patient-derived xenografts reveals targetable vulnerabilities for difficult-to-treat tumors. Commun Biol. 2020;3(1):1–15. doi: 10.1038/s42003-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong SB, et al. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PloS One. 2010;5(12):e15793. doi: 10.1371/journal.pone.0015793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maric G, et al. Glycoprotein non-metastatic b (gpnmb): A metastatic mediator and emerging therapeutic target in cancer. Onco Targets Ther. 2013;6:839–852. doi: 10.2147/OTT.S44906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vázquez CL, Colombo MI. Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol. 2009;452:85–95. doi: 10.1016/S0076-6879(08)03606-9. [DOI] [PubMed] [Google Scholar]
- 40.Cairns RA, et al. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 41.Lugano R, et al. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci. 2020;77(9):1745–1770. doi: 10.1007/s00018-019-03351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talks KL, et al. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157(2):411–421. doi: 10.1016/S0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perera RM, et al. MiT/TFE family of transcription factors, lysosomes, and cancer. Annu Rev Cancer Biol. 2019;3:203–222. doi: 10.1146/annurev-cancerbio-030518-055835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bost F, Kaminski L. The metabolic modulator PGC-1α in cancer. Am J Cancer Res. 2019;9(2):198–211. [PMC free article] [PubMed] [Google Scholar]
- 45.Aggarwal V, et al. Role of reactive oxygen species in cancer progression: molecular mechanisms and recent advancements. Biomolecules. 2019;9(11):735. doi: 10.3390/biom9110735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kauffman EC, et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol. 2014;11(8):465–475. doi: 10.1038/nrurol.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eichner LJ, et al. Genetic analysis reveals AMPK is required to support tumor growth in murine Kras-dependent lung cancer models. Cell Metab. 2019;29(2):285–302. doi: 10.1016/j.cmet.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bos R, et al. Levels of hypoxia-inducible factor-1α during breast carcinogenesis. J Natl Cancer Inst. 2001;93(4):309–314. doi: 10.1093/jnci/93.4.309. [DOI] [PubMed] [Google Scholar]
- 50.Giatromanolaki A, et al. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer. 2001;85(6):881–890. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steingrímsson E, et al. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development. 1998;125(23):4607–4616. doi: 10.1242/dev.125.23.4607. [DOI] [PubMed] [Google Scholar]
- 52.Gad S, et al. Mutations in BHD and TP53 genes, but not in HNF1β gene, in a large series of sporadic chromophobe renal cell carcinoma. B J Cancer. 2007;96(2):336–340. doi: 10.1038/sj.bjc.6603492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khoo SK, et al. Inactivation of BHD in sporadic renal tumors. Cancer Res. 2003;63(15):4583–4587. [PubMed] [Google Scholar]
- 54.Schmidt LS, Linehan WM. FLCN: The causative gene for Birt-Hogg-Dubé syndrome. Gene. 2018;640:28–42. doi: 10.1016/j.gene.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pradella LM, et al. Where Birt-Hogg-Dubé meets Cowden syndrome: mirrored genetic defects in two cases of syndromic oncocytic tumours. Eur J Hum Genet. 2013;21(10):169–1172. doi: 10.1038/ejhg.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolger AM, et al. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, et al. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(1):128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paquet ER, Hallett MT. Absolute assignment of breast cancer intrinsic molecular subtype. J Natl Cancer Inst. 2015;107(1):dju357. doi: 10.1093/jnci/dju357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.