Abstract
The complete plastome sequence of Amaryllis belladonna L. is assembled and annotated. This is the type species of Amaryllis and therefore the type of the family Amaryllidaceae and as such, is important to document the phylogenetic position of the family. The plastome sequence has a length of 158,145 bp, with the large single copy (LSC) regions comprising 85,963 bp, the small single copy (SSC) 18634 bp and two identical inverted repeats (IR) regions each of 26,774 bp. Phylogenetic analysis fully resolved Amaryllis in a clade with Crinum L. in the Amaryllidoideae, as expected, with the Allioideae as a sister group. Agapanthus (Agapanthoideae) is a sister to the other two subfamilies in the Amaryllidaceae. The phylogenetic tree produced corresponds to previous topologies based on plastome molecular markers including matK, ndhF and rbcL. This is the first paper reporting the whole plastome comparison of the type genera of all three subfamilies in the Amaryllidaceae.
Keywords: Amaryllis, Amaryllidoideae, Cape Province, geophyte, ornamental plant
The asparagalean family Amaryllidaceae J. St.-Hil. consists of three subfamilies: Agapanthoideae Endl. (one genus with nine species), Allioideae Herb. (13 genera with 795 species) and Amaryllidoideae (75 genera with ∼900 species; Meerow et al. 2020). The family has a cosmopolitan distribution, with centers of diversity for the Amaryllidoideae in the Mediterranean Basin, South Africa, and South America (Rønsted et al. 2012). The relationship between the subfamilies: (Agapanthoideae, Allioideae, Amaryllidoideae) is well-established based on plastid DNA data (Fay et al. 2000; Pires et al. 2006; Seberg et al. 2012; Givnish et al. 2018), however, the relationship between genera within the Amaryllidoideae remains elusive (Meerow et al. 2003; Meerow et al. 2006; Garcia et al. 2014). Amaryllis belladonna is the type species of the genus Amaryllis L. and consequently the type of the family Amaryllidaceae. Amaryllis comprises two species, both native to the Cape Province in South Africa. Amaryllis belladonna, also known as the ‘belladonna lily’, is a widely planted ornamental of economic importance (Tallini et al. 2017) and may have antiplasmodial uses (Cho et al. 2018). Amaryllis paradisicola Snijman is a rare species from Richtersveld National Park (Snijman and Williamson 1998) comprising, possibly, around 2000 individuals (Snijman et al. 2016). Here we report the complete plastome sequence of A. belladonna to contribute to the bioinformatic and systematic knowledge of the Amaryllidaceae.
Scape tissue was collected in silica gel from A. belladonna blooming at the Royal Horticultural Society’s Garden, Wisley, UK (51.312695° N, 0.476724° W). A herbarium voucher was deposited at WSY (WSY0150055; contact: Yvette Harvey, YvetteHarvey@rhs.org.uk). Total genomic DNA was extracted using the Qiagen DNeasy Plant Mini Kit (QIAGEN, Manchester, UK). Library development and Illumina HiSeq 150 bp PE sequencing were completed by Novogene Company Limited (Cambridge, UK).
Fast-Plast v1.2.8 (McKain and Wilson 2017) and NovoPlasty v3.7.0 (Dierckxsens et al. 2017) were used to assemble the plastome. The Bowtie reference index was built using Asparagales plastomes included in Fast-Plast. The complete plastome of Narcissus poeticus L. (MH706763; Könyves et al. 2018) was used as the starting seed for the NovoPlasty assembly. The memory was limited to 6 Gb. The complete plastome was annotated against the published Narcissus poeticus L. plastome (MH706763; Könyves et al. 2018) using Geneious Prime (v2020.2.5; https://www.geneious.com/). The A. belladonna plastome sequence was aligned to 20 published Asparagales plastome sequences including Hyacinthoides non-scripta (L.) Chouard ex Rothm. (MN824434, Asparagaceae; Garnett et al. 2020) and Xanthorrhoea preissii Endl. (KX822774, Asphodelaceae) as outgroups using the default settings with MAFFT v7.450 (Katoh et al. 2002; Katoh and Standley 2013) in Geneious Prime. Poorly aligned regions of the total alignment were excluded using trimAI v1.2 with default settings and converted to a FASTA file using readAI v1.2 (Capella-Gutiérrez et al. 2009). A maximum likelihood estimation was performed with RAxML v8.2.11 (Stamatakis 2014) in Geneious Prime using the model GTR + G + I and 1000 bootstrap replicates.
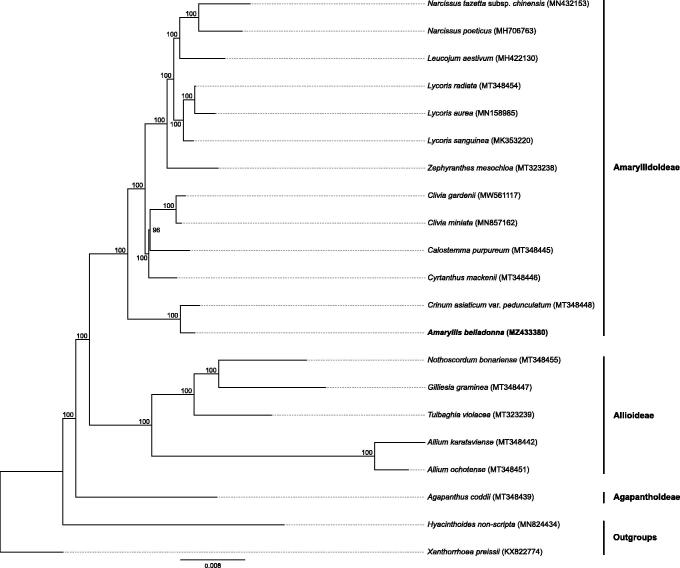
The plastome sequence of A. belladonna (MZ433380) is 158,145 bp in length, comprising the LSC (85,963 bp), SSC (18,634 bp) and two inverted repeat regions each 26,774 bp. The plastome contains 86 protein-coding, 38 tRNA and eight rRNA genes. Of these eight protein-coding genes, eight tRNA genes and four rRNA genes are duplicated in the inverted repeats. There were high levels of gene synteny between A. belladonna and other published Amaryllidoideae plastomes (Könyves et al. 2018; Li et al. 2018; Zhang et al. 2020). The phylogenetic tree produced corresponds to previous topologies based on plastome molecular markers including matK (Ito et al. 1999; Rønsted et al. 2012; Chen et al. 2013), ndhF and rbcL (Chen et al. 2013). Comparison within Amaryllidoideae samples confirms that this subfamily has a lower substitution rate than Allioideae.
The position of A. belladonna in the phylogenetic tree is congruent with previous papers (Ito et al. 1999; Rønsted et al. 2012; Chen et al. 2013) and the substitution rate modeled on the tree is similar to that for other Amaryllidoideae, and lower than that in Allioideae (Figure 1). As the type species of the family Amaryllidaceae and the subfamily Amaryllidoideae, this is a new and important reference in the plastome-based phylogeny of the group.
Figure 1.
RaxML phylogenetic tree based on 21 asparagalean complete plastome sequences. Bootstrap support values are shown at each branch and GenBank accession numbers are given in brackets, subfamilies of the samples are shown on the right. Text in bold shows the plastome assembled in this study.
Funding Statement
T
his study was funded by the Royal Horticultural Society and SCENARIO DTP.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The annotated plastome sequence is available on GenBank on NCBI (https://www.ncbi.nlm.nih.gov) under accession number MZ433380. Raw sequence data were deposited in SRA (BioProject: PRJNA743246); SRA: SRR15031247; BioSample: SAMN20014353).
References
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAL: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Kim D-K, Chase MW, Kim J-H.. 2013. Networks in a large-scale phylogenetic analysis: reconstructing evolutionary history of Asparagales (Lilianae) based on four plastid genes. PLoS One. 8(3):e59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N, Du Y, Valenciano AL, Fernández-Murga ML, Goetz M, Clement J, Cassera MB, Kingston DG.. 2018. Antiplasmodial alkaloids from bulbs of Amaryllis belladonna Steud. Bioorg Med Chem Lett. 28(1):40–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MF, Rudall PJ, Sullivan S, Stobart KL, De Bruijn AY, Reeves G, Qamaruz-Zaman F, Hong WP, Joseph J, Hahn WJ, et al. 2000. Phylogenetic studies of Asparagales based on four plastid regions. In: Wilson KL, Morrison DA, editors. Melbourne, Australia: Monocots: Systematics and Evolution, CSIRO; p. 360–371. [Google Scholar]
- Garcia N, Meerow AW, Soltis DE, Soltis PS.. 2014. Testing deep reticulate evolution in amaryllidaceae tribe hippeastreae (Asparagales) with ITS and chloroplast sequence data. Syst Bot. 39(1):75–89. [Google Scholar]
- Garnett GJL, Könyves K, Bilsborrow J, David J, Culham A.. 2020. The complete plastome of Hyacinthoides non-scripta (L.) Chouard ex Rothm. (Asparagaceae). Mitochondrial DNA B Resour. 5(1):1003–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ, Zuluaga A, Spalink D, Gomez MS, Lam VKY, Saarela JM, Sass C, Iles WJD, de Sousa DJL, Leebens-Mack J, Pires JC, et al. 2018. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am J Bot. 105(11):1888–1910. [DOI] [PubMed] [Google Scholar]
- Ito M, Kawamoto A, Kita Y, Yukawa T, Kurita S.. 1999. Phylogenetic relationships of Amaryllidaceae based on matK sequence data. J Plant Res. 112(2):207–216. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könyves K, Bilsborrow J, David J, Culham A.. 2018. The complete chloroplast genome of Narcissus poeticus L. (Amaryllidaceae: Amaryllidoideae). Mitochondrial DNA Part B. 4(2):3364–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M-DL, Wu X, Wu J-J, Zhou X, Wang R-H, Qi Z-C.. 2018. Characterization of the complete chloroplast genome of summer snowflake (Leucojum aestivum, Amaryllidaceae). Mitochondrial DNA B Resour. 3(2):1069–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKain M, Wilson M.. 2017. Fast-Plast: rapid de novo assembly and finishing for whole chloroplast genomes. https://github.com/mrmckain/Fast-Plast.
- Meerow AW, Francisco-Ortega J, Kuhn DN, Schnell RJ.. 2006. Phylogenetic relationships and biogeography within the Eurasian clade of Amaryllidaceae based on plastid ndhF and nrDNA ITS sequences: lineage sorting in a reticulate area? 31(1):42–60. [Google Scholar]
- Meerow AW, Gardner EM, Nakamura K.. 2020. Phylogenomics of the Andean tetraploid clade of the American Amaryllidaceae (subfamily Amaryllidoideae): unlocking a polyploid generic radiation abetted by continental geodynamics. Front Plant Sci. 11:582422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerow AW, Lehmiller DJ, Clayton JR.. 2003. Phylogeny and biogeography of Crinum L. (Amaryllidaceae) inferred from nuclear and limited plastid non-coding DNA sequences. Bot J Linnean Soc. 141(3):349–363. [Google Scholar]
- Pires C, Maureira I, Givnish T, Systma K, Seberg O, Peterson G, Davis J, Stevenson D, Rudall P, Fay M, et al. 2006. Phylogeny, genome size, and chromosome evolution of asparagales. ALISO. 22(1):287–304. [Google Scholar]
- Rønsted N, Symonds MRE, Birkholm T, Christensen SB, Meerow AW, Molander M, Molgaard P, Petersen G, Rasmussen N, van Staden J, et al. 2012. Can phylogeny predict chemical diversity and potential medicinal activity of plants? A case study of Amaryllidaceae. BMC Evol Biol. 12:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seberg O, Petersen G, Davis JI, Pires JC, Stevenson DW, Chase MW, Fay MF, Devey DS, Jørgensen T, Sytsma KJ, et al. 2012. Phylogeny of the Asparagales based on three plastid and two mitochondrial genes. Am J Bot. 99(5):875–889. [DOI] [PubMed] [Google Scholar]
- Snijman DA, Van Wyk PCV, Raimondo D, von Staden L.. 2016. Amaryllis paradisicola. The IUCN red list of threatened species 2016: e.T103633495A104109638; [accessed 2021 July 16]. 10.2305/IUCN.UK.2016-3.RLTS.T103633495A104109638.en [DOI]
- Snijman DA, Williamson G.. 1998. A new species of Amaryllis from the Richtersveld, South Africa. Bothalia. 28(2):192–196. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini LR, de Andrade JP, Kaiser M, Viladomat F, Nair JJ, Zuanazzi JAS, Bastida J.. 2017. Alkaloid constituents of the Amaryllidaceae plant Amaryllis belladonna L. Molecules. 22(9):1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang T, Shu X, Wang N, Zhuang W, Wang Z.. 2020. Complete chloroplast genomes and comparative analyses of L. chinensis, L. anhuiensis, and L. aurea (Amaryllidaceae). IJMS. 21(16):5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The annotated plastome sequence is available on GenBank on NCBI (https://www.ncbi.nlm.nih.gov) under accession number MZ433380. Raw sequence data were deposited in SRA (BioProject: PRJNA743246); SRA: SRR15031247; BioSample: SAMN20014353).