Abstract
Background
We evaluated the efficacy of the Siemens SARS-CoV-2 Total Antibody assay (CV2T) and IgG assay (CV2G) that can detect antibodies against the receptor binding domain of S antigen in patients with COVID-19 in a Tokyo metropolitan area.
Methods
Sensitivity and antibody levels were examined by CV2T and CV2G on Dimension EXL 200 using 236 serum samples obtained from 79 RT-PCR confirmed COVID-19 patients at multiple time points and were compared with disease severity by the World Health Organization criteria. The assay specificity was evaluated using samples collected before the COVID-19 pandemic.
Results
The sensitivity of CV2T and CV2G were low (16.7–21.4%) in days 0–6 and increased to 43.8–52.5% in days 7–13 and to 80.8–90.0% in days 14–20. The seroprevalences persisted after day 21 to days past 42 regardless of disease severity. In every day grouping, mean antibody levels were higher in severe cases than in mild cases with a significant difference in days 14–20 and days 20–27. The specificity was 97.9 % (95% CI; 92.8–99.8) for CV2T and 99.0 % (95% CI; 94.6–100) for CV2G.
Conclusions
Our results indicate a high specificity and high sensitivity at 14 days of CV2T and CV2G as antibody detection assays.
Keywords: COVID-19, SARS-CoV-2, Serology, Antibody testing
COVID-19, SARS-CoV-2, Serology, Antibody testing.
1. Introduction
A serious respiratory illness caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), referred to as Coronavirus disease 2019 (COVID-19), spread globally, and the World Health Organization (WHO) declared a pandemic in March 2020. The real-time reverse transcriptase polymerase chain reaction (RT-PCR) test, which detects SARS-CoV-2 viral RNA, was developed to confirm the diagnosis of COVID-19 [1]. However, sensitivity of RT-PCR tests can be affected by viral loads in specimens, timing of sample collection during the disease course, and the oligo target of RT-PCR. In fact, several studies demonstrated false negative results in RT-PCR tests [2, 3, 4].
To date, various kinds of serological tests, including lateral flow assays and fully automated antibody detection devices using Chemiluminescent microparticle immunoassay or Electro-Chemiluminescent immunoassay, have been developed to evaluate the status of past infection or to support diagnosis by RT-PCR assays [5]. Many of these serological tests received Emergency Use Authorization by the U.S. Food and Drug Administration (FDA) [6]. Although the performance of these assays have claimed to be rigorously tested, including high throughput capability and high sensitivity and specificity, the clinical significance of seroprevalence remains undetermined except for epidemiological studies described in the COVID-19 Serology Surveillance Strategy by the Centers for Disease Control and Prevention (CDC) [7].
Conversely, as vaccinations using messenger RNA (mRNA) composed of full length S1-receptor binding protein (S1-RBD) and S-antigen have been rapidly developed and used in many countries [8], the measurement of IgG antibody titers against the S1-RBD gained much more attention since the titers showed a good correlation with neutralizing antibody activity [9, 10, 11, 12]. However, these studies used in-house S1-RBD ELISA to determine IgG titer which is time consuming and laborious. Thus, the development of a fully automated reliable assay targeting the S1-RBD is indispensable since it has potential for using the IgG titer as a surrogate for the neutralizing antibody assay result.
Recently, Siemens Healthcare Diagnostics developed a fully automated assay, SARS-CoV-2 Total Antibody assay and IgG assay, that can specifically detect total and IgG antibodies against S1-RBD and was recently approved for emergency use by the FDA. In this study, we evaluated the clinical performance of this antibody assay using samples collected from patients with COVID-19 with varying disease severity at Juntendo University Hospital in Tokyo, Japan.
2. Materials and methods
2.1. Patient information and sample collection
Two hundred and thirty-six (236) serum samples were collected from a total of 79 symptomatic COVID-19 patients between March and August 2020. Table 1 shows the clinical information of the patients. All patients were diagnosed with COVID-19 by RT-PCR testing. RT-PCR was performed with the Light Mix Modular SARS-CoV-2 (COVID-19) N-gene and E-gene assay (Roche Diagnosis, Tokyo, Japan) or the 2019 Novel Coronavirus Detection Kit (Shimadzu, Kyoto, Japan). All clinical information was acquired by reviewing the patient charts. Based on patients’ disease severity, they were classified into two groups: Group M, equivalent to mild (without pneumonia or hypoxia) and moderate (with signs of pneumonia and SpO2 ≥ 90% in room air) cases, and Group S, equivalent to severe (with signs of pneumonia and respiratory rate of ≥30/min or SpO2 < 90% in room air) and critical (under acute respiratory distress syndrome) cases, as defined by WHO criteria. To assess specificity of the assay, samples collected in 2017 and 2018 (i.e., pre-COVID-19) were used as negative controls. The samples were stored at -80 °C until use.
Table 1.
Clinical characteristics of patients with COVID-19.
| Disease severity | Group M |
Group S |
||
|---|---|---|---|---|
| Mild | Moderate | Severe | Critical | |
| Patient | ||||
| Number (n = 79) | 51.9% (41/79) | 26.6% (21/79) | 15.2% (12/79) | 6.3% (5/79) |
| Male (%) | 68.3% (28/41) | 61.9% (13/21) | 83.3% (10/12) | 80.0% (4/5) |
| Age, y (mean) | 23-87 (46.4) | 18-75 (47.0) | 46-84 (62.9) | 66-86 (77.0) |
| Sample number (n = 236) | ||||
| Days from symptom onset | ||||
| 0-6 | 22 | 6 | 9 | 3 |
| 7-13 | 23 | 17 | 14 | 2 |
| 14-20 | 16 | 10 | 7 | 3 |
| 21-27 | 14 | 17 | 5 | 6 |
| 28-34 | 8 | 6 | 5 | 2 |
| 35-41 | 5 | 3 | 2 | 2 |
| 42+ | 6 | 2 | 4 | 17 |
The research related to human use has complied with all the relevant national regulations and institutional policies, and was conducted in accordance with the tenets of the Helsinki Declaration. It has been approved by the Institutional Review Board (IRB) at Juntendo University Hospital (IRB #20-036). Informed consents were obtained from all individuals included in this study.
2.2. Anti-SARS-CoV-2 antibody testing
The Siemens SARS-CoV-2 Total Antibody assay (CV2T) and SARS-CoV-2 IgG (CV2G) were performed using serum samples on the Dimension EXL 200 Integrated Chemistry System (Siemens Healthcare Diagnostics, Delaware, USA) according to manufacturer instructions. These assays are homogeneous sandwich chemiluminescent immunoassays based on the Luminescent Oxygen Channeling Immunoassay (LOCI) that uses the S1-RBD as a target for antibody detection. Fluorescence signal intensities measured at a wavelength of 612 nm were directly correlated with antibody concentrations, which were indicated as antibody levels (QUAL). For semiquantitative CV2G assay, Ind is used instead of QUAL. One QUAL equal to one Ind. The limit of detection (LoD) for CV2G is 600 Ind, and assay linear range is between 600 and 140,000 Ind. However, CV2T is a qualitative assay, and the linear range and LoD have not been determined. Antibody levels greater than or equal to 1000 QAUL and 1000 Ind units were interpreted as positive [13]. According to the manufacture, 1000 QUAL equals to 12 BAU/ml for CV2T and 1000 Ind equals to 17 BAU/ml for CV2G.
2.3. Statistical analysis
RT-PCR was considered the gold standard for the detection of SARS-CoV-2, and the sensitivity for the assays was assessed using days from the onset of symptoms. Confidence intervals (CI) were calculated by the Clopper-Pearson method for sensitivity and specificity. Antibody levels between Group M and Group S were compared with Mann-Whitney U testing. Antibody levels between groups of days from onset of symptoms were compared by Kruskal-Wallis analysis. A two-tailed p value of <0.05 was considered statistically significant. Statistical analysis was performed by EZR [14].
3. Results
The precision of the antibody assay was evaluated according to the Clinical and Laboratory Standards Institute guideline EP15-A3 using QC materials consisting of positive and negative serum controls for the antibodies against S1-RBD provided by the manufacturer (Siemens Healthcare Diagnostics, Delaware, USA) and one pooled sample obtained from patients with COVID-19 at Juntendo University Hospital. Reproducibility (n = 20) was assessed by replicates in one day on one instrument. Confidence of variation (CV) was 2.2–7.0 % for CV2T and 1.3–3.8 % for CV2G. Within-laboratory reproducibility (n = 20) was assessed by duplicate assays two times a day for five days. The CV was 4.0–5.7 % for CV2T and 2.9–7.5 % for CV2G.
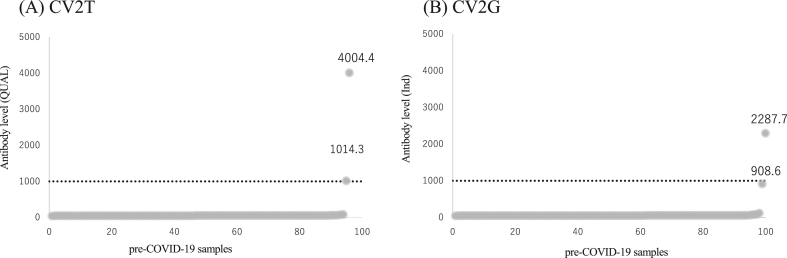
Specificity of the antibody assay was assessed using the pre-COVID-19 samples. Two out of 98 samples were detected as positive by CV2T, and 1 out of 100 samples was detected as positive by CV2G, yielding the specificity of 97.9 % (95% CI: 92.8–99.8) for CV2T and 99.0 % (95% CI: 94.6–100) for CV2G. Figure 1 shows the actual antibody levels of all negative control samples simultaneously measured by CV2T (panel A) and CV2G (panel B). One false positive sample with an antibody level of 4004.4 QUAL by CV2T showed 2287.7 Ind by CV2G. In contrast, the other false positive sample with 1014.3 QUAL by CV2T showed 908.6 Ind by CV2G, which was interpreted as negative since the cut off value was set to 1000 Ind. Nonetheless, all other negative controls showed negligible antibody levels, which validated high specificity of this assay.
Figure 1.
Distribution of antibody levels using pre-COVID-19 samples. CV2T (A) and CV2G (B) results are shown in rank order and more than 100 QUAL and 100 Ind are labeled. Cut off (1000 QUAL and 1000 Ind) is shown in dotted line.
Next, sensitivity of the antibody assay was examined using a total of 236 samples obtained from 79 patients whose RT-PCR tests were positive for SARS-CoV-2. Table 2 summarizes the seroprevalences measured in various timing from onset of symptoms. The sensitivity increased relative to the day from onset of symptoms. The seroprevalences measured by CV2T were relatively low (16.7–52.5%) in early phase (day 0–13) in both Group M and Group S. However, the seroprevalences by CV2T were relatively high (80.8–93.5%) in day 14–20 and day 21–27 in both groups. Between day 28 and 42 and higher, Group M showed 100% seroprevalence, and Group S showed 75–100% seroprevalence. The seroprevalences measured by CV2G showed similar results as measured by CV2T. The seroprevalences were relatively low in early phase and were relatively high in day 14–27. Then, Group M showed 100% seroprevalence and Group S showed 75–100% seroprevalence in day 28–42 plus.
Table 2.
Assay sensitivity of CV2T and CV2G.
| Days from symptom onset (Group M, Group S) | Sensitivity (95% CI) |
|||
|---|---|---|---|---|
| CV2T |
CV2G |
|||
| Group M | Group S | Group M | Group S | |
| 0-6 (n = 28,12) | 21.4% (8.3–41.0) | 16.7% (2.1–48.4) | 17.9% (6.1–36.9) | 16.7% (2.1–48.4) |
| 7-13 (n = 40, 16) | 52.5% (36.1–68.5) | 50.0% (24.7–75.3) | 52.5% (36.1–68.5) | 43.8% (19.8–70.1) |
| 14-20 (n = 26, 10) | 80.8% (60.6–93.4) | 90.0% (55.5–99.7) | 80.8% (60.6–93.4) | 90.0% (55.5–99.7) |
| 21-27 (n = 31, 11) | 93.5% (78.6–99.2) | 90.9% (58.7–99.8) | 93.5% (78.6–99.2) | 90.9% (58.7–99.8) |
| 28-34 (n = 14, 7) | 100% (76.8–100) | 85.7% (42.1–99.6) | 100% (76.8–100) | 85.7% (42.1–99.6) |
| 35-41 (n = 8, 4) | 100% (63.1–100) | 75.0% (19.4–99.4) | 100% (63.1–100) | 75.0% (19.4–99.4) |
| 42+ (n = 8, 21) | 100% (63.1–100) | 100% (83.9–100) | 100% (63.1–100) | 100% (83.9–100) |
Interestingly, one patient in Group S retained negative seroprevalence for all collected samples. For this patient, more than one sample was collected every week and the last sample collection was in day 37. The patient suffered from Pneumocystis pneumonia, Cytomegalovirus (CMV) infection, and deep-seated mycosis, indicating the patient was immunocompromised. When this patient was excluded from the analysis, all samples collected in Group S after day 14 had positive seroprevalence and the sensitivity reached 100%.
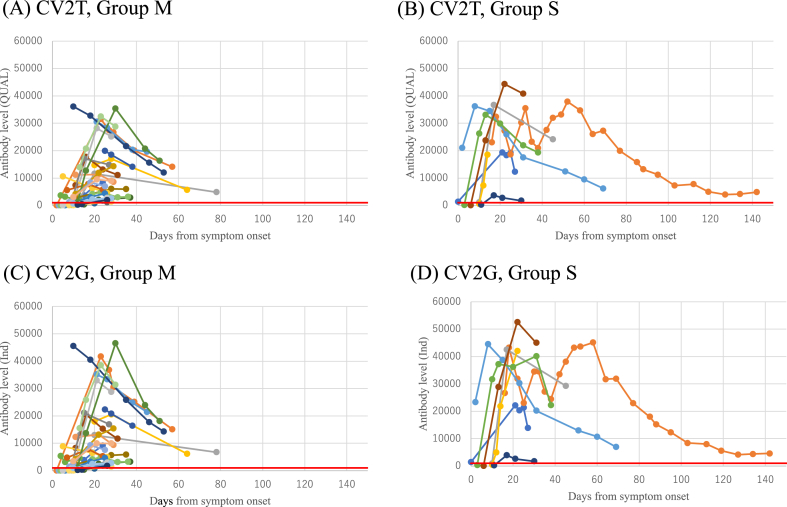
To determine dynamic changes of antibodies in the same patients, antibody levels of the seroconverted patients who had more than three consecutive samples were plotted as a function of days from onset of symptoms. Figure 2 shows that antibody levels measured by either CV2T or CV2G increased in the early phase of the infection and gradually decreased but remained positive in our observation period in both groups (Figure 2A-D). To assess the difference of mean antibody levels among the days from onset of symptoms, Kruskal-Wallis analysis was performed, and a significant difference was observed among the days from the onset of symptoms (p < 0.001). Post hoc analysis using Steel-Dwass analysis demonstrated a significant difference in mean antibody levels in days 0–6 when compared to later days. There was also a significant difference in mean antibody levels in groups of day 7–13 when compared to other days. However, no significant difference was observed in mean antibody levels in groups later than days 14–20.
Figure 2.
Chronological changes of antibody levels. Antibody levels of individuals who had more than three results are plotted by disease severity and by assays; (A) Group M, CV2T (B) Group S, CV2T (C) Group M, CV2G (D) Group S, CV2G. Red line indicates the cut off (1000 QUAL and 1000 Ind).
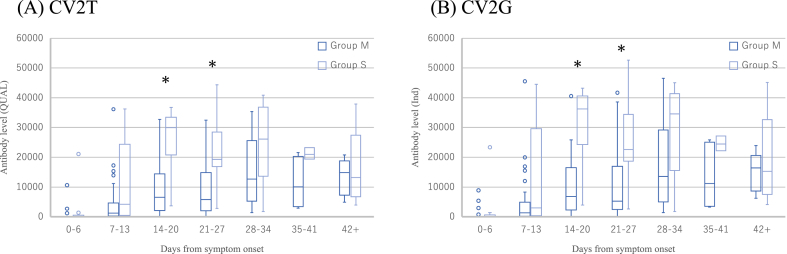
Figure 3 shows comparisons of mean antibody levels between Group M and Group S in each day groups to assess the correlation between disease severity and antibody levels. The mean antibody levels were significantly higher in Group S than Group M in group day 14–20 and 21–27 by both CV2T (Figure 3A) and CV2G (Figure 3B).
Figure 3.
Antibody responses to SARS-CoV-2. CV2T (A) and CV2G (B) antibody levels against SARS-CoV-2 in patients compared by the disease severity at different times from symptom onset are presented in box plot. Asterisks indicate p < 0.05.
4. Discussion
This study was conducted to evaluate the efficacy of the Siemens SARS-CoV-2 Total Antibody assay and IgG assay using 236 serum samples from 79 patients in a university hospital in Tokyo, Japan. Although several Siemens assays have been developed, the Dimension using the LOCI method measuring IgG has not been reported [15, 16, 17, 18].
Due to the pressing need for assays detecting antibody against SARS-CoV-2, many kits have emerged and applied for the Emergency Use Authorization (EUA) by the FDA. However, little clinical information of the assays authorized for emergency use is known. Thus, it is crucial for clinical laboratories to evaluate the assay performance declared by the vendor before using EUA products. Thus, we first validated the assay precision. According to the package insert, the CVs were set to be under 10% and under 12% for repeatability and within-laboratory reproducibility, respectively. Our results were lower than this and confirmed the manufacturer's claims.
In the determination of seroprevalences, most patients were seropositive at days 14–20 and seropositivity persisted till days 42 plus. Similar to the previous studies, one immunocompromised patient failed to seroconvert [19, 20]. This may indicate that detectable amounts of antibodies cannot be produced in immunocompromised patients.
The sensitivity after 14 days from onset of symptoms in our study nearly matched the package insert (CV2T: 100% and CV2G: 100%) [13]. However, the sensitivity reported by the manufacturer was higher in days 0–6 (CV2T: 66.67%; CV2G: 61.9%) and days 7–13 (CV2T: 91.30%; CV2G: 92.3%) compared to our results. A study using the Dimension Vista that shares the same technologies with Dimension EXL 200 in the U.S. reported a sensitivity of 78% by CV2T after 14 days from onset of symptoms, which that is lower than our study [18]. This inconsistency might occur due to the locality or the character diversity of the sample such as ethnicity, age, gender, and underlying diseases.
In the detection of SARS-CoV-2 antibody, cross reaction with other coronaviruses, such as severe acute respiratory syndrome coronavirus (SARS) and Middle East respiratory syndrome coronavirus (MERS), has been reported [21]. Moreover, cross reaction with CMV has also been reported [22]. However, upon reviewing the patients’ charts, we did not find evidence of previous exposure to SARS or MERS, which are quite rare disease in Japan. Using the pre-COVID-19 samples in the specificity assay, our study showed high specificity in both CV2T (97.9%) and CV2G (99.0%), which is essential for detecting antibodies. However, our specificity results were lower than the Zilla et al. study which had high (100%) specificity using 184 samples in the total antibody assay using the Siemens Vista [18]. The manufacturer also claims 99.86% specificity for CV2T and 100% specificity for CV2G using 1529 and 1509 healthy samples collected before COVID-19 pandemic, respectively. Although these discordances may be due to difference in samples, the difference may be negligible.
Comparing Group M and Group S, the antibody levels were significantly higher in Group S in the second and third weeks. These results are similar to a study by Long et al., which compared severe and non-severe groups from three hospitals in China although significant difference was obtained only in days 8–14 [23]. Several studies also demonstrated positive correlations between disease severity and antibody levels [9, 17, 24, 25] while others did not [20, 26]. These contradicting results may have occurred due to difference in sample size, the target for antibody detection (S protein or N protein or S1-RBD), or the genetic diversity of patients.
The results of CV2T and CV2G are not comparable because the CV2G is semi-quantitative assay, and the CV2T is qualitative assay. In our study, the CV2G assay seems to yield higher antibody levels than the CV2T assay in both Group S and Group M. However, further investigation was not able to perform because of the lack of reference material.
The CDC recommend serology testing for suspectable COVID-19 patients who are PCR negative and at least two weeks post onset of symptoms or for seroprevalence survey [27]. Importantly, the CDC also pointed out that antibody assays with positive result targeting the S antigen and negative result targeting other antigen suggest previous infection or reactions to vaccinations [27]. COVID-19 patients develop several types of antibodies against viral proteins, and as time passes the antibodies decrease and may become seronegative. Thus, negative N-specific antibodies do not deny post exposure. If a person has antibodies against N, E, or M proteins, not only S proteins, it would indicate post exposure to SARS-CoV-2. On the other hand, S-specific antibodies can be developed by exposure to SARS-CoV-2 or vaccination because most vaccines are expected to elicit S-specific antibodies. Hence, the assessment of optimal timing to measure S-specific antibodies that can act as neutralizing antibodies, not N-specific antibodies, is essential to evaluate effectiveness of the vaccination.
Concordant with the previous studies [18, 23, 28], our data showed that sensitivity reached to almost 100% in days 14–20. Also, a statistical analysis demonstrated antibody levels significantly increased during the first 14 days. Chew et al. also reported antibody levels were higher in the second week after onset of symptoms compared to the early phase [29]. Taken together, two to three weeks can be suitable timing for detecting seroconversion [9, 10, 11, 12].
This study has some limitations: 1) the present study was performed in a single university hospital, and number of samples were relatively small; 2) since this was a retrospective study, the sampling time varies by patients. Thus, we could not track the change of antibody level in all day groups; 3) clarification of the onset of symptoms was merely dependent upon the clinical charts, which was subjective and might have discrepancy with true onsets of viral infection; and 4) we have not performed neutralizing assay since it requires live virus and BSL-3 equipment. Thus, we were not able to determine whether the detected antibodies act as neutralizing antibodies. Some studies have demonstrated that the avidity assay detects the binding strength of antibodies as an alternative to neutralizing assay in determining protective humoral immunity [30, 31], which requires further investigation; 5) the sole use of Dimension EXL 200 is another potential limitation. There are several assays to detect SARS-CoV-2 S1-RBD-specific antibodies with different measurement principles [15, 16, 17, 18], which can lead to different results.
In conclusion, the Siemens CV2T and CV2G could detect the S1-RBD specific antibodies against SARS-CoV-2 at approximately 14 days after onset with a high sensitivity. Though current use of this assay as a screening tool is limited as described in the CDC recommendations, it may be useful to determine the post vaccine effect and the necessity of booster shots since they have a potential to detect neutralizing antibodies. However, a complete understanding of the antibody kinetics of these two assays is required for use as a predictor of vaccine response. This requires larger samples from various regions and prospective studies over longer timeframes, such as before and after vaccination.
Declarations
Author contribution statement
Gene Igawa: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Takamasa Yamamoto: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Yuna Baba, Konomi Shinozuka, Maiko Yuri: Performed the experiments.
Mitsuru Wakita: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Shigeki Misawa, Takashi Miida: Contributed reagents, materials, analysis tools or data.
Tomohiko Ai: Analyzed and interpreted the data; Wrote the paper.
Yoko Tabe: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank all health-care ware workers involved in the diagnosis and treatment of COVID-19 in the Juntendo University Hospital.
References
- 1.Benzigar M.R., Bhattacharjee R., Baharfar M., Liu G. Current methods for diagnosis of human coronaviruses: pros and cons. Anal. Bioanal. Chem. 2020:2311–2330. doi: 10.1007/s00216-020-03046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Y., Long L., Zhang D., Yuan T., Cui S., Yang P., et al. Potential false-negative nucleic acid testing results for severe acute respiratory syndrome coronavirus 2 from thermal inactivation of samples with low viral loads. Clin. Chem. 2020;66(6):794–801. doi: 10.1093/clinchem/hvaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Wang C., Han M., Ye J., Gao Y., Liu Z., et al. Discrimination of false negative results in RT-PCR detection of SARS-CoV-2 RNAs in clinical specimens by using an internal reference. Virol. Sin. 2020;35(6):758–767. doi: 10.1007/s12250-020-00273-8. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espejo A.P., Akgun Y., Al Mana A.F., Tjendra Y., Millan N.C., Gomez-Fernandez C., et al. Review of current advances in serologic testing for COVID-19. Am. J. Clin. Pathol. 2020;154(3):293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. https://pubmed.ncbi.nlm.nih.gov/32729549 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.COVID-19 serology surveillance Strategy. https://www.cdc.gov/coronavirus/2019-ncov/covid-data/serology-surveillance/index.html [Internet]. Available from:
- 8.Chung Y.H., Beiss V., Fiering S.N., Steinmetz N.F. Covid-19 vaccine frontrunners and their nanotechnology design. ACS Nano. 2020;14(10):12522–12537. doi: 10.1021/acsnano.0c07197. [DOI] [PubMed] [Google Scholar]
- 9.Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020 Dec 7;5(54) doi: 10.1126/sciimmunol.abe0240. https://pubmed.ncbi.nlm.nih.gov/33288645 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A.J., et al. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5(48):1–14. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suthar M.S., Zimmerman M.G., Kauffman R.C., Mantus G., Linderman S.L., Hudson W.H., et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep. Med. 2020;1(3):100040. doi: 10.1016/j.xcrm.2020.100040. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salazar E., Kuchipudi S.V., Christensen P.A., Eagar T., Yi X., Zhao P., et al. Convalescent plasma anti–SARS-CoV-2 spike protein ectodomain and receptor-binding domain IgG correlate with virus neutralization. J. Clin. Invest. 2020;130(12):6728–6738. doi: 10.1172/JCI141206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.EUA authorized serology test performance. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance [Internet]. Available from:
- 14.Kanda Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 2013;48(3):452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irsara C., Egger A.E., Prokop W., Nairz M., Loacker L., Sahanic S., et al. Clinical validation of the Siemens quantitative SARS-CoV-2 spike IgG assay (sCOVG) reveals improved sensitivity and a good correlation with virus neutralization titers. Clin. Chem. Lab. Med. 2021;59(8):1453–1462. doi: 10.1515/cclm-2021-0214. [DOI] [PubMed] [Google Scholar]
- 16.Hörber S., Soldo J., Relker L., Jürgens S., Guther J., Peter S., et al. Evaluation of three fully-automated SARS-CoV-2 antibody assays. Clin. Chem. Lab. Med. 2020;58(12):2113–2120. doi: 10.1515/cclm-2020-0975. [DOI] [PubMed] [Google Scholar]
- 17.Trabaud M.-A., Icard V., Milon M.-P., Bal A., Lina B., Escuret V. Comparison of eight commercial, high-throughput, automated or ELISA assays detecting SARS-CoV-2 IgG or total antibody. J. Clin. Virol. 2020;132:104613. doi: 10.1016/j.jcv.2020.104613. https://pubmed.ncbi.nlm.nih.gov/32942137 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zilla M., Wheeler B.J., Keetch C., Mitchell G., McBreen J., Wells A., et al. Variable performance in 6 commercial SARS-CoV-2 antibody assays may affect convalescent plasma and seroprevalence screening. Am. J. Clin. Pathol. 2020:1–11. doi: 10.1093/ajcp/aqaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei J., Zhao J., Han M., Meng F., Zhou J. SARS-CoV-2 infection in immunocompromised patients: humoral versus cell-mediated immunity. J. Immunother. Cancer. 2020 Jul;8(2) doi: 10.1136/jitc-2020-000862. https://pubmed.ncbi.nlm.nih.gov/32727811 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phipps W.S., SoRelle J.A., Li Q.Z., Mahimainathan L., Araj E., Markantonis J., et al. SARS-CoV-2 antibody responses do not predict COVID-19 disease severity. Am. J. Clin. Pathol. 2020;154(4):459–465. doi: 10.1093/ajcp/aqaa123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okba N.M.A., Müller M.A., Li W., Wang C., Geurtsvankessel C.H., Corman V.M., et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S.Y., Lee Y.L., Lin Y.C., Lee N.Y., Liao C.H., Hung Y.P., et al. Multicenter evaluation of two chemiluminescence and three lateral flow immunoassays for the diagnosis of COVID-19 and assessment of antibody dynamic responses to SARS-CoV-2 in Taiwan. Emerg. Microb. Infect. 2020;9(1):2157–2168. doi: 10.1080/22221751.2020.1825016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 24.Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17(7):773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [Internet] [DOI] [PubMed] [Google Scholar]
- 26.Oved K., Olmer L., Shemer-Avni Y., Wolf T., Supino-Rosin L., Prajgrod G., et al. Multi-center nationwide comparison of seven serology assays reveals a SARS-CoV-2 non-responding seronegative subpopulation. E Clin. Med. 2020;29:100651. doi: 10.1016/j.eclinm.2020.100651. [Internet] Available from: https://pubmed.ncbi.nlm.nih.gov/33235985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Interim guidelines for COVID-19 antibody testing. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html [Internet]. [cited 2021 May 8]. Available from:
- 28.Ng D.L., Goldgof G.M., Shy B.R., Levine A.G., Balcerek J., Bapat S.P., et al. SARS-CoV-2 seroprevalence and neutralizing activity in donor and patient blood. Nat. Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-18468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chew K.L., Tan S.S., Saw S., Pajarillaga A., Zaine S., Khoo C., et al. Clinical evaluation of serological IgG antibody response on the Abbott Architect for established SARS-CoV-2 infection. Clin. Microbiol. Infect. 2020;26(9) doi: 10.1016/j.cmi.2020.05.036. https://pubmed.ncbi.nlm.nih.gov/32531475 [Internet] 1256.e9-1256.e11. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer G. The potential significance of high avidity immunoglobulin G (IgG) for protective immunity towards SARS-CoV-2. Int. J. Infect. Dis. 2021;106:61–64. doi: 10.1016/j.ijid.2021.01.061. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldo E., Gaspari E De. Avidity assay to test functionality of anti-SARS-Cov-2 antibodies. Vaccine. 2020 doi: 10.1016/j.vaccine.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.