Abstract
For three human blood culture isolates of beta-hemolytic streptococci with Lancefield's serogroup A antigen, phylogenetic analysis of the 16S rRNA genes confirmed biochemical identification as Streptococcus dysgalactiae subsp. equisimilis. Genes encoding M or M-like proteins, which are considered to be major virulence determinants in streptococci, were detected in all of these strains. Our data clearly demonstrate that for beta-hemolytic streptococci, the species assignment should not be based on the results of serogrouping alone.
Streptococci were first serogrouped by Rebecca C. Lancefield in 1933 on the basis of specific cell wall-associated carbohydrates (16). The term group A streptococcus has been used generally as a synonym for Streptococcus pyogenes despite the fact that minute-colony-forming species of the “Streptococcus milleri” group may also rarely possess Lancefield's serogroup A antigen (12, 18). Moreover, Bert and Lambert-Zechovsky (3) have recently published a case report of recurrent bacteremia due to a biochemically identified strain of Streptococcus dysgalactiae subsp. equisimilis exhibiting Lancefield's serogroup A antigen. This clinical isolate was kindly provided by Bert and Lambert-Zechovsky for further investigation and assigned streptococcal culture collection no. AC-2074. The present report describes the phylogenetic analysis and molecular characterization of blood culture isolates of S. dysgalactiae subsp. equisimilis belonging to Lancefield's serogroup A.
(This work has been presented in part at the 9th European Congress of Clinical Microbiology and Infectious Diseases, Berlin, Germany, 21 to 24 March 1999 [4a].)
Case 1.
A 78-year-old man presented to the hospital with acute onset of headache, fever of 40°C, and chills. His medical history included liver cirrhosis due to chronic hepatitis C, with signs of portal hypertension. On physical examination, local erythema and tenderness of his right foot were noted and the diagnosis of an erysipelas was made. A minor skin lesion acquired 4 days prior to admission probably served as the site of entry for bacteria. Blood cultures were positive for beta-hemolytic streptococci, and antimicrobial therapy with a standard dose of intravenous amoxicillin was started. The patient's condition improved, and he was discharged 6 days later on an oral regimen to complete a 10-day course of amoxicillin. One blood culture isolate was referred to the National Reference Center for Streptococci, Aachen, Germany, and assigned streptococcal culture collection no. AC-2713.
Case 2.
A 72-year-old woman who was undergoing diagnostic workup for an indeterminate paraspinal mass (chronic inflammatory lesion versus osteochondroma versus osteosarcoma) was referred for symptoms consistent with acute tonsillitis. Blood was obtained, and blood cultures grew beta-hemolytic streptococci. The patient was treated according to the results for in vitro susceptibility testing with cefuroxime. Twenty-four days later, the patient had fully recovered. This clinical isolate was assigned streptococcal culture collection no. AC-2832.
Methods.
Lancefield's group-specific streptococcal carbohydrates were extracted from an overnight culture in Todd- Hewitt broth according to the hot-formamide technique of Fuller (8). Following extraction, serogroup was determined by the Ouchterlony immunodiffusion technique with rabbit antisera against the antigens A, C, G, and A-variant (14, 17). Antigen-antibody precipitation lines were visualized by Coomassie blue staining. In addition, trypsinized bacterial suspensions were tested for agglutination with the lectin from Dolichos biflorus (Biotrend, Cologne, Germany) in order to detect the group C streptococcal antigen (15, 20, 23). The isolates were biochemically identified with the Rapid ID 32 Strep API system (bioMérieux, Marcy-l'Etoile, France).
Streptococcal genomic DNA was prepared according to standard protocols (2). The genomes of the strains were hybridized with an oligonucleotide probe specific for S. pyogenes (Accuprobe; Gen-Probe Incorporated, San Diego, Calif.). Nearly the entire 16S rRNA gene was amplified by PCR, and the resulting products were purified with the Quiaquick PCR purification kit (Qiagen, Heiden, Germany) and sequenced on an ABI 373 automated DNA sequencer (Applied Biosystems, Weiterstadt, Germany) by following the protocol of Hiraishi (10). The sequence editor DCSE, version 2.54 (5), was used for editing and concatenation and for multiple alignment of the three strains' 16S ribosomal DNA (rDNA) sequences with those of the following strains deposited in the EMBL database: S. pyogenes ATCC 12344 (accession no. AB002521); S. agalactiae ATCC 13813 (accession no. U02908); Streptococcus dysgalactiae subsp. dysgalactiae ATCC 43078, serogroup C (accession no. AB002485), and ATCC 27957, serogroup C (accession no. AB002484); S. dysgalactiae subsp. equisimilis LMG 16026, serogroup C (accession no. AB008926); and S. pneumoniae MAFF 911410 (accession no. AB002522). The 16S rDNA sequences of the three strains were also aligned with 16S rDNA sequences obtained in our laboratory from the following strains of the National Reference Center's collection: S. pyogenes AC-2892; S. agalactiae AC-2581; S. dysgalactiae subsp. dysgalactiae GCS-1338, serogroup C; S. dysgalactiae subsp. equisimilis ATCC 35666, serogroup C; and S. dysgalactiae subsp. equisimilis ATCC 12394, serogroup G. Phylogenetic studies were performed by using the neighbor-joining method based on a dissimilarity matrix corrected for multiple mutations per site, using the algorithm of Juke and Cantor. Confidence values for individual branches were determined by bootstrap analysis based on 1,000 bootstrap trees that were generated from resampled characters. All calculations were performed with the software package TREECON, version 1.3b (24).
The presence of genes encoding M- or M-like proteins was determined by PCR with “all M” primers (19). The resulting products were purified and sequenced. The nucleotide sequence encoding the N-terminal hypervariable portion of the M protein was subjected to similarity searches against the National Institutes of Health DNA database (1).
Remarks and conclusion.
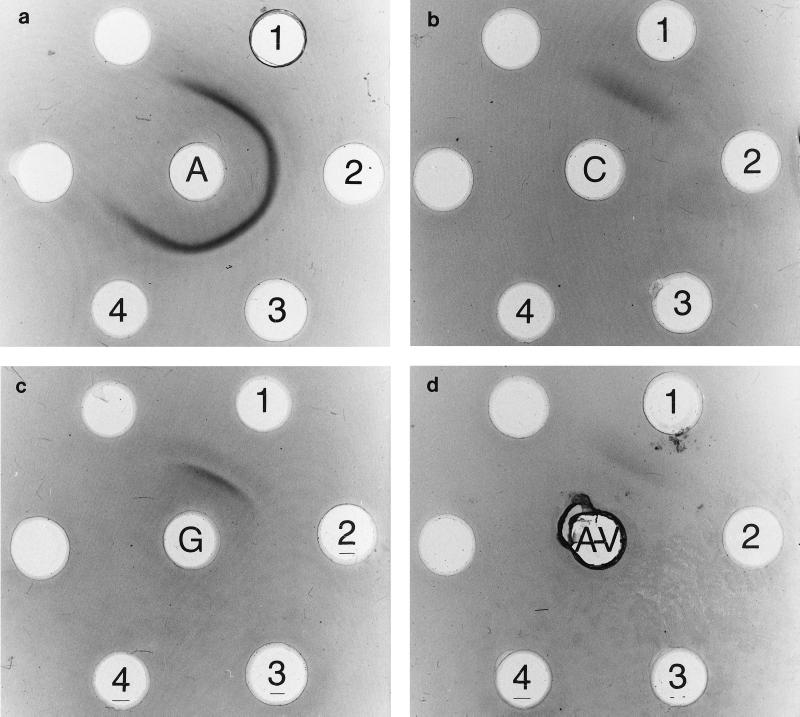
It was shown unequivocally that all three isolates exhibit the serogroup A antigen but not serogroup C, G, or A-variant antigens. Results of the Ouchterlony immunodiffusion are shown in Fig. 1. The N-acetylgalactosamine specificity of the group C carbohydrate antigen was not detected by agglutination with the lectin from D. biflorus for any of the three strains tested. Isolates AC-2713 and AC-2832 were biochemically identified as S. dysgalactiae subsp. equisimilis (API code 1512 2041 110; 99.5% identity; T = 1.0). As published previously for isolate AC-2074 (3), our results revealed the following characteristic properties to discriminate group A S. dysgalactiae subsp. equisimilis from S. pyogenes: production of β-glucuronidase, fermentation of ribose, and the absence of pyrrolidonylarylamidase production. A negative Voges-Proskauer reaction provides a means to differentiate between the large-colony-forming S. dysgalactiae subsp. equisimilis and the minute-colony-forming species of the S. milleri group. Hybridization with the Accuprobe oligonucleotide probe specific for S. pyogenes was not detected for any of the three strains tested.
FIG. 1.
Ouchterlony immunodiffusion of cell wall carbohydrate extracts (outer wells) tested with rabbit antisera against the antigens A, C, G, and A-variant (central wells). 1, serogroup-specific control antigen; 2, AC-2074; 3, AC-2713; 4, AC-2832.
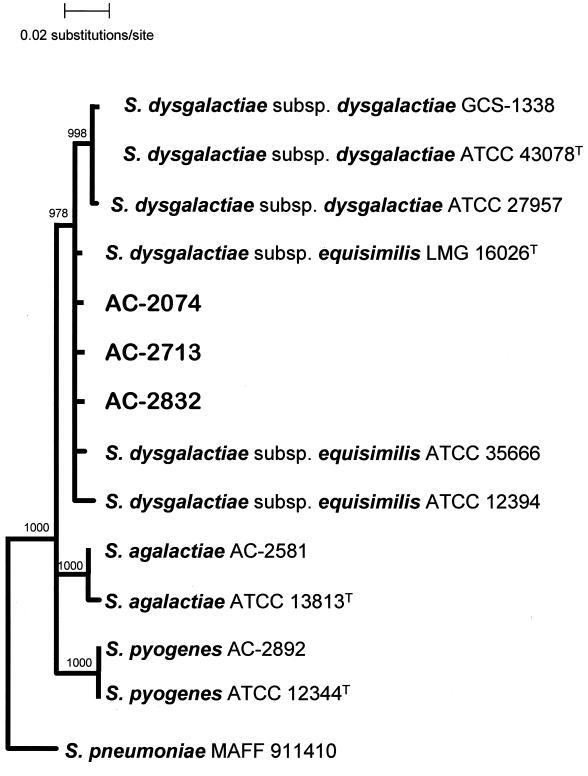
A rooted phylogenetic distance tree inferred by analysis of 16S rDNA sequences is shown in Fig. 2. S. pneumoniae was arbitrarily chosen as the outgroup taxon. All three blood culture isolates studied clustered with the type strain of S. dysgalactiae subsp. equisimilis, LMG 16026, as a sister group to S. dysgalactiae subsp. dysgalactiae in the phylogenetic tree. The phylogenetic analyses revealed a substantial evolutionary distance between S. pyogenes and the S. dysgalactiae subsp. equisimilis blood culture isolates exhibiting the Lancefield serogroup A antigen.
FIG. 2.
Rooted phylogenetic tree inferred by analysis of 16S rDNA sequences of the three blood culture isolates AC-2074, AC-2713, and AC-2832 and other members of the pyogenic streptococci by the neighbor-joining method. Bootstrap values of ≥975 are displayed; nodes with a support of <975 are drawn unresolved.
For the three strains studied, genes (emm or emmL) encoding M- or M-like proteins were detected. The respective nucleotide sequences of strains AC-2713 and AC-2832 revealed a high degree of similarity (>80% identity for a portion of at least 200 bp within the 5′ region of the respective gene) to the genotype b emmL gene described previously for serogroup G S. dysgalactiae subsp. equisimilis (21); the emm or emmL gene sequence of strain AC-2074 showed a high degree of similarity to the emm41 gene of S. pyogenes.
Pyogenic streptococci have been divided conventionally on the basis of their Lancefield group antigen, habitat, pathogenicity, and biochemical characteristics (7, 16). With the availability of automated DNA sequencing and a rapidly growing database, the analysis of 16S rRNA sequences has become one of the most powerful tools for phylogenetic analysis and species identification. The finding that the 16S rDNA sequences of all three strains studied are closely related to that of the type strain of S. dysgalactiae subsp. equisimilis suggests that biochemical methods, like the API system, are an appropriate diagnostic approach for the identification of this species. The observation of a substantial evolutionary distance between S. dysgalactiae subsp. equisimilis and S. pyogenes is in agreement with results of previous phylogenetic analyses of beta-hemolytic streptococci (13) and supports the application of an oligonucleotide probe specific for S. pyogenes in the identification of beta-hemolytic streptococci exhibiting Lancefield's serogroup A antigen. These data clearly show that for species assignment of beta-hemolytic streptococci, the determination of the serogroup is insufficient as the sole diagnostic tool, as outlined previously for group G streptococci (9).
In humans, S. dysgalactiae subsp. equisimilis may colonize the skin, throat, and genital tract. Recently, this species has been isolated with an increasing frequency as the cause of various human infections, such as cellulitis, pharyngitis, sepsis, meningitis, and endocarditis (6). Just as most patients with infections due to S. dysgalactiae subsp. equisimilis exhibiting serogroup C or G antigen have underlying diseases, predominantly malignancies (4, 22), the three patients diagnosed with infections due to group A S. dysgalactiae subsp. equisimilis also suffered from compromising underlying diseases. The finding of two additional patients with bacteremia due to S. dysgalactiae subsp. equisimilis belonging to Lancefield's serogroup A and the detection of emm or emmL genes encoding a major virulence factor in all three strains tested provide evidence that strains of S. dysgalactiae subsp. equisimilis of Lancefield's serogroup A are probably as pathogenic as those exhibiting Lancefield's serogroup C or G antigen. Therefore, it can be concluded that beta-hemolytic streptococci that possess the serogroup A antigen but are negative for the Voges-Proskauer reaction and do not produce pyrrolidonylarylamidase can be found in serious human infections and may be identified as S. dysgalactiae subsp. equisimilis.
Acknowledgments
We thank F. Bert (Clichy, France), M. Jacobs (Dillingen, Germany), and C. Jochum (Püttlingen, Germany) for their cooperation and the referral of the strains to the National Reference Laboratory for Streptococci. We appreciate the excellent technical assistance of M. Breuer-Werle and B. Weidenhaupt.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1990. [Google Scholar]
- 3.Bert F, Lambert-Zechovsky N. Analysis of a case of recurrent bacteraemia due to group A Streptococcus equisimilis by pulsed-field gel electrophoresis. Infection. 1997;25:250–251. doi: 10.1007/BF01713156. [DOI] [PubMed] [Google Scholar]
- 4.Bradley S F, Gordon J J, Baumgartner D D, Marasco W A, Kauffman C A. Group C streptococcal bacteremia: analysis of 88 cases. Rev Infect Dis. 1991;13:270–280. doi: 10.1093/clinids/13.2.270. [DOI] [PubMed] [Google Scholar]
- 4a.Brandt C, Haase G, Schnitzler N, Zbinden R, Lutticken R. Classification of blood culture isolates of Streptococcus dysgalactiae subspecies equisimilis possessing Lancefield's group A antigen. Clin Microbiol Infect. 1999;5(Suppl. 3):57. doi: 10.1128/jcm.37.12.4194-4197.1999. . (Abstract.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Rijk P, De Wachter R. DCSE v 2.54, an interactive tool for sequence alignment and secondary structure research. Comput Appl Biosci. 1993;9:735–740. doi: 10.1093/bioinformatics/9.6.735. [DOI] [PubMed] [Google Scholar]
- 6.Efstratiou A. Outbreaks of human infections caused by pyogenic streptococci of Lancefield groups C and G. J Med Microbiol. 1989;29:207–219. doi: 10.1099/00222615-29-3-207. [DOI] [PubMed] [Google Scholar]
- 7.Efstratiou A, Colman G, Hahn G, Timoney J F, Boeufgras J M, Monget D. Biochemical differences among human and animal streptococci of Lancefield group C or G. J Med Microbiol. 1994;41:145–148. doi: 10.1099/00222615-41-2-145. [DOI] [PubMed] [Google Scholar]
- 8.Fuller A T. The formamide method for the extraction of polysaccharides from haemolytic streptococci. Br J Exp Pathol. 1938;19:130–139. [Google Scholar]
- 9.Haase G, Schnitzler N. Lancefield serogrouping alone is insufficient for species assignment of streptococci. Clin Infect Dis. 1997;25:941. doi: 10.1086/597672. [DOI] [PubMed] [Google Scholar]
- 10.Hiraishi H. Direct automated sequencing of the 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett Appl Microbiol. 1993;15:210–213. doi: 10.1111/j.1472-765x.1992.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 11.Inoue M, Eifuku-Koreeda H, Kitada K, Takamatsu-Matsushita N, Okada Y, Osano E. Serotype variation in Streptococcus anginosus, S. constellatus and S. intermedius. J Med Microbiol. 1998;47:435–439. doi: 10.1099/00222615-47-5-435. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs J A, Pietersen H G, Stobberingh E E, Soeters P B. Streptococcus anginosus, Streptococcus constellatus and Streptococcus intermedius. Clinical relevance, hemolytic and serologic characteristics. Am J Clin Pathol. 1995;104:547–553. doi: 10.1093/ajcp/104.5.547. [DOI] [PubMed] [Google Scholar]
- 13.Kawamura Y, Hou X G, Sultanan F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 14.Krause R M, McCarty M. Variation in the group-specific carbohydrate of group C hemolytric streptococci. J Exp Med. 1962;116:131–140. doi: 10.1084/jem.116.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kühnemund O, Köhler W, Prokop O. Untersuchungen zur Struktur des C-Streptokokken-agglutinins aus Dolichos biflorus. Hoppe-Seyler's Z Physiol Chem. 1968;349:1434–1436. [PubMed] [Google Scholar]
- 16.Lancefield R C. A serological differentiation of human and other groups of hemolytic streptococci. J Exp Med. 1933;57:571–595. doi: 10.1084/jem.57.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCartey M, Lancefield R C. Variation in the group-specific carbohydrate of group A streptococci. I. Immunochemical studies on the carbohydrates of variant strains. J Exp Med. 1955;102:11–28. doi: 10.1084/jem.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piscitelli S C, Shwed J, Schreckenberger P, Danziger L H. Streptococcus milleri group: renewed interest in an elusive pathogen. Eur J Clin Microbiol Infect Dis. 1992;11:491–498. doi: 10.1007/BF01960802. [DOI] [PubMed] [Google Scholar]
- 19.Podbielski A, Melzer B, Lütticken R. Application of the polymerase chain reaction to study the M protein(-like) gene family in β-hemolytic streptococci. Med Microbiol Immunol. 1991;180:213–227. doi: 10.1007/BF00215250. [DOI] [PubMed] [Google Scholar]
- 20.Schaufuss P, Lämmler C, Blobel H. Rapid differentiation of streptococci isolated from cows with mastitis. J Clin Microbiol. 1986;24:1098–1099. doi: 10.1128/jcm.24.6.1098-1099.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnitzler N, Podbielski A, Baumgarten G, Mignon M, Kaufhold A. M or M-like protein gene polymorphisms in human group G streptococci. J Clin Microbiol. 1995;33:356–363. doi: 10.1128/jcm.33.2.356-363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skogberg K, Simonen H, Renkonen O V, Valtonen V V. Beta-hemolytic streptococcal septicaemia: a clinical study. Scand J Infect Dis. 1988;220:119–125. doi: 10.3109/00365548809032427. [DOI] [PubMed] [Google Scholar]
- 23.Slifkin M, Gil G M. Identification of group C streptococcal antigen extracts with lectin-bound polystyrene particles. J Clin Microbiol. 1984;19:83–84. doi: 10.1128/jcm.19.1.83-84.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van de Peer Y, De Wachter R. Treecon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:559–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]