Abstract
Undifferentiated mesenchymal tumors arising from the inner lining (intima) of large arteries are classified as intimal sarcomas (ISA) with MDM2 amplification as their molecular hallmark. Interestingly, undifferentiated pleomorphic sarcomas (UPS) of the heart have recently been suggested to represent the cardiac analog of ISA due to morphological overlap and high prevalence of MDM2 amplifications in both neoplasms. However, little is known about ISAs and cardiac UPS without MDM2 amplifications and molecular data supporting their common classification is sparse. Here, we report a series of 35 cases comprising 25 ISAs of the pulmonary artery, one ISA of the renal artery and 9 UPS of the left atrium. Tumors were analyzed utilizing the Illumina Infinium MethylationEPIC BeadChip array, enabling copy number profile generation and unsupervised DNA methylation analysis. DNA methylation patterns were investigated using t-distributed stochastic neighbor embedding (t-SNE) analysis. Histologically, all ISAs and UPS of the left atrium resembled extra-cardiac UPS. All cases exhibited highly complex karyotypes with overlapping patterns between ISA and UPS. 29/35 cases showed mutually exclusive amplifications in the cell-cycle associated oncogenes MDM2 (25/35), MDM4 (2/35), and CDK6 (2/35). We further observed recurrent co-amplifications in PDGFRA (21/35), CDK4 (15/35), TERT (11/35), HDAC9 (9/35), and CCND1 (4/35). Sporadic co-amplifications occurred in MYC, MYCN, and MET (each 1/35). The tumor suppressor CDKN2A/B was frequently deleted (10/35). Interestingly, DNA methylation profiling (t-SNE) revealed an overlap of ISA and cardiac UPS. This “ISA” methylation signature was distinct from potential histologic and molecular mimics. In conclusion, our data reveal MDM4 and CDK6 amplifications in ISAs and UPS of the left atrium, lacking MDM2 amplification. We further report novel co-amplifications of various oncogenes, which may have therapeutic implications. Finally, the genetic and epigenetic concordance of ISAs and UPS of the left atrium further supports a shared pathogenesis and common classification.
Subject terms: Diagnostic markers, Cancer
Introduction
Intimal sarcoma (ISA) is an exceedingly rare undifferentiated sarcoma that arises in the pulmonary artery and less frequently in the aorta or its branches [1]. ISAs behave highly aggressive with a mean patients’ survival ranging from 5 to 18 months [1]. Patients with ISA are mostly of middle age at diagnosis and typically present with non-specific symptoms, which sometimes masquerades as thromboembolic disease. Thus, patients are often diagnosed in an advanced disease stage. Furthermore, ISAs are often reported to be resistant to conventional chemotherapy [2].
First recognized by Mandelstamm as pulmonary artery sarcoma from an autopsy in 1923, case reports and small case series have been published for the following decades [3]. The term “intimal” was coined with reference to the attributes that these sarcomas arise from the subendothelial space of arteries, form polypoidal and endoluminal protrusions and spread laterally along the intima of large arteries before they infiltrate beyond the adventitia. However, it has also been noted that sarcomas other than ISA may occasionally exhibit such “intimal” growth pattern [4, 5].
Histologically, ISAs are composed of spindle shaped, pleomorphic or epithelioid cells often resembling soft tissue correlates like undifferentiated pleomorphic sarcoma (UPS), myxofibrosarcoma or epithelioid angiosarcoma. By definition, ISAs lack specific lineage differentiation, although myofibroblastic and rarely osteogenic or chondroid differentiation may occur [5–7].
Molecular studies on pulmonary artery ISAs revealed a high frequency of MDM2 amplifications, accompanied by co-amplifications of CDK4 and PDGFRA [8–15]. In 2014, Neuville et al. discovered MDM2 amplifications in a large proportion of UPS of the heart. Interestingly, many of the MDM2 amplified cardiac UPS presented with histologic features resembling ISA [13]. The authors proposed the concept that these cardiac UPS may represent ISA [16]. However, concerns rose with regards to the non-specific occurrence of MDM2 amplifications in a broad range of tumor types [10]. Moreover, genome-wide copy number analysis has been performed only in a few pulmonary artery ISAs, whereas ISAs of more uncommon sites such as the aorta have not been analyzed to date (Table 1). It was concluded that reclassifying these cardiac UPS as ISA could be premature, besides that fact that the term “intima” would be inappropriate for these tumors from an anatomic point of view [17, 18]. Thus, the 4th edition of the WHO classification of tumors of the heart differentiates the multiple subtypes of cardiac UPS by histopathology, but mentions ISA as a synonym or alternative designation [19, 20]. Thus, the relationship between arterial and cardiac ISAs remains incompletely understood.
Table 1.
Studies applying genome-wide copy number variation analysis in intimal sarcomas.
| Study | Year | Analyzed cases | Location | Method | Relevant findings |
|---|---|---|---|---|---|
| Bode-Lesniewska et al. | 2001 | 8 | Pulmonary artery | CGH | MDM2 amp |
| Zhao et al. | 2002 | 8 | Pulmonary artery | aCGH | MDM2 amp, PDGFRA amp |
| Sebenik et al. | 2005 | 12/14 | Aorta and branches | CGH | Complex karyotyp |
| Zhang et al. | 2007 | 1 | Heart | Karyotyping | MDM2 amp, CDK4 amp |
| Dewaele et al. | 2010 | 8/21 | Pulmonary artery (n = 5) and heart (n = 3) | aCGH | MDM2 amp, PDGFRA amp |
| Neuville et al. | 2014 | 5/100 | Heart | aCGH | MDM2 amp |
| Ito et al. | 2017 | 1 | Heart | aCGH | MDM2 amp, PDGFRA amp |
| Roszik et al. | 2019 | 13 | Unknown | GENIE database | MDM2 amp, TERT amp |
aCGH microarray-based comparative genomic hybridization, amp amplified.
High density DNA methylation arrays provide a powerful tool for robust molecular tumor classification [21, 22]. DNA methylation profiling in sarcomas has defined subtype-specific sarcoma signatures, even within seemingly morphological homogenous entities that would otherwise evade a definite histologic diagnosis. Likewise, DNA methylation profiling has shown morphological heterogeneous tumors to constitute a single molecular subtype [23–29]. Furthermore, data of these high-density DNA methylation arrays allow genome-wide mapping of copy number variations.
Herein, we comprehensively characterize a cohort of 26 ISAs and 9 UPS of the left atrium by genome-wide copy number analysis and DNA methylation profiling. We sought to further define their molecular alterations and determine whether they share a DNA methylation signature that segregates them from potential histologic mimics.
Material and methods
Sample selection
We collected a cohort of 26 ISAs and 9 UPS of the left atrium from different patients, containing 10 previously published cases [8, 9]. Samples were retrieved from the Institute of Pathology of the University Zürich (Switzerland), from the Department of Pathology of the St Antonius Hospital Nieuwegein (the Netherlands), the Institute of Pathology of the University Hospital Heidelberg (Germany), the Department of Pathology of the Memorial Sloan Kettering Cancer Center in New York (United States of America), the Department of Pathology of the Radboud University Medical Center in Nijmegen (the Netherlands), the Dermatopathology Bodensee in Friedrichshafen (Germany) and the Institute of Pathology of the Sozialstiftung Hospital in Bamberg (Germany). Basic clinical information of the investigated cases is provided in Supplementary Table 1.
Diagnoses were established according to the guidelines of the WHO classification for soft tissue and bone tumors (5th edition) and for tumors of the lung, pleura, thymus and heart (4th edition) [1, 19]. Accordingly, none of the study cases showed a definable line of differentiation. The study was performed in concordance with the guidelines set forth by the local ethics committee of the University of Heidelberg and in accordance with the Declaration of Helsinki.
DNA extraction and quantification
DNA of all tumors was extracted from formalin-fixed paraffin-embedded (FFPE) tissue samples. All tumors included in this study had sufficient tumor material available to prevent extraction of neighboring benign tissue. Areas with highest available tumor content (≥70%) were chosen for extraction of DNA. The Maxwell® 16 FFPE Plus LEV DNA Kit was applied on the automated Maxwell device (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Extracted DNA was quantified using the QuantiFast SYBR Green PCR Kit (Qiagen, Duesseldorf, NW, Germany). A minimum of 100 ng DNA was extracted in every case and provided for subsequent array-based DNA methylation analysis.
Genome-wide DNA methylation data generation and pre-processing
The total DNA input suitable for DNA methylation profiling ranges from a minimum of 10 ng to 500 ng [30]. In this study, all 35 samples reached a total DNA input of ≥100 ng and therefore were subjected to the Illumina Infinium MethylationEPIC BeadChip array (Illumina, San Diego, USA) analysis at the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ) Heidelberg. To exclude low-quality samples from the cohort, the on-chip quality metrics of all samples were checked and compared to a set of 7,500 pairs of IDAT-files [22]. All 35 samples passed this quality control check. DNA methylation data were normalized by performing background correction and dye bias correction (shifting of negative control probe mean intensity to zero and scaling of normalization control probe mean intensity to 20000, respectively). Probes targeting sex chromosomes, probes containing multiple single nucleotide polymorphisms and those that could not be uniquely mapped were removed. Human reference genome (hg19) was used for the analysis of multi-site mapping. Probes from the EPIC array were excluded if the predecessor Illumina Infinium 450k BeadChip did not cover them, thereby making data generated by both 450k and EPIC feasible for subsequent analyses. In total, 438370 probes were kept for analysis.
Copy number analysis
Copy number plots were generated on methylation array data using the R package ‘conumee’ after additional baseline correction (https://github.com/dstichel/conumee). Copy number variants were identified by manual inspection as previously described [31]. Thresholds for the identification of amplifications and homozygous deletions were derived from the difference of the baseline. Gains/amplifications usually are above a log2 value of 0.4 and losses/deletions usually are below a log2 value of 0.4. Low tumor cell content or subclonal alterations may reduce the amplitude deviation.
Fluorescence In Situ Hybridization
Additional FISH assays were performed for assessing gene copy alterations. FISH on interphase nuclei from FFPE 4 μm sections was performed using custom-designed probes of bacterial artificial chromosomes flanking the target genes CDK6 (7q21.2) and MDM4 (1q32.1). An amplification was defined as the presence of >10 signals (ratio to control probe >10) or tight clustered signals characteristic of homogeneous staining regions.
Unsupervised DNA methylation analysis
We used t-distributed stochastic neighbor embedding (t-SNE) analysis, a method enabling dimensionality reduction and visualization of clusters to detect methylation clusters. The algorithm was performed using the 10,000 most variable probes with a perplexity of 10 and 3000 iterations. Methylation groups were tested for stability by varying the number of the most variable probes and perplexities. Methylation data from 78 samples previously published in part comprising non-radiation /–UV induced angiosarcomas of soft tissue or viscera (n = 6), inflammatory myofibroblastic tumors (n = 7), leiomyosarcomas of (venous) vessels (n = 12), conventional high grade osteosarcomas (n = 13), low grade osteosarcomas, MDM2 amplified (n = 6), extraskeletal osteosarcomas (n = 4), retroperitoneal well-/de-differentiated liposarcomas, MDM2 amplified (n = 13), angiomatoid fibrous histiocytomas (n = 9) and extra-cardiac UPS (n = 8) were used for comparison [23–29].
Results
Study cohort and histopathology
The study included 25 cases with ISAs arising in the pulmonary artery and one case with an ISA of the renal artery. The sex ratio was balanced (male-female ratio, 13:13). Patient’s age at presentation ranged from 30 to 83 years, with a median age of 58 years. Furthermore, the study included 5 female and 4 male patients with UPS of the left atrium, among them one young patient with a metastatic bone lesion in the humerus, which was analyzed here. Their age at presentation ranged from 18 to 67 years, with a median age of 38 years. The patient’s characteristics are summarized in Table 2.
Table 2.
Patient characteristics and histologic features.
| Variables | Artery (n = 26) | Heart (n = 9) |
|---|---|---|
| Sex | ||
| Male | 13 | 4 |
| Female | 13 | 5 |
| Median Age in years (range) | 58 (30–83) | 38 (18–67) |
| Histological subtype | ||
| UPS-like | 23 | 9 |
| Malignant IMT-like | 1 | 0 |
| AFH-like | 1 | 0 |
| Myxofibrosarcoma | 1 | 0 |
Morphologically, all 35 cases, some at least focally, showed ISA features characterized by endoluminal growth, fibrin layering with tumor overgrowth and intimal spread (Fig. 1). Overall, the cellularity was variable within tumors. The predominant tumor architecture was loose and pattern-less, although storiform areas and collagenized stroma were recognizable in some cases. The tumor cells appeared mostly spindle shaped (Fig. 2a) and rarely epithelioid (Fig. 2b). Nuclear pleomorphism was evident in all cases. Some case showed prominent bleeding residues and two cases presented with dystrophic calcifications (Fig. 2c). Three cases exhibited patterns reminiscent of distinct soft tissue sarcoma subtypes other than undifferentiated (pleomorphic) sarcoma. One case (ID 141642) presented with myxoid areas with low cellularity, but tumor cell condensation around vessels, resembling myxofibrosarcoma (Fig. 2d). One case (ID 141634) showed pseudoangiomatous spaces filled with blood and surrounded by tumor cells, a pattern that to some extend resembled an angiomatoid fibrous histiocytoma (Fig. 2e). Another case (ID 129604) showed prominent plasma cell aggregates and scattered eosinophils within an otherwise hypocellular tumor stroma, resembling inflammatory myofibroblastic tumor (Fig. 2f).
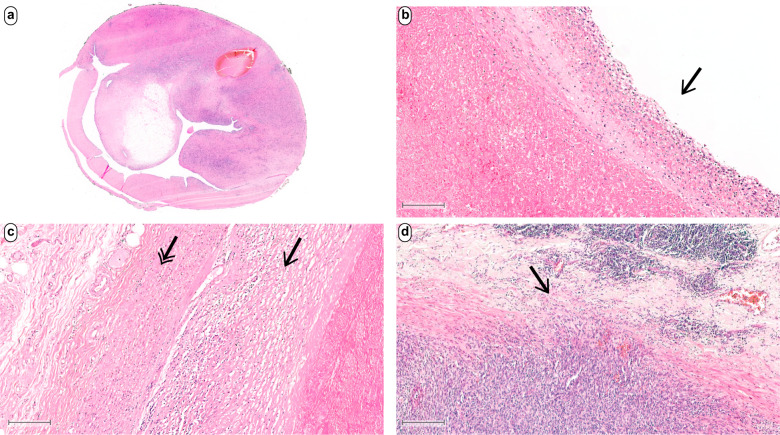
Fig. 1. Examples of characteristic architectural patterns in intimal sarcomas.
At low magnification this case illustrates the endoluminal growth of a polypoid tumor within an artery (a). Intimal sarcomas typically overgrow fibrin layers attached at the vessel wall (b). Tumor cells spread lateral within the intimal space (c). Some cases show focal infiltration and penetration of the tunica media (d). Scale bars equal 200 µm.
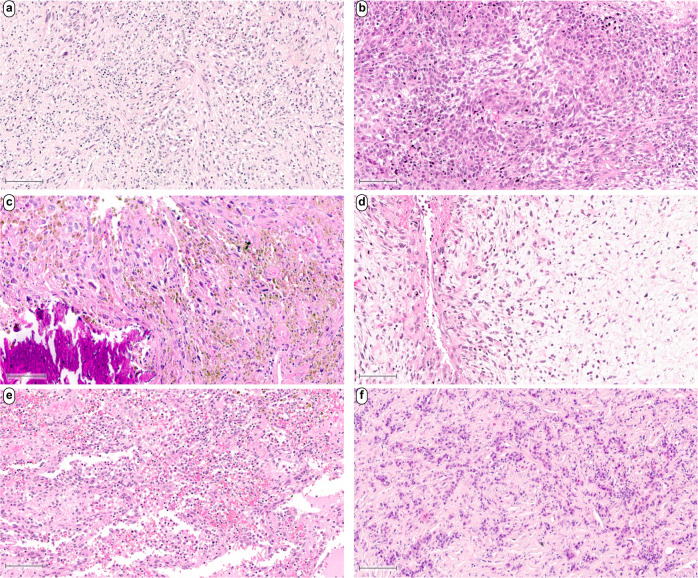
Fig. 2. Histologic features in intimal sarcomas.
Intimal sarcomas often exhibited a loose storiform growth pattern (a). Single cases showed an epithelioid cytomorphology with tumor cells focally forming diffuse sheets and solid areas (b). Some cases showed focal stromal sclerosis, hemosiderin deposits and dystrophic calcifications (c). Case 141642 exhibited a prominent myxoid stroma with elongated, thin-walled vessels and increased perivascular tumor cell density (d). Case 141634 showed variable-sized pseudocystic spaces containing homogeneous eosinophilic material or blood. These spaces are lined by tumor cells. Hemosiderin is present (e). Case 129604 showed a prominent component of aggregating plasma cells (f). Scale bars equal 100 µm.
Recurrent gene amplifications beyond MDM2 and PDGFRα
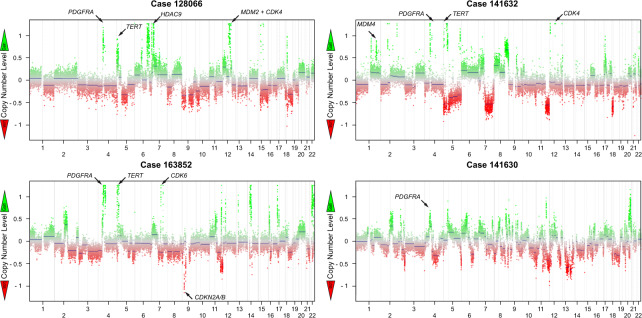
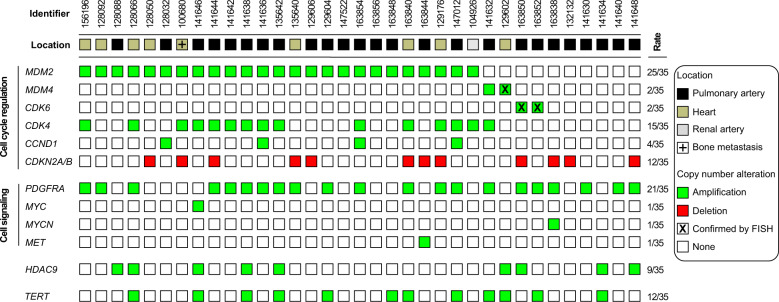
We next analyzed the copy number profiles for amplifications and deletions (Fig. 3). ISAs and UPS of the left atrium showed complex karyotypes with overlapping patterns. Copy number analysis unveiled mutually exclusive amplifications in the cell cycle regulating genes MDM2 (25/35), MDM4 (2/35), and CDK6 (2/35). MDM4 and CDK6 amplifications, where possible, were confirmed by FISH analysis (Fig. 4). Copy number variations in these pathways included co-amplifications of CDK4 (15/35) and CCND1 (4/35) and recurrent homozygous deletions in CDKN2A/B (10/35). Amplifications in signaling pathways most frequently involved receptor tyrosine kinase (RTK) PDGFRA (21/35) followed by MET, MYC, and MYCN (each 1/35). Furthermore, we observed recurrent amplifications in TERT (12/35) and HDAC9 (9/35). The copy number variations are summarized in Fig. 5.
Fig. 3. Copy number profiles in intimal sarcomas.
Exemplary copy number profiles of intimal sarcomas carrying a 12q14-q15 amplification including MDM2 and CDK4 (upper left), carrying a 1q32.1 amplification including MDM4 (upper right), carrying a 7q21.2 amplification including CDK6 (lower left) and one example case lacking one of these gene amplifications (lower right).
Fig. 4. MDM4 and CDK6 amplifications demonstrated by FISH.

2-color FISH for CDK6 (green signal) or MDM4 (red signal) in MDM2 balanced intimal sarcomas. The upper image (case 163852) shows tight clustered green signals (CDK6) and almost balanced red signals (MDM4). The lower image (case 129602) shows the opposite constellation with abundant red signals (MDM4) and almost balanced green signals (CDK6).
Fig. 5. Summary of copy number variations in intimal sarcomas.
Shown are the the most prominent copy number alterations identified. The color code is indicated in the figure.
Common DNA methylation signature in intimal sarcomas and undifferentiated pleomorphic sarcomas of the left atrium
We next analyzed DNA methylation profiles by t-SNE analysis (Fig. 6). ISAs and UPS of the left atrium formed a unique methylation cluster when compared to potential histopathologic and molecular mimics, thereby demonstrating a distinct “ISA” methylation signature. Interestingly, the sample of a bone metastasis of an ISA overlaid with primary ISA samples, which illustrates the stability of this “ISA” methylation signature. Finally, it is important to note that the aforementioned mimics also constituted subtype specific clusters in the t-SNE plot.
Fig. 6. Unsupervised DNA methylation analysis of intimal sarcomas, undifferentiated pleomorphic sarcomas of the left atrium and potential mimics.
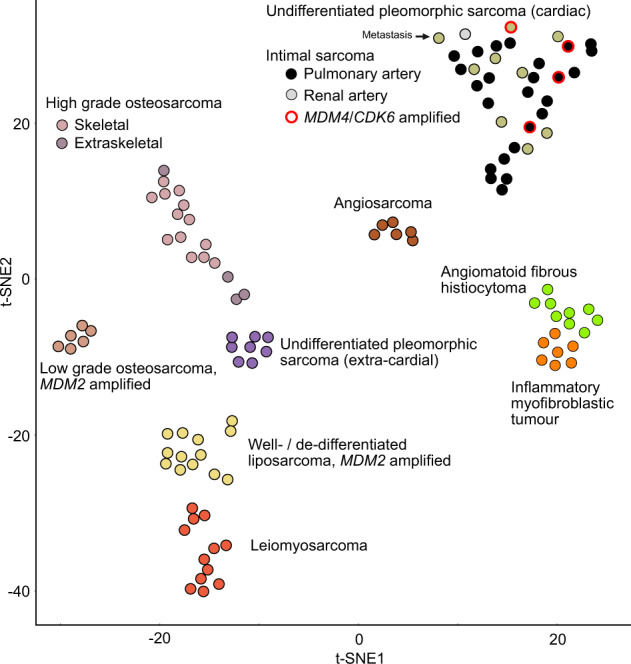
Unsupervised t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis of DNA methylation data from intimal sarcomas. The study cohort was compared with methylation data of prototypical soft tissue sarcomas that may histologically or molecularly mimic intimal sarcomas.
Discussion
In this study we assessed the molecular profiles of ISAs and UPS of the left atrium using genome-wide copy number profiling and unsupervised DNA methylation analysis. ISAs and UPS of the left atrium showed highly complex karyotypes. Our analysis revealed MDM4 and CDK6 amplifications in ISAs and cardiac UPS lacking MDM2 amplifications. Furthermore, their epigenetic patterns were highly overlapping. Thus, our data argue in favor for the proposed concept by Neuville and colleagues that ISAs and UPS of the left atrium may constitute a common entity [13, 16].
Previous studies on ISAs and cardiac sarcomas have used array-CGH analysis only in a small number of samples and tested their findings on larger cohorts by fluorescence in situ hybridization analysis. These studies revealed complex karyotypes with highly recurrent amplifications of MDM2 [8–15]. With this approach, however, there is a risk that less frequent copy number variations can be missed. In our study, all 35 cases were tested for genome-wide copy number variations. We could confirm MDM2 amplifications in the vast majority of ISAs and additionally identified previously unreported, mutually exclusive MDM4 and CDK6 amplifications in ISAs and UPS of the left atrium. MDM4 and CDK6 immunohistochemistry were strongly positive in cases with such amplifications (data not shown). However, we also noticed a marked MDM4 and CDK6 expression in cases with balanced MDM4 and CDK6 locus. Therefore, we consider MDM4/CDK6 immunohistochemistry not suitable as surrogate for detecting MDM4/CDK6 amplifications.
MDM2 and CDK6 are critical components in the p53 and RB signaling pathway, respectively. These pathways are frequently disrupted in sarcomas with complex karyotypes, probably because subunits of those pathways are also involved in maintaining chromosome integrity [32]. CDK6 and CDK4 form a complex that suppresses RB1, a key element in the RB pathway. Notably, the co-amplification of CCND1 and the recurrent deletion of CDKN2A/B also contribute to the disruption of the RB pathway [33]. MDM4 is best characterized for repressing p53 transactivation activity and p53 translational regulation in normal cells under stress [34]. Because of the high frequency of amplifications in these cell cycle pathways, they appear to contribute to the pathogenesis in ISA and UPS of the left atrium.
We also observed highly recurrent amplifications of PDGFRA, as previously reported [12]. Other RTKs were only rarely amplified, e.g., MET. Copy number alterations were rarely observed in the oncogenes MYC and MYCN. Furthermore, we observed frequent TERT amplifications, which confer unlimited proliferation potential to cancer cells through telomere length maintenance. TERT amplifications have been shown to confer the highest telomerase activity among tumors [35]. Finally, we detected HDAC9 amplifications in ISAs. Higher expression of HDAC9 has been associated with poor prognosis in different cancer types [36]. These oncogene amplifications in ISAs and UPS of the left atrium may qualify patients for targeted therapies [37, 38]. However, it remains to be determined whether targeted therapies can lead to a treatment response in ISAs [2].
Some cases lacking MDM2, MDM4, or CDK6 amplifications remain ambiguous by copy number profiling. It is conceivable that these cases harbor alternative driver mutations, although ISAs show a relative low mutation rate overall [15]. In our study, cases lacking MDM2, MDM4, or CDK6 amplifications often carried PDGFRA amplifications. However, PDGFRA amplifications are among the most common genetic alterations in cancer. Accordingly, we observed PDGFRA amplifications also in 5/12 leiomyosarcomas (data not shown). Hence, the detection of PDGFRA amplifications alone is non-specific for diagnosing ISA [39]. To some extent, the same concerns apply for MDM2/4 amplifications. ISAs of uncommon sites such as the retroperitoneum, where dedifferentiated liposarcoma is always a strong consideration, are prone to be misdiagnosed, especially in conjunction with an underlying MDM2 amplification [40–42]. Thus, clinical correlation is paramount for the diagnosis of ISAs and the isolated use of established molecular markers alone may not be helpful in this context.
We provide evidence that DNA methylation profiling may be valuable in increasing the diagnostic accuracy of these rare tumors. Our methylation analysis led to the identification of a common methylation fingerprint in ISAs and cardiac UPS. We assume this “ISA” methylation signature to be specific. Accordingly, relevant differential diagnoses, e.g., dedifferentiated liposarcomas and extra-cardiac UPS showed distinct methylation signatures compared to ISA [43, 44]. Notably, these methylation signatures remain stable over the timeline of the disease, as previously reported in other tumor entities [45]. We could detect the ISA methylation signature even in the bone metastasis of a cardiac UPS. In view of the specificity and stability of this “ISA” DNA methylation signature, DNA methylation profiling may be a valuable ancillary biomarker in ISAs and UPS of the left atrium, especially in cases lacking MDM2, MDM4 or CDK6 amplifications or occurring at unusual sites.
Supplementary information
Acknowledgements
We thank the Microarray Unit of the Genomics and Proteomics Core Facility, German Cancer Research Center (DKFZ), for providing excellent methylation services.
Funding
The work was supported by the German Cancer Aid (grant 70112499). Open Access funding enabled and organized by Projekt DEAL.
Author contributions
CK conceptualized and coordinated the project. Pathology review and copy number variation analysis was performed by CK. CK, DS, and AvD supervised array-based analysis. JKB, FKFK, DTWJ, SMP, CEH, SF, AS, RB, TM, ML, DB, CRA, JVG, UF, BB, and GM provided tumor samples and corresponding metadata. CK wrote the original draft, which was reviewed and approved of by all authors.
Data availability
The data are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Ethics Approval and Consent to Participate
The patients’ consent was waived due to the retrospective nature of this study and minimal risk to the subjects. The study was performed in accordance with the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Andreas von Deimling, Gunhild Mechtersheimer.
Supplementary information
The online version contains supplementary material available at 10.1038/s41379-021-00874-y.
References
- 1.WHO Classification of Tumours of Soft Tissue and Bone. 5th edn, Vol. 3 (2020).
- 2.Frezza AM, Assi T, Lo Vullo S, Ben-Ami E, Dufresne A, Yonemori K, et al. Systemic treatments in MDM2 positive intimal sarcoma: a multicentre experience with anthracycline, gemcitabine, and pazopanib within the World Sarcoma Network. Cancer. 2020;126:98–104. doi: 10.1002/cncr.32508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandelstamm, M. Über primäre Neubildungen des Herzens. Virchows Arch. 1923:43–54.
- 4.Hottenrott G, Mentzel T, Peters A, Schroder A, Katenkamp D. Intravascular (“intimal”) epithelioid angiosarcoma: clinicopathological and immunohistochemical analysis of three cases. Virchows Arch. 1999;435:473–478. doi: 10.1007/s004280050430. [DOI] [PubMed] [Google Scholar]
- 5.Staats P, Tavora F, Burke AP. Intimal sarcomas of the aorta and iliofemoral arteries: a clinicopathological study of 26 cases. Pathology. 2014;46:596–603. doi: 10.1097/PAT.0000000000000182. [DOI] [PubMed] [Google Scholar]
- 6.Burke AP, Virmani R. Sarcomas of the great vessels. A clinicopathologic study. Cancer. 1993;71:1761–73. doi: 10.1002/1097-0142(19930301)71:5<1761::AID-CNCR2820710510>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 7.Tavora F, Miettinen M, Fanburg-Smith J, Franks TJ, Burke A. Pulmonary artery sarcoma: a histologic and follow-up study with emphasis on a subset of low-grade myofibroblastic sarcomas with a good long-term follow-up. Am J Surg Pathol. 2008;32:1751–61. doi: 10.1097/PAS.0b013e31817d7fd0. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J, Roth J, Bode-Lesniewska B, Pfaltz M, Heitz PU, Komminoth P. Combined comparative genomic hybridization and genomic microarray for detection of gene amplifications in pulmonary artery intimal sarcomas and adrenocortical tumors. Genes Chromosomes Cancer. 2002;34:48–57. doi: 10.1002/gcc.10035. [DOI] [PubMed] [Google Scholar]
- 9.Bode-Lesniewska B, Zhao J, Speel EJ, Biraima AM, Turina M, Komminoth P, et al. Gains of 12q13-14 and overexpression of mdm2 are frequent findings in intimal sarcomas of the pulmonary artery. Virchows Arch. 2001;438:57–65. doi: 10.1007/s004280000313. [DOI] [PubMed] [Google Scholar]
- 10.Sebenik M, Ricci A, Jr., DiPasquale B, Mody K, Pytel P, Jee KJ, et al. Undifferentiated intimal sarcoma of large systemic blood vessels: report of 14 cases with immunohistochemical profile and review of the literature. Am J Surg Pathol. 2005;29:1184–93. doi: 10.1097/01.pas.0000159774.70288.7d. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Macdonald WD, Erickson-Johnson M, Wang X, Jenkins RB, Oliveira AM. Cytogenetic and molecular cytogenetic findings of intimal sarcoma. Cancer Genet Cytogenet. 2007;179:146–149. doi: 10.1016/j.cancergencyto.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Dewaele B, Floris G, Finalet-Ferreiro J, Fletcher CD, Coindre JM, Guillou L, et al. Coactivated platelet-derived growth factor receptor {alpha} and epidermal growth factor receptor are potential therapeutic targets in intimal sarcoma. Cancer Res. 2010;70:7304–14. doi: 10.1158/0008-5472.CAN-10-1543. [DOI] [PubMed] [Google Scholar]
- 13.Neuville A, Collin F, Bruneval P, Parrens M, Thivolet F, Gomez-Brouchet A, et al. Intimal sarcoma is the most frequent primary cardiac sarcoma: clinicopathologic and molecular retrospective analysis of 100 primary cardiac sarcomas. Am J Surg Pathol. 2014;38:461–469. doi: 10.1097/PAS.0000000000000184. [DOI] [PubMed] [Google Scholar]
- 14.Ito Y, Maeda D, Yoshida M, Yoshida A, Kudo-Asabe Y, Nanjyo H, et al. Cardiac intimal sarcoma with PDGFRbeta mutation and co-amplification of PDGFRalpha and MDM2: an autopsy case analyzed by whole-exome sequencing. Virchows Arch. 2017;471:423–428. doi: 10.1007/s00428-017-2135-x. [DOI] [PubMed] [Google Scholar]
- 15.Roszik, J, Khan, A, Conley, AP, Livingston, JA, Groisberg, R, Ravi, V et al. Unique aberrations in intimal sarcoma identified by next-generation sequencing as potential therapy targets. Cancers (Basel). 2019:11. [DOI] [PMC free article] [PubMed]
- 16.Neuville A, Coindre JM. Cardiac sarcoma with MDM2 amplification. Am J Surg Pathol. 2014;38:1449. doi: 10.1097/PAS.0000000000000275. [DOI] [PubMed] [Google Scholar]
- 17.Maleszewski JJ, Tavora F, Burke AP. Do “intimal” sarcomas of the heart exist? Am J Surg Pathol. 2014;38:1158–1159. doi: 10.1097/PAS.0000000000000271. [DOI] [PubMed] [Google Scholar]
- 18.Burke A, Tavora F. The 2015 WHO Classification of Tumors of the Heart and Pericardium. J Thorac Oncol. 2016;11:441–52. doi: 10.1016/j.jtho.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 19.WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. 4th edn, Vol. 7 (2015).
- 20.Watson, R, Frye, J, Trieu, M & Yang, MX Primary undifferentiated pleomorphic cardiac sarcoma with MDM2 amplification presenting as acute left-sided heart failure. BMJ Case Rep2018 (2018). [DOI] [PMC free article] [PubMed]
- 21.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–74. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koelsche C, Schrimpf D, Stichel D, Sill M, Sahm F, Reuss DE, et al. Sarcoma classification by DNA methylation profiling. Nat Commun. 2021;12:498. doi: 10.1038/s41467-020-20603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koelsche C, Mynarek M, Schrimpf D, Bertero L, Serrano J, Sahm F, et al. Primary intracranial spindle cell sarcoma with rhabdomyosarcoma-like features share a highly distinct methylation profile and DICER1 mutations. Acta Neuropathol. 2018;136:327–37. doi: 10.1007/s00401-018-1871-6. [DOI] [PubMed] [Google Scholar]
- 24.Weidema ME, van de Geer E, Koelsche C, Desar IME, Kemmeren P, Hillebrandt-Roeffen MHS, et al. DNA methylation profiling identifies distinct clusters in angiosarcomas. Clin Cancer Res. 2020;26:93–100. doi: 10.1158/1078-0432.CCR-19-2180. [DOI] [PubMed] [Google Scholar]
- 25.Ten Broek RW, Koelsche C, Eijkelenboom A, Mentzel T, Creytens D, Vokuhl C, et al. Kaposiform hemangioendothelioma and tufted angioma - (epi)genetic analysis including genome-wide methylation profiling. Ann Diagn Pathol. 2020;44:151434. doi: 10.1016/j.anndiagpath.2019.151434. [DOI] [PubMed] [Google Scholar]
- 26.Rohrich M, Koelsche C, Schrimpf D, Capper D, Sahm F, Kratz A, et al. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol. 2016;131:877–87. doi: 10.1007/s00401-016-1540-6. [DOI] [PubMed] [Google Scholar]
- 27.Kommoss FKF, Stichel D, Schrimpf D, Kriegsmann M, Tessier-Cloutier B, Talhouk A, et al. DNA methylation-based profiling of uterine neoplasms: a novel tool to improve gynecologic cancer diagnostics. J Cancer Res Clin Oncol. 2020;146:97–104. doi: 10.1007/s00432-019-03093-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koelsche C, Hartmann W, Schrimpf D, Stichel D, Jabar S, Ranft A, et al. Array-based DNA-methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod Pathol. 2018;31:1246–56. doi: 10.1038/s41379-018-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fittall MW, Lyskjaer I, Ellery P, Lombard P, Ijaz J, Strobl AC, et al. Drivers underpinning the malignant transformation of giant cell tumour of bone. J Pathol. 2020;252:433–40. doi: 10.1002/path.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hovestadt V, Remke M, Kool M, Pietsch T, Northcott PA, Fischer R, et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol. 2013;125:913–916. doi: 10.1007/s00401-013-1126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capper D, Stichel D, Sahm F, Jones DTW, Schrimpf D, Sill M, et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018;136:181–210. doi: 10.1007/s00401-018-1879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research Network. Electronic address, e. d. s. c. & Cancer Genome Atlas Research, N. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–965 e928. [DOI] [PMC free article] [PubMed]
- 33.Nebenfuehr S, Kollmann K, Sexl V. The role of CDK6 in cancer. Int J Cancer. 2020;147:2988–95. doi: 10.1002/ijc.33054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haupt S, Mejia-Hernandez JO, Vijayakumaran R, Keam SP, Haupt Y. The long and the short of it: the MDM4 tail so far. J Mol Cell Biol. 2019;11:231–44. doi: 10.1093/jmcb/mjz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan X, Larsson C, Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019;38:6172–83. doi: 10.1038/s41388-019-0872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–52. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 37.Taylor BS, Barretina J, Maki RG, Antonescu CR, Singer S, Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;11:541–57. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–21. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Dievel J, Sciot R, Delcroix M, Vandeweyer RO, Debiec-Rychter M, Dewaele B, et al. Single-center experience with intimal sarcoma, an ultra-orphan, commonly fatal mesenchymal malignancy. Oncol Res Treat. 2017;40:353–359. doi: 10.1159/000476036. [DOI] [PubMed] [Google Scholar]
- 40.Le Guellec S, Chibon F, Ouali M, Perot G, Decouvelaere AV, Robin YM, et al. Are peripheral purely undifferentiated pleomorphic sarcomas with MDM2 amplification dedifferentiated liposarcomas? Am J Surg Pathol. 2014;38:293–304. doi: 10.1097/PAS.0000000000000131. [DOI] [PubMed] [Google Scholar]
- 41.Coindre JM, Mariani O, Chibon F, Mairal A, De Saint Aubain Somerhausen N, Favre-Guillevin E, et al. Most malignant fibrous histiocytomas developed in the retroperitoneum are dedifferentiated liposarcomas: a review of 25 cases initially diagnosed as malignant fibrous histiocytoma. Mod Pathol. 2003;16:256–62. doi: 10.1097/01.MP.0000056983.78547.77. [DOI] [PubMed] [Google Scholar]
- 42.Coindre JM, Hostein I, Maire G, Derre J, Guillou L, Leroux A, et al. Inflammatory malignant fibrous histiocytomas and dedifferentiated liposarcomas: histological review, genomic profile, and MDM2 and CDK4 status favour a single entity. J Pathol. 2004;203:822–30. doi: 10.1002/path.1579. [DOI] [PubMed] [Google Scholar]
- 43.Basturk O, Weigelt B, Adsay V, Benhamida JK, Askan G, Wang L, et al. Sclerosing epithelioid mesenchymal neoplasm of the pancreas - a proposed new entity. Mod Pathol. 2020;33:456–67. doi: 10.1038/s41379-019-0334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koelsche C, Stichel D, Griewank KG, Schrimpf D, Reuss DE, Bewerunge-Hudler M, et al. Genome-wide methylation profiling and copy number analysis in atypical fibroxanthomas and pleomorphic dermal sarcomas indicate a similar molecular phenotype. Clin Sarcoma Res. 2019;9:2. doi: 10.1186/s13569-019-0113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27:728–43. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available from the corresponding author upon request.