Highlights
-
•
A novel anti-ADAM17 monoclonal antibody, D8P1C1, has been developed.
-
•
D8P1C1 inhibits the proteolysis of peptide substrates by ADAM17.
-
•
D8P1C1 inhibits the proliferation of cancer cells and tumor growth inhibition in vivo.
-
•
D8P1C1 preferentially recognizes ADAM17 on cancer cells.
-
•
Negative stain EM analysis reveals that D8P1C1 binds to the ADAM17 protease domain.
Keywords: ADAM17, Monoclonal antibody, EGFR signaling, Cancer therapy
Abbreviations: MP, Metalloprotease domain; D, Disintegrin domain; C, Cysteine rich domain; EGFR, Epidermal growth factor receptor; TNBC, Triple negative breast cancer; MMP, Matrix metalloproteinase; HRP, Horseradish peroxidase; VH, Variable heavy chain; VL, Variable light chain; EM, Electron microscopy
Abstract
ADAM17 is upregulated in many cancers and in turn activates signaling pathways, including EGFR/ErbB, as well as those underlying resistance to targeted anti-EGFR therapies. Due to its central role in oncogenic pathways and drug resistance mechanisms, specific and efficacious monoclonal antibodies against ADAM17 could be useful for a broad patient population with solid tumors. Hence, we describe here an inhibitory anti-ADAM17 monoclonal antibody, named D8P1C1, that preferentially recognizes ADAM17 on cancer cells. D8P1C1 inhibits the catalytic activity of ADAM17 in a fluorescence-based peptide cleavage assay, as well as the proliferation of a range of cancer cell lines, including breast, ovarian, glioma, colon and the lung adenocarcinoma. In mouse models of triple-negative breast cancer and ovarian cancer, treatment with the mAb results in 78% and 45% tumor growth inhibition, respectively. Negative staining electron microscopy analysis of the ADAM17 ectodomain in complex with D8P1C1 reveals that the mAb binds the ADAM17 protease domain, consistent with its ability to inhibit the ADAM17 catalytic activity. Collectively, our results demonstrate the therapeutic potential of the D8P1C1 mAb to treat solid tumors.
Introduction
ADAM proteases induce shedding of a variety of membrane-anchored proteins implicated in both health and diseases, notably cytokines, growth factors, chemokines as well as regulators of neurological processes, inflammation and cancer [1], [2], [3]. ADAM proteases consist of an N-terminal pro-sequence followed by metalloprotease (M), disintegrin (D), cysteine-rich (C), transmembrane and cytoplasmic domains [4]. The substrate specificity is not imparted by a typical substrate cleavage signature but relies on noncatalytic interactions between the substrate and the ADAM D and C domains [5, 6]. ADAM17, also known as TNF-alpha converting enzyme (TACE), is activated in many cancers, including breast, ovarian, colon and prostate, and in turn triggers oncogenic pathways, including EGFR/ErbB and those underlying resistance to targeted anti-EGFR therapies [1]. Signaling by the EGFR/erbB/HER family is important in regulating proliferation, survival, differentiation and motility during normal development, while dysregulation can promote oncogenesis [7]. The proteolytic release of cell-surface tethered erbB ligands by ADAM17 is a key regulatory switch to trigger EGFR/erbB signaling, which initiates the downstream autocrine signaling that drives tumor progression [8]. Indeed, the erbBs display de-regulated signaling in many human cancers due to overexpression and/or mutations, including in some of the most intractable and common cancers, with EGFR (and HER2) prominent in ovarian, breast, lung and colon cancer, amongst others. In addition, ADAM17-mediated cleavage of erbB2/erbB4 in tumors is associated with constitutive receptor activity and poor prognosis and undermines receptor-targeted therapies [7, 8]. It is anticipated that due to the central role of ADAM17 in oncogenic pathways and drug resistance mechanisms, specific and efficacious monoclonal antibodies against ADAM17 can be useful for a broad patient population with solid tumors [9], [10], [11]. Towards this goal, we generated and characterized an inhibitory anti-ADAM17 monoclonal antibody (mAb) that preferentially recognizes ADAM17 on cancer cells and inhibits solid tumor growth in cell-based and animal-based models.
Materials and methods
Cell lines
All the cell lines described here, except the colon cancer line LIM1215, were purchased from American Type Culture Collection (ATCC). The triple-negative breast cancer (TNBC) cell line MDA-MB-231 was cultured in Dulbecco's Modified Eagle Medium (DMEM), 10% Fetal Bovine Serum (FBS), 1% Penicillin/Streptomycin (P/S) and 2 mM l-Glutamine. The HER2-overexpressing breast cancer cell line SKBR-3 was grown in McCoy's 5a, 10% FBS and 1% P/S. The ovarian high-grade serous carcinoma cell line OVCAR-3 was cultured in RPMI-1640, 10% FBS, 1% P/S, 10 mM HEPES and 0.2 units/ml Insulin while CaOV-3 and SKOV-3 were grown in DMEM, 10% FBS and 1% P/S. The lung adenocarcinoma cell line HCC-827 was cultured in RPMI-1640, 10% FBS and 1% P/S. The glioma cell line U-87 MG was grown in Eagle's Minimum Essential Medium, 10% FBS and 1% P/S. The LIM1215 colon cancer cell line, derived from a patient with inherited nonpolyposis colorectal cancer [12], was purchased from Cell Bank Australia and cultured in RPMI1640 with 2 mM l-Glutamine, 25 mM HEPES, 10% FBS, 0.6 µg/ml Insulin, 1 µg/ml Hydrocortisone and 10 uM Thiolglycerol. The cell lines were subcultured according to instruction manuals and tested for mycoplasma contamination before use.
Expression and purification of the human ADAM17 extracellular domain (ECD)
The human ADAM17 cDNA was a gift from Dr. Carl Blobel, Hospital for Special Surgery, New York. The construct comprising of 20–655 amino acids was generated by PCR amplification and cloned into a custom-made pMA152a baculovirus vector [13]. pMA152a is based on the pAcGP67B vector (BD Biosciences) with an incorporated removable Fc-tag (human). Secreted recombinant protein was produced by baculovirus-infected Hi5 insect cell following the protocol provided by BD Biosciences. The C-terminal Fc-tag was used to facilitate protein-A affinity chromatography and removed by thrombin cleavage afterwards. SD-200 size-exclusion chromatography (GE Biosciences) was used to obtain the final purified recombinant protein.
Generation and affinity maturation of monoclonal antibodies binding to the ADAM17 ECD
An ADAM17 ECD construct comprising residues 215–655 was used as an antigen to pan a large naïve human Fab library [14]. Briefly, a naïve human Fab phage display library, constructed from peripheral blood B cells of healthy donors, was used for selection of Fab binders against purified, soluble, ADAM17 ECD conjugated to magnetic beads (Dynabeads M-270 epoxy; DYNAL Inc., New Hyde Park, N.Y.). Amplified libraries of 1012 phage displayed Fabs were incubated with varying concentrations of ADAM17 ECD for four rounds of panning. Between the rounds, the beads were washed thoroughly to remove non-specifically bound phage. The bead-bound phage was mixed with TG1 cells for 1 hour at 37 °C, and the phage was amplified from the infected cells. Randomly picked clones from the infected TGI cells after four rounds of panning were grown in 2YT medium containing 100 µg/ml carbenicillin, 0.2% glucose in 96-well plates by using the automated BioRobotics BioPick colony picking system (Genomic Solutions, Ann Arbor, MI). After the bacterial cultures reached an optical density of 0.5 (at 600 nm), helper phage M13K07 at a multiplicity of infection (MOI) of 10 and kanamycin at 50 µg/ml (final concentration), were added to the medium, and the plates were further incubated at 30 °C overnight in a shaker at 250 rpm. The phage supernatants were then used to identify clones with high binding affinities towards ADAM17 ECD in ELISA-based assays. Two clones, D5 and D8, that bound to ADAM17 ECD with a signal to noise ratio of at least 1 (at A450 nm), were selected for expression and purification. The VH and VL of the selected clones were sequenced and plasmids were extracted from these clones for transformation of HB2151 cells. A single colony was picked from the plate containing freshly transformed cells, inoculated into 200 ml 2YT medium containing 100 µg/ml ampicillin and 0.2% glucose, and incubated at 37 °C with shaking at 250 rpm. When the culture OD reached 0.90 at 600 nm, isopropyl-d-thiogalactopyranoside at a 0.5 mM final concentration was added and the culture was further incubated overnight at 30 °C. The bacterial pellet was collected after centrifugation at 8000 g for 20 min and resuspended in phosphate-buffered saline pH 7.4, (PBS) containing 0.5 mU polymixin B (Sigma-Aldrich, St. Louis, MO). After 30 min incubation the bacterial slurry was centrifuged at 25,000 g for 25 min at 4 °C, and the supernatant was used for Fab purification with a protein G column (Sigma-Aldrich, St. Louis, MO). For conversion of D5 and D8 Fabs to IgG1 format, the heavy and light chains were amplified and recloned in the PDR12 vector for whole immunoglobulin-G1 (IgG1) expression. The construct was then transfected into the Freestyle 293 expression system. Whole IgG1 was purified from the culture supernatant using protein G matrix.
The affinity maturation of the D8 and D5 antibodies was carried out by Lake Pharma, Inc. using targeted mutagenesis. After 3 rounds of selection, the affinity matured clones were highly enriched, and the affinity improvement was confirmed using parental D5 and D8 mAbs as references. The matured clones had 10-fold or better improvement in binding affinity over the parental clones and bound to human ADAM17 with Kds of 50–80 pM (Lake Pharma, Inc.). The EC50 values and sequencing results were used to select a panel of five affinity matured clones for further studies (See Supplementary Table S1). The MED13622 (MedImmune/AstraZeneca) anti-ADAM17 mAb was provided by the Tri-Institutional Therapeutics Discovery Institute (Memorial Sloan Kettering, The Rockefeller University and Weill Cornell Medical College, New York).
Fluorescent peptide cleavage assay
The purified ADAM17(ECD)-binding mAbs D8P1C1 and D5P2A11, as well as the mAb MED13622, were buffer-exchanged into 25 mM Tris, pH 9.0, containing 2 μM ZnCl2, and 0.005% (w/v) Brij-35. ADAM17-antibody complexes were formed at a 1:1 molar ratio prior to the assay. The assay was carried out by mixing 50 μM of a fluorogenic peptide substrate Mca-PLAQAV-Dpa (R&D Systems Cat# ES003) with ADAM17-antibody complexes at 37 °C and monitoring the progress of the enzymatic reaction by fluorescence emission (excitation 320 nm and emission 405 nm) over a time course of 1 h using a SpectraMax M5. ADAM17(ECD) alone was used as a positive control. The substrate peptide was derived from TNFalpha and contains a highly fluorescent 7-methoxycoumarin group and a quencher group, 2,4-dinitrophenyl. ADAM17 cleaves the amide bond between the fluorescent and the quencher group causing an increase in fluorescence (R&D Systems, Cat Number ES003). [15, 16].
Negative staining em analysis
Protein complex of ADAM17 ectodomain with D8P1C1 Fab was prepared by mixing the two proteins at a 1:1 molar ratio, then diluted with a buffer containing 10 mM HEPES, pH 7.4, 150 mM NaCl, adsorbed to a freshly glow-discharged carbon-coated copper grid, washed three times with the same buffer, and stained three times with 0.75% uranyl formate. Images were collected at a magnification of 57,000 using EPU on a Thermo Fisher Talos F200C microscope equipped with a 4k x 4k CETA 16 M camera and operated at 200 kV. The pixel size was 2.5 Å for the CETA camera. Image processing, particles picking, reference-free 2D classification, 3D reconstruction and refinement were performed using cryoSPARC. The structure was visualized using Chimera.
Alamar Blue cell viability assay
Cells (breast/ovarian/colon/glioma/lung-adenocarcinoma) were harvested in the log phase of growth (after 3 days of culturing). The cell count was determined and was adjusted to 5 × 104 cells/ml. The cells were allowed to adhere and grow for 24 h in 96-well cell culture plates, treated with test agents, in this case, the affinity matured ADAM17 mAbs D8P1C1, D5P2A11 and their parental clones D8 and D5. The cells were allowed to grow for additional 38 h. Cells not treated with mAbs were used as a control. Alamar Blue (Bio-RAD Laboratories) (10% of the well volume) was added aseptically. Cultures containing Alamar Blue were incubated for 6 hrs and cell proliferation was measured spectrophotometrically by absorbance at 570 and 600 nm. Cell viability was calculated using the following formula:
Percentage difference between treated and control cells
O1 = molar extinction coefficient (E) of oxidized alamarBlue® (Blue) at 570 nm, O2 = E of oxidized alamarBlue® at 600 nm, A1 = absorbance of test wells at 570 nm, A2 = absorbance of test wells at 600 nm, P1 = absorbance of positive growth control well (cells plus alamarBlue® but no test agent) at 570 nm, P2 = absorbance of positive growth control well (cells plus alamarBlue® but no test agent) at 600 nm [17].
Cell-based ELISA assays to gage the binding of anti-ADAM17 mAb D8P1C1 to cancer cell lines
Cellular ELISA [18] was performed to gage the binding of the anti-ADAM17 mAb D8P1C1, relative to the binding of the MED13622 mAb, to ADAM17 expressed on the cell surface of cancer cell lines (breast, ovarian, colon, glioma, adenocarcinoma), as well as to HEK293 cells and HEK293 cells transfected with full-length human ADAM17. Briefly, 5 × 104 cells/well were immobilized on 96-well ELISA plates (Greiner bio-one) with 1% paraformaldehyde for 2 hrs at 37 °C. The plate was washed thrice with PBS and blocked for 2 hrs at room temperature with 4% non-fat dry milk. The anti-ADAM17 mAbs were added in varying concentrations. Mouse mAb conjugated to HRP and recognizing human IgG was used as a secondary antibody (Abcam) and color was developed using the TMB substrate kit (Thermo Scientific). The data was recorded at 450 nm.
In vivo antitumor efficacy studies
All cells were grown in monolayer culture, harvested by trypsinization, and implanted subcutaneously into the right flank of 6- to 8-week-old female athymic nude mice for MDA-MB-231, and NSG mice for SKOV-3 (n = 5). Approximately 10 million cells (MDA-MB-231 or SKOV-3) were injected per mouse. Mice were randomized into 5 mice per group. When tumors reached 100 to 150 mm3, the anti-ADAM17 mAb D8P1C1, prepared by diluting with sterile PBS, was injected intraperitoneally, twice a week for four weeks, according to body weight. Sterile PBS was used as a control. Tumor volume was determined by external caliper and calculated by the modified ellipsoidal formula: V = ½ (Length × Width2). Antitumor efficacy was calculated as (1-dT/dC) X100, where dT is the final tumor volume minus the starting tumor volume from the treatment group and dC is the final tumor volume minus the starting tumor volume of the control group [19]. Error bars were calculated as SEM. The mouse body weight and general health were monitored daily, and the experiments were carried out in accordance with Association for Assessment and Accreditation of Laboratory Animal Care and MSKCC Institutional Animal Care and Use Committee guidelines.
Statistical analysis
The data from all in vitro and cell-based assays described, including Alamar blue, fluorescent peptide cleavage and ELISA-based assays, are representative of triplicate determinations and two independent experiments. Unpaired two-tailed Student's t-test was performed to determine the significance of the results.
Results
The recombinant ADAM17 ectodomain is monomeric in solution
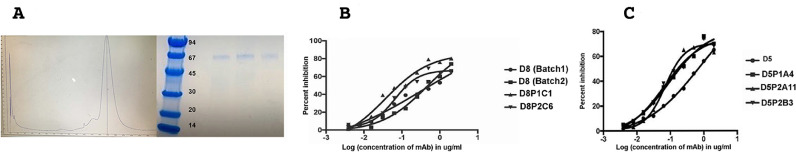
The ectodomain (ECD) of ADAM17 (residues 20–655) was cloned in a custom-made vector, pMA152a [13] for baculovirus expression. The PCR amplified product was fused to the envelope surface glycoprotein gp67 signal sequence and the ADAM17(ECD) was purified from the culture supernatant using protein A Sepharose and SD-200 (size-exclusion chromatography). ADAM17(ECD) elutes as a monomer with a native molecular weight of 65 kDa. The prodomain is cleaved during secretion of the protein into the culture medium. The purified ADAM17(ECD) recombinant protein was N-terminally sequenced confirming that the final product (amino acid residues 215–655) consists of the metalloprotease (MP), disintegrin (D) and cysteine rich (C) domains, Fig. 1A. The matured (lacking the pro domain) ECD was used as an antigen to raise a panel of ADAM17 specific monoclonal antibodies.
Fig. 1.
Antigen Expression and mAb Affinity Maturation. (A) SD-200 size-exclusion chromatography and SDS-PAGE profile of the purified ADAM17(ECD) construct. The molecular weights (kDa) of the protein standards are indicated. The three SDS-PAGE samples represent the three peak fractions from the SD-200 elute. The ADAM17(ECD) protein elutes as a monomer on the gel filtration chromatography column with a native molecular weight of 65 kDa. (B, C) Cell proliferation assays with MDA-MB-231 cells treated with five affinity matured clones of D5 and D8. The results represent mean of triplicate determinations and two independent experiments, and graphs show the effect of treatment of mAbs on MDA-MB-231 cells relative to the control (described in the Materials and Methods section, for IC50 and R-square values see Supplementary Table S1). D8 (Batch 1) was expressed and purified from Free Style™ 293 F cells and D8 (Batch 2) was expressed and purified from CHO cells.
Generation of anti-ADAM17 monoclonal antibodies
Construction of a large naïve phage-displayed Fab library was achieved through one-step cloning: SfiI was used as the cloning enzyme, since it can keep almost all the antibody gene repertoire intact during the cloning procedure. To fully cover each of the human antibody genes, carefully selected primers, matching the conserved N- and C-terminal region of the heavy- and light-chains, were used for the amplification of each of the gene families separately [14]. Two Fab's, D8 and D5, were selected based on their binding specificities and affinities towards the ADAM17 ECD (using ELISA). Cross-reactivity with closely related ADAM's, including ADAM10 and ADAM19 were also evaluated by ELISA-based screening. The selected Fabs specifically recognize the human and mouse ADAM17 ectodomain and did not bind to ADAM10 or ADAM19 (data not shown). They were then reformatted to a human IgG1 framework. Affinity maturation of D8 and D5 were carried out by Lake Pharma using targeted mutagenesis. A panel of 5 affinity matured clones for D8 and D5 were selected based on sequencing results (selecting distinct clones), EC50 and Kd values.
Two affinity matured anti-ADAM17 antibodies, D8P1C1 and D5P2A11, efficiently inhibit the proliferation of a triple-negative breast cancer cell line
ADAM17-dependent EGFR ligand (TGF-alpha) shedding is an important proliferative signal in breast cancer and ADAM17 is known to play a key role in the progression of triple-negative breast cancer (TNBC) [20]. Indeed, it has been shown that small molecule inhibitors to ADAM17 block invasion, migration and colony formation of several triple-negative breast cancer cell lines [21]. We, therefore, evaluated the anti-tumor potential of the five affinity-matured anti-ADAM17 mAbs (see above) in an Alamar blue cell-based assay using the TNBC cell line MDA-MB-231 that is HER2 negative and EGFR positive [22]. The results reveal that D8P1C1 and D5P2A11 are the most effective clones in inhibiting the proliferation of the TNBC cell line with IC50 values of 0.037 and 0.069 µg/ml, respectively (Fig. 1B, C, Supplementary Table S1). Based on these experiments, we selected D8P1C1 and D5P2A11 for further studies. The IC50 values of the five affinity matured clones and their parental counterparts, D8 and D5, are shown in Table S1. Noteworthy, the IC50 values of D8P1C1 and D5P2A11 are 5 to 10-fold lower than the parental D8 and D5 clones.
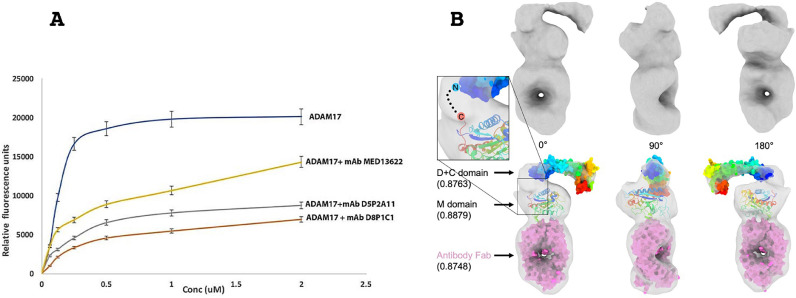
D8P1C1 binds to the catalytic domain of ADAM17 and inhibits its proteolytic activity
In a fluorescent peptide cleavage assay, the mAb D8P1C1 significantly inhibits the catalytic activity of ADAM17, Fig. 2A. At the concentrations used, D8P1C1 is twice as effective as the MedImmune/AstraZeneca mAb MED13622 in inhibiting substrate cleavage. We next carried out negative-stain EM analysis with ADAM17(ECD) bound to a Fab fragment of D8P1C1. The D8P1C1 Fab fragment was generated by restricted papain cleavage of the whole IgG. The ADAM17(ECD)/Fab complex was prepared by mixing the two components in 1:1 molar ratio. Both 2D class averaging and 3D reconstruction show detailed features corresponding to the catalytic, disintegrin and cysteine rich ADAM domains, as well as the bound Fab, Fig. 2B. In the calculated 3D EM density map, we docked the X-ray crystal structures of the ADAM10 disintegrin and cysteine rich domains (PDB ID 5L0Q) [23], as well as that of the ADAM17 catalytic domain (PDB ID 3EWJ) [24]. These results show that the Fab D8P1C1 binds to the catalytic domain of ADAM17 (Fig. 2B and Supplementary Fig. S1). Further studies are underway to obtain atomic-resolution structure of the complex.
Fig. 2.
The affinity matured D8P1C1 and D5P2A11 clones bind the ADAM17 metalloprotease domain and inhibit its proteolytic activity. (A) FRET-based peptide cleavage assays. The data represent mean of triplicate determinations and two independent experiments. Maximum dispersion was within 10% of the mean value. ADAM17-antibody complexes were formed at 1:1 molar ratio prior to the assay, and the assay was carried out in the presence of 50 μM of a fluorogenic peptide as described in the Materials and Methods section. Representative results of at least three independent experiments show effect of mAbs on substrate cleavage relative to the control (ADAM17 without the mAbs); mean ± SEM; P<0.001 by unpaired two-tailed Student's t-test, (ADAM17 without mAb vs ADAM17 with mAb). (B) Negative stain EM analysis and 3D reconstruction of the ADAM17 extracellular domain bound to a Fab derived from D8P1C1 shown in three perpendicular angles. The molecular volume rendering shows the electron density map (surface in gray) of the ADAM17/antibody complex (top), with crystal structures (bottom) of D + C domain (surface in rainbow color), M domain (cartoon in rainbow color) and antibody Fab (surface in magenta) [23, 24] fitted inside. Rainbow color follows the scheme of N-terminus in blue and C-terminus in red. Volume fitting correlation scores are shown in the parenthesis. A hypothetic linker between C-terminus of M domain and N-terminus of D + C domain is shown as dash line in the inset.
D8P1C1 and D5P2A11 inhibit the proliferation of multiple EFGR-overexpressing cancer cell lines
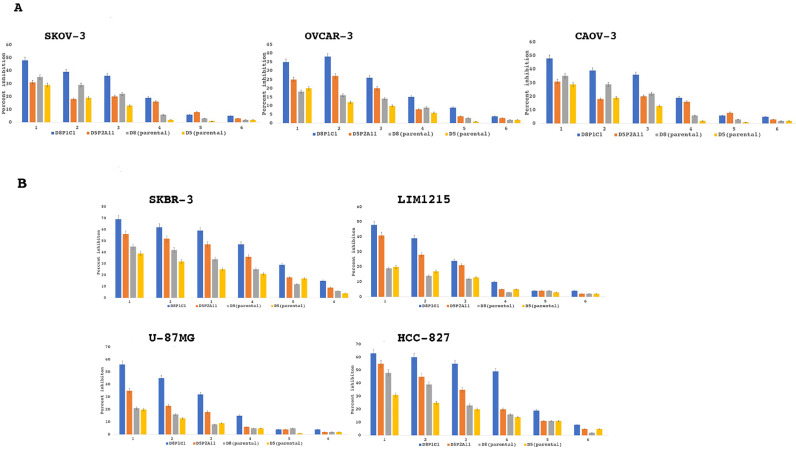
We next performed cell viability assays with other cancer cell lines, including OVCAR-3, SKOV-3, CAOV-3 (ovarian) and SKBR-3 (breast), U87-MG (glioma), LIM1215 (colon) and HCC-827 (non-small cell lung cancer or NSCLC), Fig. 3A, B. These cell lines are known to overexpress EGFR/HER2 [22, 25]. The ovarian cancer cell lines OVCAR-3 and CAOV-3 represent high-grade serous carcinoma (HGSC) type, while SKOV-3 belongs to the non-HGSC type [25]. Though high-grade serous carcinoma is the most prevalent amongst ovarian cancer patients, the non-serous types are known to migrate and invade more aggressively [25]. The Alamar blue assays show that the two anti-ADAM17 mAbs (D8P1C1 and D5P2A11) cause varying degree of proliferation inhibition, Fig. 3A, B. Specifically, they are more efficient in inhibiting the HER2-dependent breast cancer cell line SKBR-3 and the EGFR-expressing lines HCC-827, LIM1215 and U87-MG, as compared to the ovarian cancer cell lines. For instance, at a concentration of 20 µg/ml, D8P1C1 caused 70% and 63% proliferation inhibitions of SKBR-3 and HCC-827, respectively, compared to 35% inhibition of OVCAR-3.
Fig. 3.
Alamar blue cell viability assays with multiple cancer cell lines. Percent growth inhibition is show for ovarian (SKOV-3, CAOV-3, OVCAR-3), breast (SKBR-3), colon (LIM1215), glioma (U87-MG) and non-small cell lung cancer (HCC-827) tumor cell lines treated with D8P1C1, D5P2A11, D8 or D5. The data represent mean of triplicate determinations and two independent experiments, and the bar plots show the effect of treatment of mAbs on cancer cells relative to the control, mean ± SEM (described in Materials and Methods).1: 20 µg/ml; 2: 10 µg/ml; 3: 5 µg/ml; 4: 2.5 µg/ml; 5: 1.25 µg/ml; 6: 0.625 µg/ml.
D8P1C1 specifically recognizes ADAM17 on cancer cells
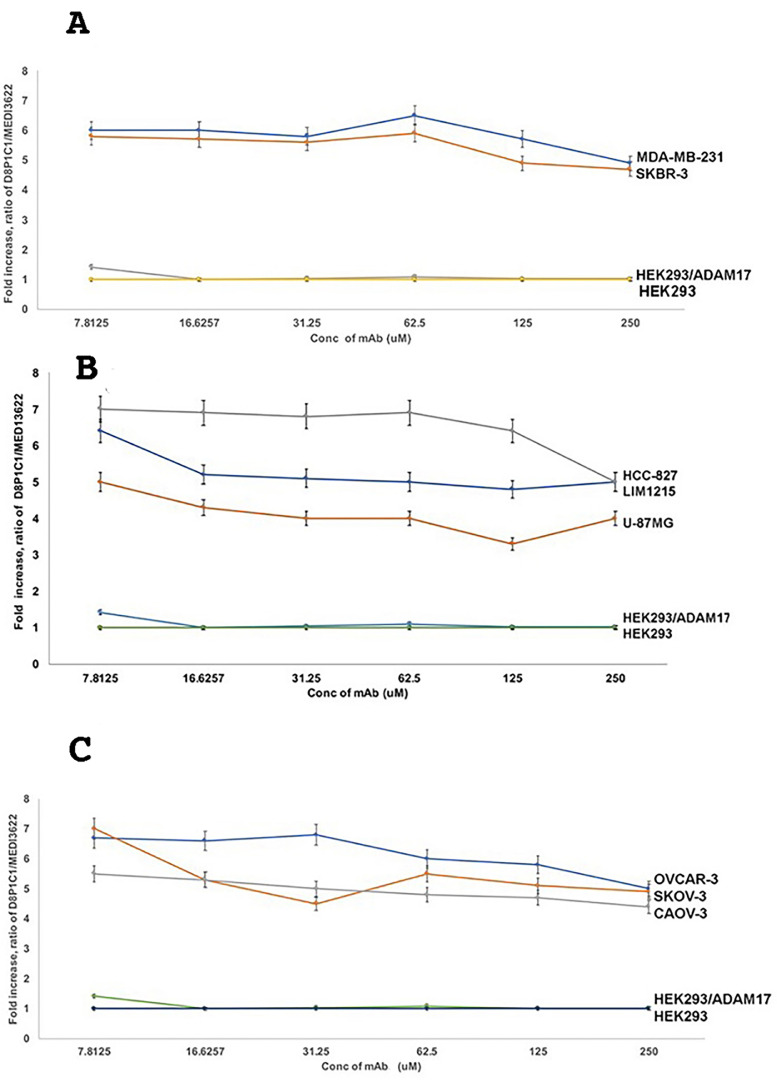
The ADAM activity is proposed to be regulated via transitions between two distinct conformations, “active” or “open” and “latent” or “closed” [26] supported by “open” and “closed” conformations observed in the crystal structures of related snake venom metalloproteinases [27]. These two radically different conformations are proposed to result from alternative disulfide-bonding arrangements of the D + C region [27, 28]. Previously, our studies with ADAM10 and the ADAM10-specific mAb 8C7 highlighted how 8C7 recognizes the active conformation of ADAM10 prevalent in tumors, as opposed to the inactive/latent ADAM10 conformation present in normal tissues [23]. To assess whether D8P1C1 also preferentially recognizes the tumor-specific ADAM17 conformation, we measured its binding to several human tumor cell lines relative to the binding of a commercial mAb (MED13622, MedImmune/AstraZeneca) that is not conformation specific. The cell-based ELISA [18] results with TNBC, ovarian, glioma, colon and non-small cell cancer cell lines document that D8P1C1 indeed preferentially binds to ADAM17 expressed on human tumor-derived cells, Fig. 4, as compared to ADAM17 expressed on HEK293 cells. We estimate that D8P1C1 binds to tumor-expressed ADAM17 approximately six-fold better than to ADAM17 expressed on HEK293 cells.
Fig. 4.
D8P1C1 preferentially binds to the activated, tumor-specific conformation of ADAM17. Cellular ELISA was performed to gage the binding of D8P1C1, relative to the binding of the control MED13622 mAb, to ADAM17 expressed on the cell surface of cancer cell lines: (A) breast, (B) colon, glioma and non-small cell lung, (C) ovarian, as well as HEK293 cells and HEK293 cells transfected with ADAM17. MED13622 binds equally well to the activated (tumor-associated) and the autoinhibited conformation of ADAM17. The graphs show the D8P1C1/MED13622 signal ratio relative to the D8P1C1/MED13622 signal ratio observed in the untransfected HEK293 cells. Specifically on the Y axes is plotted the value of
Where A(D8P1C1-HEK) is the signal for D8P1C using the untransfected HEK293 cells; A(MED13622-HEK) is the signal for MED1362 using the untransfected HEK293 cells; A(D8P1C1-HEK) is the signal for D8P1C using the cells that are being evaluated; A(MED13622-HEK) is the signal for MED1362 using the cells being evaluated. D8P1C1 binds to ADAM17 on tumors approximately six-fold better than to ADAM17 on HEK2923 cells. The data represent triplicate determinations and two independent experiments, mean ± SEM; P < 0.001 by unpaired two-tailed Student's t-test (cancer cell lines vs HEK293 cells).
D8P1C1 caused tumor growth inhibition of the TNBC cell line MDA-MB-231 and the ovarian cancer cell line SKOV-3 in mouse xenograft studies
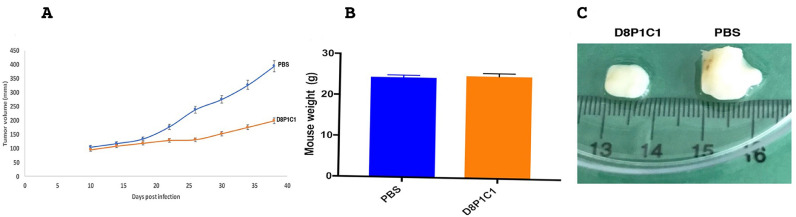
We evaluated the anti-tumor potential of D8P1C1 in vivo mouse models of TNBC using MDA-MB-231 cells. Administration of D8P1C1 at a dose of 15 mg/kg results in 78% percent tumor growth inhibition (Fig. 5). There were no toxicity effects in animals, gauged from no loss in mouse weight or presence of visible diarrhea, Fig. 5B, despite D8P1C1 binding equally well to human and mouse ADAM17 in ELISA-based assays (data not shown). This confirms that, although ADAM17 is present on a variety of cells, the inhibitory D8P1C1 mAb selectively targets tumors, without significant side effects or toxicity.
Fig. 5.
Anti-tumor effect of D8P1C1 in a triple-negative breast cancer xenograft model. 6–8 weeks old female athymic nude mice (n = 5) were used. 10 million MDA-MB-231 cells per mouse were introduced and each mouse was injected with D8P1C1 (i.p.) at a dose of 15 mg/kg, biweekly for 4 weeks. Sterile PBS was used as a control. Graphs show mean ± SEM, P< 0.001 by unpaired two-tailed Student's t-test, (mAb D8P1C1 vs PBS). D8P1C1 causes 78% tumor growth inhibition (A) with no loss in mouse weight (B) although it binds both human and mouse ADAM17. Panel (C) shows representative tumors excised from the treated and untreated animals.
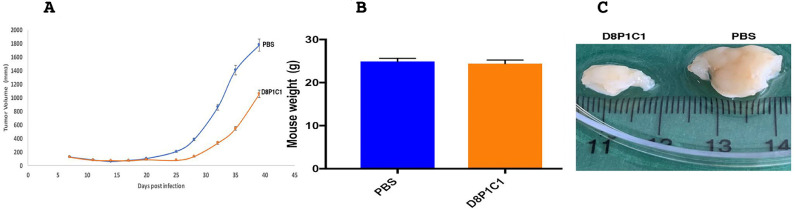
Finally, in an ovarian cancer xenograft model with SKOV-3 cells, D8P1C1 causes 45% tumor growth inhibition at a higher dose of 60 mg/kg, which we selected to assess toxicity at high concentrations. As above, no discernible toxicity effects were observed (Fig. 6).
Fig. 6.
Xenograft model using SKOV-3 ovarian cancer cells implanted into female NSG mice. The anti-ADAM17 mAb D8P1C1 causes 45 percent tumor growth inhibition at a dose of 60 mg/kg, administered bi-weekly for 4 weeks (A). 6–8 weeks old NSG mice (n = 5) were chosen. 10 million SKOV-3 cells per mouse were used and each mouse was injected with D8P1C1 (i.p.) at a dose of 60 mg/kg, biweekly for 4 weeks. Sterile PBS was used as a control. Graphs show mean ± SEM, P < 0.01 by unpaired two-tailed Student's t-test (mAb D8P1C1 vs PBS). Treatment with D8P1C1 did not affect mouse weight (B) although it binds both human and mouse ADAM17. Panel (C) shows representative tumors excised from the treated and untreated animals.
Discussion
ADAM17 is a member of the ADAM family of metalloproteinases. It cleaves over 80 substrates that are involved in tumor progression, inflammation and neurological diseases. In addition to erbB ligands as discussed above, substrates include Jagged1 [29], Glypican [30] and the C-MET receptor [31] that are primarily responsible for metastasis, cell proliferation and maintenance of stem cell populations in a wide variety of solid and hematological cancers [21]. ADAM17 also cleaves substrates, such as Neogenin [32], Syndecan-4 [33] and Glycoprotein VI, which augment inflammatory pathways [34]. It has been documented that ADAM17 sheds the Fc receptor CD16 that is present on peripheral blood natural killer (NK) cells and over-activation of ADAM17 dampens the effector functions of NK cells by downregulating the cell surface expression of CD16. ADAM17 antagonists play a major role in restoring the effector functions of NK cells [35]. Furthermore, ADAM17 is upregulated or mutated in many cancers (Fig. S2). Consequently, ADAM17 is considered to be an attractive target for therapeutic intervention. Several small molecule inhibitors targeting the catalytic domain of ADAM17 have been previously developed, such as Apratastat (Wyeth pharmaceuticals), DPC 333 (Bristol-Myers SquibbCompany) and INCB7839 (Incyte corporation), but they failed in clinical trials due to their toxicity and lack of specificity [24]. This is mainly because the inhibitors also target a close homolog of ADAM17, ADAM10, and some essential matrix metalloproteinases, such as MMP2, MM12, MMP15 [24]. Further chemical modifications of these compounds are in progress. A quinoline-derivative of the hydoxamate-based inhibitors DPC-33 and Apratastat was shown to inhibit proliferation of human keratinocytes with an IC50 of 3 nM and has been selected as a clinical candidate for psoriasis. Non-hydroxamate and non-zinc binding inhibitors of ADAM17 have also been investigated in cell-based assays but none has been tested in clinical trials [36]. Thus, monoclonal antibodies targeting ADAM17 are suggested to provide a better arsenal to deter cancer progression. To date, two anti-ADAM17 inhibitory mAbs, D1(A12) and MED13622, have shown moderate to high efficacy in vitro and in vivo tumor models of triple-negative breast, head and neck squamous cell epithelial, as well as ovarian and esophageal, cancers [37], [38], [39]. Binding and modeling studies indicate that D1(A12) binds to both the catalytic and noncatalytic ADAM17 domains while MED13622 binds a unique hairpin loop in the ADAM17 metalloprotease domain. This loop is absent in other ADAMs, ADAMTSs and MMPs explaining the basis for the mAb's specificity [40].
Here we describe the generation and anti-tumor potential of a new anti-ADAM17 mAb, D8P1C1. D8P1C1 inhibits proliferation of a host of cancer cell lines, including TNBC and HER2-overexpressing breast, ovarian, glioma, colon and non-small cell lung cancer lines. Though the inhibitory effect is modest in ovarian, glioma and colon cancer cells, it is significant in the case of the two breast cancer cell lines MDA-MB-231 (TNBC) and SKBR-3 (HER2 overexpressing), as well as the non-small cell lung cancer cell line HCC-827 that has an acquired mutation in the tyrosine kinase domain (E746-E750) [41]. Importantly, in an in vivo xenograft model of TNBC, D8P1C1 causes 78% tumor growth inhibition. On the other hand, in an in vivo xenograft model of SKOV3 (ovarian cancer) the mAb exhibits moderate inhibition. This could be attributed to the prevalence of the MUC-16 signaling pathway, which promotes progression of ovarian cancers independently of the EGFR-ADAM17 signaling events [41]. MUC-16 binds to adhesion molecules, including mesothelins and gelectins, and activates the AKT/ERK pathways contributing to the adverse outcomes in ovarian cancer [42].
Unlike the previously described anti-ADAM17 mAbs, D8P1C1 specifically recognizes ADAM17 expressed on cancer cell lines. This suggests that the mAb D8P1C1 selectively binds to an activated conformation of ADAM17 prevalent on cancer cells [23, 26]. An in vitro fluorescent peptide cleavage assay reveals that D8P1C1 acts via inhibiting the ADAM17 catalytic activity, and a negative-stain EM structure of the D8P1C1/ADAM17 complex, derived from 3D-reconstruction, shows that D8P1C1 binds to the ADAM17 proteinase domain. The catalytic inhibition studies also document that D8P1C1 is more efficient in blocking peptide cleavage by ADAM17 than the MedImmune/AstraZeneca mAb MED13622 mAb. It is known that TNBCs are not sensitive to targeted therapeutics used for HER2-positive and ERα-positive tumors [43]. Likewise, in the case of non-small cell lung cancer, the currently used anti-EGFR therapeutic monoclonal antibodies, including Cetuximab and Panitumumab, although effective in killing the cancer cells, exhibit severe side effects, including rash and diarrhea. The observation that D8P1C1 is tumor-conformation-specific and exhibits no discernible toxicity effect in animal models, validates future efforts to develop D8P1C1-based reagents for treatment of TNBC, NSCLC and solid tumors at large.
Declaration of Competing Interest
The authors declare that they have no competing interest.
Acknowledgments
Acknowledgements
Our studies are supported by the National Institutes of Health [R21CA185930 and R01NS038486 to D.B.N]; The Experimental Therapeutics Center of Memorial Sloan-Kettering, support from Mr. William H. and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research [2018–2020 to D.B.N]; Functional Genomics Initiative [2019–2020 to D.B.N.]
Author contribution
Nayanendu Saha: conceptualization, writing original draft, investigation. Kai Xu, Zhongyu Zhu, Yan Xu: data curation, analysis, investigation, editing, methodology. Dorothea Robev and Teja Kalidindi: data curation. Elisa de Stanchina, Naga Vara Kishore Pillarsetty, Juha Himanen and Dimiter S Dimitrov: formal analysis of data, review and editing. Dimitar B. Nikolov: supervison, formal analysis, review and writing, funding acquisition.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101265.
Appendix. Supplementary materials
References
- 1.Murphy G. The ADAMS: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 2.Dreymueller D., Uhlig S., Ludwig A. ADAM-family metalloproteinases in lung inflammation: potential therapeutic targets. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2015;308(4):L325–L343. doi: 10.1152/ajplung.00294.2014. [DOI] [PubMed] [Google Scholar]
- 3.Pruessmeyer J., Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin. Cell Dev. Biol. 2009;20(2):164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Seals D.F., Courtneidge SA S.A. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17(1):7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 5.Janes P.W., Saha N., Barton W.A., Kolev M.V., Wimmer-Kleikamp S.H., Nievergall E., Blobel C.P., Himanen J.P., Lackmann M., Nikolov D.B. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 2005;123(2):291–304. doi: 10.1016/j.cell.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Smith K.M., Gaultier A., Cousin H., Alfandari D., White J.M., Simone D.W. The cysteine-rich domain regulates ADAM protease function in vivo. J. Cell Biol. 2002;159(5):893–902. doi: 10.1083/jcb.200206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarden Y., Sliwkowski M.X. Untangling the ErbB signaling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 8.Blobel C.P., Carpenter G., Freeman M. The role of protease activity in ErbB biology. Exp. Cell Res. 2009;315(4):671–682. doi: 10.1016/j.yexcr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drag M., Salvesen G.S. Emerging principles in protease- based drug discovery. Nat. Rev. Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turk B. Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Dis. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 11.DasGupta S., Murumkar P.R., Giridhar R., Yadav R.M. Current perspective of TACE inhibitors: A review Bioorganic & Medicinal Chemistry. 2009;17:444–459. doi: 10.1016/j.bmc.2008.11.067. [DOI] [PubMed] [Google Scholar]
- 12.Whitehead R.H., Macrae F.A., St John D.J., Ma J. A colon cancer cell line (LIM1215) derived from a patient with inherited nonpolyposis colorectal cancer. J. Natl. Cancer Inst. 1985;74(4):759–765. [PubMed] [Google Scholar]
- 13.Xu K., Rajashankar K.R., Chan Y.P., Himanen J.P., Broder C.C., Nikolov D.B. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl Acad. Sci. 2008;105(29):9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z., Dimitrov D.S. Construction of a Large Naïve Human Phage-Displayed Fab Library through One-step cloning methods. Mol Bio. 2009;525:129–142. doi: 10.1007/978-1-59745-554-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Primakoff P., Myles D.G. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000;16:83–87. doi: 10.1016/s0168-9525(99)01926-5. [DOI] [PubMed] [Google Scholar]
- 16.Black R.A., Becherer J.D., et al. In: Handbook of Proteolytic Enzymes. Barret A.J., et al., editors. Academic Press; San Diego: 1998. in Tumor Necrosis Factor alpha -Converting Enzyme; p. 1315. [Google Scholar]
- 17.O’Brien J., Wilson I., Orton T., Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cytotoxicity. Eur. J. Biochem. 2000;267(17):5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 18.Smith D D., Cohick C.B., Lindsley H.B. Optimization of Cellular ELISA for Assay of Surface Antigens on Human Synoviocytes. BioTechniques. 1997;22:952–957. doi: 10.2144/97225rr03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rios-Doria J., Sabol D., Chesebrough J., Stewart D., Xu L., Tammali R., Cheng L., Du Q., Schifferli K., Rothstein R., Leow C.C., Heidbrink-Thompson J., Jin X X., Gao C., Friedman J., Wilkinson B., Damschroder M., Pierce A.J., Hollingsworth R.E., Tice D.A., Michelotti E.F. A Monoclonal Antibody to ADAM17 Inhibits Tumor Growth by Inhibiting EGFR and Non-EGFR-Mediated Pathways. Mol. Cancer Ther. 2015;14:1637–1649. doi: 10.1158/1535-7163.MCT-14-1040. [DOI] [PubMed] [Google Scholar]
- 20.Giricz O., Calvo V., Peterson E.A., Abouzeid C.M., Kenny P.A. TACE-dependent TGF-alpha shedding drives triple-negative breast cancer cell line. Int. J. Cancer. 2013;133(11):2587–2595. doi: 10.1002/ijc.28295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss M.L., Minod D. Recent Advances in ADAM17 Research: a Promising Target for Cancer and Inflammation. Mediators Inflamm. 2017 doi: 10.1155/2017/9673537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subik K., Lee J.F., Baxter L., Strzepek T., Costello D., Xing L., Hung M.C., Bonfiglio T., Hicks D.G., Tang P. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl) 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- 23.Atapattu L., Saha N., Chheang C., Eissman M.F., Xu K., Vail M.E., Hi L., Llerena C., Liu Z., Horvay K., Abud H.E., Kusebauch U., Moritz R.L., Ding B.S., Cao Z., Rafii S., Ernst M., A.M Scott, Nikolov D.B., Lackman M., Janes P.W. An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth. J. Exp. Med. 2016;213(9):1741–1757. doi: 10.1084/jem.20151095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss M.L., Sklair-Tavron L., Nudelman R. Drug insight: tumor necrosis factor-converting enzyme as a pharmaceutical target for rheumatoid arthritis. Nat. Clin. Pract. Rheumatol. 2008;4(6):300–309. doi: 10.1038/ncprheum0797. [DOI] [PubMed] [Google Scholar]
- 25.Potts A.H., Dawson J.C., Herrington C.S. Ovarian cancer cell lines derived from non-serous carcinomas migrate and invade more aggressively than those derived from high-grade serous carcinomas. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-41941-4. article number: 5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha N., Robev D., Himanen J.P., Nikolov D.B. ADAM proteases: emerging role and targeting of the non-catalytic domains. Cancer Lett. 2019;467:50–57. doi: 10.1016/j.canlet.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeda S., Igarashi T., Mori H., Araki S. Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold. The EMBO J. 2006;25:2388–2396. doi: 10.1038/sj.emboj.7601131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dusterhoft S., Jung S., Hung C.W., Tholey A., Sonnichsen F.D., Grotzinger J., Lorenzen I. Membrane-proximal domain of a disintegrin and metalloprotease-17 represents the putative molecular switch of its shedding activity operated by protein-disulfide isomerase. J. Am. Chem. Soc. 2013;135(15):5776–5781. doi: 10.1021/ja400340u. [DOI] [PubMed] [Google Scholar]
- 29.Li D., Masiero M., Banham A.H., Harris A.L. The notch ligand JAGGED1 as a target for anti-tumor therapy. Front. Oncol. 2014;4:254. doi: 10.3389/fonc.2014.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawahara R., Granato D.C., Yokoo S., Domingues R.R., Trindade D.M., Leme A.F. Mass spectrometry-based proteomics revealed glypican-1 as a novel ADAM17 substrate. J. Proteomics. 2017:pp151. doi: 10.1016/j.jprot.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 31.Chalupsky K., Kanchev I., Zbodakova O., Buryova H., Jirouskova M., Korinek V., Gregor M., Sedlacek R. ADAM10/17-dependent release of soluble c-Met correlates with hepatocellular damage. Folia Biol. (Krakow) 2013;59(2):76–86. [PubMed] [Google Scholar]
- 32.Okamura Y., Kohmura E., Yamashita T. TACE cleaves neogenin to desensitize cortical neurons to the repulsive guidance molecule. Neurosci. Res. 2011;71(1):63–70. doi: 10.1016/j.neures.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Pruessmeyer J., Martin C., Hess F.M., Schwarz N., Schmidt S., Kogel T., Hoettecke N., Schmidt B., Sechi A., Uhlig S., Ludwig A. A disintegrin and metalloproteinase 17 (ADAM17) mediates inflammation-induced shedding of syndecan-1 and -4 by lung epithelial cells. J. Biol. Chem. 2010;285(1):555–564. doi: 10.1074/jbc.M109.059394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bender M., Hofmann S., Stegner D., Chalaris A., Bosl M., Braun A., Scheller J., Rose-John S., Nieswandt B. Differentially regulated GPVI ectodomain shedding by multiple platelet-expressed proteinases. Blood. 2010;116(17):3347–3355. doi: 10.1182/blood-2010-06-289108. [DOI] [PubMed] [Google Scholar]
- 35.Romee R., Foley B., Lenvik T., Wang Y., Zhang B., Ankario D., Luo X., Cooley S., Verneris M., Walcheck B., Miller J. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17) Blood. 2013;121(18):3599–3608. doi: 10.1182/blood-2012-04-425397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calligaris M., Cuffaro D., Bonelli S., Spanò D.P., Rossello A., Nuti E., Scilabra S.D. Strategies to Target ADAM17 in Disease: from Its Discovery to the iRhom Revolution. Molecules. 2021;26(4):944. doi: 10.3390/molecules26040944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards F.M., Tape C.J., Jodrell D.I., Murphy G. Anti-tumour effects of a specific anti-ADAM17 antibody in an ovarian cancer model in vivo. PLoS ONE. 2012;7(7):e40597. doi: 10.1371/journal.pone.0040597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tape C.J., Willems S.H., Dombernowsky S.L., Stanley P.L., Fogarasi M., Ouwehand W., McCafferty J., Murphy G. Cross-domain inhibition of TACE ectodomain. Proc. Natl. Acad. Sci. U. S. A. 2011;108(14):5578–5583. doi: 10.1073/pnas.1017067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y., Benaich N., Tape C., Kwok H.F., Murphy G. Targeting the sheddase activity of ADAM17 by an anti-ADAM17 antibody D1 (A12) inhibits head and neck squamous cell carcinoma cell proliferation and motility via blockage of bradykinin induced HERs transactivation. Int. J. Biol. Sci. 2014;10:702–714. doi: 10.7150/ijbs.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng L., Cook K., Xu L., Cheng L., Damschroder M., Gao C., Wu H., Dall'Acqua W.F. Molecular basis for the mechanism of action of an anti-TACE antibody. Mabs. 2016;8(8):1598–1605. doi: 10.1080/19420862.2016.1226716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler D.L., Dunn E.F., Harari P.M. Understanding resistance to EGFR inhibitors—Impact on future treatment strategies. Nat. Rev. Clin. Oncol. 2010;(7):493. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S., Batra S.K. Understanding unique attributes of MUC16 (CA1215): potential implications in targeted therapy. Cancer Res. 2015;75(22):5337–5353. doi: 10.1158/0008-5472.CAN-15-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Won K.-.A., Spruck C. Triple negative breast cancer therapy: current and future perspectives. Int. J. Oncol. 2020;57(6):1245–1261. doi: 10.3892/ijo.2020.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.