Abstract
Temperature fluctuation is one of the most frequent threats to which organisms are exposed in nature. The activation of gene expression programs that trigger the transcription of heat stress-protective genes is the main cellular response to resist high temperatures. In addition, reversible accumulation and compartmentalization of thermosensitive proteins in high-order molecular assemblies are emerging as critical mechanisms to ensure cellular protection upon heat stress. Here, we summarize representative examples of membrane-less intracellular bodies formed upon heat stress in yeasts and human cells and highlight how protein aggregation can be turned into a cytoprotective mechanism.
Introduction
High temperature induces the unfolding and exposure of hydrophobic stretches in thermosensitive proteins, which can establish non-native intra- and inter-molecular interactions leading to aggregation into high-order protein assemblies (Chiti and Dobson 2006; Kammerer et al. 2004; Tyedmers et al. 2010). Furthermore, a significant part of the proteome contains intrinsically disordered domains (IDD) (Oldfield and Dunker 2014; Uversky 2017). IDDs usually show high structural flexibility and, under environmental perturbations, such as high temperature, they can acquire new folding states that make them more prone to establish interactions with other proteins. This leads to phase separation and the concentration of molecules in intracellular condensates (Alberti and Hyman 2021; Dyson and Wright 2005; Fomicheva and Ross 2021; Franzmann and Alberti 2019; Uversky and Dunker 2010). The dynamic properties of IDDs also contribute to the formation of protein aggregates under more extreme stress conditions (Kim et al. 2013; Molliex et al. 2015; Patel et al. 2015).
Cells respond to protein folding stress by activating the heat stress response (HSR), a transcriptional program that induces the expression of heat stress-response proteins (HSPs) such as chaperones and other cytoprotective factors, which boost the refolding of damaged proteins or their clearance by proteolytic mechanisms (Richter et al. 2010; Verghese et al. 2012). However, conditions of high proteotoxic stress that overcome the protein quality-control system may result in persistent protein aggregates which have been classically linked to a pervasive cellular decline and the development of degenerative and age-related diseases (pathological aggregation) (Diaz-Villanueva et al. 2015; Hipp et al. 2014; Klaips et al. 2018; Schneider and Bertolotti 2015). Interestingly, the accumulation of proteins in aggregates has also been more recently described as an organized and reversible process that displays cytoprotective functions (physiological aggregation) (Audas et al. 2016; Franzmann and Alberti 2019; Marijan et al. 2019; Tyedmers et al. 2010). This protective protein aggregation results in the formation of a variety of specific membrane-less inclusion bodies or biomolecular condensates, with different cellular locations, physico-chemical properties, and functions (Gallardo et al.2020; Shav-Tal et al. 2005; Sontag et al. 2017; Tyedmers et al. 2010; van Leeuwen and Rabouille 2019; Wallace et al. 2015; Wang et al. 2018). This review summarizes current knowledge on biomolecular condensates induced by HS in yeasts and humans with a focus on the recently described nucleolar rings (NuRs) in Schizosaccharomyces pombe. We discuss the function of the Hsf1-dependent HSR in the regulation of NuRs and its role in the maintenance of cell viability in high proteotoxic conditions such as acute HS.
Stress granules and cytoplasmic protein aggregation centers.
One of the most conserved and well-described inclusion bodies are the cytoplasmic stress granules (SGs) that are rapidly formed as a response to several environmental stresses, including high temperature (Fig. 1) (Buchan and Parker 2009). SGs are biomolecular condensates containing mainly RNA-binding proteins (RBPs), translationally repressed mRNAs, translation factors, and 40S ribosomal subunits. SG formation is driven by the unfolding and promiscuous interactions of IDDs present in RNA-binding proteins (RBPs) and involves a number of post-translational protein modifications (Hofmann et al. 2021; Molliex et al. 2015). SGs are dynamic structures that contribute to the repression of protein synthesis upon stress and are considered sites of mRNA triage, sorting towards the decay or the storage of mRNAs (Alberti et al. 2017; Anderson and Kedersha 2009).
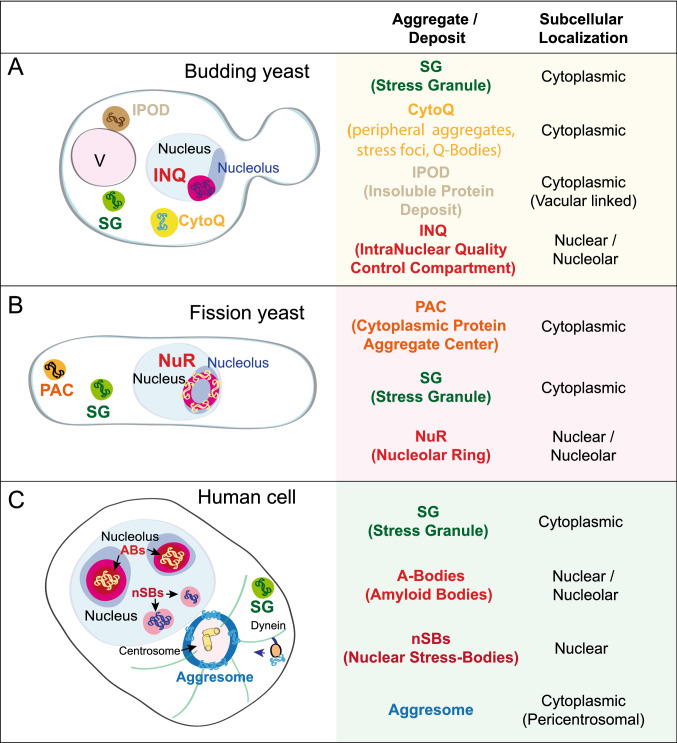
Fig. 1.
Cytoprotective aggregation centers under HS. Schematic representation of different intracellular biomolecular condensates formed in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and human cells at different cellular locations upon HS. A Biomolecular condensates in S. cerevisiae upon HS. SGs and CytoQ encompass different cytoplasmic condensates, which contain non-terminally misfolded proteins mainly destined for refolding after the stress, similar to the INQ formed in the nucleus adjacent to the nucleolus. IPODs, however, are aggregate deposits formed in the cytoplasm adjacent to the vacuole, which accumulate terminally misfolded proteins destined for degradation. B Two types of cytoplasmic condensates have been described in S. pombe; PACs and SGs. PACs form upon mild HS and accumulate non-terminally misfolded proteins whose fate is refolding. PACs have been proposed to constitute the precursors of SGs formed upon more severe stress conditions. In the nucleus, misfolded proteins accumulate at NuRs upon acute HS. NuRs form at the nucleolar region and function as reversible protein sequestering centers. Upon stress relief, NuRs dissolution is linked to the restoration of cell growth. C Upon stress conditions, human cells form different cytoplasmic deposition sites, such as SGs or aggresomes. Aggresomes represent sites of terminal aggregation of damaged proteins whose destiny is degradation by autophagic mechanisms. In the nucleus, human cells protect thermosensitive proteins by their coalescence along with lncRNAs transcribed from ribosomal intergenic spacers in nucleolar structures known as amyloid bodies (ABs). AB are reversible structures and have been shown to promote local nuclear translation during the stress. On the other hand, nSBs are nuclear assemblies of RNA and proteins formed upon HS which function by sequestering transcription-related factors facilitating the HS-induced inhibition of bulk RNA transcription
Other cytosolic biomolecular condensates termed protein aggregate centers (PACs) have been recently described to form in the fission yeast upon exposure to a mild HS (37 ℃) (Fig. 1B) (Cabrera et al. 2020). PACs are dynamic assemblies that contain chaperones and components of the translational machinery; however, misfolded proteins also accumulate at PACs and this is required to avoid their degradation during the HS. Therefore, PACs have been proposed to protect these non-terminally misfolded proteins from degradation. PACs behave as liquid-like condensates that change to a more compacted state upon incubation of cells at higher temperatures, which suggests that PACs can function as seeds for SG formation. The assembly and disassembly of SGs and PACs are regulated by Hsp70 chaperones (Boronat et al. 2021; Cabrera et al. 2020; Cherkasov et al. 2013; Kroschwald et al. 2015; Mateju et al. 2017; O'Driscoll et al. 2015; Walters et al., 2015), and the disaggregase Hsp104 is key for their dissolution (Cabrera et al. 2020; Cherkasov et al. 2013; Kroschwald et al. 2015).
Insoluble protein deposit, CytoQ, and Intranuclear quality-control compartment.
Other protein deposition centers have been observed in Saccharomyces cerevisiae as a result of HS. These include insoluble protein deposit (IPOD) and CytoQ in the cytoplasm, and intra-nuclear quality-control compartment (INQ) in the nucleus (Fig. 1A). IPODs are perivacuolar deposits constituted by immobile, terminally aggregated proteins, including the amyloidogenic prion proteins Rnq1 and Sup35 (Kaganovich et al. 2008; Kumar et al. 2016), while CytoQ refers to different cytoplasmic condensates containing cytosolic non-terminally misfolded proteins. On the contrary, INQ, also termed juxtanuclear quality-control compartment (JUNQ), localize inside the nucleus, adjacent to the nucleolus, and harbor nuclear and cytosolic misfolded proteins (Kaganovich et al. 2008; Miller et al. 2015). CytoQ and INQ share similarities in their formation, which is dependent on cell-compartment-specific aggregases: the cytoplasmic small HSP (sHSP) Hsp42 for CytoQs and the nuclear aggregase Btn2 for INQs. Hsp42 and Btn2 function as sorting factors that promote the partitioning of misfolded proteins into CytoQs or INQs, respectively (Malinovska et al. 2012; Miller et al. 2015; Specht et al. 2011), and their activities are key to avoid the overload of the proteostatic capacity and, consequently, to prevent the loss of cell viability. Non-terminally unfolded proteins stored at CytoQs and INQs will be mainly targeted to the refolding pathway (Ho et al. 2019; Miller et al. 2015; Wallace et al. 2015), whereas terminally misfolded substrates accumulated at IPODs are either targeted for clearance (mainly by autophagy) or diluted by cell division (Kaganovich et al. 2008). Of note, in both cases, the Hsp70 chaperone system and the disaggregase Hsp104 participate in their sorting and re-solubilization (Gallina et al. 2015; Ho et al. 2019; Kaganovich et al. 2008; Malinovska et al. 2012; Miller et al. 2015; O'Driscoll et al. 2015).
Aggresomes, nuclear stress bodies, and amyloid bodies
Human cells have also been shown to form specialized inclusion bodies for sequestering misfolded proteins under HS. These include SGs and aggresomes in the cytoplasm, and nuclear stress bodies (nSBs) and amyloid bodies (AB) in the nucleus (Fig. 1C).
Aggresome assembly depends on the active delivery (by dynein motor complex) and the accumulation of aggresomal particles containing misfolded proteins near the centrosome (Johnston et al. 1998) (Fig. 1). Aggresomes recruit chaperones, ubiquitination enzymes, and proteasomes and, in conditions of insufficient proteasome degradation, the prolonged presence of aggresomes results in their autophagic clearance (Johnston et al. 1998; Tyedmers et al. 2010). Therefore, they are considered as garbage depositories that aid in the clearance of terminally aggregated proteins (Kawaguchi et al. 2003; Tyedmers et al. 2010).
On the other hand, ABs are formed in the nucleus by the rapid and reversible interaction between heterogeneous proteins and ribosomal intergenic non-coding RNA (rIGSRNA). rIGSRNAs are expressed in a stress-inducible manner and act as the seeding elements for AB formation in subnuclear foci, trapping, and immobilizing proteins that are characterized by their insolubility. Upon stress relief, ABs are disaggregated in an HSP70-dependent manner and their components return to their normal localization (Audas et al. 2016, 2012). Interestingly, a new and unexpected function has been recently ascribed to ABs as solid-like condensates that support nuclear translation of Hsf1 targets during acidosis and HS (Theodoridis et al. 2021).
Another subnuclear foci that is found exclusively in primates upon heat and chemical stresses are the nuclear stress bodies (nSBs) (Biamonti and Vourc'h 2010). nSB formation is initiated by binding of Hsf1 to pericentric tandem repeats of satellite III (Sat 3). Hsf1-dependent transcription of Sat 3 transcripts promotes the binding and sequestration of transcription factors and RBPs to these long non-coding RNAs (lncRNAs). Sat3 transcripts tend to stay associated with the transcription site, forming subnuclear foci that concentrate and sequester several nuclear factors required for transcription, mainly at the 9q12 locus (Biamonti and Vourc'h 2010). nSBs do not seem to colocalize with Hsf1 canonical targets of the HSR, suggesting that nSB formation is not required for the transcription of HSR genes (Jolly et al. 1997). However, nSB assembly is required to sustain the HS-induced transcriptional repression and to maintain cell viability after the HS (Goenka et al. 2016) and, therefore, they have been proposed to function as sequestering centers which facilitate the downregulation of general transcription during the stress.
Nucleolar rings
In fission yeast, acute heat stress (42 ℃), which prevents bulk protein synthesis and blocks general mRNA metabolism (Cabrera et al. 2020; Gallardo et al. 2020; Ribeiro et al. 1997), results in a dramatic reorganization of the nucleus. These nuclear architectural changes include nucleolar contraction and the formation of ring-shaped nucleolar aggregation centers, named Nucleolar Rings (NuR) (Gallardo et al. 2020). NuRs assemblies contain a wide and functionally divergent group of nuclear factors, including factors involved in mRNA processing and export, nuclear pore complex (NPC) components, mRNA, chromatin-associated factors, and cell cycle regulators. The formation of these assemblies is triggered by HS-induced unfolding and aggregation of proteins and RNAs (Gallardo et al. 2020). While the aggregation of cell cycle factors at NuRs could contribute to the arrest of cell division observed under this extreme temperature, the rearrangement of the NPC and mRNA machinery could be responsible for the block of housekeeping mRNA metabolism and export and for the formation of a reservoir of messenger ribonucleoprotein (mRNPs) particles, which would be ready for export once stress conditions cease. This rearrangement bears similarities with the compositional changes of mRNPs under heat stress described for other organisms, including budding yeast and higher eukaryotes (Bond 1988; Bracken and Bond 1999; Hochberg-Laufer et al. 2019; Kay et al. 1987; Lutz et al. 1988; Mahl et al. 1989; Mayrand and Pederson, 1983; Sadis et al. 1988). In fact, in S. cerevisiae, heat stress leads to the uncoupling of several RBPs from the mRNA and their accumulation in nuclear foci, in a manner dependent on the nucleoporins Mlp1 and Mlp2. This reorganization has been proposed to prevent the quality control and export of regular mRNAs, while promoting the export of HS mRNAs (Carmody et al. 2010; Zander et al. 2016; Zander and Krebber 2017).
The study of NuRs has shed light on the dynamic nature of these stress-induced assemblies. NuRs are formed by the rapid aggregation of multiple nuclear proteins, which remain mostly immobile, while the stress persists. However, when the stress is relieved, NuRs disaggregate and their multiple components relocate back to their functional localization (Gallardo et al. 2020). This shows that fission yeast cells can modulate protein aggregation, turning deleterious protein aggregates into cytoprotective protein sequestering centers which contribute to maintain cellular homeostasis.
Hsf1 is the main transcription factor involved in the activation of the HSR (Akerfelt et al. 2010). Upon incubation of fission yeast cells at 42 ℃, Hsf1 rapidly accumulates in the nucleoplasm; however, it does not localize at NuRs. Interestingly, once the heat stress is over, cell growth recovery tightly correlates with a peak in Hsf1 expression (Gallardo et al. 2020). This delayed Hsf1 upregulation is also observed in S. cerevisiae during the recovery from severe heat shock (Yamamoto et al. 2008). Furthermore, a partial Hsf1 depletion significantly delays NuRs dissolution and cell growth reinitiation during the recovery from acute HS. This suggests that Hsf1 expression during the acute HS is limited, and that a burst in newly synthesized Hsf1 during the recovery period is required to trigger the refolding of most heat-denatured proteins in order to reactivate cellular metabolism (Gallardo et al. 2020; Yamamoto et al. 2008).
Hsf1 activation leads to the expression of HSPs, such as chaperones and disaggregases (Akerfelt et al. 2010). These proteins localize at aggregates and are responsible for aggregate dissolution once the stress is over. In addition, chaperones are commanders in the triage of aggregated proteins for recycling or degradation during the recovery from HS (Balchin et al. 2016; Escusa-Toret et al. 2013; Hartl et al. 2011; Malinovska et al. 2012; Mogk et al. 2015; Vabulas et al. 2010). Consistently, NuRs accumulate several HSPs, such as the Hsp70 homologs Ssa1 and Ssa2, and the disaggregase Hsp104.
Importantly, Hsp104 is required for NuR aggregate dissolution, for the relocalization of NuRs components, and for the resumption of cell growth after acute HS (Gallardo et al. 2020). This is in agreement with previous studies that have demonstrated that Hsp104, in concert with the chaperones Hsp70/Hsp40, functions in the reassembly of small nuclear RNPs after HS (Bracken and Bond 1999; Cherkasov et al. 2013; Haslbeck et al. 2005). Interestingly, a recent study in budding yeast shows that the Hsp70/Hsp40 chaperone partners aid in the solubilization of intra-nuclear inclusions formed after incubation of the cells at 42 ℃, independently on Hsp104. In these conditions, Hsp40 and Hsp104 compete for protein disaggregation, targeting proteins towards turnover or refolding, respectively (den Brave et al. 2020). Metazoans lack Hsp104 orthologues and the disaggregase activity is accomplished by Hsp70 and Hsp110, among others (Mogk et al. 2015; Rampelt et al. 2012; Shorter 2011).
In conclusion, fission yeast NuRs are reversible molecular assemblies rapidly formed upon acute heat stress by the aggregation of nuclear and nucleolar factors along with RNA. Their formation correlates with nucleolar contraction and the inhibition of cell growth and their Hsp104-dependent dissolution is required for cell growth restoration once the heat stress is relieved. Therefore, NuRs can be considered emergency storage deposits for thermolabile proteins and its assembly might contribute to the inhibition of nuclear functions under acute HS.
Concluding remarks
As discussed above, HS elicits the formation of a variety of high-order-molecular assemblies, which are formed both in the cytoplasm and the nucleus, by the promiscuous interaction of misfolded proteins, IDD-containing proteins and usually RNA. Although the functions of all these intracellular protein deposits are still under investigation, they share a common role as protective mechanisms preserving cellular homeostasis during stress conditions and promoting cell survival. The concentration of proteins in biomolecular condensates contributes to inactivate unneeded or energetically consuming cellular functions and avoids the interference of misfolded proteins with stress-response cellular pathways. The local recruitment and concentration of HSPs at these condensates facilitates the refolding or clearance of stress-sensitive proteins and can also promote the efficiency of essential processes, such as translation in the case of ABs.
Understanding how chaperones modulate and reverse protein aggregation is of prime importance, since the formation of amyloid deposits has been linked to the development of a variety of degenerative diseases, such as Amyotrophic Lateral Sclerosis, Alzheimer's, Huntington’s, or Parkinson's disease, among others (Chiti and Dobson 2017). In addition, recent advances have shown that aging is associated with progressive exhaustion of the proteostatic capacity of cells and the formation of pathogenic aggresomes, which compromises multiple cellular processes (Hipp et al. 2019, 2014; Moreno-Blas et al. 2018; Morimoto 2008; Thiruvalluvan et al. 2020). Therefore, achieving a better knowledge of what factors and mechanisms are involved in the aggregation and disaggregation processes would be extremely valuable for future therapeutic strategies of age-related diseases. In this sense, yeasts as model organisms represent a valuable tool for genetic or drug screening purposes to find new modulators that could potentially alleviate pathologic aggregation in humans.
Acknowledgements
We would like to thank Daga’s lab members and our colleagues at the CABD for helpful discussions.
Funding
This work was supported by the Ministerio de Ciencia, Innovación y Universidades (Grant: PGC2018-099849-B-I00 to R.R. Daga) and Junta de Andalucía-FEDER-UPO (grant: UPO-1264663 to S.S.P.).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Silvia Salas-Pino, Email: ssalpin@upo.es.
Rafael R. Daga, Email: rroddag@upo.es
References
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol. 2021;22:196–213. doi: 10.1038/s41580-020-00326-6. [DOI] [PubMed] [Google Scholar]
- Alberti S, Mateju D, Mediani L, Carra S. Granulostasis: protein quality control of RNP granules. Front Mol Neurosci. 2017;10:84. doi: 10.3389/fnmol.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Audas TE, Audas DE, Jacob MD, Ho JJ, Khacho M, Wang M, Perera JK, Gardiner C, Bennett CA, Head T, Kryvenko ON, Jorda M, Daunert S, Malhotra A, Trinkle-Mulcahy L, Gonzalgo ML, Lee S. Adaptation to stressors by systemic protein amyloidogenesis. Dev Cell. 2016;39:155–168. doi: 10.1016/j.devcel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016 doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 2010;2(6):a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988;7:3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronat S, Cabrera M, Hidalgo E. Spatial sequestration of misfolded proteins as an active chaperone-mediated process during heat stress. Curr Genet. 2021;67:237–243. doi: 10.1007/s00294-020-01135-2. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Bond U. Reassembly and protection of small nuclear ribonucleoprotein particles by heat shock proteins in yeast cells. RNA. 1999;5:1586–1596. doi: 10.1017/s1355838299991203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Boronat S, Marte L, Vega M, Perez P, Ayte J, Hidalgo E. Chaperone-facilitated aggregation of thermo-sensitive proteins shields them from degradation during heat stress. Cell Rep. 2020;30(7):2430–2443. doi: 10.1016/j.celrep.2020.01.077. [DOI] [PubMed] [Google Scholar]
- Carmody SR, Tran EJ, Apponi LH, Corbett AH, Wente SR. The mitogen-activated protein kinase Slt2 regulates nuclear retention of non-heat shock mRNAs during heat shock-induced stress. Mol Cell Biol. 2010;30:5168–5179. doi: 10.1128/MCB.00735-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol. 2013;23:2452–2462. doi: 10.1016/j.cub.2013.09.058. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- den Brave F, Cairo LV, Jagadeesan C, Ruger-Herreros C, Mogk A, Bukau B, Jentsch S. Chaperone-mediated protein disaggregation triggers proteolytic clearance of intra-nuclear protein inclusions. Cell Rep. 2020;31(9):107680. doi: 10.1016/j.celrep.2020.107680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Villanueva JF, Diaz-Molina R, Garcia-Gonzalez V. Protein folding and mechanisms of proteostasis. Int J Mol Sci. 2015;16:17193–17230. doi: 10.3390/ijms160817193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Escusa-Toret S, Vonk WI, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol. 2013;15:1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomicheva A, Ross ED. From prions to stress granules: defining the compositional features of prion-like domains that promote different types of assemblies. Int J Mol Sci. 2021 doi: 10.3390/ijms22031251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann TM, Alberti S. Protein phase separation as a stress survival strategy. Cold Spring Harb Perspect Biol. 2019;11(6):034058. doi: 10.1101/cshperspect.a034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo P, Real-Calderon P, Flor-Parra I, Salas-Pino S, Daga RR. Acute heat stress leads to reversible aggregation of nuclear proteins into nucleolar rings in fission yeast. Cell Rep. 2020;33(6):108377. doi: 10.1016/j.celrep.2020.108377. [DOI] [PubMed] [Google Scholar]
- Gallina I, Colding C, Henriksen P, Beli P, Nakamura K, Offman J, Mathiasen DP, Silva S, Hoffmann E, Groth A, Choudhary C, Lisby M. Cmr1/WDR76 defines a nuclear genotoxic stress body linking genome integrity and protein quality control. Nat Commun. 2015;6:6533. doi: 10.1038/ncomms7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka A, Sengupta S, Pandey R, Parihar R, Mohanta GC, Mukerji M, Ganesh S. Human satellite-III non-coding RNAs modulate heat-shock-induced transcriptional repression. J Cell Sci. 2016;129:3541–3552. doi: 10.1242/jcs.189803. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem. 2005;280:23861–23868. doi: 10.1074/jbc.M502697200. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Kasturi P, Hartl FU. The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol. 2019;20:421–435. doi: 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- Ho CT, Grousl T, Shatz O, Jawed A, Ruger-Herreros C, Semmelink M, Zahn R, Richter K, Bukau B, Mogk A. Cellular sequestrases maintain basal Hsp70 capacity ensuring balanced proteostasis. Nat Commun. 2019;10:4851. doi: 10.1038/s41467-019-12868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg-Laufer H, Schwed-Gross A, Neugebauer KM, Shav-Tal Y. Uncoupling of nucleo-cytoplasmic RNA export and localization during stress. Nucleic Acids Res. 2019;47:4778–4797. doi: 10.1093/nar/gkz168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Kedersha N, Anderson P, Ivanov P. Molecular mechanisms of stress granule assembly and disassembly. Biochim Biophys Acta Mol Cell Res. 2021;1868(1):118876. doi: 10.1016/j.bbamcr.2020.118876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Morimoto R, Robert-Nicoud M, Vourc'h C. HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J Cell Sci. 1997;110(Pt 23):2935–2941. doi: 10.1242/jcs.110.23.2935. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer RA, Kostrewa D, Zurdo J, Detken A, Garcia-Echeverria C, Green JD, Muller SA, Meier BH, Winkler FK, Dobson CM, Steinmetz MO. Exploring amyloid formation by a de novo design. Proc Natl Acad Sci U S a. 2004;101:4435–4440. doi: 10.1073/pnas.0306786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kay RJ, Russnak RH, Jones D, Mathias C, Candido EP. Expression of intron-containing C. elegans heat shock genes in mouse cells demonstrates divergence of 3' splice site recognition sequences between nematodes and vertebrates, and an inhibitory effect of heat shock on the mammalian splicing apparatus. Nucleic Acids Res. 1987;15:3723–3741. doi: 10.1093/nar/15.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaips CL, Jayaraj GG, Hartl FU. Pathways of cellular proteostasis in aging and disease. J Cell Biol. 2018;217:51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Nawroth PP, Tyedmers J. Prion aggregates are recruited to the insoluble protein deposit (IPOD) via myosin 2-based vesicular transport. PLoS Genet. 2016;12(9):e1006324. doi: 10.1371/journal.pgen.1006324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz Y, Jacob M, Fuchs JP. The distribution of two hnRNP-associated proteins defined by a monoclonal antibody is altered in heat-shocked HeLa cells. Exp Cell Res. 1988;175:109–124. doi: 10.1016/0014-4827(88)90259-5. [DOI] [PubMed] [Google Scholar]
- Mahl P, Lutz Y, Puvion E, Fuchs JP. Rapid effect of heat shock on two heterogeneous nuclear ribonucleoprotein-associated antigens in HeLa cells. J Cell Biol. 1989;109:1921–1935. doi: 10.1083/jcb.109.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell. 2012;23:3041–3056. doi: 10.1091/mbc.E12-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marijan D, Tse R, Elliott K, Chandhok S, Luo M, Lacroix E, Audas TE. Stress-specific aggregation of proteins in the amyloid bodies. FEBS Lett. 2019;593:3162–3172. doi: 10.1002/1873-3468.13597. [DOI] [PubMed] [Google Scholar]
- Mateju D, Franzmann TM, Patel A, Kopach A, Boczek EE, Maharana S, Lee HO, Carra S, Hyman AA, Alberti S. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36:1669–1687. doi: 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrand S, Pederson T. Heat shock alters nuclear ribonucleoprotein assembly in Drosophila cells. Mol Cell Biol. 1983;3:161–171. doi: 10.1128/mcb.3.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SB, Ho CT, Winkler J, Khokhrina M, Neuner A, Mohamed MY, Guilbride DL, Richter K, Lisby M, Schiebel E, Mogk A, Bukau B. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015;34:778–797. doi: 10.15252/embj.201489524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Kummer E, Bukau B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front Mol Biosci. 2015;2:22. doi: 10.3389/fmolb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Blas D, Gorostieta-Salas E, Castro-Obregon S. Connecting chaperone-mediated autophagy dysfunction to cellular senescence. Ageing Res Rev. 2018;41:34–41. doi: 10.1016/j.arr.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll J, Clare D, Saibil H. Prion aggregate structure in yeast cells is determined by the Hsp104-Hsp110 disaggregase machinery. J Cell Biol. 2015;211:145–158. doi: 10.1083/jcb.201505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro MJ, Reinders A, Boller T, Wiemken A, De Virgilio C. Trehalose synthesis is important for the acquisition of thermotolerance in Schizosaccharomyces pombe. Mol Microbiol. 1997;25:571–581. doi: 10.1046/j.1365-2958.1997.4961856.x. [DOI] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Sadis S, Hickey E, Weber LA. Effect of heat shock on RNA metabolism in HeLa cells. J Cell Physiol. 1988;135:377–386. doi: 10.1002/jcp.1041350304. [DOI] [PubMed] [Google Scholar]
- Schneider K, Bertolotti A. Surviving protein quality control catastrophes–from cells to organisms. J Cell Sci. 2015;128:3861–3869. doi: 10.1242/jcs.173047. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE. 2011;6(10):e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag EM, Samant RS, Frydman J. Mechanisms and functions of spatial protein quality control. Annu Rev Biochem. 2017;86:97–122. doi: 10.1146/annurev-biochem-060815-014616. [DOI] [PubMed] [Google Scholar]
- Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoridis PR, Bokros M, Marijan D, Balukoff NC, Wang D, Kirk CC, Budine TD, Goldsmith HD, Wang M, Audas TE, Lee S. Local translation in nuclear condensate amyloid bodies. Proc Natl Acad Sci U S A. 2021 doi: 10.1073/pnas.2014457118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiruvalluvan A, de Mattos EP, Brunsting JF, Bakels R, Serlidaki D, Barazzuol L, Conforti P, Fatima A, Koyuncu S, Cattaneo E, Vilchez D, Bergink S, Boddeke E, Copray S, Kampinga HH. DNAJB6, a key factor in neuronal sensitivity to amyloidogenesis. Mol Cell. 2020;78(2):346–358. doi: 10.1016/j.molcel.2020.02.022. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol. 2010;2(12):a004390. doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen W, Rabouille C. Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic. 2019;20:623–638. doi: 10.1111/tra.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J, Abrams J, Wang Y, Morano KA. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev. 2012;76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, Airoldi EM, Pan T, Budnik BA, Drummond DA. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell. 2015;162:1286–1298. doi: 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RW, Muhlrad D, Garcia J, Parker R. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA. 2015;21:1660–1671. doi: 10.1261/rna.053116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tao X, Jacob MD, Bennett CA, Ho JJD, Gonzalgo ML, Audas TE, Lee S. Stress-induced low complexity RNA activates physiological amyloidogenesis. Cell Rep. 2018;24(7):1713–1721. doi: 10.1016/j.celrep.2018.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Maeda Y, Ikeda A, Sakurai H. Regulation of thermotolerance by stress-induced transcription factors in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:783–790. doi: 10.1128/EC.00029-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander G, Krebber H. Quick or quality? How mRNA escapes nuclear quality control during stress. RNA Biol. 2017;14:1642–1648. doi: 10.1080/15476286.2017.1345835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander G, Hackmann A, Bender L, Becker D, Lingner T, Salinas G, Krebber H. mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature. 2016;540:593–596. doi: 10.1038/nature20572. [DOI] [PubMed] [Google Scholar]