Abstract
In recent years, the incidence and the mortality rate of cervical cancer have been gradually increasing, becoming one of the major causes of cancer-related death in women. In particular, patients with advanced and recurrent cervical cancers present a very poor prognosis. In addition, the vast majority of cervical cancer cases are caused by human papillomavirus (HPV) infection, of which HPV16 infection is the main cause and squamous cell carcinoma is the main presenting type. In this study, we performed screening of differentially expressed genes (DEGs) based on The Cancer Genome Atlas (TCGA) database and GSE6791, constructed a protein–protein interaction (PPI) network to screen 34 hub genes, filtered to the remaining 10 genes using the CytoHubba plug-in, and used survival analysis to determine that RPS27A was most associated with the prognosis of cervical cancer patients and has prognostic and predictive value for cervical cancer. The most significant biological functions and pathways of RPS27A enrichment were subsequently investigated with gene set enrichment analysis (GSEA), and integration of TCGA and GTEx database analyses revealed that RPS27A was significantly expressed in most cancer types. In this study, our analysis revealed that RPS27A can be used as a prognostic biomarker for HPV16 cervical cancer and has biological significance for the growth of cervical cancer cells.
Keywords: RPS27A, HPV type 16 cervical cancer, HPV16, cervical cancer, squamous cell carcinoma
Introduction
Cervical cancer (cervical squamous cell carcinoma and endocervical adenocarcinoma, CESC) is the fourth most common cancer in women, after breast, colorectal, and lung cancers (1). In the last 2 years, the incidence and the mortality of cervical cancer have been increasing globally, with more than 600,000 new cases and nearly 350,000 deaths in 2020 (2, 3). The age of onset of cervical cancer is “bimodal” and is concentrated in women in their 30s and 40s (4), and about 85% of cervical cancer deaths occur in less developed and developing countries due to medical conditions (5). Patients with early-stage (IB–IIA) cervical cancer overwhelmingly show a trend of good prognosis after receiving appropriate treatment (6), but the prognosis of patients with advanced and recurrent cervical cancer remains poor (7). In addition, cervical cancer is metastatic, with the most frequent site being the bone, and the median survival time after diagnosis is only 7–12 months (8).
Of the 22 million new cancer cases caused by an infection in 2018, up to 690,000 were affected by human papillomavirus (HPV) (9). HPV is a double-stranded DNA virus, and most types of HPV infections are cleared by autoimmunity. However, a few types of HPV viruses can transform infected cells into malignant tumor cells (10). HPV infection is transmitted through sexual contact (early-age sexual intercourse and multiple sexual partners are both high-risk factors for HPV infection), and persistent HPV infection is the most important factor in the development of cervical cancer (11). HPV testing is the primary modality for cervical cancer screening and can significantly reduce the risk of death from cervical cancer (12). In addition, broad-spectrum HPV vaccination is an effective way to prevent the development of cervical cancer (13, 14). To date, three HPV vaccines have been licensed for use: the bivalent HPV virus-like particle vaccine (2vHPV), the quadrivalent HPV virus-like particle vaccine (4vHPV), and the nonavalent HPV virus-like particle vaccine (9vHPV), which can prevent 70% of cervical cancers worldwide (15).
More than 40 HPV virus types colonize the genital tract, 15 of which are associated with cervical cancer, and HPV16 is one of the most virulent genotypes (16). High-risk HPV16 is associated with genital and oropharyngeal cancers (17) and approximately 50% of cases of squamous cell carcinoma (SCC), the most frequent type of cervical cancer (18). Women who have been persistently infected with HPV16 for 2 years have a high probability of developing precancerous lesions within the next 5 years (19), and persistent HPV16 infection is the most important factor leading to the recurrence of high-grade cervical intraepithelial neoplasia (CIN) after treatment in patients with HPV infection (20).
In this study, based on the analysis of The Cancer Genome Atlas (TCGA) database and Gene Expression Omnibus (GEO) database, we aimed to screen the pivotal genes through the screening of differentially expressed genes (DEGs) and the construction of a protein–protein interaction (PPI) network, identify the key gene by analyzing the relationship between the high and low expressions of the pivotal genes and the survival of cervical cancer patients, use this key gene to predict the survival of cervical cancer patients, and analyze the main functional pathways of the key gene using gene set enrichment analysis (GSEA) to determine the prognostic biomarkers for cervical cancer caused by HPV16 infection.
Information and Methods
Data Sources
All clinical information and gene expression-related matrix data related to cervical cancer were obtained from TCGA database, GEO database (GSE6791) (https://www.ncbi.nlm.nih.gov/geo/), and the Genotype–Tissue Expression (GTEx) database. TCGA included 12 HPV16-positive samples and 294 HPV16-negative samples, GSE6791 included eight HPV16-positive samples and three HPV16-negative samples, and para cancer tissue data were obtained from GTEx.
DEG Screening and Functional Pathway Enrichment Analysis
DEGs were screened for HPV16-related genes using the R limma package, and volcano plots were plotted by the R package ggplot2 with log2FC = 0.3785 and p < 0.05 as the screening conditions. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional pathway enrichment analyses were performed on the screened DEGs using the R package clusterProfiler to explore the biological characteristics of DEGs. Venn diagrams were drawn to identify overlaps between the DEGs in TCGA and GSE6791.
Selection and Identification of Hub Genes
DEGs were imported into the STRING database (https://string-db.org/) to construct a PPI network, visualized by Cytoscape (version 3.8.2), and pivotal genes were screened by degree sorting using the CytoHubba plugin. The performance of each pivotal gene was observed in TCGA and GSE6791 databases, and the expression levels of the pivotal genes in cervical cancer, cervical SCC, and cervical adenocarcinoma tissues were compared with those in normal tissues. Kaplan–Meier curves were plotted to observe the overall survival (OS) of high and low expressions of pivotal genes, and the genes most associated with the prognosis of cervical cancer patients were selected as key genes.
Validation of Pivotal Genes
The expression levels of key genes in the different clinical stages were analyzed using the R package ggplot2, and the relationship between the key genes and the different clinical stages, including prognosis, of cervical cancer patients was analyzed by the R package survival. Subsequently, the subject operating curve (receiver operating characteristic, ROC) was plotted to assess the diagnostic value of the expression levels of the key genes for HPV16 positivity. P < 0.05 was considered statistically significant.
Single Gene Set Enrichment Analysis
GSEA is a method for analyzing gene expression data to assess pathway enrichment in transcriptional data (21). The median gene expression was used as a grouping condition, and the biological functions associated with the hub genes in HPV16 were analyzed using the R package clusterProfiler by matching mutual species with the functions in the R package msigdbr. The screening conditions were p < 0.05 and false discovery rate (FDR) < 0.2. The presentation was visualized using gseaplot2, a function in the R package clusterProfiler.
Pan-Cancer Analysis
The data of 33 cancers and normal tissues were obtained from TCGA database and GTEx database to analyze the expression difference between the key genes in cancer and para cancer and observe the association of the gene with other cancers.
Results
Screening of DEGs
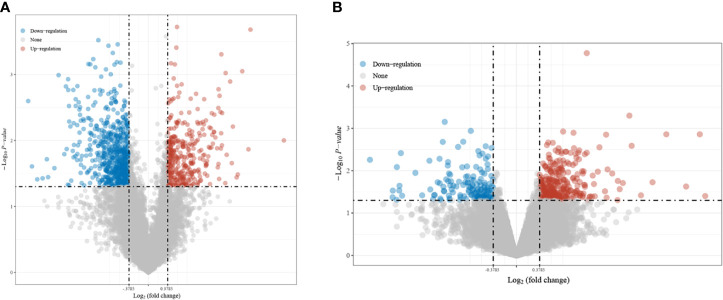
In this study, 1,069 DEGs were screened from TCGA, among which 362 genes were upregulated and 707 genes were downregulated ( Figure 1A ). Four hundred and forty-six DEGs were screened from GSE6791, of which 302 genes were upregulated and 144 genes were downregulated ( Figure 1B ).
Figure 1.
Screening results of the differentially expressed genes (DEGs). (A) intersecting genes in TCGA and GSE6791. (B) Screened from the differential genes in The Cancer Genome Atlas (TCGA).
GO/KEGG Enrichment Analysis of DEGs
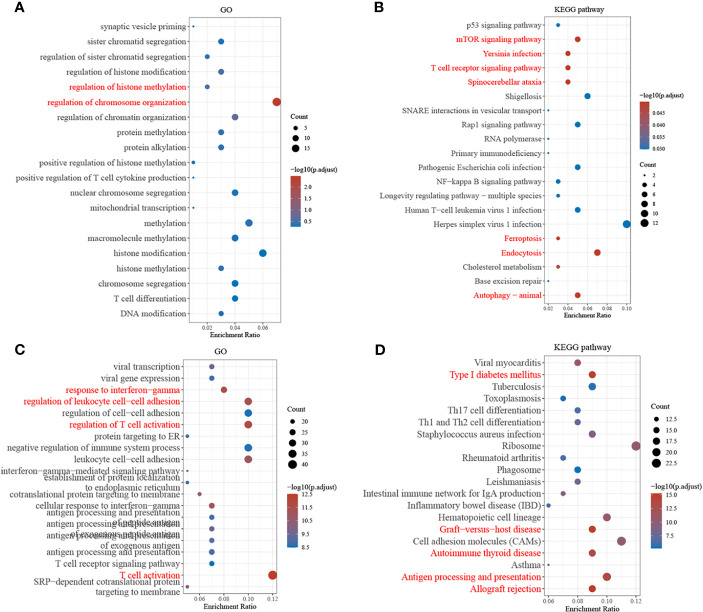
The top 20 GO terms and KEGG pathways with the most enriched upregulated genes in DEGs were listed by p-value, as shown in Figure 2 . In the GSE6791 database, DEGs were mainly enriched in GO functions such as regulation of chromosome organization, histone, and methylation regulation and in KEGG pathways such as the ribosome, tumor necrosis factor signaling pathway, and iron death. In TCGA database, the DEGs were mainly enriched in GO functions such as T-cell activation and regulation, response to interferon−gamma, and regulation of leukocyte cell–cell adhesion and in KEGG pathways such as transplant rejection, antigen outgrowth, and presentation.
Figure 2.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of the differentially expressed genes (DEGs). (A, B) Screening of GO terms and KEGG pathway for DEGs from GSE6791. (C, D) Screening of GO terms and KEGG pathway for the differential genes from The Cancer Genome Atlas (TCGA).
Screening of Pivotal Genes
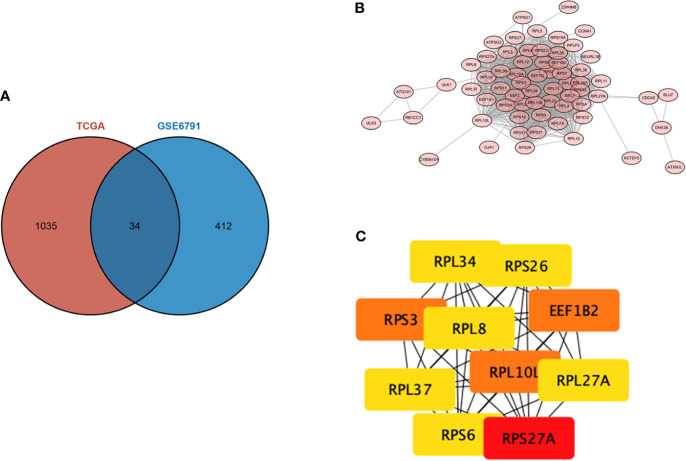
The intersection of the DEGs screened from TCGA and GSE6791 was taken, and a total of 34 overlapping genes were screened, as shown in the Wayne diagram ( Figure 3A ). To analyze the interactions between DEGs, we constructed a PPI using the STRING database and obtained the genes with an integrated score of >0.4 ( Figure 3B ), among which the top 10 pivotal genes in degree ranking were RPS27A, RPS3, EEF1B2, RPL10L, RPL27A, RPL34, RPS6, RPS26 RPL8, and RPL37 ( Figure 3C ).
Figure 3.
Protein–protein interaction (PPI) network of the differentially expressed genes (DEGs). (A) intersecting genes in TCGA and GSE6791. (B) PPI constructed using the STRING database. (C) Degree ranking of the top 10 potential key genes.
RPS27A Is a Key Gene
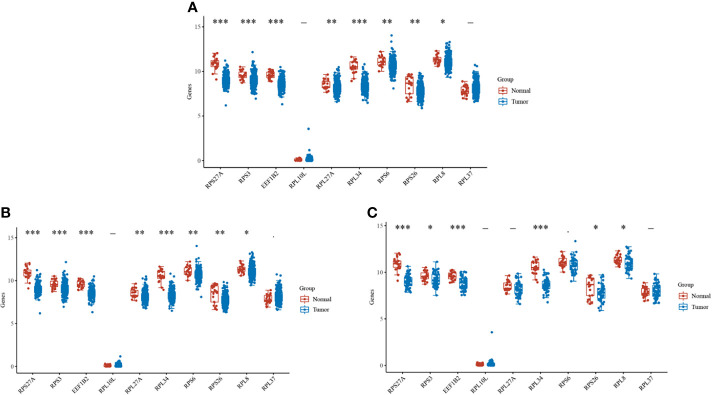
Cervical cancer was divided into cervical SCC and cervical adenocarcinoma. By observing the difference in the expression levels of key genes in the different types of cervical cancer and normal tissues, we found that most genes, such as RPS27A, RPS3, and EEF1B2, were significantly expressed in cervical cancer ( Figure 4 ). Subsequently, Kaplan–Meier analysis was performed on 10 pivotal genes, and only RPS27A was significantly associated with the prognosis of cervical cancer ( Table 1 ). Therefore, RPS27A was designated as a pivotal gene.
Figure 4.
Expressions of the 10 key genes in different cervical cancer types. (A–C) Differences in the expressions of the key genes in cervical cancer, cervical phospho-cellular carcinoma, cervical adenocarcinoma, and normal tissues, respectively. *P < 0.05; **P < 0.01; ***P < 0.001.
Table 1.
Summary of the Kaplan–Meier curve data for the 10 key genes.
| Genes | p-value | HR | Low 95%CI | High 95%CI |
|---|---|---|---|---|
| RPS27A | 0.015882274 | 1.791330669 | 1.115359057 | 2.876979882 |
| RPS3 | 0.158973292 | 1.397120997 | 0.877287386 | 2.224980218 |
| EEF1B2 | 0.26483894 | 1.301492142 | 0.818991291 | 2.068253733 |
| RPL10L | 0.416411021 | 0.824521873 | 0.517726551 | 1.313118512 |
| RPL27A | 0.6639216 | 1.10829083 | 0.696995342 | 1.76229092 |
| RPL34 | 0.377287049 | 1.232092918 | 0.77521818 | 1.958226726 |
| RPS6 | 0.720160167 | 1.088208158 | 0.685267336 | 1.728080319 |
| RPS26 | 0.508204272 | 1.169684167 | 0.735239992 | 1.860836006 |
| RPL8 | 0.099889699 | 1.477767994 | 0.928054852 | 2.353091781 |
| RPL37 | 0.2883298 | 1.285697073 | 0.808499074 | 2.044550223 |
Diagnostic and Prognostic Value of RPS27A Expression Level for HPV16
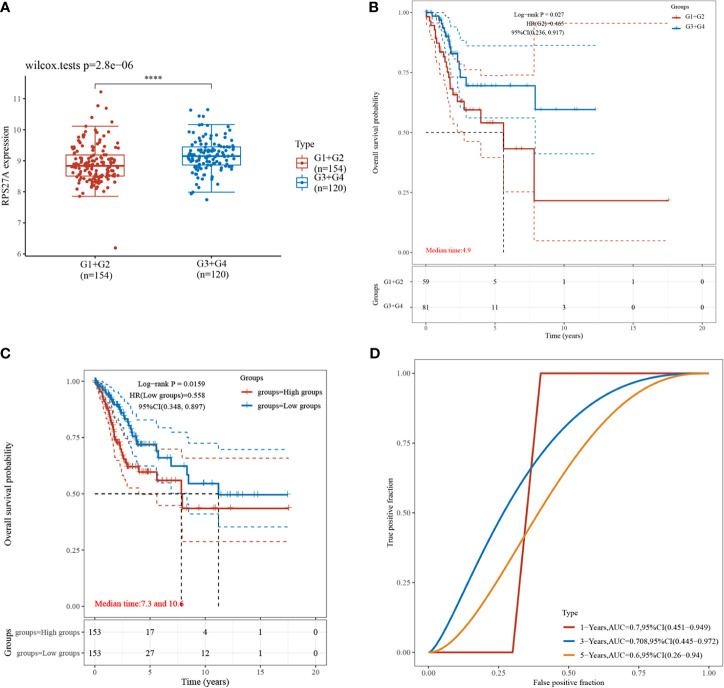
Validation of RPS27A revealed significant differences in its expression during the different clinical stages of cervical cancer ( Figure 5A ), and a high RPS27A expression was associated with poorer prognosis in patients with advanced cervical cancer (p = 0.0023) ( Figure 5B ). In addition, survival analysis showed that cervical cancer patients with a high RPS27A expression had worse prognosis ( Figure 5C ), and RPS27A expression was associated with poorer prognosis in patients with HPV16-positive cervical cancer at 1 year (AUC = 0.7, 95%CI = 0.451–0.949), 3 years (AUC = 0.708, 95%CI = 0.445–0.972), and 5 years (AUC = 0.6, 95%CI = 0.26–0.94), which were predictive of prognostic survival ( Figure 5D ).
Figure 5.
Clinical expression comparison and prognostic analysis of RPS27A. (A) Comparison of the expressions in different clinical stages, ****P < 0.0001. (B) Prognostic impact of a high RPS27A expression in clinical analysis. (C) Kaplan–Meier curve distribution of high and low RPS27A expressions. (D) Receiver operating characteristic (ROC) curve of the different survival times of RPS27A.
Biological Characteristics of RPS27A
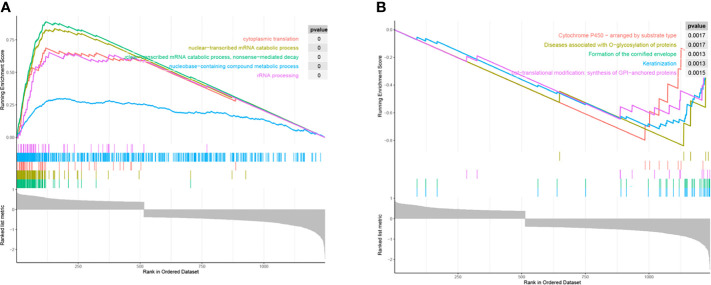
The results of GSEA showed that RPS27A was mainly enriched in GO functions such as cytoplasmic translation, nuclear–transcriptional mRNA catabolic processes, and ribosomal RNA (rRNA) processing ( Figure 6 ). It was also associated with cytochrome P450 (CYP450) arrangement by substrate type, keratinized envelope formation, post-translational modifications: GPI-anchored protein synthesis, and other biological pathways.
Figure 6.
Functional mining of RPS27A. (A) Gene Ontology (GO) terms of RPS27A. (B) Biological pathway of RPS27A in the Reactome gene set.
RPS27A Is Significantly Expressed in Most Tumours
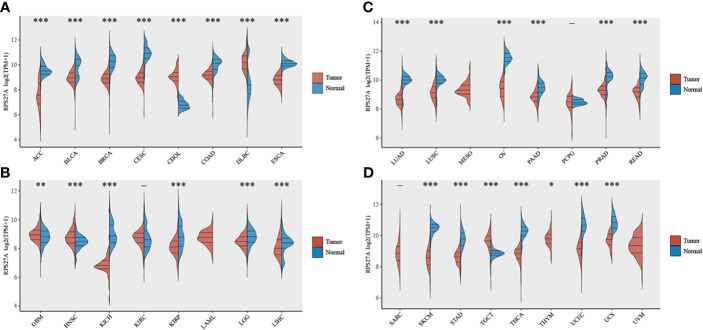
As shown in Figure 7 , RPS27A expression was significantly associated with the majority of tumors, such as adrenocortical carcinoma (ACC; p < 0.001), bladder urothelial carcinoma (BLCA; p < 0.001), breast invasive carcinoma (BRCA; p < 0.001), and CESC (p < 0.001).
Figure 7.
Expression levels of RPS27A in different tumor tissues. (A–D) Differences in RPS27A expressions in 33 different tumor tissues and normal tissues in The Cancer Genome Atlas (TCGA). *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
HPV infection is a major cause of cervical carcinogenesis, and repeated infections with the same species of HPV genotypes have a synergistic effect in inducing cervical carcinogenesis (22). HPV encodes two oncoproteins, E6 and E7, and their sustained expression promotes cervical carcinogenesis (23). In particular, HPV16 infection shows a high prevalence in cervical cancer cases, with most infections manifesting without symptoms (24). This study aimed to discover the genes associated with HPV16 cervical cancer and to provide biomarkers for the diagnosis, treatment, and prognosis of cervical cancer.
Ribosomal protein S27A (RPS27A), the only key gene screened in this study associated with prognostic survival in cervical cancer patients, belongs to the ribosomal protein S27AE family. It is a component of the ribosomal 40S subunit and is involved in the encoding of the ubiquitin carboxyl terminus (25). RPS27A is an RNA-binding protein that performs extra-ribosomal functions, including ribosome biosynthesis and post-translational modification processes (26). It has been documented that RPS27A is a direct transcriptional target of p53. It is overexpressed in DNA damage and in kidney, breast, and colon cancers (27) and has roles in promoting proliferation, regulating cell cycle progression, and inhibiting apoptosis (28). In addition, RPS27A is involved in the progression of several diseases or cancers: it may be a potential target for Epstein–Barr virus (EBV)-induced LMP1-positive cancer cells (29), its upregulated expression promotes colorectal cancer cell growth and inhibits apoptosis (30), it is involved in the pathogenesis of diabetic pancreatic ductal adenocarcinoma (PDAC) (25), and it is also one of the pathway links that promote the proliferation of HPV immortalized cervical epithelial cells (H8), which can promote cervical carcinogenesis (26). In the present study, a high expression of RPS27A could lead to poor prognosis in patients with advanced cervical cancer, serve as a prognostic survival predictor in patients with HPV16-positive cervical cancer, and act as an oncogene in the development of HPV16 cervical cancer.
GSEA showed that RPS27A is also associated with GO functions such as cytoplasmic translation, nuclear–transcriptional mRNA catabolic processes, and rRNA processing, all of which are associated with ribosomes. Ribosome biogenesis is a tightly regulated cellular process that begins in the nucleolus and is subsequently processed into rRNA (31). When disrupted during ribosome biogenesis, it can differentially promote cell cycle arrest, senescence, or apoptosis (32). In recent years, the role of ribosomes in carcinogenesis has been extensively validated, linking their involvement in cell cycle regulation and p53 activation to cancer progression (33). For example, RPS19, RPS21, and RPS24 can be used as biomarkers for prostate cancer (34), and ribosome dysfunction is associated with the pathogenesis of nasopharyngeal carcinoma (35) and can promote breast cancer metastasis (36).
In addition, CYP450 is also enriched for significant biological pathways. It is a large, intact membrane-conserved superfamily that includes 57 coding genes, mainly found in hepatocytes and enterocytes, involved in the metabolism of cholesterol, oestrogen, vitamin D, and arachidonic acid (37). Li et al. showed that HPV integrated genes strongly prefer the CP450 pathway (38), which is consistent with the results of the present study. Moreover, CP450 is one of the factors that predispose patients to cervical cancer. Studies by several scholars have shown that polymorphic variants in the CP450 family gene CYP1A1 can increase the risk of cervical cancer, especially in Asians (39, 40). In addition, CP450-related genes also play important roles in other cancers: CYP4Z1 is involved in regulating breast cancer progression (41), the CYP17 inhibitor prevents the growth of prostate cancer cells (42), CYP24A1, a proto-oncogene in human lung cancer, has anti-differentiation and anti-proliferative effects in human lung cancer cell lines (43), and CYP1B1 causes apoptosis in neural cancer cells by inducing melatonin (44). This shows that CP450 is important in cancer cell differentiation, proliferation, and apoptosis. The above analysis indicates that the functions between RPS27A and CP450 have overlapping parts, suggesting that RPS27A might have a synergistic effect with CP450.
Previously, comprehensive bioinformatics analysis methods, such as functional enrichment analysis, PPI network construction, and survival analysis, were used to screen DEGs using the GEO database or TCGA database to identify key genes for cancer progression. For example, Sun et al. (45) and Liu et al. (46) identified biomarkers associated with gastric cancer (GC) progression using this method. In this study, we identified the key gene for HPV16 cervical cancer as RPS27A, developed a survival prediction model to confirm its predictive ability, and performed a GSEA to investigate its functional pathway. In conclusion, this study identified RPS27A as a key gene for HPV type 16 cervical cancer using a comprehensive bioinformatics analysis approach and that it has an accurate predictive ability for patients’ prognostic survival. Although systematic bias may have arisen due to the large variation in sample size, our findings still provide therapeutic targets with clinical significance for HPV16-associated cervical cancer.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by Ningbo Medical and Health Care Brand Discipline (PPXK2018-06).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, et al. Estimates of Incidence and Mortality of Cervical Cancer in 2018: A Worldwide Analysis. Lancet Global Health (2020) 8(2):e191–203. doi: 10.1016/S2214-109X(19)30482-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 4. Darre T, Kpatcha TM, Bagny A, Maneh N, Gnandi-Piou F, Tchangai B. Descriptive Epidemiology of Cancers in Togo from 2009 to 2016. APJCP (2017) 18(12):3407–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Small J, Bacon MA, Bajaj A, Chuang LT, Fisher BJ, Harkenrider MM, et al. Cervical Cancer: A Global Health Crisis. Cancer (2017) 123(13):2404–12. doi: 10.1002/cncr.30667 [DOI] [PubMed] [Google Scholar]
- 6. Liu Y-M, Ni L-Q, Wang S-S, Lv Q-L, Chen W-J, Ying S-P. Outcome and Prognostic Factors in Cervical Cancer Patients Treated With Surgery and Concurrent Chemoradiotherapy: A Retrospective Study. World J Surg Oncol (2018) 16(1):1–7. doi: 10.1186/s12957-017-1307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charakorn C, Thadanipon K, Chaijindaratana S, Rattanasiri S, Numthavaj P, Thakkinstian A. The Association Between Serum Squamous Cell Carcinoma Antigen and Recurrence and Survival of Patients With Cervical Squamous Cell Carcinoma: A Systematic Review and meta-analysis. Gynecologic Oncol (2018) 150(1):190–200. doi: 10.1016/j.ygyno.2018.03.056 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y, Guo X, Wang G, Ma W, Liu R, Han X, et al. Real-World Study of the Incidence, Risk Factors, and Prognostic Factors Associated With Bone Metastases in Women With Uterine Cervical Cancer Using Surveillance, Epidemiology, and End Results (SEER) Data Analysis. Med Sci Monit (2018) 24:6387. doi: 10.12659/MSM.912071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global Burden of Cancer Attributable to Infections in 2018: A Worldwide Incidence Analysis. Lancet Global Health (2020) 8(2):e180–e90. doi: 10.1016/S2214-109X(19)30488-7 [DOI] [PubMed] [Google Scholar]
- 10. Manini I, Montomoli E. Epidemiology and Prevention of Human Papillomavirus. Ann Ig (2018) 30(4):28–32. doi: 10.7416/ai.2018.2231 [DOI] [PubMed] [Google Scholar]
- 11. Bedell SL, Goldstein LS, Goldstein AR, Goldstein AT. Cervical Cancer Screening: Past, Present, and Future. Sexual Med Rev (2020) 8(1):28–37. doi: 10.1016/j.sxmr.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 12. Wright TC, Stoler MH, Behrens CM, Sharma A, Zhang G, Wright TL. Primary Cervical Cancer Screening With Human Papillomavirus: End of Study Results From the ATHENA Study Using HPV as the First-Line Screening Test. Gynecol Oncol (2015) 136(2):189–97. doi: 10.1016/j.ygyno.2014.11.076 [DOI] [PubMed] [Google Scholar]
- 13. Simms KT, Steinberg J, Caruana M, Smith MA, Lew J-B, Soerjomataram I, et al. Impact of Scaled Up Human Papillomavirus Vaccination and Cervical Screening and the Potential for Global Elimination of Cervical Cancer in 181 Countries, 2020–99: A Modelling Study. Lancet Oncol (2019) 20(3):394–407. doi: 10.1016/S1470-2045(18)30836-2 [DOI] [PubMed] [Google Scholar]
- 14. Momenimovahed Z, Salehiniya H. Cervical Cancer in Iran: Integrative Insights of Epidemiological Analysis. BioMedicine (2018) 8(3):18. doi: 10.1051/bmdcn/2018080318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marth C, Landoni F, Mahner S, McCormack M, Gonzalez-Martin A, Colombo N. Cervical Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann Oncol (2017) 28:iv72–83. doi: 10.1093/annonc/mdx220 [DOI] [PubMed] [Google Scholar]
- 16. Kaliff M, Sorbe B, Mordhorst LB, Helenius G, Karlsson MG, Lillsunde-Larsson G. Findings of Multiple HPV Genotypes in Cervical Carcinoma Are Associated With Poor Cancer-Specific Survival in a Swedish Cohort of Cervical Cancer Primarily Treated With Radiotherapy. Oncotarget (2018) 9(27):18786–96. doi: 10.18632/oncotarget.24666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsang KY, Fantini M, Fernando RI, Palena C, David JM, Hodge JW, et al. Identification and Characterization of Enhancer Agonist Human Cytotoxic T-Cell Epitopes of the Human Papillomavirus Type 16 (HPV16) E6/E7. Vaccine (2017) 35(19):2605–11. doi: 10.1016/j.vaccine.2017.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McClung NM, Gargano JW, Bennett NM, Niccolai LM, Abdullah N, Griffin MR, et al. Trends in Human Papillomavirus Vaccine Types 16 and 18 in Cervical Precancers, 2008–2014. Cancer Epidemiol Prev Biomarkers (2019) 28(3):602–9. doi: 10.1158/1055-9965.EPI-18-0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao W, Liu Y, Zhang L, Ding L, Li Y, Zhang H, et al. MicroRNA-154-5p Regulates the HPV16 E7-pRb Pathway in Cervical Carcinogenesis by Targeting CUL2. J Cancer (2020) 11(18):5379. doi: 10.7150/jca.45871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smelov V, Elfström KM, Johansson AL, Eklund C, Naucler P, Arnheim‐Dahlström L, et al. Long‐Term HPV Type‐Specific Risks of High‐Grade Cervical Intraepithelial Lesions: A 14‐Year Follow‐Up of a Randomized Primary HPV Screening Trial. International J Cancer (2015) 136(5):1171–80. [DOI] [PubMed] [Google Scholar]
- 21. Powers RK, Goodspeed A, Pielke-Lombardo H, Tan A-C, Costello AC. GSEA-InContext: Identifying Novel and Common Patterns in Expression Experiments. Bioinformatics 34:i555–64. doi: 10.1093/bioinformatics/bty271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Senapati R, Nayak B, Kar SK, Dwibedi B. HPV Genotypes Co-Infections Associated With Cervical Carcinoma: Special Focus on Phylogenetically Related and non-Vaccine Targeted Genotypes. PloS One (2017) 12(11):e0187844. doi: 10.1371/journal.pone.0187844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yeo-Teh NS, Ito Y, Jha S. High-Risk Human Papillomaviral Oncogenes E6 and E7 Target Key Cellular Pathways to Achieve Oncogenesis. Int J Mol Sci (2018) 19(6):1706. doi: 10.3390/ijms19061706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. da Silva RL, da Silva Batista Z, Bastos GR, Cunha APA, Figueiredo FV, de Castro LO, et al. Role of HPV 16 Variants Among Cervical Carcinoma Samples From Northeastern Brazil. BMC Women’s Health (2020) 20(1):1–11. doi: 10.1186/s12905-020-01035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou W, Wang Y, Gao H, Jia Y, Xu Y, Wan X, et al. Identification of Key Genes Involved in Pancreatic Ductal Adenocarcinoma With Diabetes Mellitus Based on Gene Expression Profiling Analysis. Pathol Oncol Res (2021) 27:58. doi: 10.3389/pore.2021.604730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen L, Wang H. Nicotine Promotes Human Papillomavirus (HPV)-Immortalized Cervical Epithelial Cells (H8) Proliferation by Activating RPS27a-Mdm2-P53 Pathway In Vitro . Toxicol Sci (2019) 167(2):408–18. doi: 10.1093/toxsci/kfy246 [DOI] [PubMed] [Google Scholar]
- 27. Kidokoro Y, Sakabe T, Haruki T, Kadonaga T, Nosaka K, Nakamura H, et al. Gene Expression Profiling by Targeted RNA Sequencing in Pathological Stage I Lung Adenocarcinoma With a Solid Component. Lung Cancer (2020) 147:56–63. doi: 10.1016/j.lungcan.2020.06.035 [DOI] [PubMed] [Google Scholar]
- 28. Wang H, Yu J, Zhang L, Xiong Y, Chen S, Xing H, et al. RPS27a Promotes Proliferation, Regulates Cell Cycle Progression and Inhibits Apoptosis of Leukemia Cells. Biochem Biophys Res Commun (2014) 446(4):1204–10. doi: 10.1016/j.bbrc.2014.03.086 [DOI] [PubMed] [Google Scholar]
- 29. Hong S-W, Kim S-M, Jin D-H, Kim YS, Hur DY. RPS27a Enhances EBV-Encoded LMP1-Mediated Proliferation and Invasion by Stabilizing of LMP1. Biochem Biophys Res Commun (2017) 491(2):303–9. doi: 10.1016/j.bbrc.2017.07.105 [DOI] [PubMed] [Google Scholar]
- 30. Mu Q, Luo G, Wei J, Zheng L, Wang H, Yu M, et al. Apolipoprotein M Promotes Growth and Inhibits Apoptosis of Colorectal Cancer Cells Through Upregulation of Ribosomal Protein S27a. EXCLI J (2021) 20:145. doi: 10.17179/excli2020-2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu H, Kong X, Huang H, Wu W, Park J, Yun D-J, et al. STCH4/REIL2 Confers Cold Stress Tolerance in Arabidopsis by Promoting rRNA Processing and CBF Protein Translation. Cell Rep (2020) 30(1):229–42. e5. doi: 10.1016/j.celrep.2019.12.012 [DOI] [PubMed] [Google Scholar]
- 32. Turi Z, Lacey M, Mistrik M, Moudry P. Impaired Ribosome Biogenesis: Mechanisms and Relevance to Cancer and Aging. Aging (Albany NY) (2019) 11(8):2512. doi: 10.18632/aging.101922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Penzo M, Montanaro L, Treré D, Derenzini M. The Ribosome Biogenesis—Cancer Connection. Cells (2019) 8(1):55. doi: 10.3390/cells8010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arthurs C, Murtaza BN, Thomson C, Dickens K, Henrique R, Patel HR, et al. Expression of Ribosomal Proteins in Normal and Cancerous Human Prostate Tissue. PloS One (2017) 12(10):e0186047. doi: 10.1371/journal.pone.0186047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sim EU-H, Lee C-W, Narayanan K. The Roles of Ribosomal Proteins in Nasopharyngeal Cancer: Culprits, Sentinels or Both. biomark Res (2021) 9(1):1–10. doi: 10.1186/s40364-021-00311-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ebright RY, Lee S, Wittner BS, Niederhoffer KL, Nicholson BT, Bardia A, et al. Deregulation of Ribosomal Protein Expression and Translation Promotes Breast Cancer Metastasis. Science (2020) 367(6485):1468–73. doi: 10.1126/science.aay0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elfaki I, Mir R, Almutairi FM, Duhier FMA. Cytochrome P450: Polymorphisms and Roles in Cancer, Diabetes and Atherosclerosis. Asian Pac J Cancer Prevent: APJCP (2018) 19(8):2057. doi: 10.22034/APJCP.2018.19.8.2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li W, Tian S, Wang P, Zang Y, Chen X, Yao Y, et al. The Characteristics of HPV Integration in Cervical Intraepithelial Cells. J Cancer (2019) 10(12):2783. doi: 10.7150/jca.31450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wongpratate M, Settheetham-Ishida W, Phuthong S, Natphopsuk S, Ishida T. Genetic Polymorphisms of the Human Cytochrome P450 1a1 (CYP1A1) and Cervical Cancer Susceptibility Among Northeast Thai Women. Asian Pac J Cancer Prevent: APJCP (2020) 21(1):243–8. doi: 10.31557/APJCP.2020.21.1.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ding B, Sun W, Han S, Cai Y, Ren M, Shen Y. Cytochrome P450 1A1 Gene Polymorphisms and Cervical Cancer Risk: A Systematic Review and Meta-Analysis. Medicine (2018) 97(13). doi: 10.1097/MD.0000000000010210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McDonald MG, Ray S, Amorosi CJ, Sitko KA, Kowalski JP, Paco L, et al. Expression and Functional Characterization of Breast Cancer-Associated Cytochrome P450 4Z1 in Saccharomyces Cerevisiae. Drug Metab Disposition (2017) 45(12):1364–71. doi: 10.1124/dmd.117.078188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Norris JD, Ellison SJ, Baker JG, Stagg DB, Wardell SE, Park S, et al. Androgen Receptor Antagonism Drives Cytochrome P450 17A1 Inhibitor Efficacy in Prostate Cancer. J Clin Invest (2017) 127(6):2326–38. doi: 10.1172/JCI87328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li M, Li A, He R, Dang W, Liu X, Yang T, et al. Gene Polymorphism of Cytochrome P450 Significantly Affects Lung Cancer Susceptibility. Cancer Med (2019) 8(10):4892–905. doi: 10.1002/cam4.2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu Z, Tian X, Peng Y, Sun Z, Wang C, Tang N, et al. Mitochondrial Cytochrome P450 (CYP) 1B1 Is Responsible for Melatonin-Induced Apoptosis in Neural Cancer Cells. J Pineal Res (2018) 65(1):e12478. doi: 10.1111/jpi.12478 [DOI] [PubMed] [Google Scholar]
- 45. Sun C, Yuan Q, Wu D, Meng X, Wang B. Identification of Core Genes and Outcome in Gastric Cancer Using Bioinformatics Analysis. Oncotarget (2017) 8(41):70271. doi: 10.18632/oncotarget.20082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu X, Wu J, Zhang D, Bing Z, Tian J, Ni M, et al. Identification of Potential Key Genes Associated With the Pathogenesis and Prognosis of Gastric Cancer Based on Integrated Bioinformatics Analysis. Front Genet (2018) 9:265. doi: 10.3389/fgene.2018.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.