Abstract
Homeoproteins and basic helix-loop-helix (bHLH) transcription factors are known for their critical role in development and cellular differentiation. The pituitary pro-opiomelanocortin (POMC) gene is a target for factors of both families. Indeed, pituitary-specific transcription of POMC depends on the action of the homeodomain-containing transcription factor Pitx1 and of bHLH heterodimers containing NeuroD1. We now show lineage-restricted expression of NeuroD1 in pituitary corticotroph cells and a direct physical interaction between bHLH heterodimers and Pitx1 that results in transcriptional synergism. The interaction between the bHLH and homeodomains is restricted to ubiquitous (class A) bHLH and to the Pitx subfamily. Since bHLH heterodimers interact with Pitx factors through their ubiquitous moiety, this mechanism may be implicated in other developmental processes involving bHLH factors, such as neurogenesis and myogenesis.
Cell-specific transcription results from complex molecular interactions that involve the synergistic action of multiple transcription factors. Taken together, these interactions provide the molecular basis for the complex program of cell differentiation and development. Factors of two classes of transcription factors often involved in developmental processes, the homeodomain (HD) and the basic helix-loop-helix (bHLH) factors, were found to form the basis for cell-specific transcription of pituitary pro-opiomelanocortin (POMC) gene expression. We have used this system to define the molecular mechanism by which these two classes of transcription factors can synergistically interact to control transcription. More specifically, our studies reveal a specific interaction between the HD of the Pitx (Ptx) subfamily of homeobox proteins and the widely expressed class A bHLH transcription factors.
The HD is a 60-amino-acid DNA-binding domain that contains a well-conserved helix-turn-helix motif. The third helix of this motif interacts with the major groove of DNA and is responsible for sequence-specific recognition. In particular, residue 50 of the HD is responsible for subdividing the HD-containing genes into large subgroups (10). HD proteins related to Drosophila bicoid have a lysine at this position, and this residue dictates recognition of a unique DNA sequence (20, 63). In vertebrates, this subgroup includes the goosecoid, Otx, and Pitx subfamilies (8, 15). While Pitx1 was originally cloned as a pituitary transcription factor (26, 60) that is involved in transcription of many pituitary hormone genes (64), it was later found to play a wide role during early development, in agreement with its early expression in posterior lateral plate mesoderm and in the stomodeum (27). Indeed, inactivation of the Pitx1 gene impairs hind limb and mandible development (28, 59). In the stomodeum, Rathke's pouch, and definitive pituitary, Pitx1 is coexpressed with the related Pitx2 gene, and it is likely that the two genes serve partly redundant functions in these tissues (14, 27, 41). Pitx2 was first isolated as the causative gene of Rieger's syndrome (56), a craniofacial malformation that affects facial and tooth development; it was later found to be expressed in left lateral plate mesoderm and to be the effector for left-right asymmetry in the development of internal organs (4, 33, 51, 55, 68). In addition to its left-side-specific expression in lateral plate mesoderm, Pitx2 is also expressed bilaterally in the head and branchial arches, as well as in the myotome and in migrating myoblasts of the limbs (33, 51).
Whereas HD-containing genes have mostly been implicated in patterning (25), like Pitx1 for specification of hind limbs (17, 28, 59, 67) and Pitx2 in determination of laterality (21), bHLH transcription factors have been more often implicated in cell differentiation. Indeed, the four myogenic bHLH factors play critical roles at different stages of muscle cell differentiation (47, 54), and other bHLH are involved in hematopoietic differentiation (2, 57) or in neurogenesis (18, 19, 31, 32, 38). bHLH factors are characterized by a helix-loop-helix dimerization domain and a basic domain that binds specific DNA sequences (NCANNTGN) called E boxes (36, 42). The bHLH family can be subdivided into various subgroups (22). The class A bHLH are ubiquitously expressed and appear to act primarily as general transcription factors, although they play a particular role in B-lymphocyte development (69). The class A bHLH can form homodimers (58), but they also form heterodimers with class B bHLH transcription factors (30). Different bHLH dimers interact with specific E boxes that differ in their nonconserved residues (3). The class B bHLH have a tissue- or cell-restricted expression, and many members of these groups are critical for differentiation in various lineages, like muscle (47), the erythroid lineage (57), and neuronal differentiation (31).
One neurogenic bHLH, NeuroD1 (also called BETA2) (32, 45), was also shown to be important for development of endocrine cells in the pancreas and of secretin and cholecystokinin cells in the intestine (43). In neural tissues, NeuroD1 has been associated with late-differentiation events (32), and in mice, it is expressed in neurons of cranial nerves (V to XI), dorsal root ganglia, cerebral cortex, nasal epithelium, vomeronasal organ, and retina of the eye. Its expression is transient in cranial nerves and dorsal root ganglia but persistent in the other structures. It is also excluded from the mitotically active ventricular zone of the cerebral cortex and spinal cord. Neurogenin 1 and 2 are bHLH factors that precede and cause NeuroD1 expression in Xenopus laevis (38) and mice (37). It was suggested that neurogenins and NeuroD1 may act as determination and differentiation factors, respectively. They are part of a proneural gene cascade that is similar to the cascade of myogenic bHLH.
NeuroD1 is also found in endocrine cells of the pancreas, and inactivation of its gene causes a diabetic phenotype due to a failure to develop mature islets (44). By analogy with the nervous system, NeuroD1 expression is preceded by and requires the presence of neurogenin 3 in the pancreas (1, 16). In this tissue, NeuroD1-containing heterodimers are required for transcription of the insulin gene, and their action on this promoter depends on interaction with a pancreas-specific homeoprotein, Pdx1 (48, 49). Very recently, it was suggested that these factors interact physically with each other (46).
We have previously shown expression of NeuroD1 in adult pituitary corticotroph cells and we have documented its role in corticotroph-specific transcription of the POMC gene (52). We have also shown the importance of the Pitx1 transcription factor for expression of POMC (26). Interestingly, the action of NeuroD1-containing bHLH dimers on POMC transcription is entirely dependent on transcriptional interaction with Pitx1 (52, 62). However, the molecular basis of this interaction is unknown. Here we present evidence of direct physical interaction between the DNA-binding domains of bHLH and HD transcription factors and suggest that this interaction may account for the transcriptional synergism observed between them. The specificity of this interaction, which is restricted to the Pitx family and the class A ubiquitous bHLH, suggests that the mechanism revealed in the present work might be involved in other differentiated tissues where the Pitx genes are coexpressed with bHLH factors, like muscle and neurons.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
Reporter plasmids were constructed in the pXP1-luciferase vector as described previously (52). The simplified reporter plasmids were made of oligonucleotide sequences of the corresponding regions of the rat POMC promoter as described before (52). Point mutations were made using the pALTER system from Promega. The oligonucleotide sequences used for mut E boxneuro, mut Pitx1 site, and E boxneuro ⇒ E boxubi are, respectively, GCCAGGAAGGCTACCGGACGCACACAGG, CACACCAGGATTAGACTACTCTGTCCAGT, and GCCAGGAAGCCAGGTGTGCGCACACAGG (bold characters are the mutated nucleotides). The expression vectors used were described in previous work (52).
Transfection assays.
L or AtT-20 D16v cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum and maintained at 37°C and 5% CO2. L cells were transfected by the calcium phosphate coprecipitation method, and 50,000 cells were plated in 12-well plates. A total of 8 μg of total DNA was used for each transfection, performed in duplicate. Control experiments contained equivalent amounts of empty expression vector or pSP64. AtT-20 cells were transfected by using Lipofectamine (Pharmacia). Briefly, 250,000 cells were plated in 12-well plates, and 3 μg of total DNA was used for each transfection, performed in duplicate. Harvesting and luciferase analysis were described previously (26, 52).
Coimmunoprecipitation.
COS-1 cells (2.5 × 105) were transfected in a 60-mm dish with the appropriate expressing vectors (5 μg) for a total of 15 μg of DNA. The cells were harvested, centrifuged, and resuspended in 800 μl of buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EGTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], and 10 μg each of the protease inhibitors leupeptin, aprotinin, and pepstatin per ml). Cells were incubated on ice for 15 min before addition of 50 μl of NP-40 followed by vigorous vortexing. After centrifugation, the nuclear pellet was resuspended in 100 μl of buffer B (10 mM HEPES [pH 7.9], 0.1 mM EGTA, 0.4 M NaCl, 0.5 mM PMSF, 1 mM DTT, and 10 μg of each protease inhibitor as above per ml) and shaken vigorously at 4°C for 15 min. The extract was then centrifuged, and the supernatant was dialyzed against 500 ml of buffer C (10 mM Tris [pH 7.9], 150 mM NaCl, 0.3% NP-40, 0.25% bovine serum albumin [BSA], 1 mM DTT, 0.01 M NaN3) at 4°C with four changes of buffer every 2 h. The protein concentration of the extracts was estimated by the Bradford assay. Coimmunoprecipitation experiments were performed essentially as described before (11), except that 10 μg of transfected COS-1 or 200 μg of untransfected AtT-20 cell nuclear extracts was used and subjected to a preclearing step using 1 μg of purified immunoglobulin G (IgG) from a rabbit (Sigma). Anti-Pitx1a antibody (0.1 μg) (affinity purified) was used for immunoprecipitation. Pan1 was revealed by Western blotting using an anti-E2A antibody (Santa Cruz) and an anti-mouse IgG-horseradish peroxidase conjugate (Sigma). Revelation was performed by chemiluminescence as described by the manufacturer (ECL+plus; Amersham Pharmacia).
Transgenic mice.
The plasmids used in transgenic mice were described previously (62). The injected fragments were produced by BamHI-XmnI digestion and purified on an agarose gel. Transgenic mice were produced by injection of DNA fragments in eggs from C3H mice. Implantation was done in pseudopregnant CD1 mice. POMC-luciferase transgenes were identified by Southern blot, and mice heterozygous for the transgene were sacrificed for analysis. Anterior and intermediate pituitaries were dissected, immediately frozen on dry ice, and then homogenized in 200 to 300 μl of a buffer containing Tris-HCl (pH 8), 0.5% NP-40, and 0.1 M DTT. After centrifugation (5 min), half of the supernatant was analyzed for luciferase activity, and the other half was used for protein quantification using the Bradford assay.
Antibody against NeuroD1 and immunohistochemistry.
A PCR fragment encoding amino acids 122 to 165 of mouse NeuroD1 was subcloned in frame with maltose binding protein (MBP) and glutathione-S-transferase (GST) into their respective vectors. The fusion proteins were purified from Escherichia coli BL-21 with maltose and Sepharose beads according to the manufacturer's recommendations (New England Biolabs and Pharmacia Biotech, respectively). Antibodies were raised by injection of 100 μg of MBP-NeuroD1122–165 in New Zealand female rabbits; two booster injections were made at 4 and 6 weeks after the control injection. After assessment of immunological response by Western blot, the rabbits were sacrificed and serum was collected. The antiserum was purified on a GST-NeuroD1122–165 column to obtain an affinity-purified antibody preparation. Embryos were fixed in 4% paraformaldehyde, paraffin embedded, and sectioned sagitally in 5-mm slices. Immunohistochemistry was performed as previously described (52, 53) except that an amplification step was added using the Renaissance Tyramide Signal Amplification-indirect system (NEN).
Pull-down assay.
MBP-LacZ, MBP-Pitx1, MBP-Nkx2.5, MBP-Gsc, and MBP-Pitx1 deletion mutants were purified from E. coli BL-21 following the manufacturer's recommendations, and 500 ng of each fusion protein coupled to Amylose beads (New England Biolabs) was used in all assays. Pan1, NeuroD1, and luciferase were synthesized in vitro using [35S]methionine and the TnT-coupled transcription-translation rabbit reticulocyte lysate system (Promega). Pull-downs were performed as described before (11).
RESULTS
Transcriptional synergism between Pitx1 and NeuroD1 requires the E boxneuro but not NeuroD1 per se.
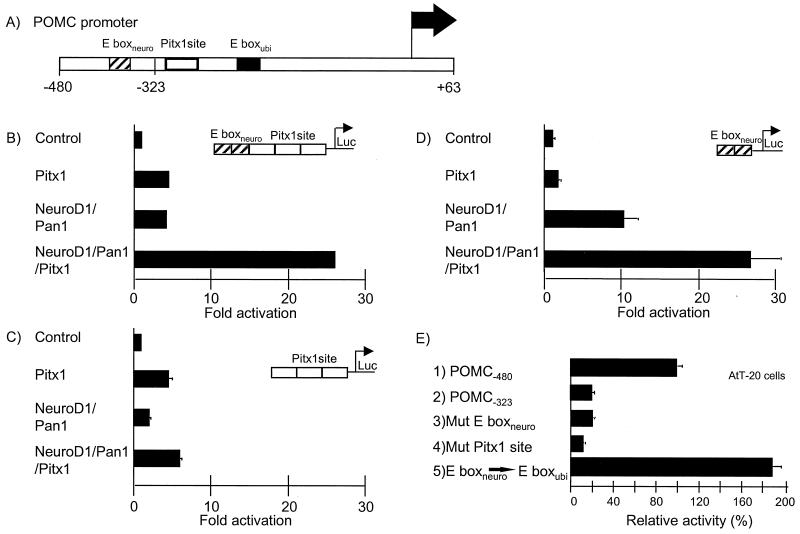
We have previously demonstrated that the corticotroph specificity of POMC transcription (Fig. 1A) depends on the action of a homeoprotein, Pitx1, and of bHLH heterodimers containing NeuroD1 (26, 52). The transcriptional synergism between these factors was reconstituted in L cell fibroblasts using simple reporters containing Pitx1 binding sites and the NeuroD1-specific E box, E boxneuro (Fig. 1B). We tested whether both binding sites are required for synergism in this simple system and found that E boxneuro is essential (Fig. 1C), whereas the Pitx1 binding site is not (Fig. 1D). Thus, DNA-bound bHLH heterodimers containing NeuroD1 and Pan1 can recruit Pitx1 to the promoter but not the reverse.
FIG. 1.
Binding site requirement for synergism between NeuroD1-Pan1 heterodimers and Pitx1. (A) Schematic representation of the rat POMC promoter (−480 to +63 bp) and relative positions of E boxneuro, Pitx1 binding site, and E boxubi. (B) Synergism between bHLH heterodimers (NeuroD1-Pan1) and homeoprotein (Pitx1) reconstituted in cotransfected L cells. A luciferase (Luc) reporter gene was used to assess transcriptional activity. The reporter contained three copies of the POMC gene Pitx1 binding site and two copies of the E boxneuro, as indicated. (C and D) Same experiments as in B but with a Pitx1 site and with the E boxneuro-containing reporter, respectively. (E) AtT-20 cells were transfected with a luciferase reporter gene driven by the intact POMC promoter (bar 1), a promoter deleted of its distal domain (bar 2), a promoter with a mutated E boxneuro (bar 3), a promoter with a mutation of the Pitx1 binding site (bar 4), or an E box specificity mutant promoter in which E boxneuro was transformed into E boxubi (bar 5). Results are the averages (± standard error of the mean [SEM]) from three sets of duplicate experiments (C, D, and E) or one representative experiment (B).
Since the POMC promoter contains two different E boxes as well as one Pitx1 binding site (26, 52), we investigated their respective roles in the intact promoter (Fig. 1E, POMC-480). Previous work had shown the bipartite organization of the upstream 300 bp of the POMC promoter (61) and the importance of the distal E box (E boxneuro, previously called DE2C) for activity of its distal half (62). In agreement with these observations, mutagenesis of the E boxneuro decreased promoter activity in POMC-expressing AtT-20 cells (Fig. 1E, Mut E boxneuro) to the same extent as deletion of the distal promoter sequences (Fig. 1E, POMC-323).
In contrast to the simple reporter used in Fig. 1, mutagenesis of the Pitx1 binding site of the intact POMC promoter had a similar effect on activity as mutation of E boxneuro (Fig. 1E, Mut Pitx site). Thus, promoter context imposes constraints on transcription factor activity that are not mimicked in simplified reporters containing multimerized regulatory elements. In this case, the E boxneuro in its natural context is not sufficient to recruit Pitx1 in the absence of the Pitx1 binding site. We have previously shown that another E box of the POMC promoter, E boxubi (CE1B), is activated upon binding of Pan1 (the rodent homologue of E47) or other ubiquitous bHLH factors, in contrast to the E boxneuro, which is only activated upon binding of NeuroD1-Pan1 heterodimers; the E boxubi itself is not bound or activated by NeuroD1-Pan1 heterodimers (52). In order to test the importance of NeuroD1 for activity of the distal E box, we mutated this E box into an E boxubi that no longer requires NeuroD1 for DNA binding and transcriptional activation (52). This mutant promoter (Fig. 1E, Mut E boxneuro⇒E boxubi) is as active as the intact promoter, suggesting that (i) NeuroD1 per se is not essential for synergism with Pitx1, (ii) it can be replaced by Pan1, and (iii) the role of NeuroD1 is to confer sequence-specific recognition.
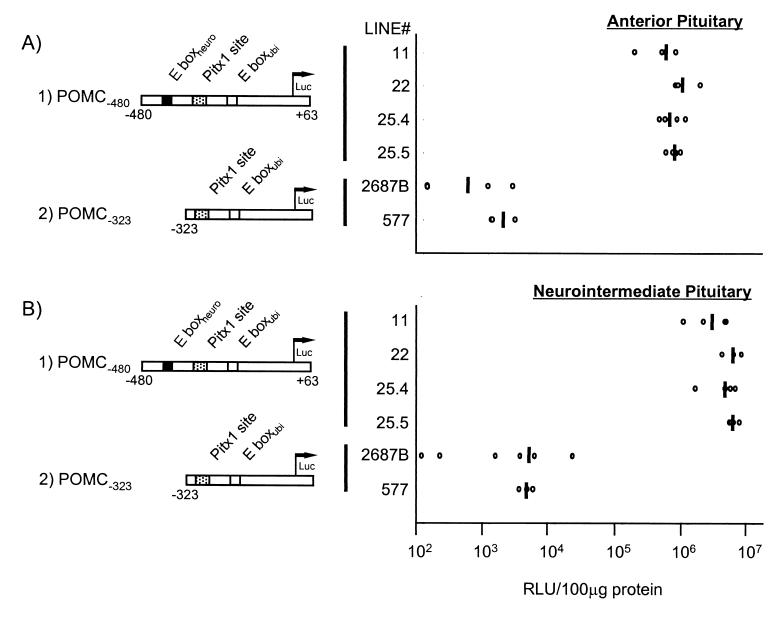
In agreement with the importance of the distal E boxneuro for promoter activity in cell culture (Fig. 1E), anterior pituitary expression of a POMC-luciferase transgene deleted of its distal sequences was found to be at least 1,000 times less active than a transgene driven by the intact POMC promoter (Fig. 2A). This may even constitute an underestimate of the activity of the deleted sequences, since five of seven POMC−323-luc lines tested did not express the transgene at detectable levels; in contrast, the POMC−480-luc transgene was expressed in four of five lines tested. This great in vivo dependence on distal promoter sequences (by comparison to activity in cultured cells) was also observed in the intermediate pituitary (Fig. 2B). Thus, the distal POMC promoter plays a critical role in pituitary expression; prior work had indicated that the activity of this promoter domain absolutely requires an active E boxneuro (52, 62).
FIG. 2.
Pituitary expression of POMC-luciferase transgenes. Two different transgenes driven by either the intact POMC promoter (POMC−480) or a deleted promoter (POMC−323) were assessed for luciferase activity in pituitary tissues. POMC−323 transgenic mice lines (2687B and 577) show a 1,000-fold-reduced activity in both anterior (A) and neurointermediate pituitary (B) compared to POMC−480 lines 11, 22, 25.4, and 25.5. Every open circle represents luciferase activity in an individual mouse, and the bar is the calculated average. Results are expressed in relative light units (RLU) and standardized according to the amount of protein.
NeuroD1 expression is restricted to corticotroph cells during pituitary development.
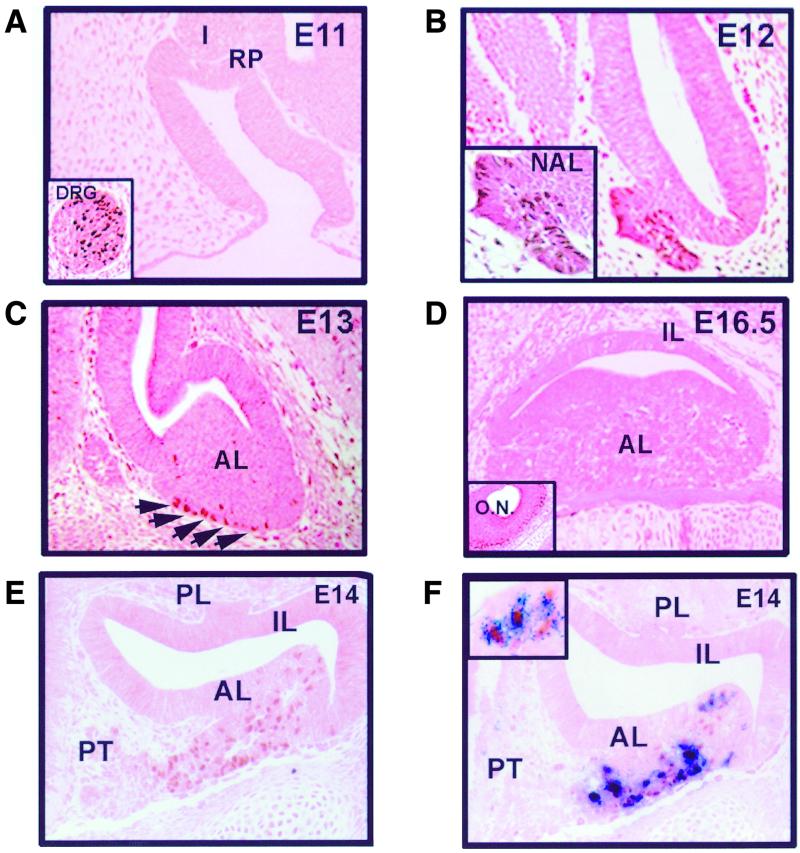
The unique role played by NeuroD1 in sequence-specific recognition of the distal E boxneuro (52) suggests that this bHLH factor may be an important developmental regulator of POMC expression. In order to assess this possibility, we followed NeuroD1 expression during pituitary development using a NeuroD1-specific antiserum. Before corticotroph differentiation, at E11 of mouse development, NeuroD1 is not expressed in Rathke's pouch but is expressed in dorsal root ganglia (Fig. 3A), as previously found by in situ hybridization (32).
FIG. 3.
Correlation of NeuroD1 and POMC expression during pituitary development. (A) NeuroD1 immunoreactivity (brown) is not detected by immunohistochemistry in Rathke's pouch (RP) or infudibulum (I) at E11, but it is present in nuclei of dorsal root ganglia cells (DRG, inset). (B to D) NeuroD1 was detected in the nascent anterior lobe (NAL) at E12 (B) and in the ventral portion of the anterior lobe (AL) indicated by arrows at pituitary E13 (C), but no expression could be detected at E16.5 in anterior lobe (D), although it was still expressed in olfactory neurons (O.N., inset). (E) At E14, expression is seen in anterior lobe but not in pars tuberalis (PT), intermediate lobe (IL), or posterior lobe (PL). (F) Coimmunohistochemistry against ACTH (blue) and NeuroD1 (brown) was performed on E14. ACTH-positive cells are also positive for NeuroD1. There are a few NeuroD1-positive nuclei in this section that do not appear positive for ACTH; however, the same cells were ACTH positive on the consecutive section (not shown). Higher magnification (inset) shows coexpression of ACTH and NeuroD1 in individual cells.
NeuroD1 expression started at E12 within the region of the nascent anterior lobe of the pituitary (Fig. 3B). At E13, NeuroD1-positive cells were observed within the ventral portion of the anterior lobe (Fig. 3C). From E16 onward, NeuroD1 was no longer detected in the anterior lobe, but olfactory neurons still expressed it (Fig. 3D), as reported before (32). The onset of this pituitary expression correlated well with in situ hybridization (data not shown). However, NeuroD1 transcripts were detected (52) in the adult pituitary, but no proteins could be detected (data not shown). Coimmunohistochemistry experiments performed on E14 pituitaries revealed that all NeuroD1-positive cells (brown) were also adrenocorticotropin (ACTH) positive (blue) (Fig. 3E and F and data not shown). Thus, NeuroD1 appeared just before POMC around E12 to E12.5, and its pituitary expression is entirely restricted to ACTH-producing cells.
Pan1 interacts physically with Pitx1.
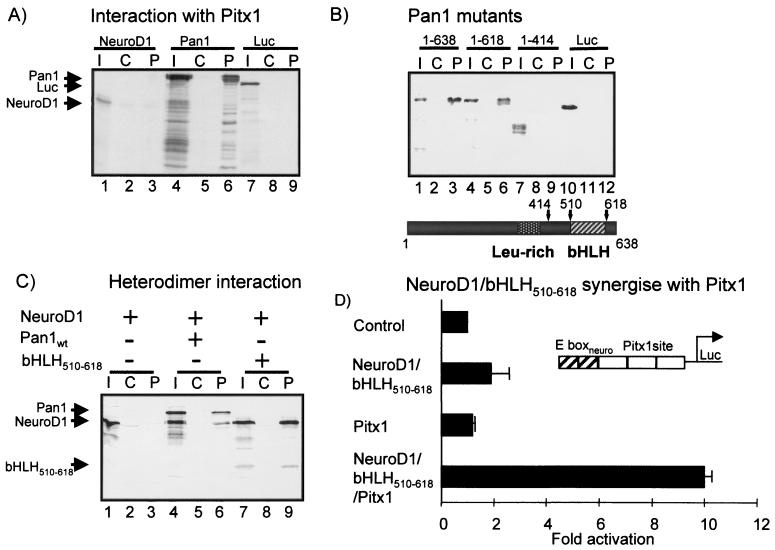
The activity of the POMC promoter E boxneuro⇒E boxubi mutant (Fig. 2A) suggested that ubiquitous bHLH factors could mediate the synergistic interaction with Pitx1. To test the possibility of a physical interaction between Pitx1 and Pan1 or NeuroD1, pull-down assays were performed using purified MBP-Pitx1 and in vitro-produced [35S]methionine labeled NeuroD1 and Pan1 (Fig. 4A, lanes 1 and 4). In the presence of purified MBP-LacZ and MBP-Pitx1 (Fig. 4A, lanes 2 and 3), no interaction with NeuroD1 could be detected. However, Pan1 was retained by an MBP-Pitx1 column (about 20%) and not by an MBP-LacZ column (Fig. 4A, lanes 4 to 6). The demonstration of an interaction between Pan1 (but not NeuroD1) and Pitx1 is in accordance with the effect of the E box specificity mutation (Fig. 2A) and suggests that Pan1 mediates the synergistic interaction with Pitx1.
FIG. 4.
Direct interaction between Pan1 bHLH domain and Pitx1 homeodomain. (A) Pan1 but not NeuroD1 can interact with Pitx1 in vitro. A pull-down assay was set up using resin-bound control (C) and MBP-Pitx1 (P) together with 35S-labeled NeuroD1, Pan1, or luciferase (Luc) synthesized in vitro. The arrows point to the main protein products. Input (I) is 20% of the amount used in lanes C and P. (B) Deletion of Pan1 bHLH domain impairs Pitx1 interaction. Two C-terminal deletions of Pan1 were produced. One removed the last 20 amino acids (1–618), and another deleted 224 amino acids (1–414), which removed the bHLH domain. Pull-down was done as in panel A except that the input was 10%. (C) NeuroD1-Pan1 heterodimers interact with Pitx1 through Pan1 bHLH510–618. NeuroD1 (N1) and Pan1 (P1) or its bHLH domain (bHLH510–618) were cosynthesized in vitro. (D) bHLH510–618 is sufficient to form an active NeuroD1 heterodimer with conserved synergistic capacity with Pitx1 in transfected L cells.
bHLH domain of Pan1 is essential for Pitx1 interaction.
We next identified the Pan1 domain responsible for Pitx1 interaction by using two different Pan1 C-terminal truncations in the pull-down assay. The first is a 20-amino-acid deletion (mutant 1–618) which migrates slightly faster than the full-length Pan1 (1–638) (Fig. 4B, lanes 4 and 1), and the second mutation is a deletion of the bHLH domain (1 to 414) (Fig. 4B lane 7). The 1–618 mutant did not interact with MBP-LacZ (Fig. 4B, lane 5) but interacted with MBP-Pitx1 at the same level as full-length Pan1 (Fig. 4B, lanes 6 and 3). However, mutant 1–414, which had been deleted of its bHLH domain but still contained its leucine-rich region, did not have the capacity to interact with MBP-Pitx1 (Fig. 4B, lane 9). Thus, the last 204 amino acids that mainly constitute the bHLH domain of Pan1 are required to mediate Pitx1 interaction.
Heterodimers formed between NeuroD1 and Pan1 can interact with Pitx1.
We tested the capacity of NeuroD1-Pan1 heterodimers to interact with Pitx1. As shown above, NeuroD1 is not capable of interaction with Pitx1 (Fig. 4C, lanes 1 to 3). However, when NeuroD1 is cosynthesized with Pan1, the resulting heterodimers are retained by MBP-Pitx1 (Fig. 4C, lanes 4 to 6). A mutant containing solely the Pan1 bHLH domain (amino acids 512 to 618) can form heterodimers with NeuroD1, and these heterodimers also interacted with Pitx1 (Fig. 4C, lanes 7 to 9). These data extend the mapping of the bHLH domain by C-terminal deletion (Fig. 4B) and indicate that a small bHLH polypeptide of 97 amino acids is sufficient for dimer formation with NeuroD1 and for interaction with Pitx1. The same bHLH polypeptide was tested for its ability to form active dimers and to synergize with Pitx1 in transfection experiments (Fig. 4D). By comparison to the data in Fig. 1B using the full-length Pan1, it can be concluded that the short Pan1 bHLH domain is entirely sufficient to form active heterodimers with NeuroD1 and to mediate the synergistic interaction with Pitx1.
Pitx1 HD is sufficient for Pan1 interaction.
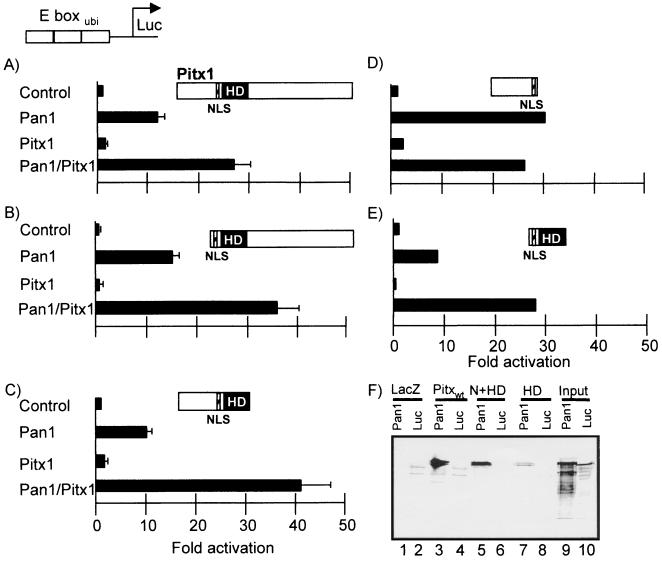
In order to define the region of Pitx1 involved in synergism with Pan1, a set of Pitx1 deletion mutants (65) were tested using the simplest reporter system available: the different Pitx1 mutants were cotransfected in L cells with a reporter containing trimers of the E boxubi upstream of the minimal POMC promoter. This reporter requires only Pan1 homodimers for activation and is not a target for Pitx1, and thus enhancement of transcriptional activity can be ascribed to an in vivo interaction between Pan1 homodimers and Pitx1 or its deletion mutants. Pan1 expression activated the E boxubi reporter about 12-fold, and Pitx1 on its own did not affect its activity (Fig. 5A). However, when both were present, activity was stimulated 28-fold (Fig. 5A, Pan1/Pitx1). In agreement with previous data, this suggests that Pitx1 interacts with Pan1 in vivo. Deletion of the N- or C-terminal domain of Pitx1 (Fig. 5B and C) did not affect its ability to enhance Pan1-dependent activity. However, the N-terminal domain on its own did not show synergism with Pan1 (Fig. 5D), but the HD did (Fig. 5E). In order to verify whether the HD is also sufficient for in vitro interaction with Pan1, MBP-Pitx1 deletion mutants were produced and used in pull-down assays. Two truncated Pitx1 proteins containing the HD with or without the N terminus were fused to MBP and tested for Pan1 interaction. Both mutants were capable of in vitro interaction with Pan1 (Fig. 5F, lanes 5 and 7, respectively) but not with MBP-lacZ (lanes 6 and 8). Thus, the HD seems sufficient and essential for Pan1 interaction (Fig. 5F) and for transcriptional synergism with Pan1 (Fig. 5E). A similar transcriptional synergism was observed with another ubiquitous bHLH, ITF2 (data not shown).
FIG. 5.
Pitx1 HD can interact in vivo with Pan1. Transient transfections were performed in L cells using a luciferase reporter containing three copies of E boxubi. Transfections were done with mammalian expression vectors for the indicated cDNAs. Various deletion mutants of Pitx1 (A) were used, including an N-terminal deletion (B), a C-terminal deletion (C), the N terminus (D), and the HD (E). These mutants were previously shown to be expressed at similar levels (65). NLS, nuclear localization signal. The results are the average (± SEM) of at least two independent experiments performed in duplicate. (F) The Pitx1 HD is sufficient for interaction with Pan1. The following MBP fusion proteins were used for interaction with Pan1: LacZ as a control; MBP-Pitx1 (Pitxwt); and N+HD and HD are the N-terminal region and/or the Pitx1 HD fused to MBP.
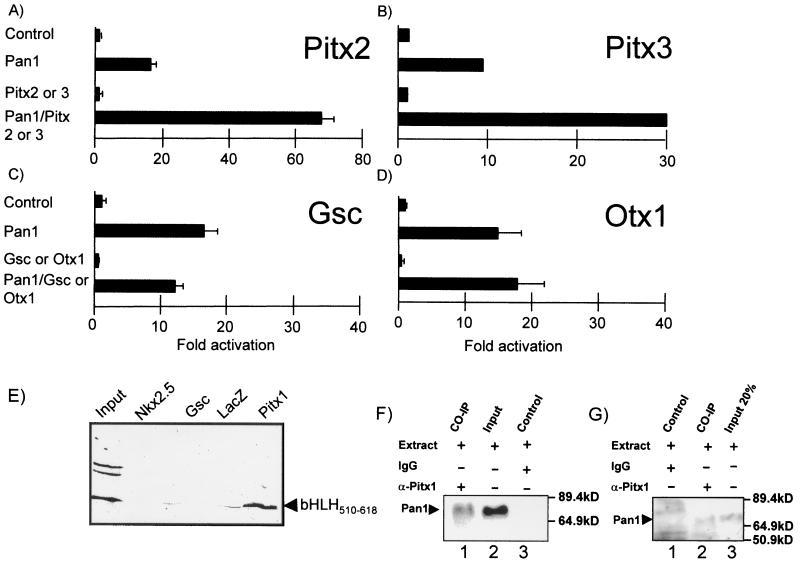
Pan1 interaction is restricted to the Pitx subfamily.
We tested whether other members of the bicoid-related family of HD could interact with Pan1. Compared to Pitx1, Pitx2 and Pitx3 have highly homologous HD, but Gsc and Otx1 are more distantly related. As for Pitx1, Pan1-dependent activity was enhanced by Pitx2 or Pitx3 (Fig. 6A and B). In contrast, Gsc and Otx1 were without effect when cotransfected with Pan1 (Fig. 6C and D). To correlate this specificity with in vitro interaction, pull-down assays were used to show that the small Pan1 bHLH domain (Fig. 4C) does not interact with MBP-Gsc or MBP-Nkx2.5 (another HD protein) (Fig. 6E), in accordance with in vivo experiments (Fig. 6C and data not shown). Thus, Pan1 can only interact with members of the Pitx subfamily. To confirm that Pitx1 and Pan1 can interact in vivo, nuclear extracts from COS cells overexpressing Pitx1 and Pan1 (Fig. 6F) or from normal AtT-20 cells (Fig. 6G) were subjected to coimmunoprecipitation. Western blot analysis of the immunoprecipitates revealed Pan1 (Fig. 6F, lane 1, and Fig. 6G, lane 2) in extracts immunoprecipitated with a Pitx1 antiserum (αPitx1) but not in control IgG-treated extracts (Fig. 6F, lane 3, and 6G, lane 1). Thus, Pan1 and Pitx1 interact as efficiently in vivo as in vitro.
FIG. 6.
Pan1 interaction is specific to the Pitx family. In experiments performed as described in the legend to Fig. 5, Pitx2 (A) and Pitx3 (B) show the same synergism as Pitx1 (Fig. 5A) but not goosecoid (Gsc, C) or Otx1 (D). Results are the average (± SEM) of at least two experiments performed in duplicate. (E) Gsc and Nkx2.5 homeoproteins do not interact with Pan1 in vitro. Pull-down assays were performed with the small in vitro-synthesized bHLH domain of Pan1 and purified MBP-Nkx2.5, MBP-Gsc, MBP-LacZ, and MBP-Pitx1. Input was 10% of the amount used. (F) An anti-Pitx1 antibody coimmunoprecipitates Pan1. Nuclear extracts were prepared from COS-1 cells transfected with Pitx1- and Pan1-expressing plasmids, and 10 μg of extracts was used for coimmunoprecipitation (CO-IP), input, and control samples. (G) Coimmunoprecipitation of Pan1 with Pitx1 from nuclear extracts of untreated AtT-20 cells. The experiment was performed as in panel F except that 200 μg of extracts was used for each sample.
DISCUSSION
We have demonstrated lineage-restricted expression of NeuroD1 in the pituitary; indeed, NeuroD1 is expressed only in corticotroph cells, and its expression begins just prior to POMC (Fig. 3). Thus, NeuroD1 is the only corticotroph-restricted factor known so far. This neurogenic bHLH has otherwise been implicated in neuronal differentiation and pancreas development (32, 39, 44, 45). We have presented evidence supporting a direct protein-protein interaction mechanism by which NeuroD1 heterodimers can take part in cell-specific transcription of pituitary POMC. This interaction between bHLH and HD transcription factors is generally relevant, since the direct interaction between these proteins involves the ubiquitous class A dimerization partner of NeuroD1. The role of NeuroD1 in this system is to ensure the specificity of E box recognition.
NeuroD1 and POMC transcription.
We have shown lineage-restricted pituitary expression of NeuroD1 in corticotroph cells with an onset at E12 that is consistent with the onset of POMC expression (Fig. 3). The transient presence of NeuroD1 protein in corticotroph cells between E12 and E16 of mouse development would be consistent with the involvement of other neurogenic bHLH in corticotroph cells before or after NeuroD1. Indeed, this might be analogous to the sequential involvement of neurogenins 1 and 2 and of NeuroD1 during neuronal differentiation (37). NeuroD1 is only one of the many neurogenic bHLH related to Drosophila atonal that are implicated in neuronal differentiation (31). Other factors of the NeuroD or neurogenin subfamily may also form heterodimers with class A ubiquitous bHLH and exhibit DNA-binding specificity similar to that of NeuroD1 heterodimers. Thus, different neurogenic bHLH could act sequentially as heterodimers on the E boxneuro of the POMC promoter. In such a model, the AtT-20 cells would be representative of the E12 to E16 developmental window during corticotroph differentiation, since all their E boxneuro binding activity contains NeuroD1 (52).
NeuroD1 heterodimers are essential for efficient transcription of the POMC gene in AtT-20 cells (52). Mutagenesis of their target site, the E boxneuro, has the same deleterious effect as deletion of the entire distal region of the promoter or mutagenesis of the Pitx1 binding site (Fig. 1E) or, for that matter, deletion of the central region of the promoter that contains the Pitx1 binding site (61, 62). Hence, the E boxneuro is an essential element of the POMC enhancer. The highly specific binding and activation of this target by NeuroD1 heterodimers (52) indicates that the role of NeuroD1 in promoter activation is to ensure selective recognition of E boxneuro. In vivo, transgenic promoter studies (Fig. 2) reveal that the role of the enhancer region containing E boxneuro is quantitatively much more important than suggested from transfection studies (Fig. 1E). However, NeuroD1 itself may not be required sensu strictu for transcription. Indeed, replacement of E boxneuro by an E boxubi sequence restored promoter activity, indicating that the requirement is for bHLH dimers rather than for NeuroD1 itself (Fig. 1E). This is consistent with the mechanism for interaction between bHLH and Pitx described in the present work, but it also suggests that other bHLH with a DNA-binding specificity similar to that of NeuroD1 may substitute for NeuroD1, for example, later in development, after NeuroD1 protein is no longer detected.
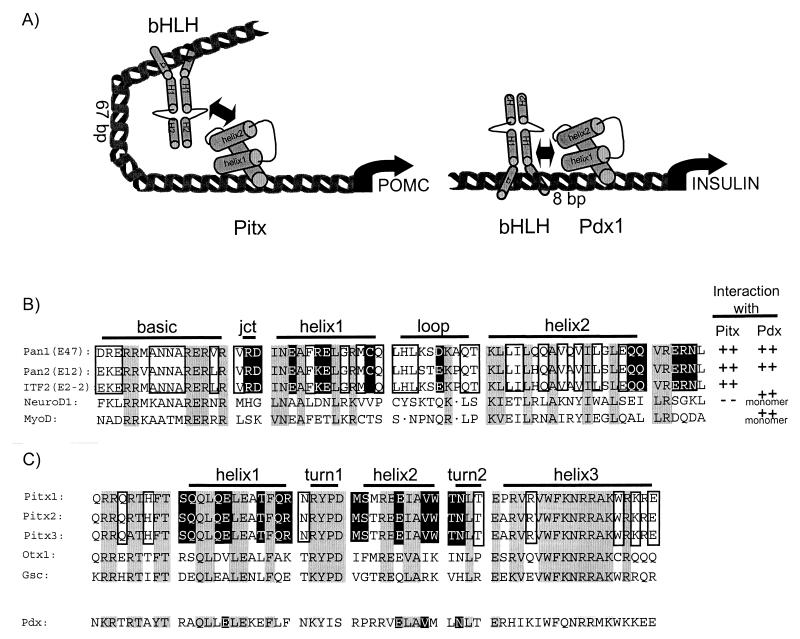
Within the native POMC promoter, the E boxneuro is separated from the Pitx binding site by 67 bp (62). This spacing suggests that the interaction between DNA-bound bHLH dimers and Pitx proteins is not like that of factors bound to neighboring DNA sites, which may interact with each other through side surfaces, as for example the Hox and Pbx homeoproteins (5, 7, 35, 50). The interaction between bHLH factors and Pdx1 identified in the insulin promoter is another example of proteins bound to adjacent DNA sites (46). This interaction results in weakly cooperative DNA binding, as illustrated in Fig. 7A. In contrast, the distance between E boxneuro and the Pitx binding site on the POMC promoter suggests that interaction between these proteins occurs through surfaces that are opposite to the DNA-interacting surface, as suggested by the model in Fig. 7A. In this model, the bending of promoter DNA required to bring Pitx and bHLH proteins together results in very different contact surfaces between the proteins compared to their respective orientations when bound to adjacent sites on DNA. Differences in bHLH-HD interaction on the POMC and insulin promoters are further highlighted by the unique specificity of each interaction, as discussed below.
FIG. 7.
Specificity of interactions between Pitx HD and bHLH dimers. (A) Schematic drawing representing the Pitx HD and bHLH dimers bound to their respective sites, which are separated by 67 bp in the POMC promoter. In view of the distance between these sites, it is likely that promoter DNA is bent in order to allow interaction between these proteins. In contrast, bHLH factors also interact with the HD of another factor, Pdx1, on the insulin promoter. In this case, the binding sites for these two factors are almost adjacent (separated by 8 bp). (B) Alignment of the bHLH domains of class A bHLH that interact with Pitx and/or Pdx1 and of NeuroD1 and MyoD1, which were shown to interact with Pdx1 but not with Pitx1 (NeuroD1). The protein interaction data are summarized on the left of the sequence alignment; the Pitx data are from this study, and the Pdx data are from Ohneda et al. (46). Pdx1 is inferred to interact in vitro with monomers of NeuroD1 and MyoD, since these class B bHLH are not thought to form homodimers (22, 45). Pitx1 does not interact in vitro with NeuroD1 but will interact with heterodimers that contain NeuroD1 and the Pan1 bHLH domain (Fig. 4C). In the sequence alignment, amino acid residues that are conserved in all five bHLH domains are shown in gray shading. Boxed residues are specific to the class A bHLH and are involved in maintenance of bHLH tertiary structure or dimerization. Residues that face towards the outside of the bHLH domain and that constitute the best candidates for Pitx interaction are shown in reverse type (white on black). This analysis was based on modeling the various bHLH sequences using the X-ray structure of the E47 bHLH as a reference (12). (C) Homology between the HD of the bicoid-related HD proteins and Pdx1. Shading is used as in panel B to indicate conserved residues between all HD (gray shading), conserved Pitx residues that are involved in maintenance of tertiary structure (boxes), and residues that are specific to the Pitx HD and that are accessible on the surface of the HD for protein-protein interaction (reverse type). This analysis was based on the X-ray structure of the Antennapedia HD-DNA complex (13).
We have documented the Pan1-Pitx1 physical interaction in different ways: the in vitro pull-down assay was useful to map interaction domains, and this analysis correlated well with transcriptional synergism (Fig. 4). In addition, we could show transcriptional synergism even in the absence of DNA binding by Pitx1 (Fig. 1D and 5) but not the reverse, i.e., in the absence of DNA binding by bHLH dimers (Fig. 1C). Most importantly, we showed by coimmunoprecipitation that Pitx1 and Pan1 are present in a common complex in vivo whether these proteins are overexpressed in heterologous cells (Fig. 6F) or not, as in POMC-expressing AtT-20 cells (Fig. 6G). We have previously shown that transcription of the POMC enhancer depends on multiple regulatory elements, including the E boxneuro and Pitx binding site (61, 62) (Fig. 1E). The cognate transcription factors appear to be present together in a multiprotein complex (Fig. 6G) that may associate with the POMC promoter (Fig. 7A); this protein complex may also contain other POMC transcription factors.
Specificity of bHLH-HD interactions.
The interaction between bHLH and HD factors appears to be quite specific with regard to the HD. The HD of Pitx1, Pitx2, and Pitx3 are nearly identical to each other, and it is not surprising that all three should interact with Pan1. However, their closest relatives, the Otx and Gsc HD, do not share this property (Fig. 6 and 7C) despite the fact that they bind the same target DNA sequence (8, 9). The HD of another subfamily, Nkx2.5, was also found to be incapable of interaction with Pan1. Although no other HD has significant homology with the Pitx HD, it cannot be excluded that other members of the HD family might interact with Pan1. Indeed, the pancreas-specific HD protein Pdx1 exerts transcriptional synergism with Pan1 on the rat insulin promoter (48), and these proteins interact directly in vitro (46). However, the bHLH-HD interaction between Pan1 and Pdx1 appears to be different from that of Pan1 with Pitx1. Indeed, whereas Pdx1 can interact just as well in vitro with the class A bHLH Pan1 (E47) as with class B bHLH like NeuroD1, MyoD, and Mash1 but not Tal1 (46), Pitx1 only interacts with the class A bHLH and not with NeuroD1 (Fig. 4A and C). Furthermore, this specificity highlights another more important difference: since NeuroD1 does not form homodimers (45), its in vitro binding with Pdx1 suggests that the Pdx1-bHLH interaction requires only a bHLH monomer. In striking contrast, NeuroD1 on its own does not interact with Pitx1 (Fig. 4A and C) but will form part of a Pitx1 complex when heterodimerized with Pan1 or its bHLH domain (Fig. 4C); these observations suggest that, in contrast to Pdx1, Pitx homeoproteins may interact only with bHLH dimers, although definite proof of this would require experiments with dimerization-deficient mutants of a bHLH, like Pan1, that interacts with both Pdx1 and Pitx1.
The bHLH domains involved in interaction with Pitx and/or Pdx proteins have been aligned to reveal similarities and differences (Fig. 7B). This comparison shows few amino acid residues (shaded in gray) that are conserved between class A (Pan1, Pan2, E47, E12, ITF2, and E2-2) and class B bHLH (NeuroD1 and MyoD); a subset of these are involved in maintaining the integrity of the bHLH tertiary structure (12), and others could be involved in the interaction with Pdx1. Class A bHLH residues that are unique to this class and that may interact with Pitx proteins were subdivided into two groups, those that are involved in tertiary structure or dimer formation (boxed) and that are less likely to be available for Pitx interaction, and those that face the outside surface of the bHLH domain (reverse type, white on black) and that constitute the best candidates for Pitx interaction. These later residues are mostly found in helix 1 and after helix 2 (Fig. 7B). When a similar analysis was performed on related HDs that do and do not interact with class A bHLH, a subset of residues primarily found in helixes 1 and 2 of the Pitx HD were identified (Fig. 7C). Only four of those are conserved in Pdx1, again highlighting the differences between the Pitx and Pdx HDs. Thus, in considering amino acid homologies in these domains from the perspective of the bHLH domain as well as the HD domain, the specificity of their interaction is much more stringent for Pitx and class A bHLH than for Pdx and both class A and B bHLH.
bHLH factors were previously shown to interact with other transcription factors, some of which have HD domains. However, none of these interactions were found to involve the HD itself. Indeed, bHLH factors interact with HD factors lmx1.1 (lmx1a) and lmx1.2 (lmx1b) and with LIM-only proteins like LMO2 (23, 29, 66). In both cases, the interaction is between the LIM domain, a zinc finger-like structure, and the bHLH domain. Recently, an in vitro interaction between myogenic bHLH heterodimers and HD proteins of the Pbx/Meis family was reported (24). However, this interaction is very different from the bHLH-Pitx interaction reported here. The Pbx/Meis factors interact with an amino acid motif that is present N-terminal of the myogenic factor bHLH domain; this motif is similar to the tryptophan-containing motif recognized by Pbx/Meis in Hox HD proteins (5, 6, 35, 50). In summary, the interaction between bHLH and Pitx HD proteins is unique in that (i) it is specific to class A bHLH, (ii) when class B bHLH are involved, their recruitment is through dimerization with Pitx-interacting class A bHLH, and (iii) it is restricted to the Pitx subfamily.
Developmental code involving HD and bHLH factors.
HD genes have been involved in patterning, specification of segmental identity, and sometimes cellular differentiation. Numerous roles have been assigned to Pitx factors, including hind limb specification (Pitx1), left-right asymmetry (Pitx2), neuronal differentiation (Pitx3), eye development (Pitx2 and Pitx3), and craniofacial development (Pitx1 and Pitx2). Even though tissue-restricted bHLH factors have not been implicated in all these systems, ubiquitous bHLH factors like Pan1 are expressed in all these structures, and therefore a Pan1-Pitx interaction would be possible in many of these tissues. However, coexpression of these genes is not a sufficient criterion for significant physical interaction. Indeed, promoter context is likely to play a critical role in cross-talk between these transcription factors. We have preliminary data to support the hypothesis of a bHLH-Pitx interaction involving myogenic bHLH. These observations suggest that interactions between Pitx and bHLH factors might be of general relevance. In muscle, both Pitx1 and Pitx2 can be expressed, as shown for Pitx1 in muscles of the posterior half of the body (27) and for Pitx2 in myotomes, myoblasts, and muscles (33, 51). Hence, interactions between Pitx factors that may be involved in muscle patterning (33, 34) and myogenic bHLH that are essential for muscle differentiation (40) may represent one mechanism by which molecular cross-talk is achieved between these two families of developmental regulators. Similarly, neuronal development might involve interactions between Pitx and bHLH factors at the level of common target genes.
ACKNOWLEDGMENTS
We are grateful to Ming Tsai, François Guillemot, and Harold Weintraub for providing the BETA2, NeuroD1, and MyoD plasmids, respectively. Daniel Durocher was helpful in pull-down experiments. The efficient secretarial assistance of Lise Laroche was much appreciated.
G.P. is a recipient of a Terry Fox Studentship of the National Cancer Institute of Canada. This work was funded by the National Cancer Institute of Canada, supported with funds provided by the Canadian Cancer Society.
REFERENCES
- 1.Apelqvist A, Sommer L, Beatus P, Anderson D J, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 2.Aplan P D, Nakahara K, Orkin S H, Kirsch I R. The SCL gene product: a positive regulator of erythroid differentiation. EMBO J. 1992;11:4073–4081. doi: 10.1002/j.1460-2075.1992.tb05500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 4.Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe L A, Nowotschin S, Viebahn C, Haffter P, Kuehn M R, Blum M. The homeobox gene Ptx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development. 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- 5.Chan S K, Jaffe L, Capovilla M, Botas J, Mann R S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 6.Chang C P, Brocchieri L, Shen W F, Largman C, Cleary M L. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C P, Shen W F, Rozenfeld S, Lawrence H J, Largman C, Cleary M L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 8.Drouin J, Lamolet B, Lamonerie T, Lanctôt C, Tremblay J J. The Ptx family of homeodomain transcription factors during pituitary development. Mol Cell Endocrinol. 1998;140:31–36. doi: 10.1016/s0303-7207(98)00026-4. [DOI] [PubMed] [Google Scholar]
- 9.Drouin J, Lanctôt C, Tremblay J J. La famille Ptx des facteurs de transcription à homéodomaine. Medecine/Sciences. 1998;14:335–339. [Google Scholar]
- 10.Duboule D. Guidebook to the homeobox genes. Oxford, U.K: Sambrook & Tooze Publications; 1994. [Google Scholar]
- 11.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellenberger T, Fass D, Arnaud M, Harrison S C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 13.Fraenkel E, Pabo C O. Comparison of X-ray and NMR structures for the Antennapedia homeodomain-DNA complex. Nat Struct Biol. 1998;5:692–697. doi: 10.1038/1382. [DOI] [PubMed] [Google Scholar]
- 14.Gage P J, Camper S A. Pituitary homeobox 2, a novel member of the bicoid-related family of homeobox genes, is a potential regulator of anterior structure formation. Hum Mol Genet. 1997;6:457–464. doi: 10.1093/hmg/6.3.457. [DOI] [PubMed] [Google Scholar]
- 15.Gage P J, Suh H, Camper S A. The bicoid-related Pitx gene family in development. Mamm Genome. 1999;10:197–200. doi: 10.1007/s003359900970. [DOI] [PubMed] [Google Scholar]
- 16.Gradwohl G, Dierich A, LeMeur M, Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham A, McGonnell I. Limb development: farewell to arms. Curr Biol. 1999;9:R368–R370. doi: 10.1016/s0960-9822(99)80229-2. [DOI] [PubMed] [Google Scholar]
- 18.Guillemot F, Joyner A L. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech Dev. 1993;42:171–185. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- 19.Guillemot F, Lo L C, Johnson J E, Auerbach A, Anderson D J, Joyner A L. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 20.Hanes S D, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 21.Harvey R P. Links in the left/right axial pathway. Cell. 1998;94:273–276. doi: 10.1016/s0092-8674(00)81468-3. [DOI] [PubMed] [Google Scholar]
- 22.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson J D, Zhang W, Rudnick A, Rutter W J, German M S. Transcriptional synergy between LIM-homeodomain proteins and basic helix-loop-helix proteins: the LIM2 domain determines specificity. Mol Cell Biol. 1997;17:3488–3496. doi: 10.1128/mcb.17.7.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoepfler P S, Bergstrom D A, Uetsuki T, Dac-Korytko I, Sun Y H, Wright W E, Tapscott S J, Kamps M P. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx-Meis1/Prep1. Nucleic Acids Res. 1999;27:3752–3761. doi: 10.1093/nar/27.18.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 26.Lamonerie T, Tremblay J J, Lanctôt C, Therrien M, Gauthier Y, Drouin J. PTX1, a bicoid-related homeobox transcription factor involved in transcription of pro-opiomelanocortin (POMC) gene. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 27.Lanctôt C, Lamolet B, Drouin J. The bicoid-related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development. 1997;124:2807–2817. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- 28.Lanctôt C, Moreau A, Chamberland M, Tremblay M L, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- 29.Larson R C, Lavenir I, Larson T A, Baer R, Warren A J, Wadman I, Nottage K, Rabbitts T H. Protein dimerization between LMO2 (RBTN2) and TAL1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 1996;15:1021–1027. [PMC free article] [PubMed] [Google Scholar]
- 30.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 31.Lee J E. Basic helix-loop-helix genes in neural development. Curr Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 32.Lee J E, Hollenberg S M, Snider L, Turner D L, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 33.Logan M, Pagán-Westphal S M, Smith D M, Paganessi L, Tabin C J. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- 34.Logan M, Tabin C J. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science. 1999;283:1736–1739. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- 35.Lu Q, Kamps M P. Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing Hox proteins: proposal for a model of a Pbx1-Hox-DNA complex. Mol Cell Biol. 1996;16:1632–1640. doi: 10.1128/mcb.16.4.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma P C, Rould M A, Weintraub H, Pabo C O. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 37.Ma Q, Fode C, Guillemot F, Anderson D. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Q F, Kintner C, Anderson D J. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 39.Miyata T, Maeda T, Lee J E. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molkentin J D, Olson E N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mucchielli M L, Martinez S, Pattyn A, Goridis C, Brunet J F. Otlx2, an Otx-related homeobox gene expressed in the pituitary gland and in a restricted pattern in the forebrain. Mol Cell Neurosci. 1996;8:258–271. doi: 10.1006/mcne.1996.0062. [DOI] [PubMed] [Google Scholar]
- 42.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 43.Mutoh H, Fung B P, Naya F J, Tsai M J, Nishitani J, Leiter A B. The basic helix-loop-helix transcription factor beta2/neurod is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci USA. 1997;94:3560–3564. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naya F J, Huang H P, Qiu Y, Mutoh H, DeMayo F J, Leiter A B, Tsai M J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naya F J, Stellrecht C M M, Tsai M J. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 46.Ohneda K, Mirmira R G, Wang J H, Johnson J D, German M S. The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol Cell Biol. 2000;20:900–911. doi: 10.1128/mcb.20.3.900-911.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olson E N, Klein W H. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 48.Peers B, Leonard J, Sharma S, Teitelman G, Montminy M R. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol Endocrinol. 1994;8:1798–1806. doi: 10.1210/mend.8.12.7708065. [DOI] [PubMed] [Google Scholar]
- 49.Peshavaria M, Henderson E, Sharma A, Wright C V, Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol Cell Biol. 1997;17:3987–3996. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phelan M L, Rambaldi I, Featherstone M S. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piedra M E, Icardo J M, Albajar M, Rodriguez-Rey J C, Ros M A. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell. 1998;94:319–324. doi: 10.1016/s0092-8674(00)81475-0. [DOI] [PubMed] [Google Scholar]
- 52.Poulin G, Turgeon B, Drouin J. NeuroD1/BETA2 contributes to cell-specific transcription of the POMC gene. Mol Cell Biol. 1997;17:6673–6682. doi: 10.1128/mcb.17.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raphael S J, Apel R L, Asa S L. Brief report: detection of high-molecular-weight cytokeratins in neoplastic and non-neoplastic thyroid tumors using microwave antigen retrieval. Modern Pathol. 1995;8:870–872. [PubMed] [Google Scholar]
- 54.Rawls A, Olson E N. MyoD meets its maker. Cell. 1997;89:5–8. doi: 10.1016/s0092-8674(00)80175-0. [DOI] [PubMed] [Google Scholar]
- 55.Ryan A K, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura I, Tsukui T, de la Peña J, Sabbagh W, Greenwald J, Choe S, Norris D P, Robertson E J, Evans R M, Rosenfeld M G, Izpisúa Belmonte J C. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- 56.Semina E V, Reiter R, Leysens N J, Alward W L, Small K W, Datson N A, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel B U, Carey J C, Murray J C. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 57.Shivdasani R A, Mayer E L, Orkin S H. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 58.Sun X-H, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- 59.Szeto D P, Rodriguez-Esteban C, Ryan A K, O'Connell S, Liu R, Kioussi C, Gleiberman A S, Izpisua-Belmonte J C, Rosenfeld M G. Role of the bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szeto D P, Ryan A K, O'Connell S M, Rosenfeld M G. P-OTX: a PIT-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Therrien M, Drouin J. Pituitary pro-opiomelanocortin gene expression requires synergistic interactions of several regulatory elements. Mol Cell Biol. 1991;11:3492–3503. doi: 10.1128/mcb.11.7.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Therrien M, Drouin J. Cell-specific helix-loop-helix factor required for pituitary expression of the pro-opiomelanocortin gene. Mol Cell Biol. 1993;13:2342–2353. doi: 10.1128/mcb.13.4.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 64.Tremblay J J, Lanctôt C, Drouin J. The pan-pituitary activator of transcription, Ptx-1 (pituitary homeobox1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. MolEndocrinol. 1998;12:428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 65.Tremblay J J, Marcil A, Gauthier Y, Drouin J. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J. 1999;18:3431–3441. doi: 10.1093/emboj/18.12.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wadman I A, Osada H, Grutz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weatherbee S D, Carroll S B. Selector genes and limb identity in arthropods and vertebrates. Cell. 1999;97:283–286. doi: 10.1016/s0092-8674(00)80737-0. [DOI] [PubMed] [Google Scholar]
- 68.Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina E V, Murray J C, Hamada H, Noji S. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- 69.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]