Abstract
Objectives
Develop an individualised prognostic risk prediction tool for predicting the probability of adverse COVID-19 outcomes in patients with inflammatory bowel disease (IBD).
Design and setting
This study developed and validated prognostic penalised logistic regression models using reports to the international Surveillance Epidemiology of Coronavirus Under Research Exclusion for Inflammatory Bowel Disease voluntary registry from March to October 2020. Model development was done using a training data set (85% of cases reported 13 March–15 September 2020), and model validation was conducted using a test data set (the remaining 15% of cases plus all cases reported 16 September–20 October 2020).
Participants
We included 2709 cases from 59 countries (mean age 41.2 years (SD 18), 50.2% male). All submitted cases after removing duplicates were included.
Primary and secondary outcome measures
COVID-19 related: (1) Hospitalisation+: composite outcome of hospitalisation, ICU admission, mechanical ventilation or death; (2) Intensive Care Unit+ (ICU+): composite outcome of ICU admission, mechanical ventilation or death; (3) Death. We assessed the resulting models’ discrimination using the area under the curve of the receiver operator characteristic curves and reported the corresponding 95% CIs.
Results
Of the submitted cases, a total of 633 (24%) were hospitalised, 137 (5%) were admitted to the ICU or intubated and 69 (3%) died. 2009 patients comprised the training set and 700 the test set. The models demonstrated excellent discrimination, with a test set area under the curve (95% CI) of 0.79 (0.75 to 0.83) for Hospitalisation+, 0.88 (0.82 to 0.95) for ICU+ and 0.94 (0.89 to 0.99) for Death. Age, comorbidities, corticosteroid use and male gender were associated with a higher risk of death, while the use of biological therapies was associated with a lower risk.
Conclusions
Prognostic models can effectively predict who is at higher risk for COVID-19-related adverse outcomes in a population of patients with IBD. A free online risk calculator (https://covidibd.org/covid-19-risk-calculator/) is available for healthcare providers to facilitate discussion of risks due to COVID-19 with patients with IBD.
Keywords: COVID-19, inflammatory bowel disease, statistics & research methods
Strengths and limitations of this study.
Our study includes data from an international cohort with a wide range of ages including paediatric patients.
The use of regularised regression methods for prediction allowed us to consider a wide range of potential predictors in a statistically sound way.
The data for this study comes from a voluntary registry, and the differences between the registry population and the general population of patients with inflammatory bowel disease (IBD) are unknown.
The models were validated using a test data set from the same registry and have not yet been validated in an external cohort of individuals with IBD.
Our methods are associational, not causal—when using the online risk calculator, healthcare providers should not use it to answer ‘what-if’ questions (eg, how an individual’s risk would change if they altered the medications they were taking) which are inherently causal questions.
Introduction
Since the onset of the COVID-19 pandemic, almost 50 million cases have been reported globally. Many countries, including the USA, are reporting record numbers of new cases as of November 2020.1 While the majority of cases are mild, patients with at least one comorbidity are at higher risk of adverse outcomes, including hospitalisation, respiratory failure or death.2 3 Risk calculators can facilitate shared decision making between patients and healthcare providers,4 and such tools have been created to predict death due to COVID-19 in US patients 65 years and older,5 to determine hospitalisation risk6 and to guide early vaccine allocation.
Patients with inflammatory bowel disease (IBD) are prescribed immunosuppressive medications such as corticosteroids, immunomodulators, biological therapies and Janus-kinase inhibitors, which are linked with a higher risk of viral infection.7 8 Demographics, comorbidities, medication use, geographic region and other factors may increase the risk for COVID-19-related complications among patients with IBD.9 10 To help healthcare providers and patients navigate these myriad potential risk factors, we developed and validated penalised multivariable logistic regression models for predicting the probability of hospitalisation, intensive care unit (ICU) admission and death due to COVID-19 in patients with IBD. We used an international registry of 2709 patients with IBD with COVID-19 from 59 countries. We also developed a free, publicly available personalised risk calculator using the final models that is available online (https://covidibd.org/covid-19-risk-calculator/). Reporting follows Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis guidelines.11
Methods
Source of data
The Surveillance Epidemiology of Coronavirus Under Research Exclusion for Inflammatory Bowel Disease (SECURE-IBD) database (www.covidibd.org) is an international registry to study outcomes of COVID-19 in paediatric and adult patients with IBD.12 SECURE-IBD is a voluntary registry with ongoing data collection where healthcare providers can report cases of COVID-19 in patients with IBD, confirmed by PCR or antibody testing. Healthcare providers are instructed to report cases of severe outcomes after a minimum of 7 days from the onset of symptoms and after a sufficient time has passed to observe the disease course through the resolution of acute illness or death. In the event that a patient’s status changed after submission, reporters are instructed to re-report and contact the research team. Reporters were not explicitly informed of what data could be used as predictors or outcomes, but being a voluntary registry, reporters were not blinded. A fuller account of the data collection is given in Brenner et al.13
Patient
Patient and professional organisations representing many countries were engaged in planning the registry and the data collection, promoting the registry and disseminating results from studies using the SECURE-IBD database. A list of the organisations involved is available in online supplemental table 1.
bmjopen-2021-049740supp001.pdf (1.5MB, pdf)
Participants
We included all patients reported to the registry from 13 March 2020, the data collection start date, through 20 October 2020. For model development, a training sample consisting of 85% of the entire surveillance data set available as of 15 September 2020 was used. The random split was done using stratified random sampling based on an ordinal version of the outcome. The test data set consisted of the remaining 15% of the data available on 15 September, plus all of the additional cases reported to the registry between 16 September 2020 and 20 October 2020. We added the entirety of the last month of data to the test data set in order to provide a more honest assessment of our model’s performance in an environment that is changing over time.
We reported means and SD for continuous variables, counts for categorical variables and proportions for binary variables. We reported the missing data for all variables. We did not include p values in our descriptive tables following the Strengthening the Reporting of Observational Studies in Epidemiology guidelines.14
Outcomes
We examined three primary outcomes: (1) hospitalisation or death (Hospitalisation+), (2) ICU admission, mechanical ventilation or death (ICU+) and (3) death due to COVID-19 related causes (Death). Patients may experience multiple outcomes. All outcomes were reported by the patient’s healthcare provider at the time of the case report.
Predictors
As our aim was to create models and a risk stratification tool intended to allow physicians to inform patients of their risk before presenting with COVID-19, we restricted our attention to predictors that would be available during a routine consultation. COVID-19 presenting symptoms and information about the COVID-19 treatment received were therefore not included in this analysis. All predictors were reported by the patients’ healthcare provider.
A full description of the predictors is available in online supplemental table 2. Demographic predictors included age, country of residence, state of residence (for US cases), gender, race and ethnicity. Racial indicators included white, black and Asian. American Indian and Pacific Islander indicators were excluded due to low prevalence. Multi-racial patients belong to multiple categories. As only one patient had reported a gender other than male or female, only two genders were considered in the analysis. Due to the nature of reporting, ethnicity, gender and race should be interpreted as provider-perceived race and gender. Assessing race and ethnicity is important for identifying potential health inequities in COVID-19 related outcomes. For cases from US states with very low prevalence in the registry, a more general geographic predictor (census region or census division) was used in place of the state itself. Clinical predictors included height, weight, body mass index (BMI, study derived), IBD diagnosis (Crohn’s disease, ulcerative colitis or IBD unspecified) and IBD disease activity as defined by physician global assessment. We included indicators for the following a priori defined medication classes: biologicals (including antitumour necrosis factor (anti-TNF), anti-interleukin 12 (anti-IL-2) and anti-integrin agents), 5-aminosalicylates/sulfasalazine, immunomodulators (6MP, azathioprine, methotrexate), corticosteroids (prednisone, budesonide and other oral/parenteral steroids) and Janus kinase inhibitors (tofacitinib). We also included indicators for subclasses of biologicals (eg, anti-TNF) at the time of COVID-19 diagnosis. Additionally, dosage information was included for prednisone, 6-mercaptopurine and azathioprine.
For categorical (including binary) predictors without a meaningful reference level, all levels were included in the model. Quadratic terms were considered for all continuous covariates. Interactions were considered based on a combination of subject matter expert advice and a minimum threshold of thirty observations for every cell for interactions involving two binary predictors.
Missing data
Multiple imputation of the covariates and outcomes was performed using multivariate imputation by chained equations to address missing data.15 16 A total of 30 imputed data sets were created. Imputation was performed separately on the training and test data to prevent inducing dependence between the training and the test data through the imputation models. For transformed variables that are derived from other covariates (eg, BMI from height and weight), we imputed the missing root variables and then created the transformed variable to ensure that the relationship between the transformed variable and its inputs was preserved.
Table 1 includes the level of missing data in each of the covariates included in the analysis. Medication variables, clinical descriptions of disease and severity, location, age, and gender all had very low levels of missingness, ranging from 0% to under 5%. There were three covariates with a moderate amount of missing data—height, weight and ethnicity were missing in approximately 20% of patients.
Table 1.
Main characteristics of COVID-19 inflammatory bowel disease patients in the study*
| Characteristics | Training data (n=2009) | Test data (n=700) | Overall (n=2709) |
| Age, mean (SD), years | 42.2 (18.2) | 38.7 (17.4) | 41.2 (18.0) |
| Gender, n (%) | |||
| Female | 982 (48.9%) | 344 (49.1%) | 1326 (48.9%) |
| Male | 998 (49.7%) | 341 (48.7%) | 1339 (49.4%) |
| Other | 1 (0.0%) | 0 (0.0%) | 1 (0.0%) |
| Asian†, n (%) | 112 (5.6%) | 38 (5.4%) | 150 (5.5%) |
| Black†, n (%) | 138 (6.9%) | 39 (5.6%) | 177 (6.5%) |
| White†, n (%) | 1603 (79.8%) | 547 (78.1%) | 2150 (79.4%) |
| Hispanic/Latino, n (%) | 350 (17.4%) | 115 (16.4%) | 465 (17.2%) |
| Missing | 375 (18.7%) | 120 (17.1%) | 495 (18.3%) |
| BMI, mean (SD) | 26.0 (6.50) | 25.2 (6.26) | 25.8 (6.44) |
| Missing | 398 (19.8%) | 106 (15.1%) | 504 (18.6%) |
| Current smoker, n (%) | 61 (3.0%) | 25 (3.6%) | 86 (3.2%) |
| Disease type, n (%) | |||
| Crohn’s disease | 1115 (55.5%) | 401 (57.3%) | 1516 (56.0%) |
| Ulcerative colitis | 854 (42.5%) | 278 (39.7%) | 1132 (41.8%) |
| Cardiovascular disease, n (%) | 133 (6.6%) | 43 (6.1%) | 176 (6.5%) |
| Diabetes, n (%) | 117 (5.8%) | 30 (4.3%) | 147 (5.4%) |
| Hypertension, n (%) | 243 (12.1%) | 67 (9.6%) | 310 (11.4%) |
| Count of comorbidities, mean (SD) | 0.544 (0.938) | 0.479 (0.879) | 0.527 (0.923) |
| Biological therapy, n (%) | 1203 (59.9%) | 437 (62.4%) | 1640 (60.5%) |
| Tumour necrosis factor inhibitor, n (%) | 796 (39.6%) | 313 (44.7%) | 1109 (40.9%) |
| Anti-integrin, n (%) | 213 (10.6%) | 72 (10.3%) | 285 (10.5%) |
| IL-12/23 inhibitor, n (%) | 187 (9.3%) | 47 (6.7%) | 234 (8.6%) |
| 5-Aminosalicylates, n (%) | 636 (31.7%) | 200 (28.6%) | 836 (30.9%) |
| Sulfasalazine, n (%) | 64 (3.2%) | 24 (3.4%) | 88 (3.2%) |
| Mesalamine, n (%) | 561 (27.9%) | 175 (25.0%) | 736 (27.2%) |
| Immunomodulators, n (%) | 459 (22.8%) | 140 (20.0%) | 599 (22.1%) |
| Methotrexate, n (%) | 81 (4.0%) | 24 (3.4%) | 105 (3.9%) |
| Azathioprine or 6-mercaptopurine, n (%) | 367 (18.3%) | 110 (15.7%) | 477 (17.6%) |
| Corticosteroids, n (%) | 209 (10.4%) | 65 (9.3%) | 274 (10.1%) |
| Budesonide, n (%) | 59 (2.9%) | 17 (2.4%) | 76 (2.8%) |
| Oral or parenteral steroids, n (%) | 154 (7.7%) | 49 (7.0%) | 203 (7.5%) |
| Janus kinase inhibitors (tofacitinib), n (%) | 30 (1.5%) | 9 (1.3%) | 39 (1.4%) |
| Hospitalisation+, n (%) | 499 (24.8%) | 138 (19.7%) | 637 (23.5%) |
| Missing | 37 (1.8%) | 17 (2.4%) | 54 (2.0%) |
| ICU+, n (%) | 133 (6.6%) | 36 (5.1%) | 169 (6.2%) |
| Missing | 49 (2.4%) | 22 (3.1%) | 71 (2.6%) |
| Death, n (%) | 57 (2.8%) | 12 (1.7%) | 69 (2.5%) |
| Missing | 33 (1.6%) | 14 (2.0%) | 47 (1.7%) |
*Missingness is only reported for predictors with >5% missing values, and for all outcomes. Only comorbidities with an overall prevalence above 5% are shown in this table. For a complete list of characteristics and number and percentage of missing values, see online supplemental table 3.
†Individuals can be assigned to more than one physician-reported racial group (white, black, Asian).
BMI, body mass index; ICU, intensive care unit; IL, interleukin.
Statistical analysis
The 10-fold cross-validation deviance averaged across each of the imputed data sets was used to decide between the least absolute shrinkage and selection operator (LASSO), ridge or elastic net penalties and to choose the value of the regularisation parameter.17 18 Separate logistic regression models were fit for each of the outcomes. Smoothing splines for continuous covariates,19 a multinomial model, Group LASSO20 and Sparse Group LASSO,21 were all investigated as potential methods, but the performance improvement in terms of the cross-validation deviance was not sufficient to justify the additional complexity and computational time. The non-parametric resampling bootstrap was used to generate 1000 samples, and for each bootstrap sample, the same sampled participants were used across the 30 imputed data sets.22 A total of 30 000 (30×1000) fitted models were created. Predicted probabilities were created by averaging the predictions from all models. We used the sample mean of the bootstrap distribution of the predicted probabilities for the final predicted probability and the percentiles of the bootstrap distribution to find the 90% CI for the risk estimate. Risk groups were not created.
To assess the performance of the resulting predictions, we created receiver operator characteristic (ROC) curves and calculated the corresponding area under the curve (AUC) using the held-out test data set for the imputed data sets.22 We provide two graphical summaries of the resulting models: (1) a summary of the sign distribution showing, across bootstrap replications and imputed data sets, the proportion of estimated associations which were negative (better outcomes), zero or positive and (2) box plots of the estimated effect on the log-odds scale. We opted to show box plots instead of CIs to highlight the exploratory nature of the results for individual predictors. LASSO estimates are biased, and the lack of a priori hypotheses makes statistical significance testing inappropriate. We first defined a set of contrasts in order to make meaningful comparisons while accounting for the second-order terms in the model rather than report results for every parameter. The contrast matrices are available on a public repository (https://github.com/KosorokLab/CovidIBDRiskCalc) in a CSV format.
Predictions are averaged over bootstrap replications and imputed data sets, and so there is no single set of model coefficients to report. Because the logistic link function is non-linear, the predicted probability from averaging over predictions from each fitted model does not equal the predicted probability from averaging over coefficients across the models. Additionally, averaging the model coefficients would result in none of the coefficients being equal to zero unless that coefficient is equal to zero in every fitted model. Instead of reporting a misleading model summary, we opted to make all the model coefficients available online (https://github.com/KosorokLab/CovidIBDRiskCalc).
Software
The analysis was conducted using R V.4.0.2 and the tidyverse, glmnet, glmnetUtils, mice, magrittr, future and pROC packages.16 23–29 The online calculator was created using shiny.30 The most recent draw for the Carolina Pick 4 lottery (https://nclottery.com/Pick4) at the time of analysis was used for the random number generation seed. The code used to conduct the analysis is available on GitHub (https://github.com/KosorokLab/CovidIBDRiskCalc). This does not include the study data, but the estimated model coefficients are available.
Results
Participants
A total of 2709 patients were reported to the registry, split into a training set of 2009 patients and a test data set of 700 patients. The test data set was comprised 366 from the 15% split and 334 patients added to the registry after model fitting and before manuscript submission. Table 1 provides demographic, clinical, medication and outcome descriptive summaries for the training set, test set and the whole sample. A total of 633 (24%) patients were hospitalised, 137 (5%) were admitted to the ICU or intubated and 69 (3%) patients died. The cohort has 1076 (40%) patients from the USA, with the rest coming from a variety of other countries summarised in table 1.
Model performance
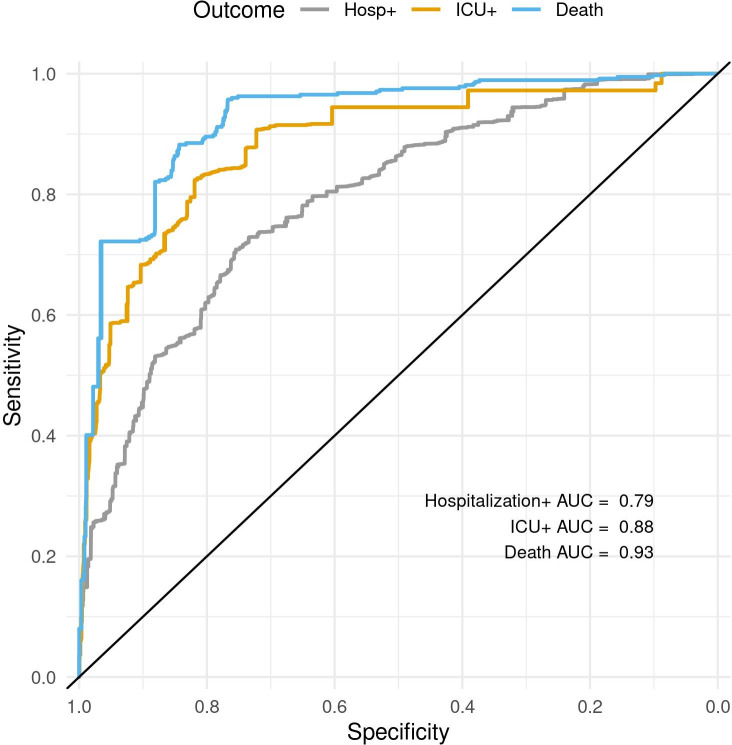
The models have excellent discrimination, with an AUC and associated 95% CI estimated on the test data set averaged over the imputations of 0.79 (0.75 to 0.83) for Hospitalisation+, 0.88 (0.82 to 0.95) for ICU+ and 0.94 (0.89 to 0.99) for Death. The receiver operator character curves are shown in figure 1.
Figure 1.
ROC curves for Hospitalisation+, ICU+ and Death showing the models’ sensitivities as a function of their specificity (axis reversed). AUC, area under the curve; ICU, intensive care unit; ROC, receiver operator characteristic.
Predictors of hospitalisation, intensive care and death
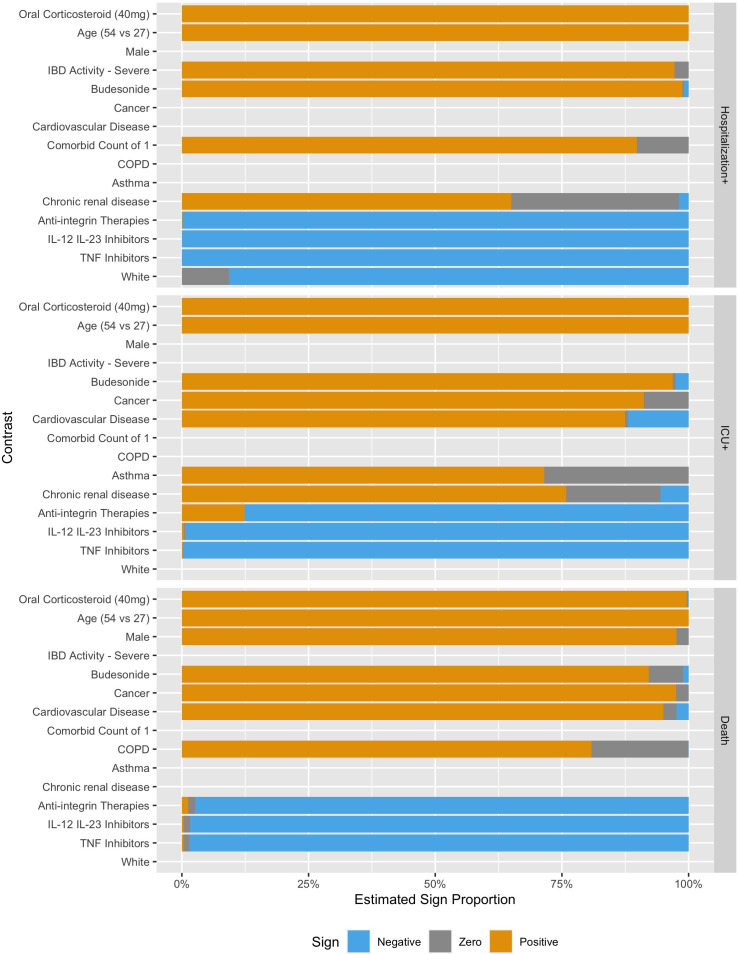
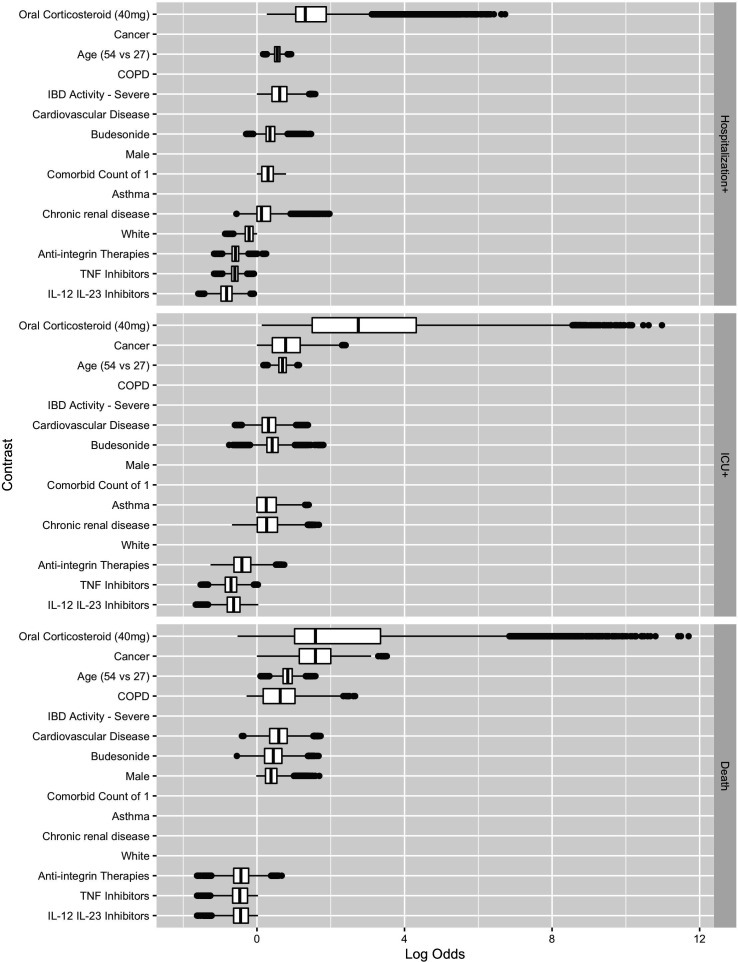
Figures 2 and 3 show the estimated coefficient sign distribution and the effect on the log-odds scale for the ten contrasts most strongly associated with each outcome, respectively. Consistent with other studies on risk factors for hospitalisation and death, we find older age, male gender and comorbidities to be associated with worse outcomes due to COVID-19.3 31 White race is associated with a lower risk of Hospitalisation+ in 89.2% of our replications but is not consistently selected in the models for ICU+ (30%) or Death (10%). These plots, not restricted to the top ten effects, are available in the supplement for all demographic, clinical and medication predictors (online supplemental figures 1 and 2), for countries (online supplemental figures 3 and 4) and for US regions (online supplemental figures 5 and 6).
Figure 2.
Estimated contrast sign distribution showing the proportion of times the estimated association was positive, negative or zero. IBD, inflammatory bowel disease; COPD, chronic obstructive pulmonary disease; IL, interleukin; TNF, tumour necrosis factor.
Figure 3.
Estimated contrast effect size distribution box plots where the effects are shown on the log-odds scale. COPD, chronic obstructive pulmonary disease; IBD, inflammatory bowel disease; IL, interleukin; TNF, tumour necrosis factor.
Corticosteroids are associated with a higher risk of Hospitalisation+, ICU+ and Death. Oral corticosteroid use is the most important predictor, in terms of the magnitude of the absolute value of the coefficient, for Hospitalisation+, ICU+ and Death (figure 3). Biological medicines are associated with a lower risk of Hospitalisation+, ICU+ and Death, with integrin antagonists having directionally smaller effects than TNF antagonists or IL-12/23 inhibitors.
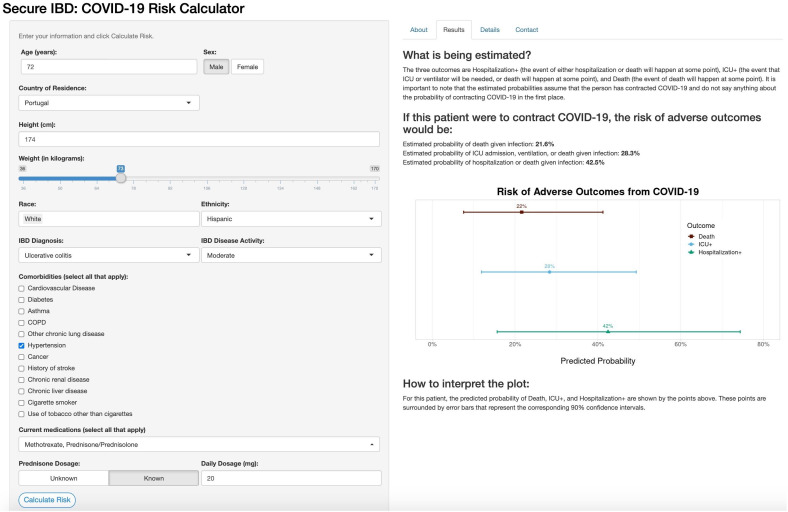
Online risk tool
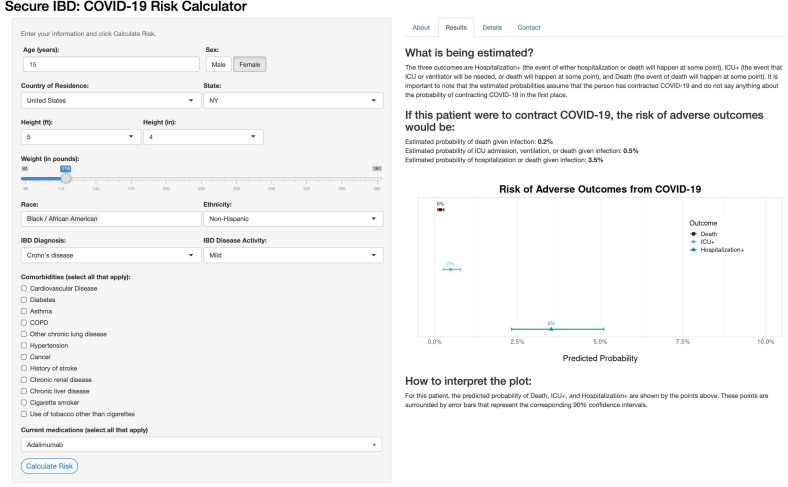
The online risk calculator where physicians can enter their patient’s information and receive predictions from our models is freely available online (http://shiny.bios.unc.edu/secure-ibd-risk-calc/). The SECURE-IBD COVID-19 Risk Calculator was designed for physicians to use during consultations with their patients and includes detailed clinical characteristics, including demographics, disease diagnosis information, comorbidities and current medications. Daily dosage may optionally be entered for certain medications. The output of the risk calculator numerically and visually summarises the patient’s probabilities of adverse outcomes and associated prediction intervals among the three nested outcomes discussed earlier. Figures 4 and 5 display the results for two example patients and their associated probabilities (and 90% CIs) of adverse outcomes if they were to contract COVID-19. The interactive application could provide a reliable basis for distinguishing between high-risk and low-risk patients to aid in personalising clinical guidance on decisions about precautions, returning to normal activities and vaccination.
Figure 4.
Online risk prediction tool example for a patient with below-average predicted risk. Young age, gender and a lack of comorbidities contribute to a lower-than-average risk of adverse COVID-19 outcomes. IBD, inflammatory bowel disease; ICU, intensive care unit.
Figure 5.
Online risk prediction tool example for a patient with above-average predicted risk. Older age, prednisone dosage and hypertension were the major contributors to increased risk, with ethnicity having a small positive association with increased risk. IBD, inflammatory bowel disease; ICU, intensive care unit.
Discussion
We developed and validated risk prediction models for hospitalisation, intensive care stay and death resulting from COVID-19 in patients with IBD using data from 2709 cases from 59 countries reported through an international voluntary registry.12 We made a free online risk calculator using these models (https://covidibd.org/covid-19-risk-calculator/) for healthcare providers to facilitate discussion of risks due to COVID-19 with their patients with IBD.4 The interactive application could provide a reliable basis for distinguishing between high-risk and low-risk patients to aid in personalising clinical guidance on decisions about precautions, returning to normal activities and vaccination.
Other COVID-19-related risk prediction tools have focused on predicting hospital course based on clinical data captured at the time of admission,6 and predicting mortality among US patients aged 65 years and older.5 Our risk tool is unique in at least three ways. First, we focus on a specialised population of patients with IBD, a chronic, immune-mediated condition frequently treated with immune suppressive medications and often affected by other comorbidities. Second, our model focuses on predictors that are known before a patient were to contract COVID-19 and thus can be used to inform lifestyle or treatment decisions to prevent infection or downstream complications. Finally, we examine a broader range of outcomes than tools focused solely on mortality. Our work can serve as a model for other disease areas, and our code is publicly available and could be adapted for similar online risk tools in other settings or populations.
Strong associations with worse adverse COVID-19 outcomes were oral corticosteroids, older age, comorbidities, gender and non-white physician-reported race (for Hospitalisation+). Caution must be used when interpreting penalised regression results because the coefficients are biased, but the results for oral corticosteroids were particularly dramatic. Compared with not taking an oral corticosteroid, taking a daily dose equivalent of 40 mg of prednisone was associated with 10 times greater adjusted odds of death. Biological therapies were associated with a lower risk of adverse COVID-19 outcomes, with small differences between the subcategories of biological therapies. Compared with not taking a biological therapy, TNF inhibitors were associated with an adjusted OR of 0.62 for death. In contrast to earlier studies using this database,13 32 we did not find a consistent association between 5-aminosalicylates and a higher risk of adverse outcomes; depending on the imputation and bootstrap replication, the sign would often change from positive to negative.
The worldwide collaboration that enabled this study and the detailed clinical data reported by physicians or trained medical staff is an important strength of this study. The machine learning approach allowed us to consider a wide variety of potential associations with adverse outcomes, and we examined multiple adverse COVID-19-related adverse outcomes enabling preliminary comparisons between risks. Certain comorbidities, including chronic obstructive pulmonary disease (COPD), cardiovascular disease (CVD), and cancer, were not as strongly associated with Hospitalisation+ as they were for death. In contrast, severe IBD disease activity was an important predictor of Hospitalisation+ but was not consistently associated with a higher risk of ICU+ or Death.
Limitations
The data for this study comes from a voluntary registry, and the registry population may differ in unknown ways from the general population of patients with IBD. Reported cases may under-represent both low-risk asymptomatic cases and severely ill patients who may be hospitalised at an outside hospital or die without their healthcare provider’s knowledge. Model development and validation were conducted using data from the same registry, and validation in an independent cohort of patients with IBD will be an important future direction. Our results are associational, not causal—when using the online risk calculator, healthcare providers should not use it to answer ‘what-if’ questions (eg, how an individual’s risk would change if they altered the medications they were taking) which are inherently causal questions.33 While the registry has a wealth of clinical data, it does not collect granular data on many social determinants of health. Additionally, insurance status is not collected, which, for patients in the USA, likely factors into the decision making of whether to visit a hospital. Finally, we cannot compare the risk of adverse COVID-19 outcomes in patients with IBD to that in the general population.
Conclusions
This prognostic model can effectively predict which patients with IBD may be at higher risk for COVID-19-related morbidity. The free and publicly available (https://covidibd.org/covid-19-risk-calculator/) risk calculator should facilitate patient–provider discussions regarding the individualised risk of COVID-19 based on patient and treatment-related factors. As COVID-19 cases continue to rise in the USA and the rest of the world, this tool will be important in assisting physicians in identifying high-risk patients with downstream clinical implications. This tool can inform public health efforts to promote rational vaccine allocation and could help providers target their outreach to higher-risk patients. We believe this approach can also serve as a model for risk stratification in other chronic diseases.
Supplementary Material
Footnotes
Twitter: @kushalshah96, @Minxin Lu
Contributors: MK conceptualised and acquired funding for the study. XZ curated the data. The investigation including data collection and analysis was conducted by JS, KSS, ML, XZ, RU, EJB, MA, J-FC, MK and MRK. The formal analysis, software programming, validation, visualisations and preparation of the original draft were done by JS, KSS and ML. The modelling methods were determined by JS, KSS, ML and MRK. The execution of the project was supervised by MK and MRK. The draft was reviewed and edited by JS, KSS, ML, XZ, RU, EJB, MA, J-FC, MK and MRK.
Funding: This work was funded by the Helmsley Charitable Trust (2003-04445), National Center for Advancing Translational Sciences (UL1TR002489), a T32DK007634 (EJB) and a K23KD111995-01A1 (RCU). Additional funding was provided by Pfizer, Takeda, Janssen, Abbvie, Eli Lilly, Genentech, Boehringer Ingelheim, Bristol Myers Squibb, Celltrion and Arenapharm.
Competing interests: JS, KSS, ML, XZ, EJB, MA and MRK report no conflicts of interest. RU has served as a consultant and/or advisory board member for Bristol Myers Squibb, Eli Lilly, Janssen, Pfizer and Takeda. He has received research support from AbbVie, Boehringer Ingelheim and Pfizer. He is supported by a Career Development Award from the National Institutes of Health (K23KD111995‐01A1). J-FC reports receiving research grants from AbbVie, Janssen Pharmaceuticals and Takeda; receiving payment for lectures from AbbVie, Amgen, Allergan, Inc. Ferring Pharmaceuticals, Shire and Takeda; receiving consulting fees from AbbVie, Amgen, Arena Pharmaceuticals, Boehringer Ingelheim, Celgene Corporation, Celltrion, Eli Lilly, Enterome, Ferring Pharmaceuticals, Genentech, Janssen Pharmaceuticals, Landos, Ipsen, Medimmune, Merck, Novartis, Pfizer, Shire, Takeda, Tigenix, Viela bio; and hold stock options in Intestinal Biotech Development and Genfit. MK has consulted for Abbvie, Janssen, Pfizer and Takeda, is a shareholder in Johnson & Johnson, and has received research support from Pfizer, Takeda, Janssen, Abbvie, Lilly, Genentech, Boehringer Ingelheim, Bristol Myers Squibb, Celtrion and Arenapharm.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. We are committed to sharing our data with the international research community. Data requests are reviewed by our SECURE-IBD team including the International Advisory Committee to ensure data will be used in a scientifically and ethically sound way. The data request form and additional information can be found online at https://covidibd.org/sharing-secure-ibd-data/. Data collection is ongoing, and data beyond the current study’s data may be available at the time of the request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
The UNC-Chapel Hill Office for Human Research Ethics has determined that the storage and analysis of deidentified data for this project does not constitute human subjects research as defined under federal regulations (45 CFR 46.102 and 21 CFR 56.102) and does not require IRB approval.
References
- 1.World Health Organization . WHO coronavirus disease (COVID-19) Dashboard, 2020. Available: https://covid19.who.int
- 2.Poblador-Plou B, Carmona-Pírez J, Ioakeim-Skoufa I, et al. Baseline chronic comorbidity and mortality in laboratory-confirmed COVID-19 cases: results from the PRECOVID study in Spain. Int J Environ Res Public Health 2020;17:5171. 10.3390/ijerph17145171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson EJ, Walker AJ, Bhaskaran K, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020;584:430–6. 10.1038/s41586-020-2521-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lloyd-Jones DM, Braun LT, Ndumele CE, et al. Use of risk assessment tools to guide decision-making in the primary prevention of atherosclerotic cardiovascular disease: a special report from the American heart association and American College of cardiology. Circulation 2019;139. 10.1161/CIR.0000000000000638 [DOI] [PubMed] [Google Scholar]
- 5.Dun C, Walsh C, Bae S. A machine learning study of 534023 Medicare beneficiaries with COVID-19: implications for personalized risk prediction. medRxiv 2020. [Google Scholar]
- 6.Jehi L, Ji X, Milinovich A, et al. Development and validation of a model for individualized prediction of hospitalization risk in 4,536 patients with COVID-19. PLoS One 2020;15:e0237419. 10.1371/journal.pone.0237419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fakhoury M, Negrulj R, Mooranian A, et al. Inflammatory bowel disease: clinical aspects and treatments. J Inflamm Res 2014;7:113–20. 10.2147/JIR.S65979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin SC, Cheifetz AS. The use of complementary and alternative medicine in patients with inflammatory bowel disease. Gastroenterol Hepatol 2018;14:415–25. [PMC free article] [PubMed] [Google Scholar]
- 9.Torres J, Mehandru S, Colombel J-F, et al. Crohn’s disease. The Lancet 2017;389:1741–55. 10.1016/S0140-6736(16)31711-1 [DOI] [PubMed] [Google Scholar]
- 10.Ungaro R, Mehandru S, Allen PB, et al. Ulcerative colitis. The Lancet 2017;389:1756–70. 10.1016/S0140-6736(16)32126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moons KGM, Altman DG, Reitsma JB, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015;162:W1–73. 10.7326/M14-0698 [DOI] [PubMed] [Google Scholar]
- 12.Rubin DT, Abreu MT, Rai V, et al. Management of patients with Crohn's disease and ulcerative colitis during the coronavirus Disease-2019 pandemic: results of an international meeting. Gastroenterology 2020;159:6–13. 10.1053/j.gastro.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner EJ, Ungaro RC, Gearry RB, et al. Corticosteroids, but not TNF antagonists, are associated with adverse COVID-19 outcomes in patients with inflammatory bowel diseases: results from an international registry. Gastroenterology 2020;159:481–91. 10.1053/j.gastro.2020.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandenbroucke JP, von Elm E, Altman DG, et al. Strengthening the reporting of observational studies in epidemiology (STROBE): explanation and elaboration. PLoS Med 2007;4:e297. 10.1371/journal.pmed.0040297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin DB. Multiple imputation after 18+ years. J Am Stat Assoc 1996;91:473–89. 10.1080/01621459.1996.10476908 [DOI] [Google Scholar]
- 16.van Buuren S, Groothuis-Oudshoorn K. Mice: multivariate imputation by chained equations in R. J Stat Softw 2010:1–68. [Google Scholar]
- 17.Tibshirani R. Regression shrinkage and selection via the LASSO. Journal of the Royal Statistical Society: Series B 1996;58:267–88. [Google Scholar]
- 18.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B 2005;67:301–20. 10.1111/j.1467-9868.2005.00503.x [DOI] [Google Scholar]
- 19.Hastie T, Tibshirani R, Wainwright M. Statistical learning with sparsity: the lasso and generalizations. CRC press 2015.
- 20.Meier L, Van De Geer S, Bühlmann P. The group LASSO for logistic regression. Journal of the Royal Statistical Society: Series B 2008;70:53–71. 10.1111/j.1467-9868.2007.00627.x [DOI] [Google Scholar]
- 21.Simon N, Friedman J, Hastie T, et al. A Sparse-Group LASSO. Journal of Computational and Graphical Statistics 2013;22:231–45. 10.1080/10618600.2012.681250 [DOI] [Google Scholar]
- 22.Efron B, Tibshirani R. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Statistical Science 1986;1:54–75. 10.1214/ss/1177013815 [DOI] [Google Scholar]
- 23.Team RC . R: A language and environment for statistical computing. Vienna, Austria: Team RC, 2013. [Google Scholar]
- 24.Wickham H, Averick M, Bryan J, et al. Welcome to the Tidyverse. Journal of Open Source Software 2019;4:1686. 10.21105/joss.01686 [DOI] [Google Scholar]
- 25.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Microsoft, Ooi H . glmnetUtils: Utilities for “Glmnet.”, 2020. Available: https://CRAN.R-project.org/package=glmnetUtils
- 27.Milton Bache S, Wickham H, Henry L. magrittr: a Forward-Pipe operator for R, 2020. Available: https://CRAN.R-project.org/package=magrittr
- 28.Bengtsson H. Future: unified parallel and distributed processing in R for everyone, 2020. Available: https://CRAN.R-project.org/package=future
- 29.Robin X, Turck N, Hainard A, et al. pROC: display and analyze ROC curves, 2020. Available: https://CRAN.R-project.org/package=pROC
- 30.Chang W, Cheng J, Allaire JJ, et al. Shiny: web application framework for R, 2020. Available: https://CRAN.R-project.org/package=shiny
- 31.Kim L, Garg S, O’Halloran A, et al. Risk factors for intensive care unit admission and in-hospital mortality among hospitalized adults identified through the US coronavirus disease 2019 (COVID-19)-associated hospitalization surveillance network (COVID-NET). Clinical Infectious Diseases 2021;72:e206–14. 10.1093/cid/ciaa1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ungaro RC, Brenner EJ, Gearry RB, et al. Effect of IBD medications on COVID-19 outcomes: results from an international registry. Gut 2021;70:725–32. 10.1136/gutjnl-2020-322539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernán MA, Robins JM. Chapter 1 a definition of causal effect. In: Causal Inference: What If 2020;311. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2021-049740supp001.pdf (1.5MB, pdf)
Data Availability Statement
Data are available upon reasonable request. We are committed to sharing our data with the international research community. Data requests are reviewed by our SECURE-IBD team including the International Advisory Committee to ensure data will be used in a scientifically and ethically sound way. The data request form and additional information can be found online at https://covidibd.org/sharing-secure-ibd-data/. Data collection is ongoing, and data beyond the current study’s data may be available at the time of the request.