Abstract
CREB binding protein (CBP) is a 270-kDa nuclear protein required for activated transcription of a large number of cellular genes. Although CBP was originally discovered through its interaction with phosphorylated CREB (pCREB), it is utilized by a multitude of cellular transcription factors and viral oncoproteins. Both CREB and the tumor suppressor p53 have been shown to directly interact with the KIX domain of CBP. Although coactivator competition is an emerging theme in transcriptional regulation, we have made the fortuitous observation that protein kinase A-phosphorylated CREB strongly enhances p53 association with KIX. Phosphorylated CREB also facilitates interaction of a p53 mutant, defective for KIX binding, indicating that CREB functions in a novel way to bridge p53 and the coactivator. This is accomplished through direct interaction between the bZIP domain of CREB and the amino terminus of p53; a protein-protein interaction that is also detected in vivo. Consistent with our biochemical observations, we show that stimulation of the intracellular cyclic AMP (cAMP) pathway, which leads to CREB phosphorylation, strongly enhances both the transcriptional activation and apoptotic properties of p53. We propose that phosphorylated CREB mediates recruitment of CBP to p53-responsive promoters through direct interaction with p53. These observations provide evidence for a novel pathway that integrates cAMP signaling and p53 transcriptional activity.
CREB binding protein (CBP) is a large pleiotropic cellular coactivator protein critical to the execution of virtually all known cellular programs, including cell growth, differentiation, the integration of both signal-dependent and -independent cellular responses, and apoptosis. CBP and its sister protein p300 are highly conserved in multicellular organisms from Caenorhabditis elegans to mammals and have been shown to have profound effects on somatic differentiation during early embryogenesis (2, 20, 33, 38). CBP interacts with a multitude of structurally unrelated cellular transcription factors, promoting selective gene activation by providing communication between promoter-bound transcription factors and components of the basal transcription apparatus (34). Following promoter localization of CBP, the coactivator is also believed to directly acetylate nucleosomes, leading to transcriptional activation through localized chromatin remodeling. These acetylation events are carried out through both intrinsic and associated (P/CAF) histone acetyltransferase activities (7, 28).
CBP was originally discovered, and thus named, through its interaction with the kinase-inducible activation domain (KID) of the cellular transcription factor CREB (4, 11, 23). Protein kinase A (PKA) phosphorylation of a critical serine residue on CREB (amino acid [aa] 133) is required for complex formation between these two proteins (29). A small subdomain of CBP (aa 586 to 679), called the KIX domain, participates in protein-protein interaction with pCREB (31). KIX is composed of three interacting α helices that come together to form a hydrophobic core. A shallow groove on the surface of the KIX structure provides the site for molecular interaction with pCREB.
Although pCREB complexed with the KIX domain of CBP has been well characterized, KIX has also been shown to interact with several other proteins, including the tumor suppressor p53 (36). p53 is a transcription factor that induces cell cycle arrest or apoptosis in response to a variety of cellular stress signals, thereby preventing the transmission of genetic mutations (reviewed in reference 22). Mutations within the coding sequence of the p53 gene, leading to loss of p53 activity, have been identified in 60% of the human malignancies examined (17, 25). This statistic underscores the significance of p53 in genome surveillance and suppression of malignant transformation. The transcriptional activity of p53, which is tightly linked to its tumor suppressor function, appears to depend upon efficient recruitment of CBP to p53 target promoters. Consistent with this observation, recent studies have shown that the activation domain of p53 participates in CBP recruitment (16, 36). Together, these findings indicate that CBP plays a critical role in supporting p53-dependent transcription function.
The recent study showing p53 binding to KIX led us to ask whether both pCREB and p53 bind to KIX in a mutually exclusive manner, possibly leading to coactivator competition within the cell. From this line of investigation, we made the surprising observation that pCREB strongly facilitates p53 association with KIX. We demonstrate that phosphorylated CREB, but not unphosphorylated CREB, mediates p53 association with KIX, and that this is accomplished through protein-protein interaction between the basic leucine zipper (bZIP) domain of CREB and the amino terminus of p53. The significance of this interaction is supported by studies showing that p53 and CREB interact in vivo and that p53 function is enhanced by forskolin, a stimulator of the PKA signal transduction pathway. These data support a model where phosphorylated CREB bridges the interaction between promoter-bound p53 and CBP, furnishing an alternate mechanism of coactivator recruitment to p53-responsive genes. Finally, the evidence supports a unique form of transcription factor organization, leading to multilayered regulation of gene expression, and underscores the importance of CBP in mediating convergent signaling pathways.
MATERIALS AND METHODS
Cloning, expression and purification of recombinant proteins.
Glutathione S-transferase (GST)-KIX (CBP aa 588 to 683), His6-p53, His6-p53 (L22Q-W23S) (36), and KID (CREB aa 100 to 160) (27) were expressed and purified as previously described. pGST-CREB-His6 expression plasmid was provided by C.-Z. Giam and was expressed and purified as described (18). GST-p53 was made by PCR amplification of the p53 gene, followed by insertion into pDEST 15 for expression in Escherichia coli and pDEST 10 for use in the Bac-To-Bac baculovirus expression system (Life Technologies). Proteins were purified to near homogeneity, dialyzed against TM 0.1 M KCl buffer (50 mM Tris-HCl [pH 7.9], 100 mM KCl, 12.5 mM MgCl2, 1 mM EDTA [pH 8.0], 20% glycerol, 0.025% [vol/vol] Tween 20, 1 mM dithiothreitol), aliquoted, and stored at −70°C. p53 proteins were dialyzed against 20 mM Tris-HCl [pH 8.0]–100 mM KCl–0.5 mM EDTA–20% glycerol. PKA phosphorylation of CREB and KID was performed as previously described (15).
GST pull-down assays.
All GST pull-down experiments were performed using 12.5 μl of glutathione-agarose beads equilibrated in 0.5× Superdex buffer (1× Superdex buffer is 25 mM HEPES [pH 7.9], 12.5 mM MgCl2, 10 μM ZnSO4, 150 mM KCl, 20% [vol/vol] glycerol, 0.1% Nonidet P-40, and 1 mM EDTA). The purified GST fusion protein was incubated with the beads for 1 to 2 h at 4°C and then washed with 0.5× Superdex buffer. The second protein(s) was then added to the washed beads and incubated for 1 to 2 h (or overnight) at 4°C. The beads were washed as before, and bound proteins were eluted with sodium dodecyl sulfate (SDS) sample dyes. Bound proteins were separated by electrophoresis on a 10% or 12% SDS gel or a 10% Tris-Tricine gel, transferred to nitrocellulose, and probed with the appropriate antibody. The following antibodies were used in this study: anti-p53 (DO-1 [epitope corresponding to aa 11 to 25]; Santa Cruz Biotechnology), anti-p53 (Ab-1 [epitope corresponding to aa 371 to 380]; Calbiochem), anti-CREB (C-21 [epitope corresponding to the carboxy terminus of human CREB-1]; Santa Cruz Biotechnology), anti-phosphoserine 133-CREB (New England Biolabs), and anti-His (H-15; Santa Cruz Biotechnology).
Cell culture, transient cotransfection assays, and expression plasmids.
Jurkat T cells and Molt 4 T cells were cultured in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and penicillin-streptomycin. The H24 p53-3 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS, tetracycline (2 μg/ml), puromycin (1 μg/ml), and geneticin (250 μg/ml). The MCF7 breast cancer cells were cultured in Eagle's minimum essential medium supplemented with 2 mM l-glutamine, penicillin-streptomycin, Earle's BBS adjusted to contain 1.5 g of sodium bicarbonate per liter, a 0.1 mM concentration of nonessential amino acids, 1.0 mM sodium pyruvate, 10% FBS, and 0.01 mg of bovine insulin per ml. For transient cotransfection assays (36), cells were grown to a density of 106 cells/ml and transfected with Lipofectamine (Life Technologies, Inc.) and a constant amount of DNA for 5 h. The cells were allowed to recover for 24 h before harvest, in the absence or presence of 20 μM forskolin. Cells were lysed, and luciferase activity was measured using the Dual-Luciferase reporter assay system with a Turner Designs model TD 20-e luminometer. Luciferase activity was normalized to pRL-TK vector (Promega), which encodes the Renilla luciferase from HSV-TK promoter, as an internal control. Expression plasmids for p53 (pC53-SN3 [6]), RSV-PKA, or Gal4-p53 fusion proteins carrying p53 aa 1 to 393 (pGalp53) or aa 1 to 52 (pGalp53N) (12) have been previously described. The luciferase reporter plasmids pG13-Luc (21), pGalTK-Luc (12), and viral CRE-Luc (15) have also been described. The Bax-Luc reporter plasmid was prepared by cloning the Bax promoter (−340 to +31) upstream of the luciferase gene.
Western blots.
MCF7 cells were grown to 70% confluency, medium was removed, and fresh medium was added to the cells in the absence or presence of 20 μM forskolin, and the cells were incubated for the indicated amount of time. Cells were lysed and resuspended in SDS sample dyes. Proteins were separated on a 10% Tris-Tricine SDS-polyacrylamide gel electrophoresis and analyzed by Western blot analysis.
Northern blots.
MCF7 cells were grown to ∼70% confluency, the medium was removed and fresh medium was added to the cells in the absence or presence of 20 μM forskolin and incubated for 45 min. The cells were harvested in a solution containing 4 M guanidinium thiocyanate, 0.5% sarcosyl, and 25 mM sodium citrate, pH 7.0, and total RNA was isolated using acidic phenol extraction followed by isopropanol precipitation and resuspension in FORMAzol (Molecular Research Center, Inc.). Purified total RNA (15 μg) was separated by electrophoresis on a 1% agarose–formaldehyde-MOPS (morpholinepropane sulfonic acid) gel, transferred to Genescreen (DuPont-NEN) membrane, and cross-linked for 3 min with UV radiation at a wavelength of 320 nm. Membranes were hybridized at 43°C with 32P-labeled cDNA probes complimentary to either the p21 or GAPDH mRNAs. The membrane was washed several times, dried, and analyzed by Phosphorimager analysis.
Coimmunoprecipitation (co-IP) assays.
Molt 4 T-cell lysates were prepared in RIPA buffer (50 mM Tris-HCl [pH 8.0], 1% Triton X-100, 100 mM NaCl, 1 mM MgCl2, 2 mM benzamidine, 2 μg of leupeptin per ml, and 1 mM phenylmethylsulfonyl fluoride). Antibody-bound beads (20 μl) (Ab-1 [epitope corresponding to aa 371 to 380] [Calbiochem] or DO-1 [epitope corresponding to aa 11 to 25] [Santa Cruz Biotechnology]) were washed in RIPA buffer. Cell lysates (500 μg) were added to each antibody column, incubated overnight, and washed several times in RIPA buffer. The bound proteins were analyzed on an SDS–10% polyacrylamide gel, and detected by Western blotting using anti-CREB-1 antibody (C-21 [epitope corresponding to aa 295 to 321]; Santa Cruz Biotechnology) and anti-p53 (DO-1; Santa Cruz Biotechnology).
Apoptotic analysis.
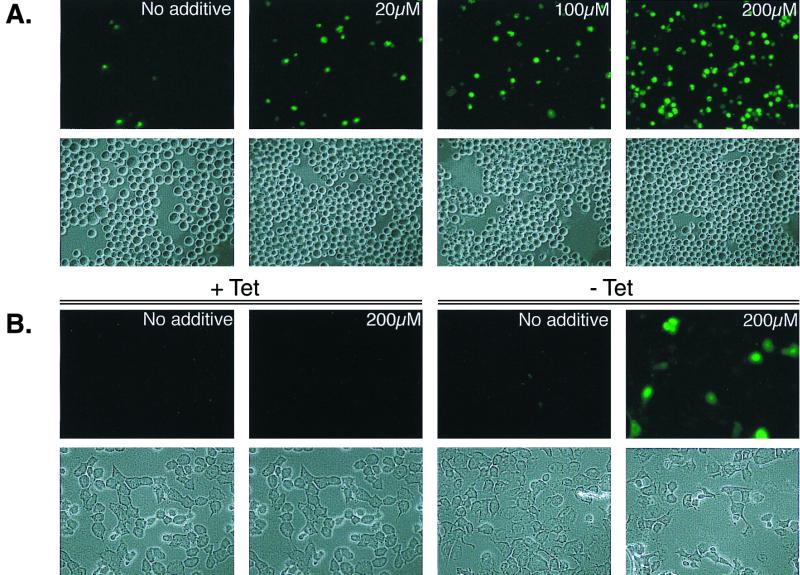
Molt 4 T-cells (5 × 105 cells) and H24 p53-3 cells (reference 10) were treated with the indicated concentrations of forskolin (or dimethyl sulfoxide carrier) for 24 h. In the p53-inducible cell line H24 p53-3, tetracycline was removed at the time of forskolin addition. Cells were harvested and treated with YO-PRO-1 (from Vybrant apoptosis assay kit no. 4; Molecular Probes). Cells were deposited on microslides coated with poly-l-lysine (Poly-Prep slides; Sigma), and the fluorescent apoptotic cells were analyzed and photographed using a fluorescence microscope.
RESULTS
pCREB enhances p53 association with the KIX domain of CBP.
In a recent structural study characterizing the phosphorylated-kinase-inducible domain of CREB bound to the hydrophobic groove on the surface of KIX, the authors noted key sequence and structural similarities between phosphorylated KID (pKID) and the activation domain of p53 (31). This led to the prediction that the hydrophobic groove on the surface of KIX may also serve as the binding site for p53. To test this hypothesis, we performed a GST pull-down competition experiment to determine whether titration of pCREB can displace p53 bound to KIX. Purified GST-KIX (aa 588 to 683) was bound to glutathione-agarose beads and incubated with recombinant, purified p53. Increasing amounts of PKA-phosphorylated CREB were added into the binding reaction mixtures, and the amount of p53 remaining bound to KIX was determined by Western blot analysis (Fig. 1A). Surprisingly, the addition of pCREB did not compete with p53 for binding to KIX but produced a dramatic increase in the amount of p53 associated with the complex (compare lanes 4 to 7). As expected, neither p53 nor pCREB bound to GST beads alone (Fig. 1A, lanes 2 and 3).
FIG. 1.
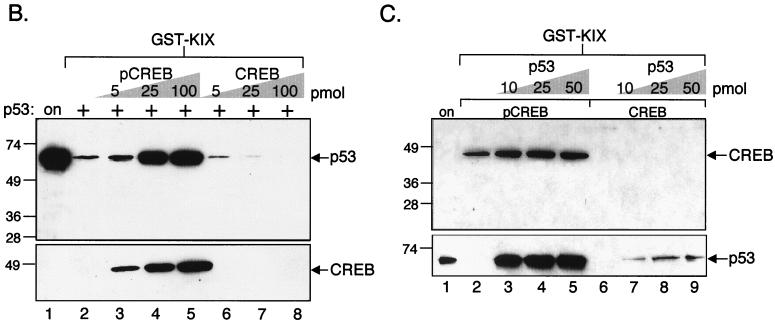
(A) CREB enhances p53 binding to the KIX domain of CBP. Purified p53 (5 pmol) was incubated with either GST alone (1 pmol) (lanes 2 and 3) or GST-KIX (aa 588 to 683) (1 pmol) (lanes 4 to 7) in the absence (−) and presence (+) of the indicated amount of purified, PKA-phosphorylated CREB. Bound p53 protein was detected by Western blot analysis. Onput protein (6%) is shown in lane 1 and protein standards (in kilodaltons) are indicated at left. (B) pCREB, but not unphosphorylated CREB, enhances p53 binding to the KIX domain of CBP. Purified p53 (5 pmol) was incubated with GST-KIX (1 pmol) in the absence or presence of the indicated amounts of either purified pCREB (lanes 3 to 5) or purified, mock-phosphorylated CREB (lanes 6 to 8). p53 (upper panel) was detected by Western blot analysis, and bound protein is indicated. The blot was reprobed using an anti-CREB antibody, and bound protein is indicated in the lower panel. Onput protein (2%) is shown in lane 1 and protein standards (in kilodaltons) are indicated at left. (C) p53 does not enhance the binding of CREB to the KIX domain of CBP. Purified, phosphorylated or mock-phosphorylated CREB (10 pmol) was incubated with GST-KIX (10 pmol) in the absence or presence of the indicated amount of purified p53 (lanes 3 to 5 and 7 to 9). CREB (upper panel) was detected by Western blot analysis. The blot was reprobed using an anti-p53 antibody, and bound protein is indicated in the lower panel. Onput protein (2%) is shown in lane 1, and protein standards (in kilodaltons) are indicated at left.
We were next interested in determining whether the increase in p53 association with KIX was specific to pCREB or if unphosphorylated CREB could also promote the interaction. To test this hypothesis, we again compared the binding of p53 to KIX following the addition of increasing amounts of either unphosphorylated or phosphorylated CREB. As shown in the GST pull-down assay in Fig. 1B (upper panel), we observed enhancement of p53 binding to KIX only in the presence of pCREB (compare lanes 2 to 5). In fact, increasing amounts of unphosphorylated CREB appeared to reduce the p53-KIX interaction in a dose-dependent manner (compare lane 2 with lanes 6 to 8). Interestingly, we observed that the enhancement of p53 association with KIX correlated precisely with pCREB binding to KIX, suggesting that both pCREB and p53 are simultaneously in complex with this region of CBP (Fig. 1B, compare lanes 3 to 5 of the upper and lower panels). Further, these data strongly support a role for a direct pCREB-KIX molecular interaction in ternary complex formation. As expected, we did not detect the binding of unphosphorylated CREB to the KIX domain (Fig. 1B, lower panel, lanes 6 to 8).
To better characterize the ternary complex containing pCREB, p53, and KIX, we performed the reciprocal experiment and tested whether p53 could also enhance pCREB binding to KIX. We performed a GST pull-down assay in which p53 was titrated into binding reaction mixtures containing GST-KIX and a constant amount of either phosphorylated or unphosphorylated CREB. As shown in Fig. 1C, p53 had only a modest effect on pCREB binding to KIX and no detectable effect on CREB binding to KIX (lanes 2 to 9). These data suggest that pCREB plays the primary role in facilitating formation of the ternary complex. This conclusion is further supported by the observation that significantly more KIX-associated p53 was detected in the presence of pCREB than in the presence of unphosphorylated CREB (Fig. 1C, lower panel, compare lanes 3 to 5 with lanes 7 to 9).
pCREB contacts KIX in the ternary complex.
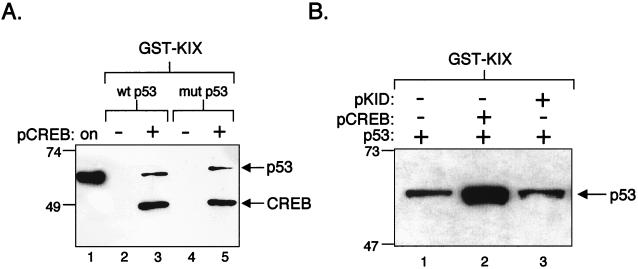
The solution structure of the pCREB-KIX complex reveals intimate molecular contacts between the hydrophobic groove of KIX and the pKID domain of CREB (31). Although we do not know where p53 binds on the surface of KIX, it seems unlikely that the hydrophobic groove could accommodate both pCREB and p53 simultaneously. The strong dependence on CREB phosphorylation in ternary complex formation suggests that pCREB directly contacts the hydrophobic groove, as previously described (31). To determine whether, in the ternary complex, p53 also directly contacts KIX, we utilized an activation domain mutant of p53 (L22Q-W23S) that we have previously shown to be defective for KIX binding (36). We reasoned that if p53 makes direct contacts with KIX in the ternary complex, then the mutant p53 should also be defective for ternary complex formation. On the other hand, if pCREB alters or abolishes the p53-KIX interaction, then we may observe enhanced binding of both wild-type and mutant forms of p53. To sort out these questions, we compared the binding of the wild-type and mutant forms of p53 to GST-KIX in the presence of pCREB. We found that both forms of p53 bound well in the ternary complex, with the binding of each molecule dependent upon pCREB (Fig. 2A, lanes 3 and 5). These data indicate that aa 22 and 23 of the p53 activation domain are not involved in ternary complex formation and support the idea that pCREB alters or abolishes the p53-KIX interaction.
FIG. 2.
(A) A p53 mutant, defective for KIX binding, binds KIX in the presence of pCREB. Purified wild-type (wt) (10 pmol) (lanes 2 and 3) or mutant (mut) (L22Q-W23S) (10 pmol) (lanes 4 and 5) p53 was incubated with GST-KIX (2 pmol) in the absence or presence of 30 pmol of purified, phosphorylated CREB. p53 and CREB were detected simultaneously by Western blot analysis using an anti-His–anti-CREB antibody mixture. Onput wt p53 (2%) is shown in lane 1, and protein standards (in kilodaltons) are indicated at left. At the exposure presented, p53 binding to KIX was not detected. (B) The PKA-phosphorylated kinase-inducible domain of CREB does not support enhanced binding of p53 to KIX. Purified p53 (10 pmol) was incubated with GST-KIX (10 pmol) in the absence or presence of 30 pmol of phosphorylated full-length CREB (lane 2) or pKID domain of CREB (lane 3). p53 was detected by Western blot analysis. Protein standards (in kilodaltons) are indicated at left.
Two possible scenarios are consistent with the data presented in Fig. 2A. In the presence of pCREB, p53 forms different contacts with KIX, or alternatively, p53 binding to KIX is abolished. The second possibility would therefore require a direct CREB-p53 interaction as a means to incorporate p53 into the ternary complex. To test this possibility, we utilized a highly truncated form of CREB that encompassed only the Ser-133 phosphorylated-kinase-inducible domain (CREB aa 115 to 148). Although this region of CREB lacks the bZIP and the Q domains, it is competent for interaction with KIX (29, 31). To determine whether pKID alone can enhance p53 association with KIX, we compared pKID in parallel with full-length pCREB in a GST-KIX pull-down assay (Fig. 2B). Interestingly, the pKID domain had no effect on p53 association with KIX (Fig. 2B, lane 3). Both pCREB and pKID were fully competent for KIX binding, as assayed in a GST-KIX pull-down reaction (data not shown). These data indicate that a region of CREB, outside of the KIX-interacting kinase-inducible domain, is required for efficient formation of the ternary complex. Furthermore, they are consistent with the idea that CREB and p53 interact directly.
Characterization of the CREB-p53 interaction in vitro and in vivo.
Based on the data presented above, we began to formulate a model where the p53-pCREB-KIX ternary complex is formed through direct pCREB interaction with the hydrophobic groove of KIX, with p53 incorporated into the coactivator complex via protein-protein interaction with CREB. To directly test for a p53-CREB interaction, we examined the binding of p53 to GST-CREB, in the absence of KIX. As shown in Fig. 3A, both wild-type and mutant p53 (L22Q-W23S) interacted equally well with GST-CREB (lanes 5 and 6). To confirm the p53-CREB interaction, we performed the reciprocal GST pull-down assay. As shown in Fig. 3B, both phosphorylated and unphosphorylated forms of full-length CREB bound equally well to GST-p53 in the absence of KIX (lanes 2 and 4). Taken together, these results indicate that p53 interacts directly with CREB, in a binding reaction that is independent of KIX. Together, these data provide direct evidence for a novel interaction between CREB and p53 and support the hypothesis that pCREB enhancement of p53 binding to KIX is mediated through a direct p53-CREB protein-protein interaction. Finally, we were interested in testing whether posttranslational modifications of p53 might affect interaction with CREB. Figure 3C shows that GST-p53 derived from baculovirus-infected Sf9 cells interacted with CREB in a manner indistinguishable from that observed with p53 derived from E. coli (compare Fig. 3C, lane 3, with Fig. 3B, lane 2). These data suggest that Sf9 cells do not introduce modifications in p53 that affect interaction with CREB. We have also shown that GST-CREB interacts with non-GST-tagged forms of p53, derived from both E. coli (Fig. 3A) and baculovirus-infected Sf9 cells (data not shown).
FIG. 3.
(A) Wild-type (wt) and mutant (mut) (L22Q-W23S) p53 directly interact with CREB in the absence of KIX. Purified wt or mutant p53 protein (10 pmol each) was incubated with either GST alone (10 pmol) (lanes 3 and 4), or GST-CREB-His6 (10 pmol) (lanes 5 and 6). Bound p53 proteins were detected by Western blot analysis. Onput protein (10%) is shown in lanes 1 and 2, and protein standards (in kilodaltons) are shown at left. (B) PKA-phosphorylated or mock-phosphorylated CREB (25 pmol) was incubated with either GST alone (100 pmol) (lanes 1 and 3) or GST-p53 (100 pmol) (lanes 2 and 4). Bound CREB was detected by Western blot analysis. Output proteins (10%) (lanes 5 and 6) and protein standards (in kilodaltons) are indicated. (C) CREB (5 pmol) was incubated with either GST alone (10 pmol) (lane 2) or GST-p53 produced and purified from baculovirus-infected Sf9 cells (10 pmol) (lane 3). Bound CREB was detected by Western blot analysis. Onput protein (10%) (lane 1) and protein standards (in kilodaltons) are indicated.
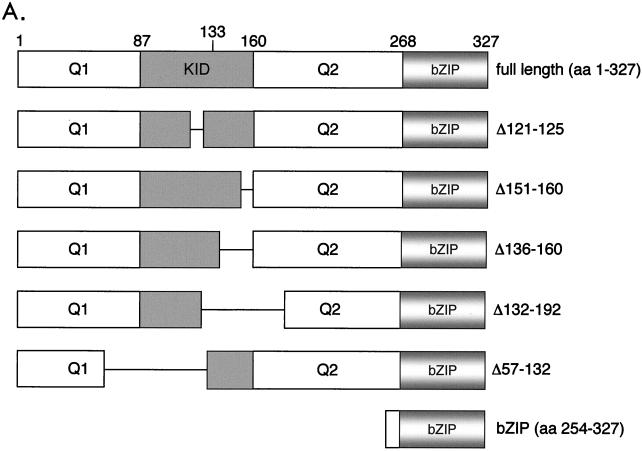
To determine which region of CREB interacts with p53, we tested a series of CREB deletion mutants in the GST pull-down assay using GST-p53. Most of the CREB deletions were centered in the region of KID and extended into both the Q1 and Q2 domains (Fig. 4A). Surprisingly, all the CREB deletion mutants tested were competent for binding to GST-p53 (Fig. 4B, lanes 1 to 6). Since all of the mutants possessed the bZIP domain (aa 254 to 327) (and a portion of Q1 and Q2), we directly tested whether bZIP alone was competent for p53 binding. Figure 4C shows that both full-length CREB and bZIP bound GST-p53 with comparable relative affinities (lanes 2, 3, 5, and 6).
FIG. 4.
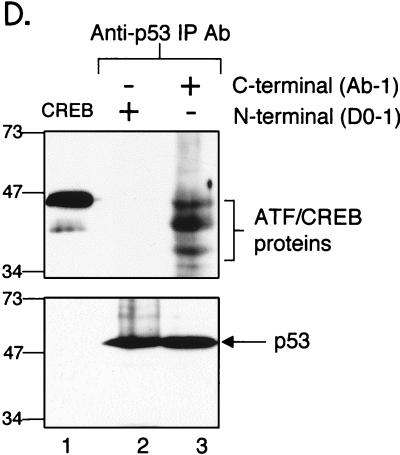
(A) Schematic of the CREB deletion mutants. (B) CREB deletion mutants are competent for p53 binding. The indicated CREB deletion mutants (10 pmol) were incubated with GST-p53 (25 pmol) (lanes 1 to 6). Bound CREB proteins were detected by Western blot analysis. Protein standards (in kilodaltons) are indicated. (C) The bZIP domain is sufficient for interaction with p53. The indicated amounts of either full-length or bZIP (aa 254 to 327) CREB were incubated with GST alone (100 pmol) (lanes 1 and 4) or GST-p53 (100 pmol) (lanes 2, 3, 5, and 6). Bound CREB was detected by Western blot analysis. Onput proteins (5 pmol of CREB, 10 pmol of bZIP) (lanes 7 and 8), and molecular mass standards (in kilodaltons) are indicated. (D) Anti-p53 immunoprecipitates ATF/CREB proteins from a T-cell lysate. The upper panel shows a CREB Western blot of the proteins bound to p53 immobilized using either the Ab-1 or DO-1 anti-p53 antibodies (lanes 2 and 3). Purified, recombinant CREB was electrophoresed as a control (lane 1). The lower panel shows a Western blot of p53 bound to the immobilized anti-p53 antibodies (lanes 2 and 3).
Although these studies establish an interaction between CREB and p53 in vitro, we were interested in determining whether this interaction could also be detected in vivo. To investigate this possibility, we performed a co-IP assay in which a carboxy terminus-reactive p53 antibody (epitope corresponding to aa 371 to 380) was immobilized on a resin. A lysate prepared from the wild-type p53-expressing T-cell line, Molt4, was then passed over the column, the column was washed, and the bound proteins were analyzed by Western blot analysis using an anti-CREB antibody. As shown in Fig. 4D (upper panel), we detected several anti-CREB immunoreactive polypeptides, ranging in molecular mass from about 35 to 45 kDa. The band with the highest molecular mass comigrated with recombinant CREB, indicating that it likely represents endogenous CREB in the Molt4 lysate (Fig. 4D, compare lanes 1 and 3). The other polypeptides are likely cross-reactive ATF/CREB family members (CREM and ATF-1). In a parallel reaction, we also performed the co-IP assay using an anti-p53 antibody reactive against the amino terminus of p53 immobilized on the resin (epitope corresponding to aa 11 to 25) and probed for bound polypeptides using the anti-CREB antibody. Interestingly, we did not detect the binding of any ATF/CREB proteins when the amino-terminal p53 antibody was used in the co-IP, suggesting that this antibody masks critical p53 amino acids that may be required for interaction with CREB (Fig. 4D, upper panel, lane 2). p53 bound equally well to both the amino-terminal and carboxy-terminal antibodies (Fig. 4D, lower panel, lanes 2 and 3). These data support the in vitro data and provide evidence for the binding of multiple ATF/CREB proteins to p53 in vivo. Furthermore, the results suggest that the amino terminus of p53 participates in the protein-protein interaction with CREB.
Activation of the PKA pathway stimulates p53 function.
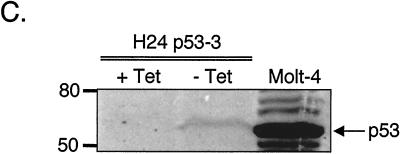
Our biochemical characterization of the ternary complex led us to address the question of whether the binding of p53 to pCREB and the formation of a complex with CBP might facilitate the biological effects of p53 in the cell. To first address this question, we were interested in testing whether forskolin, a stimulator of adenylate cyclase and the cyclic AMP (cAMP) pathway, might induce apoptosis. We treated p53-positive Molt4 T cells with increasing concentrations of forskolin, and monitored the cells undergoing apoptosis. Figure 5A shows that forskolin treatment produced a significant increase in the number of apoptotic cells in a concentration-dependent fashion. Treatment of p53-negative Jurkat T cells had no effect on cell death (data not shown). However, since these experiments did not address whether the observed apoptotic effects were directly mediated through endogenous p53, we also tested whether forskolin induced apoptosis in the p53-inducible (tet-regulated) cell line, H24 p53-3 (10). Figure 5B shows that, upon induction of p53, forskolin treatment produced a significant increase in the number of cells undergoing apoptosis. Although the effect was not as dramatic as that observed in the Molt4 T cells, Western blot analysis indicated that p53 expression was significantly lower in the H24 p53-3 cell line than in Molt4 cells (Fig. 5C).
FIG. 5.
Induction of apoptosis by forskolin. (A) Molt 4 cells were treated with the indicated concentrations of forskolin for 24 h. (B) H24 p53-3 cells (10) were removed from tetracycline-containing medium and immediately treated with forskolin for 24 h. Apoptotic cells (upper panels) were detected under fluorescent microscopy using YO-PRO-1. Lower panels correspond to bright-field microscopy of the cells presented in the upper panel. (C) Western blot analysis of H24 p53-3 extracts derived from cells grown in the presence (uninduced) (+) and absence (induced) (−) of tetracycline. Extracts from Molt4 cells were run in an adjacent lane for comparison. p53 protein was detected by Western blot analysis (DO-1).
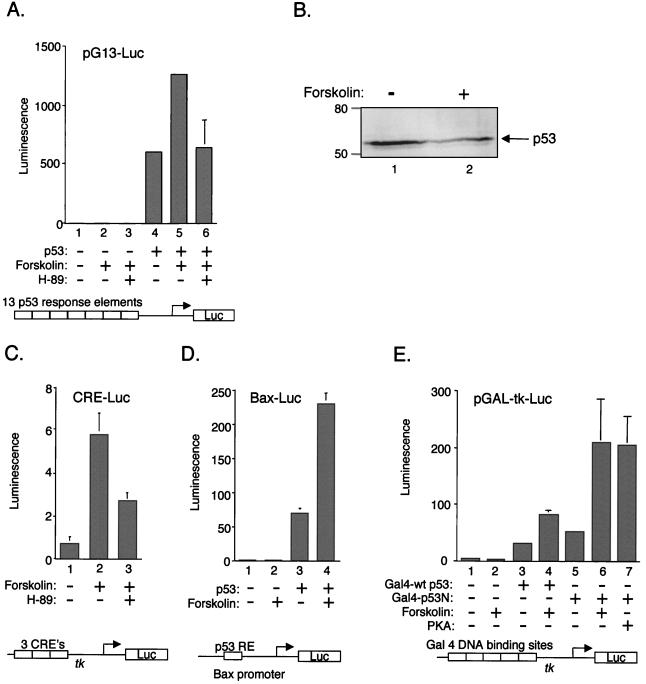
To more directly address the role of forskolin on p53 transcription function, we performed transient-transfection assays. Jurkat T cells were transiently transfected with a reporter plasmid carrying 13 copies of a p53-responsive promoter driving expression of the luciferase gene (pG13-Luc). As expected, transfection of a p53 expression plasmid dramatically stimulated luciferase expression from the p53-responsive reporter plasmid (Fig. 6A, lane 4). Treatment of the cells with 20 μM forskolin further increased p53-dependent transcriptional activity, resulting in an additional twofold stimulation. The effect of forskolin was only observed in the presence of transfected p53, suggesting that the effect was dependent upon transcriptionally competent p53 in the cell (Fig. 6A, compare lanes 2 and 5). The effect of forskolin was likely mediated through CREB (or a related factor in the cell), as there is no evidence for direct PKA phosphorylation of p53 (1). Additionally, since pG13 carries only reiterated p53 response elements upstream of a minimal promoter, it is likely that the observed stimulation by forskolin was mediated through these elements. Figure 6B shows that forskolin treatment had little or no effect on p53 levels produced in the transfection assay.
FIG. 6.
(A) Forskolin activates p53-dependent transcription in vivo. The p53-responsive pG13-Luc reporter (400 ng) was cotransfected into p53-null Jurkat T cells with the p53 expression plasmid (pC53-SN3) (400 ng), as indicated. Forskolin (20 μM) and/or H-89 (5 μM) was added (+) or not added (−) to the indicated transfection reactions. (B) Forskolin had no effect on transfected p53 protein levels. Western blot analysis of p53 derived from cells transfected with pC53-SN3 (400 ng), in the absence (−) and presence (+) of forskolin (20 μM). (C) A CREB-responsive promoter responds appropriately to forskolin and H-89. The reporter plasmid CRE-Luc (1 μg) (24) was transfected in Jurkat T cells in the presence (+) or absence (−) of forskolin (20 μM) and/or H-89 (5 μM) as indicated. (D) Forskolin activates the p53-responsive Bax promoter. The Bax-Luc reporter plasmid (400 ng) was cotransfected in Jurkat T-cells with the p53 expression plasmid (pC53-SN3) (400 ng) in the presence (+) or absence (−) of forskolin (20 μM), as indicated. (E) The amino terminus of p53 is sufficient for forskolin responsiveness. The Gal4 reporter plasmid (pGalTK-Luc) (400 ng) was cotransfected in Jurkat T cells with the Gal4-p53 expression plasmids carrying either full-length p53 (pGal4-wtp53) or the amino terminus of p53 (aa 1 to 52) (pGal4-p53N) (800 ng each) (12). Reactions were either cotransfected with the catalytic subunit of PKA (100 ng) or treated with forskolin (20 μM) as indicated. Reported values for each transient-transfection experiment are the average luminescence + the standard error (error bar) from one experiment performed in duplicate. Each experiment was performed at least twice.
Since forskolin is an activator of adenylate cyclase, we were interested in determining more specifically whether the p53-dependent transcriptional stimulation was mediated directly through PKA. To address this question, we simultaneously treated the transfected cells with both forskolin and the drug H-89, a specific pharmacological inhibitor of PKA. H-89 appeared to abolish the forskolin stimulation of pG13-Luc, reducing transcription to the level observed with transfected p53 alone (Fig. 6A, lane 6). Figure 6C shows that forskolin and H-89 were functioning correctly in the assay, as both drugs appropriately activated and inhibited transcription initiated through a minimal cAMP responsive promoter (lanes 1–3).
We were also interested in testing a natural p53-responsive promoter in the transient-transfection assay, as pG13 is an artificial promoter construct and may not behave in a physiologically appropriate fashion. To address this question, we tested the effect of forskolin on the p53-responsive Bax promoter, linked to luciferase. Figure 6D shows that expression from the Bax promoter was strongly stimulated by forskolin, in a p53-dependent manner (lanes 1 to 4).
In Fig. 4D, we present a co-IP experiment that provides evidence suggesting that the amino terminus of p53 is involved in interaction with CREB. Based on this observation, we were interested in testing whether this region of p53 is sufficient for PKA-dependent transcription in transient-transfection assays. We utilized a truncation mutant of p53, which carried only the amino-terminal 52 aa of the activation domain of p53, fused to the DNA binding domain of Gal4 (pGal4-p53N) (12). Figure 6E shows that cotransfection of Gal 4-wtp53 and Gal4-p53N similarly enhanced transcription from the Gal4-Luc reporter plasmid (compare lanes 3 and 5). This observation is not unexpected, as the amino terminus of p53 encompasses the primary activation domain of the protein. The addition of forskolin to the transfection reaction strongly enhanced Gal4-p53N-dependent transcription, with the stimulation exceeding that observed with the full-length protein (Fig. 6E, compare lanes 3 and 4 with lanes 5 and 6). Cotransfection of the catalytic subunit of PKA, which is constitutively active for CREB phosphorylation, also produced strong pGAL4-p53N stimulation comparable to that observed with forskolin (Fig. 6E, lanes 6 and 7). These data provide strong supportive evidence for an interaction between pCREB and the amino terminus of p53, resulting in PKA-dependent transcriptional stimulation. The data also indicate that p53 tetramerization is not required for interaction with pCREB.
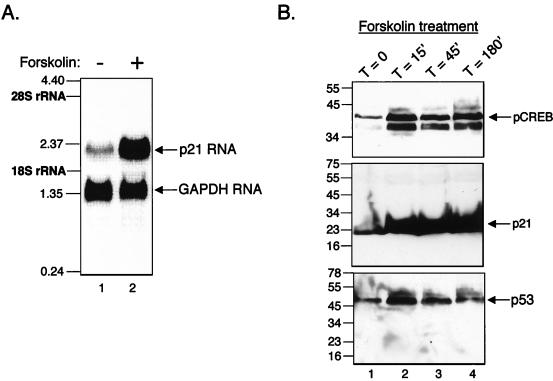
To further address the biological relevance of the cAMP pathway on p53-dependent transcription function, we tested the effect of forskolin on an endogenous p53-responsive target gene. We selected the cell cycle inhibitor gene p21 for our studies and examined p21 mRNA levels in the p53 wild-type breast cancer cell line MCF7. We selected this cell line as the p21 message was undetectable in the Molt4 T-cell lines used above. Northern blot analysis revealed that treatment of MCF7 cells with forskolin strongly stimulated p21 mRNA synthesis in vivo (fourfold) (Fig. 7A). Forskolin-induction of p21 mRNA correlated with an increase in ATF/CREB phosphorylation in vivo, as demonstrated by Western blot analysis using an antibody directed against the serine 133 phosphorylated form of CREB (Fig. 7B, upper panel). We also detected a concomitant increase in p21 protein levels, with little or no change in p53 protein levels (Fig. 7B, middle and lower panels). Interestingly, forskolin did not increase p21 protein levels in two human T-cell lymphotrophic virus type 1 infected T-cell lines, where the presence of the Tax protein inactivates the endogenous wild-type p53 (data not shown) (9, 14, 30, 36).
FIG. 7.
(A) Forskolin induces expression of the p21 gene. Shown is the northern blot analysis of total RNA (15 μg) isolated from untreated or 20 μM forskolin-treated MCF7 cells. RNA was hybridized with a p21-specific probe or a GAPDH-specific probe as a loading control. Specific transcripts are indicated. An RNA kilobase ladder and 18S and 28S rRNAs are shown at left. (B) CREB phosphorylation correlates with p21 expression. Western blot analysis of pCREB (anti-phosphoserine 133-CREB) (upper panel), p21 (gift from J. Wade Harper) (middle panel), and p53 (DO-1) (lower panel) was performed following a time course of 20 μM forskolin treatment of MCF7 cells. Protein standards (in kilodaltons) are shown at left.
DISCUSSION
Deciphering the mechanism(s) of p53 function in the cell has been one of the most intensively studied areas of modern biology. This focus derives from the critically important role p53 plays in genome surveillance and suppression of oncogenic transformation. Understanding the role of p53 as a transcription factor and the identification of p53-responsive target genes have greatly advanced our understanding of p53 function; however, many of the molecular events that contribute to p53 transcriptional activity remain elusive. The recent identification of a direct p53-CBP interaction provides insight into the transcriptional mechanisms that govern p53 function (5, 16, 26, 32, 36). In this study, we demonstrate the convergence of the PKA signaling pathway on p53 coactivator recruitment, delineating a novel mechanism of p53 coactivator utilization. We report the unexpected discovery that PKA phosphorylation of CREB at Ser133 results in p53-dependent, indirect tethering of CBP to p53-responsive genes.
Our studies support a model where the formation of a ternary complex, containing p53, pCREB, and CBP, facilitates transcriptional activation of p53 target genes. In the ternary complex, pCREB serves as a molecular bridge between p53 and CBP. Since the complex is phosphate dependent, pKID likely makes the primary, and possibly exclusive, contacts with KIX (31). Concomitantly, the bZIP domain interacts with residues located within the amino terminus of p53, without detectably interacting with the DNA. This interaction leaves the DNA binding domain of p53 available, allowing p53 to deliver the entire coactivator-containing complex to the promoters of p53-responsive genes. A schematic illustrating these interactions is shown in Fig. 8.
FIG. 8.
Model showing the ternary complex, containing CBP, pCREB, and p53, assembled on a p53-responsive promoter.
Our experiments support the idea that the ternary complex is strongly stabilized by a direct interaction between p53 and CREB. The possibility that p53 also contacts KIX in this complex cannot be excluded; however, our evidence supports the CREB-p53 interaction as the primary mode of p53 stabilization in the ternary complex. Specifically, we show that the amino terminus of p53, which carries the activation domain of the protein, is involved in direct interaction with CREB. We show that an antibody against the amino terminus of p53 blocks complex formation between p53 and CREB, and we supply evidence showing that the amino-terminal 52 aa of p53 are sufficient for mediating cAMP responsiveness in vivo. Since we demonstrate that the p53 double point mutant (L22Q-W23S) is fully active for interaction with CREB in vitro, these experiments imply that other residues within the amino terminus of p53 must participate in the interaction. Therefore, when p53 is in complex with CREB, it may be unavailable for direct coactivator interactions, as previous studies have shown that the activation domain of p53 directly contacts CBP (16, 36). Since p53 binds to DNA as a tetramer, it is possible that individual monomers participate in a combination of separable contacts with CREB and/or various domains of CBP, generating a highly complex regulatory response.
The identification of this unique ternary complex also has implications for CREB-regulated transcription, as the binding of the bZIP domain to p53 would likely abolish the DNA binding properties of the protein. Therefore, under conditions of p53 activation, pCREB may be diverted from CRE-responsive genes to p53-responsive genes. Although the physiological circumstances leading to costimulation of these pathways is not known, there is evidence that UV irradiation induces CREB phosphorylation at Ser133 (19); thus, pCREB and p53 may synergize in response to a UV-induced signal transduction pathway. More importantly, the PKA pathway of p53-activated transcription may represent an incomplete picture of this new model of p53 transcription function, as a number of other kinases, including AKT and pp90rsk, phosphorylate CREB at Ser133 (3, 8, 13, 37).
Our observation that the bZIP domain of CREB participates in the interaction with p53 raises the question of whether additional bZIP-containing transcription factors are competent for p53 binding. Evidence supporting this idea comes from the co-IP assay (Fig. 4D), where we detected up to four ATF/CREB immuno-reacting family members. It is likely that one of these proteins was the cellular transcription factor ATF-1, which shares a high degree of sequence similarity with CREB (and cross-reacts with the anti-CREB antibody). It is also possible that additional bZIP proteins, outside of the ATF/CREB family, may also bind to p53, thus expanding the repertoire of cellular proteins that may partner with p53 and regulate p53-dependent transcriptional activation. These might include components of the AP-1 complex, c-fos and c-jun, and the c/EBP family of transcription factors—each in complex with a coactivator. If this prediction is correct, it would suggest that the p53-pCREB interaction characterized herein may represent a prototype of a much more general mechanism that exerts precise control of p53 transcription function.
The observations reported in this study are very reminiscent of a recent study showing that the bZIP transcription factor CHOP participates in transcriptional activation via protein-protein interactions with the AP-1 complex (35). CHOP appears to be recruited to preexisting, DNA-bound AP-1 complexes through a direct tethering mechanism. Similar to the case with CREB, the bZIP domain of CHOP is required, and it appears to be directly involved in protein-protein interaction with the AP-1 complex. The bZIP protein ATF-6 has also been shown to function in a similarly unusual fashion (39). ATF-6 interacts directly with the activation domain of serum response factor, contributing to serum stimulation of the c-fos promoter. Although the bZIP has not been directly implicated in the ATF-6/serum response factor protein-protein interaction, it is provocative that so many bZIP proteins appear to be involved in this unusual tethering mechanism.
Together, the emerging evidence suggests that transcription factors may function in unexpected ways, assembling vertically, as well as horizontally, on target promoters, and exerting multilayered transcriptional responses. The interplay between the interacting transcription factors and their associated coactivators may serve to precisely modulate responses to external stimuli, impacting decisions that control cellular differentiation, oncogenesis, and programmed cell death.
ACKNOWLEDGMENTS
H.A.G. and I.L. contributed equally to this work.
We are grateful to C.-Z. Giam for providing the GST-CREB-His6 expression plasmid, Tom Shenk for providing the Gal4 DBD-p53 expression system, Xinbin Chen for the p53-inducible cell line, J. Wade Harper for the anti-p21 antibody, and Anne Brauweiler for construction of the CREB deletion mutants.
This work was supported by INSERM (I.L.) and NIH CA80002.
REFERENCES
- 1.Adler V, Pincus M R, Minamoto T, Fuchs S Y, Bluth M J, Brandt-Rauf P W, Friedman F K, Robinson R C, Chen J M, Wang X W, Harris C C, Ronai Z. Conformation-dependent phosphorylation of p53. Proc Natl Acad Sci USA. 1997;94:1686–1691. doi: 10.1073/pnas.94.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akimaru H, Hou D X, Ishii S. Drosophila CBP is required for dorsal-dependent twist gene expression. Nat Genet. 1997;17:211–214. doi: 10.1038/ng1097-211. [DOI] [PubMed] [Google Scholar]
- 3.Andrisani O M. CREB-mediated transcriptional control. Crit Rev Eukaryot Gene Expr. 1999;9:19–32. [PubMed] [Google Scholar]
- 4.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 5.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 6.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 7.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 8.Bonni A, Brunet A, West A, Datta S R, Takasu M A, Greenberg M E. Cell survival promoted by the Ras-MAPK singalling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 9.Cereseto A, Diella F, Mulloy J C, Cara A, Michieli P, Grassman R, Franchini G, Klotman M E. p53 functional impairment and high p21waf1/cip1 expression in human T-cell lymphotropic/leukemia virus type-I-transformed cells. Blood. 1996;88:1551–1560. [PubMed] [Google Scholar]
- 10.Chen X, Ko L J, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 11.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 12.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 13.Du K, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 14.Gartenhaus R B, Wang P. Functional inactivation of wild-type p53 protein correlates with loss of IL-2 dependence in HTLV-I transformed human T lymphocytes. Leukemia. 1995;9:2082–2086. [PubMed] [Google Scholar]
- 15.Giebler H A, Loring J E, Van Orden K, Colgin M A, Garrus J E, Escudero K E, Brauweiler A, Nyborg J K. Anchoring of CREB binding protein to the human T-cell leukemia virus type-1 promoter: a molecular mechanism of Tax transactivation. Mol Cell Biol. 1997;17:5156–5164. doi: 10.1128/mcb.17.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 17.Harris N, Brill E, Shohat O, Prokocimer M, Wolf D, Arai N, Rotter V. Molecular basis for heterogeneity of the human p53 protein. Mol Cell Biol. 1986;6:4650–4656. doi: 10.1128/mcb.6.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrod R, Tang Y, Nicot C, Lu H S, Vassilev A, Nakatani Y, Giam C-Z. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol Cell Biol. 1998;18:5052–5061. doi: 10.1128/mcb.18.9.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iordanov M, Bender K, Ade T, Schmid W, Sachsenmaier C, Engel K, Gaestel M, Rahmsdorf H J, Herrlich P. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato Y, Shi Y, He X. Neutralization of the Xenopus embryo by inhibition of p300/CREB-binding protein function. J Neurosci. 1999;19:9364–9373. doi: 10.1523/JNEUROSCI.19-21-09364.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 22.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 23.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bächinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 24.Lenzmeier B A, Giebler H A, Nyborg J K. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol Cell Biol. 1998;18:721–731. doi: 10.1128/mcb.18.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine A J, Perry M E, Chang A, Silver A, Dittmer D, Welsh D. The 1993 Walter Hubert Lecture: the role of the p53 tumour-suppressor gene in tumorigenesis. Br J Cancer. 1994;69:409–416. doi: 10.1038/bjc.1994.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 27.Mestas S P, Lumb K J. Electrostatic contribution of phosphorylation to the stability of the CREB-CBP activator-coactivator complex. Nat Struct Biol. 1999;6:613–614. doi: 10.1038/10655. [DOI] [PubMed] [Google Scholar]
- 28.Ogryzko V V, Schiltz R L, Russanove V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 29.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pise-Masison C A, Kyeong-Sook C, Radonovich M, Dittmer J, Kim S-J, Brady J N. Inhibition of p53 transactivation function by the human T-cell lymphotropic virus type 1 Tax protein. J Virol. 1998;72:1165–1170. doi: 10.1128/jvi.72.2.1165-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson J H, Montiminy M R, Wright P. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interaction. Cell. 1997;91:7441–7452. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 32.Scolnick D M, Chehab N H, Stavridi E S, Lien M C, Caruso L, Moran E, Berger S L, Halazonetis T D. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 33.Shi Y, Mello C A. CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 1998;12:943–955. doi: 10.1101/gad.12.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 35.Ubeda M, Vallejo M, Habener J F. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol Cell Biol. 1999;19:7589–7599. doi: 10.1128/mcb.19.11.7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Orden K, Giebler H A, Lemasson I, Gonzales M, Nyborg J K. Binding of p53 to the KIX domain of CREB binding protein. A potential link to human T-cell leukemia virus, type I-associated leukemogenesis. J Biol Chem. 1999;274:26321–26328. doi: 10.1074/jbc.274.37.26321. [DOI] [PubMed] [Google Scholar]
- 37.Wang J-M, Chao J-R, Chen W, Kuo M L, Yen J J-Y, Yang-Yen H-F. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol Cell Biol. 1999;19:6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao T P, Oh S P, Fuchs M, Zhou N D, Chn'g L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 39.Zhu C, Johansen F E, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]