Abstract
Background & Aims
TP53 mutations underlie Barrett’s esophagus (BE) progression to dysplasia and cancer. During BE progression, the ubiquitin ligase (E3) RNF128/GRAIL switches expression from isoform 2 (Iso2) to Iso1, stabilizing mutant p53. However, the ubiquitin-conjugating enzyme (E2) that partners with Iso1 to stabilize mutant p53 is unknown.
Methods
Single-cell RNA sequencing of paired normal esophagus and BE tissues identified candidate E2s, further investigated in expression data from BE to esophageal adenocarcinoma (EAC) progression samples. Biochemical and cellular studies helped clarify the role of RNF128-E2 on mutant p53 stability.
Results
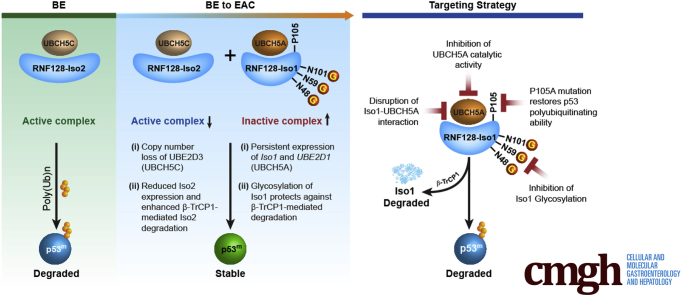
The UBE2D family member 2D3 (UBCH5C) is the most abundant E2 in normal esophagus. However, during BE to EAC progression, loss of UBE2D3 copy number and reduced expression of RNF128 Iso2 were noted, 2 known p53 degraders. In contrast, expression of UBE2D1 (UBCH5A) and RNF128 Iso1 in dysplastic BE and EAC forms an inactive E2–E3 complex, stabilizing mutant p53. To destabilize mutant p53, we targeted RNF128 Iso1 either by mutating asparagine (N48, 59, and 101) residues to block glycosylation to facilitate β-TrCP1–mediated degradation or by mutating proline (P54 and 105) residues to restore p53 polyubiquitinating ability. In addition, either loss of UBCH5A catalytic activity, or disruption of the Iso1-UBCH5A interaction promoted Iso1 loss. Consequently, overexpression of either catalytically dead or Iso1-binding–deficient UBCH5A mutants destabilized Iso1 to degrade mutant p53, thus compromising the clonogenic survival of mutant p53-dependent BE cells.
Conclusions
Loss of RNF128 Iso2–UBCH5C and persistence of the Iso1–UBCH5A complex favors mutant p53 stability to promote BE cell survival. Therefore, targeting of Iso1-UBCH5A may provide a novel therapeutic strategy to prevent BE progression.
Keywords: Barrett’s Esophagus, Esophageal Adenocarcinoma, RNF128-UBCH5 Complex, p53 Protein Stability
Abbreviations used in this paper: ANOVA, analysis of variance; BE, Barrett’s esophagus; BSA, bovine serum albumin; CpA, CpA (KR-42421) ATCC# CRL-4027 hTERT-immortalized non-dysplastic metaplasia cell line; CpD, CpD (CP-18821) ATCC# CRL-4030 hTERT-immortalized high-grade dysplasia cell line; EAC, esophageal adenocarcinoma; HBSS, Hank’s balanced salt solution; HGD, high-grade dysplasia; hg19, human reference genome; Iso, isoform; LGD, low-grade dysplasia; mRNA, messenger RNA; NDBE, nondysplastic Barrett’s esophagus; PA, Pro to Ala; RNAseq, RNA sequencing; SCFβ-TrCP1, Skp1-Cul1-F-box protein complex; SE, squamous esophagus; siRNA, small interfering RNA; UBCH5(A–D), ubiquitin-conjugating enzyme, isoform A–D; UMI, unique molecular identifier; WT, wild-type
Graphical abstract
Summary.
TP53 mutations underlie Barrett’s esophagus progression to dysplasia and cancer. Loss of UBCH5C but maintained UBCH5A expression cooperates with RNF128 Isoform 1 to protect mutant p53. Thus, targeting RNF128 Isoform 1 UBCH5A to destabilize mutant p53 may provide a therapeutic advantage.
Esophageal adenocarcinoma (EAC) is one of the most rapidly increasing cancers with frequent mutations in the tumor-suppressor gene TP53.1 We and others have shown that TP53 mutations are highly prevalent (60%–70%) in Barrett’s esophagus (BE) with high-grade dysplasia (HGD), the strongest risk factor and potential precursor to EAC.2, 3, 4, 5 TP53 mutations typically result in abnormally high nuclear p53 levels and BE cells become dependent on mutant p53 because its knock-down results in cell death.2 Strategies aimed at targeting p53 may selectively remove precursor cells before the development of EAC. However, this kind of cell-type–specific targeting has proven elusive for p53. There are many reported ubiquitin ligases that can degrade p53,6,7 including RNF128 (GRAIL),8,9 which is an E3-ubiquitin ligase that causes T-cell anergy.10 We showed that RNF128 is a critical regulator of p53 protein stability in BE through differential expression of isoform 1 (Iso1) and Iso2, which have mutually exclusive first exons.2 In tissues representing the progression from nondysplastic BE (NDBE) to EAC, we observed a significant reduction in Iso2, which efficiently degrades both wild-type and mutant p53, and an increase in Iso1 that stabilizes both forms of p53.2 The ability of RNF128 Iso2 to regulate p53 occurs via Skp1-Cul1-F-box protein complex (SCFβ-TrCP1)–mediated proteasomal degradation, whereas Iso1, stabilized by hyperglycosylation, acts to stabilize p53.2
The formation of homodimers or heterodimers of the 2 RNF128 isoforms also influences the ability of RNF128 to ubiquitinate and degrade p53 in BE cells.2 RNF128 isoforms are subject to differential regulation by glycosylation11 that might be targeted by statins, which are known to inhibit the N-linked glycosylation of specific proteins.12 In this regard, statin use is associated with a marked reduction in EAC incidence in patients with BE. Furthermore, we found that simvastatin treatment reduced colony formation of mutant p53 containing dysplastic BE cells and reduced Iso1 levels.2 These results suggest that Iso1 and Iso2 may interact differentially with specific proteins critical for their respective E3-ubiquitin ligase activities. Understanding individual E3 substrate interactions requires identifying the specific ubiquitin conjugating (E2) enzyme(s) involved in the ubiquitin transfer. Clues to the potential identity of p53 critical E2s can be found in studies into the mechanism of cadmium poisoning, which can induce nuclear accumulation of p53 in a dose-dependent manner.13 Several studies have suggested that cadmium-induced changes to p53 are the result of altered p53 degradation via tissue-specific changes in members of the UBCH5 family of E2 ubiquitin-conjugating enzymes.14,15 The 4 members, UBCH5A–D, are highly homologous (88%–92%) 147AA proteins produced from 4 discrete genes UBE2D1–4 that are highly conserved among mammals. Here, we analyze RNA sequencing (RNAseq) data from BE tissues and show UBE2D3 is a highly expressed E2 in BE and use single-cell RNAseq to show that UBE2D3 and RNF128 messenger RNAs (mRNAs) are co-expressed in NDBE cells. Herein, we examine the interaction of key UBCH5 proteins with RNF128, define the structural regions critical for catalytic activity, as well as their influence on wild-type and mutant p53 ubiquitination and stability in BE cells. Our results suggest that targeting UBCH5A catalytic activity may be a novel mechanism to cause RNF128 Iso1 degradation to impact the survival of the mutant p53-driven BE cells.
Results
Using RNAseq Data to Rank Human E2s in BE as Potential RNF128 Partners
Identifying the correct E3–E2 partnering is critical for understanding the biological context of specific ubiquitination events. Because RNF128 isoform switching resulted in inefficient mutant p53 polyubiquitination, promoting its stabilization, we sought to identify potential E2 changes that co-occur with RNF128 isoform shifts during BE progression. We first characterized mRNA expression for all known human E2 genes16, 17, 18, 19 in our previously published RNAseq expression data from a 65-sample BE and EAC tissue cohort.2 Of the 40 E2 genes, 3 (UBE2D3, UBE2E1, and UBE2I) showed good expression levels in BE (>95%; n = 52 of 54) samples with more than 32 Reads Per Kilobase of transcript per Million mapped reads (RPKM) expression (>5 log2 RPKM units), with UBE2D3 having consistently higher expression across all BE/EAC sample groups (mean, 141 RPKM vs 65 for next highest UBE2E1) (Supplementary Table 1). Although a number of E2 enzymes show varied expression patterns, several showed positive (UBE2D3, UBE2J1, and UBE2A) or negative (UBE2E1, UBE2I, UBE2D1, UBE2Q1, UBE2Q2, and UBE2W) correlations to RNF128 Iso2 (Pearson correlation, ≥0.4), but none to RNF128 Iso1. These data suggest that changes in E2 levels may associate with BE progression, along with our previously reported isoform changes to RNF128.2 In addition, yeast 2-hybrid experiments place UBE2D3 and UBE2E1, as well as UBE2D1, among the top 10 most versatile E2 enzymes in terms of E3 binding partners.16,19 Given the dependence on protein members of the UBE2D family for cadmium-induced nuclear accumulation of p53, and in particular UBCH5C from UBE2D3 and UBCH5A from UBE2D1, which appear to have opposing roles in liver cells,15 we chose to further investigate UBE2D1/UBCH5A along with those with the strongest expression (UBE2D3, UBE2E1, and UBE2I).
Co-expression of UBE2D3 and RNF128 mRNA in BE Cells
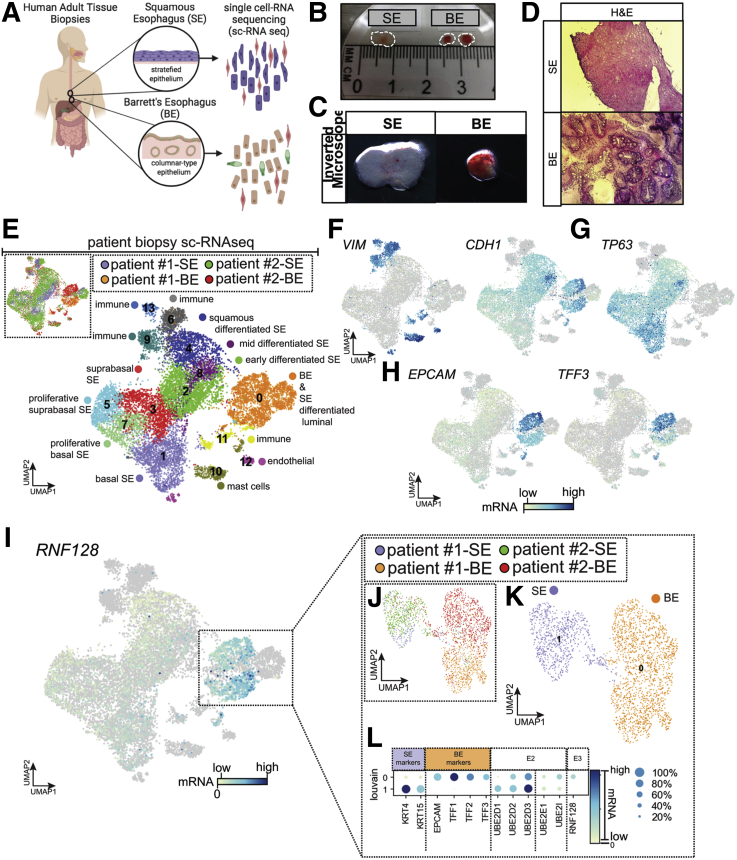
Having established UBE2D1, UBE2D3, UBE2E1, and UBE2I as candidate E2s for RNF128 in BE tissue transcriptomes, we examined the cell-type–specific expression of each relative to RNF128. Because BE and normal squamous esophagus (SE) tissues each contain a mixture of cell types, including potential islands of squamous mucosa amidst BE glandular structures, we used single-cell RNAseq to characterize the heterogenous cell types observed in NDBE with matched normal SE biopsy specimens. During the patient’s upper endoscopy procedure, we obtained fresh biopsy specimens of approximately 3 mm3 of SE and BE tissues (Figure 1A–D) from 2 patients diagnosed with NDBE. Clustering analysis showed 14 distinct transcriptomic clusters in SE (n = 2) and BE biopsy specimens (n = 2) (Figure 1E). Top cluster-specific gene (n = 5; P > .001) expression patterns were used to characterize cell types in each cluster (Figure 1F–I, Supplementary Table 2). Using known tissue characterized gene marker signatures, we identified epithelial (CDH1+) vs lamina propria clusters (VIM+). RNF128 was highly expressed in the BE-epithelial–specific cluster (C0) (Figure 1J) compared with squamous (C1, C2, C3, C4, C5, C7, and C8) or lamina propria (C6, C9, C10, C11, C12, and C13) clusters, while expression of the 4 E2 enzymes was distributed across most groups. For a more detailed assessment we extracted and reclustered the BE-specific cluster alone (C0) and observed 2 distinct subcluster types expressing either SE-specific (KRT4 and KRT15) or BE-specific (EPCAM and TFF3) markers (Figure 1J–L). Dot-plot analysis combining levels of expression and the number of cells expressing key genes again showed more general expression for each of the E2s, while RNF128 expression was present in 40%–60% of the BE cell types compared with less than 20% in SE-specific clusters (Figure 1L).
Figure 1.
Single-cell RNAseq analysis of patient-matched tissue biopsy specimens to examine co-expression patterns for RNF128 and key E2 gene mRNAs across esophageal cell type clusters. (A) Schematic (assistance from BioRender.com) of human sample biopsy specimens of matched normal SE and BE, with (B) tissue size details, (C) representative inverted microscope images, and (D) representative sections of frozen biopsy specimens from matched patient biopsy specimens of BE and SE. (E) A total of 13,332 cells were analyzed using UMAP cluster plots from SE (n = 2) and BE (n = 2) biopsy specimens. A total of 14 distinct molecular clusters were identified, with the top 200 genes per cluster listed in Supplementary Table 2. Using the top gene expression signatures composed of 5–10 characteristic genes expressed in each cluster, we define the cell type identity for each cluster. (F–H) Representative expression of known established markers in the literature for epithelial (CDH1) vs lamina propria (VIM) clusters, TP63 identifies the cell types specific to SE epithelium, and EPCAM and TFF3 are validated markers to identify metaplastic BE cells (cells with zero expression are shown in gray). We observed that cluster 0 is enriched for BE-specific cells vs clusters 1, 2, 3, 4, 5, 7, and 8, which represent SE-specific cell types. (I) RNF128 expression is higher in BE-specific clusters when compared with SE or lamina propria clusters. (J) Extraction and remapping (UMAP) of BE (cluster 0) cell types. (K) Cluster 0 vs cluster 1 identifies SE vs BE cell types. (L) Louvain analysis showing expression levels (color) vs percentage of cells expressing the gene (size of circle) shows that RNF128 is expressed specifically in the BE cluster (cluster 0; with expression of BE markers EPCAM, TFF1, TFF2, and TFF3 vs SE cluster 1; expressing stratification markers KRT4 and KRT15). We observed that E2 family member UBE2D3 is highly expressed vs UBE2D1, which has low expression in BE cluster 0.
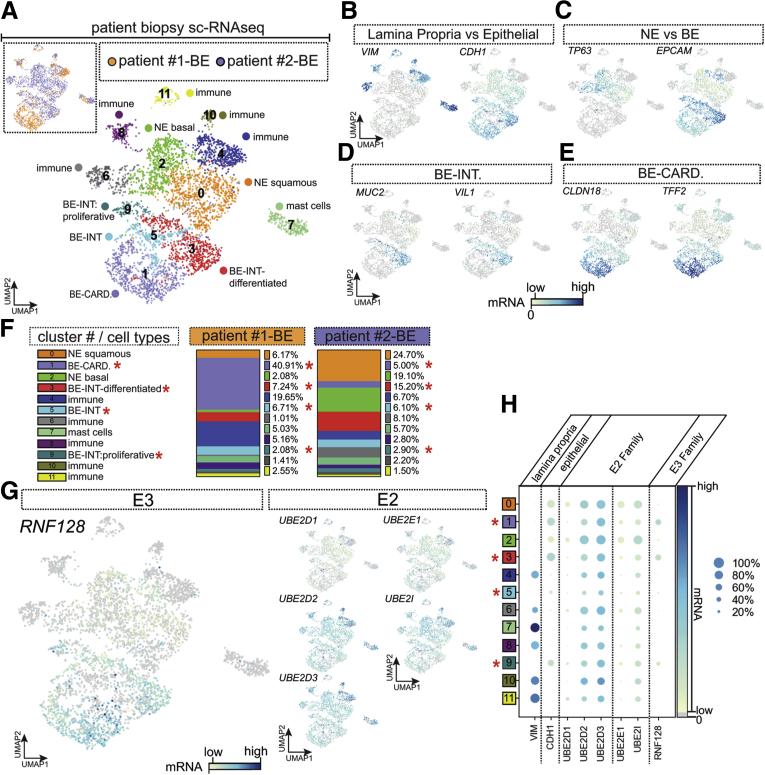
Given our focus on RNF128 as an E3 for p53 in BE cells, we independently clustered BE cell data (n = 2; final cell count, 4293) (Figure 2A), identifying 11 molecular clusters (with the top 200 genes for each outlined in Supplementary Table 3). We used known markers for stratified esophageal cells (TP63),20,21 glandular BE cells (EPCAM), goblet cells (MUC2, MUC5AC, TFF1, TFF2, and TFF3), and immune cell groups (CD247, CD3D, and CD3E). Within the overlapping but distinct columnar-epithelial clusters (C1, C3, C5, and C9) (Figure 2A–E) we could subclassify populations as either intestinal-like (MUC2, TFF3) or cardia-like (MUC5AC; TFF1, and TFF2) (Figure 2D and E) as reported previously.22 Using cell distribution plots (Figure 2F), we observed that sample 1BE was enriched with cardia-like BE cells (cluster 1: Patient 1: BE, 40.91% vs Patient 2: BE, 5.00%), while both BE samples (1:BE and 2:BE) had similar distributions of intestinal-like cells (clusters 3, 5, and 9: with 7.24:15.20, 6.71:6.10, and 2.08:2.90% in BE from Patient 1 vs BE from Patient 2, respectively). BE tissue can be enriched for either gastric or intestinal-type mucins.23, 24, 25 Thus, in cluster analyses that included both BE and SE tissues, RNF128 mRNA showed strong expression in columnar cardia cell groups, relative to other cell clusters, with particularly stark contrasts against those of squamous origin (Figures 1E and 2G and H). RNF128 expression co-localized with both MUC2- and MUC5AC-enriched components of cluster C0 (Figure 2F). A comparison of key E2 mRNA transcripts showed that, of these, UBE2D3 co-localizes with both columnar cell groups and RNF128 expression, whereas other E2 genes (including UBE2D1) show broader cell cluster expression (Figures 1I–L and 2G and H). These data show that RNF128 expression is specific to columnar epithelium, regardless of whether the BE cells have intestinal or cardia-like phenotypes, and that each of the 4 E2 mRNAs investigated co-localizes with RNF128 expression, offering the potential for interaction within BE cells.
Figure 2.
Single-cell RNA sequencing of nondysplastic BE to characterize cell types and expression patterns. (A) Patient biopsy specimens (n = 2) characterized at the single-cell level. A total of 7684 cells were analyzed for a final cell count of cell expression of 4293 for UMAP analysis. Clustering analysis showed 12 predictive distinct molecular cell clusters within the BE biopsy specimens. Cell-type classification clusters are characterized using known genes, and genes per cluster can be found in Supplementary Table 3. (B) Using known markers for lamina propria vs epithelial cells, we classified clusters based on epithelial (C0, C2, C3, C5, and C9) vs lamina propria (C4, C6, C7, C8, C10, and C11) (cells with zero expression are shown in gray). (C) Characterization of esophageal SE vs BE cell types using known markers TP63 vs EPCAM, clusters 0 and 2 are SE cells vs clusters 1, 3, 5, and 9, which express columnar-epithelial marker EPCAM (squamous cells do not express EPCAM but are positive for the basal-squamous-esophagus marker TP63). (D and E) Characterization of intestinal-type (MUC2; VIL) (clusters 3, 5, and 9) vs cardia-stomach (CLDN18; TFF2) cluster 1. (F) Quantification of cell distribution from each patient biopsy specimen into each cluster type. (G) Cell-type–specific expression levels of RNF128 (E3-ligase) vs E2-conjugating enzyme genes UBE2D1, UBE2D2, UBE2D3, UBE2E1, and UBE2I. (H) Dot-plot analysis by cluster showing normalized z-score gene expression levels (depicted by the color) vs percentage of cells expressing the gene (dot size represents the proportion of cells in each cluster expressing the marker). BE-CARD, BE cardia-like; BE-INT, intestinal-like; NE, normal (esophageal) epithelium.
Changes in UBE2D Family Members During BE Progression
UBE2D family member involvement with p53 stability appears somewhat tissue-specific, with changes in UBE2D3 and UBE2D1 showing opposite effects on p53 levels upon cadmium exposure in several model systems.15 An independent study reported that UBCH5B and UBCH5C degrade p53 relative to UBCH5A.26 In addition, high UBCH5C expression is associated with a favorable prognosis, whereas high UBCH5A appears a poor prognostic factor for esophageal and lung cancers, respectively.27,28 We next investigated the potential for BE progression-related changes in E2 mRNA levels.
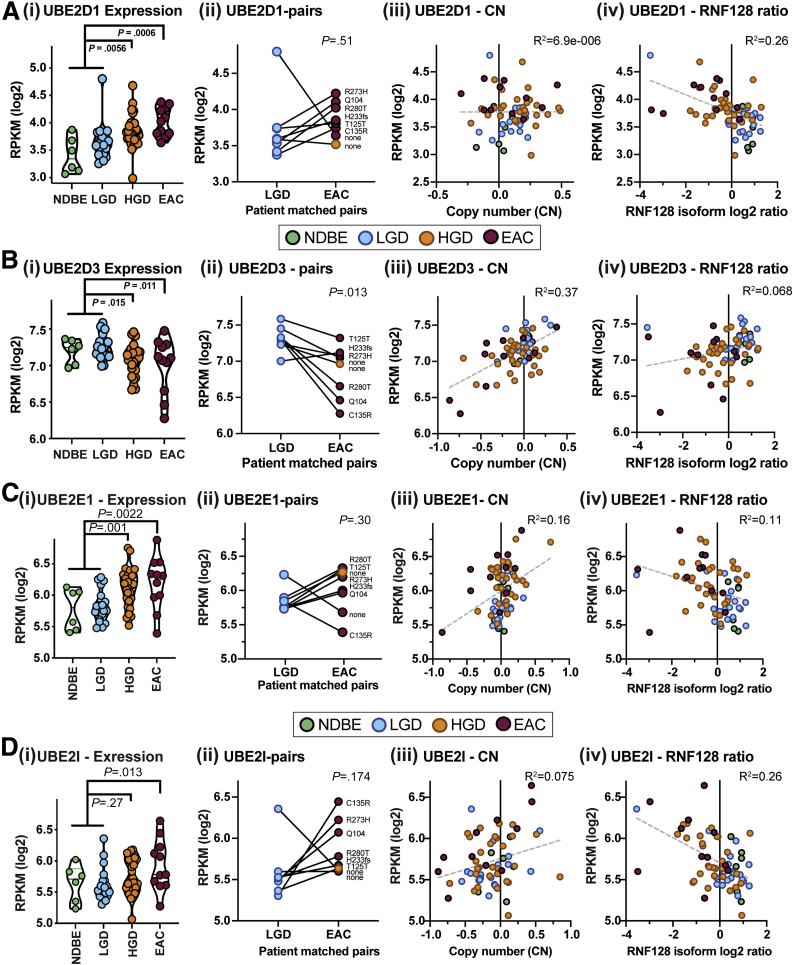
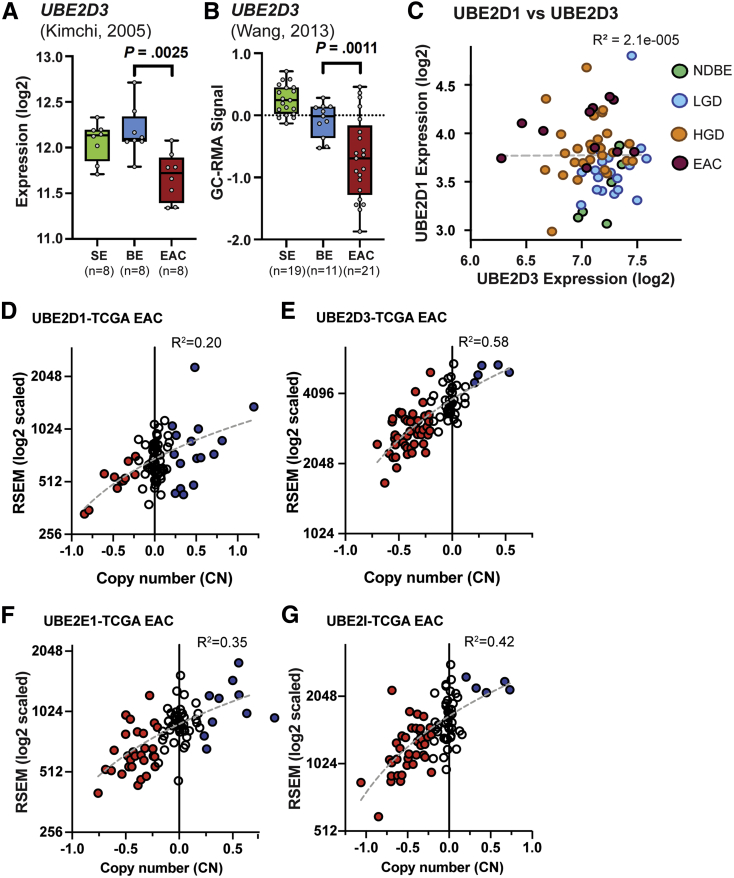
We extracted normalized expression data for UBE2D1, UBE2D3, UBE2E1, and UBE2I from our previously published BE-EAC cohort analyzed using RNAseq,2 and then applied analysis of variance (ANOVA) to compare expression levels of BE tissues containing HGD (n = 28), or EAC (n = 11), with the combined subset of NDBE only (n = 6) plus those containing low-grade dysplasia (LGD) with no histologic evidence of HGD (n = 20). These data show that UBE2D1 expression is higher in HGD (P = .0056) and EAC (ANOVA P = .0006), relative to NDBE and LGD, while UBE2D3 shows nominal group-wise expression differences in the opposite direction (P = .015 and .011 for HGD and EAC groups, respectively) (Figure 3Ai and Bi). UBE2E1 showed increased expression in HGD (P = .001) and EAC (P = .0022), while UBE2I showed a slight increase in EAC (P = .013) but not HGD (P = .27), relative to NDBE and LGD tissues (Figure 3Ci and Di). In addition, we provide nominal evidence of UBE2D3 mRNA reduction in LGD vs EAC/HGD paired t test analysis (Figure 3Bii) (n = 8; P = .013), whereas UBE2D1 and UBE2I gene expression correlate somewhat with RNF128 Iso2 mRNA levels (Figure 3Aiv and Div). Normalized Gene Expression Ombudsman data from 2 other genome-wide expression studies29,30 that used histopathology-driven selection of SE, BE, and EAC tissues showed high UBE2D3 mRNA levels in both SE and BE tissues, with reduced expression in EAC relative to BE (Welch t test P = .0025 and P = .0011 in GSE1420 and GSE26886, respectively) (Figure 4A and B). Although we saw inverse group-wise differences for UBE2D1 and UBE2D3 in our BE-related RNAseq cohort (Figure 3A and B), expression levels for the 2 genes appeared to be independent of each other across the 65 samples (Figure 4C).
Figure 3.
RNAseq-based expression analysis of top candidate E2-conjugating enzymes for RNF128. Panels show gene-based RNAseq data extracted from a 65-sample BE progression-related cohort with mRNA from NDBE (n = 6), LGD admixture (10%–100%) without HGD (n = 20), HGD admixture (10%–100%; n = 28), and EAC (n = 11) tissue samples resected from HGD or EAC patients.2 For (A) UBE2D1, (B) UBE2D3, (C) UBE2E1, and (D) UBE2I horizontal subpanels are as follows. (Ai–Di) Violin-style boxplots, with ANOVA P values for NDBE plus LGD vs HGD or EAC group comparisons. (Aii–Dii) Expression changes in a patient subset (n = 8) with matched LGD and EAC (n = 7) or HGD (n = 1), with paired t test P values for each E2 shown. (Aiii–Diii) RNAseq-based comparison of expression and expression-derived copy number estimates (using recursive median smoothing of genes mapping north and south of target loci chromosomal locations in genome version hg38) for UBE2D1 (10q21.1), UBE2D3 (4q24), UBE2E1 (3p24.2), and UBE2I (16p13.3). Pearson correlations using all 65 samples are shown. (Aiv–Div) shows expression correlations (Pearson, using all 65 samples) to the RNF128 Iso2/Iso1 log2 ratio. RPKM, Reads Per Kilobase of transcript per Million mapped reads.
Figure 4.
Confirmation of UBE2D3 changes in other BE cohorts, and RNF128 to specific E2 copy number correlations in the ESCA TCGA cohort for esophageal cancer. (A and B) Normalized deposited data from 2 GEO EAC cohorts from Kimchi et al29 (GEO series GSE1420) and Wang et al30 (GEO series GSE26886), which support the loss of UBE2D3 expression between BE and EAC, as well as the observed high UBE2D3 expression in both SE and BE. Both studies applied histopathology-driven tissue dissection and U133 Affymetrix expression array platforms, but different analytic methodologies. P values represent unequal variances t tests applied between BE and EAC groups in each cohort. (C) Lack of correlated expression between UBE2D1 and UBE2D3 across the 65 RNAseq samples in the BE progression cohort. (D–G) RNAseq by expectation maximization (RSEM) expression vs capped relative linear copy number plots in TCGA EAC samples (n = 87) from ESCA cohort31 for (D) UBE2D1, (E) UBE2D3, (F) UBE2E1, and (G) UBE2I.
Analysis of copy number changes in TCGA EAC for the same 4 Ub-conjugating E2s (Figure 4D–G), as well as the estimated copy number in our BE progression-related cohort (Figure 3Aiii-Diii), showed consistent correlations between DNA copy number change and mRNA expression levels for UBE2D3 (Pearson R2 = 0.37 and 0.58) (Figures 3Biii and 4E), less so for UBE2E1 (0.16 and 0.35, respectively) (Figures 3Ciii and 4F) and UBE2I (<0.1 and 0.42, respectively) (Figures 3Diii and 4G), but not UBE2D1 (R2 < 0.1 and 0.2, respectively) (Figures 3Aiii and 4D). These data offer a potential explanation for the reduced UBE2D3 expression levels in both HGD and EAC, with evidence of low-level copy number loss in the samples from both of these tissue sets (Figures 3Biii and 4E). It is notable that UBE2D3 (or the gene for UBCH5C) is located at chromosome 4q, and broad copy number loss on 4q is reported to be prevalent among EAC patients.32,33
RNAseq data also suggest a weak inverse correlation between UBE2D1 expression and the reduction in RNF128 isoform ratio (R2 = 0.26) (Figure 3Aiv), while changes in UBE2D3 may be linked to chromosome 4q copy number loss, which is independent of Iso2 changes (Figure 3Biv). Together, these data suggest that the independent loss of RNF128 Iso2 and UBE2D3 may represent critical events for BE progression, allowing UBCH5A-Iso1 to form an inactive E2–E3 complex that stabilizes mutant p53 (summarized in the graphical abstract).
Tighter Binding Between RNF128 Iso1 and UBCH5A Is Responsible for Reduced p53 Polyubiquitinating Ability
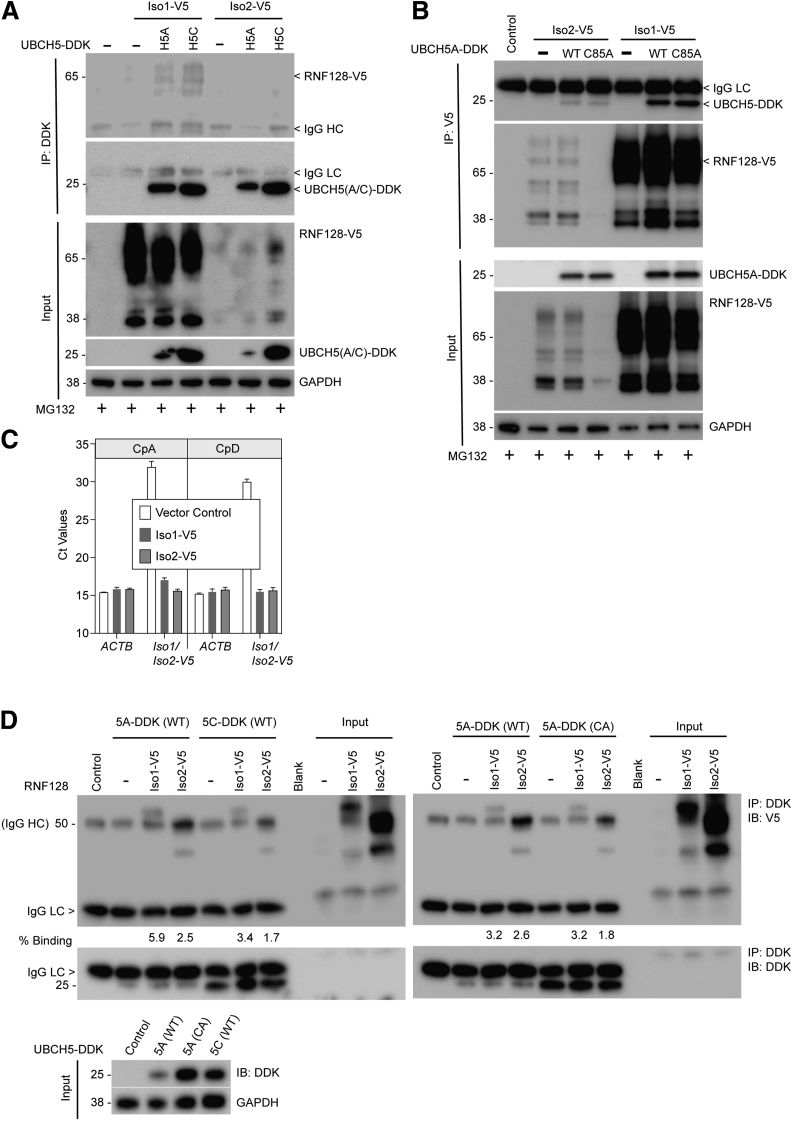
From our prior analyses2 using an in vitro ubiquitination assay platform, we noted limited p53 ubiquitination by the RNF128 Iso1 and UBCH5 complex, likely resulting in p53 stabilization. To determine the basis for the lack of catalytic activity of Iso1-UBCH5 complex on p53, we compared the binding of the 2 E3 isoforms (Iso1 and 2) with UBCH5 (A and C). As shown in Figure 5A, the interaction of Iso1 was readily detected with both E2 proteins, in contrast to their interaction with Iso2. Successful E3–E2 interactions often are too moderate or weak to facilitate easier transfer of activated ubiquitin from E2 to E3, and then to the substrate.18 Thus, a tighter E3–E2 interaction suggests suboptimal functionality, as we reported in an independent study.34 To clarify this effect on the E2 catalytic activity, we compared the Iso1 and Iso2 interaction with either the wild-type (WT) or a catalytically dead Cys85Ala (CA) mutant of UBCH5A. Our results indicate (Figure 5B) that the interaction of Iso1 with UBCH5A was tighter irrespective of E2 catalytic activity. As noted previously and seen here (Figure 5A–C), the differences in protein expression for Iso1 and Iso2 was not owing to differential transcription. We also always observed more abundant Iso1 protein expression relative to Iso2 when overexpressed in multiple BE cells.2 To address such large differences in Iso1 and Iso2 protein expression, we synthesized both isoforms using an in vitro transcription/translation system, resulting in relatively comparable quantities. When these synthesized isoforms (nonglycosylated) were subjected to interaction studies with UBCH5A or UBCH5C, we noted an approximately 2-fold stronger association of Iso1 with both UBCH5 family members compared with Iso2 (Figure 5D), which is consistent with the observations when interaction studies were performed in cells (Figure 5A and B).
Figure 5.
RNF128 Iso1 binds tighter with UBCH5 family members. (A) V5-tagged RNF128 (either Iso1 or Iso2) were co-transfected either with UBCH5A or UBCH5C as indicated in CpA cells. Twenty hours after transfection, cells were treated with proteasomal inhibitor MG132 (2 μmol/L for 4 hours) before harvest and immunoprecipitation followed by immunoblotting using indicated antibodies. (B) To test the interaction ability between RNF128 Iso1 and Iso2 with either WT or catalytic-dead (C85A) mutant UBCH5A, we performed co-transfection, immunoprecipitation, and immunoblot analysis as described earlier using the indicated antibodies. (C) Quantitative reverse-transcription polymerase chain reaction showing exogenous Iso1 or Iso2 transcript expression in CpA and CpD cells along with ACTB (housekeeping) compared with cell lines transfected with empty vector. Cycle threshold (Ct) expression values, rather than ΔΔCt ratios are presented, given the lack of RNF128 construct in the vector controls. Error bars represent the triplicate Ct range for each condition. (D) DDK-tagged UBCH5A (either WT or C85A mutant) and UBCH5C proteins were overexpressed in HEK293 cells, and 24 hours after transfection cell lysates were subjected to immunoprecipitation using FLAG-M2 beads. Thoroughly washed beads then were incubated with equal volumes of in vitro transcribed and translated V5-tagged either RNF128 Iso1 or Iso2. Unlike cells, Iso2 synthesis was approximately 1.9-fold higher compared with Iso1. Compared with input, 100-fold more amounts of Iso1 and Iso2 were used for interaction studies and the percentage binding was calculated as shown after normalization of input band intensity using ImageJ software (National Institutes of Health, Bethesda, MD). Lower panel: Expression of different UBCH5 family members in HEK293 cells. DDK, DYKDDDDK tag; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HC, Heavy Chain; IP, Immunoprecipitation; LC, Light Chain.
In RNF128 Iso1, N-terminal Glycosylation Helps Maintain Stability, Whereas the Proline Loop Is Important for Functionality
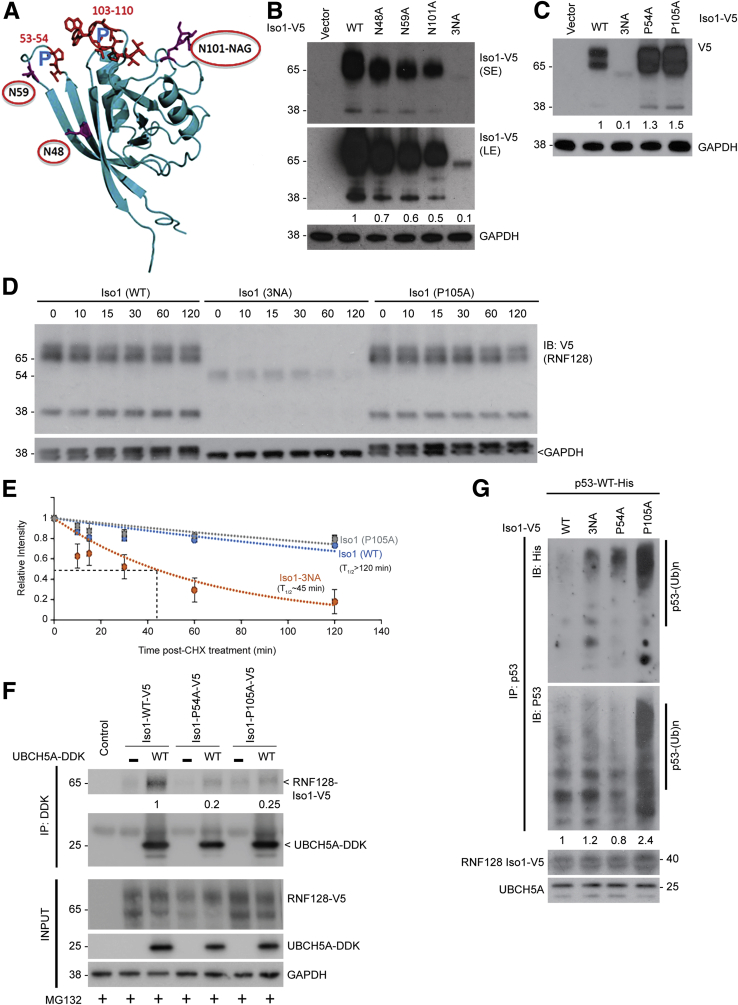
To better understand the differential binding abilities of the 2 RNF128 isoforms with UBCH5A, we analyzed the existing Iso1 crystal structure (Protein Data Bank ID: 3ICU) and identified 2 unique features absent in Iso2: 3 asparagine (N) residues at amino acid positions 48, 59, and 101, and 2 proline (P) residues at positions 54 and 105 (Figure 6A). While analyzing Iso1 glycosylation mutants, we noted lower steady-state levels of the Iso1 single mutants, which was even more pronounced in mutants in which all 3 N-residues were changed to alanine (3NA) (Figure 6B), suggesting the importance of N-terminal glycosylation in maintaining Iso1 protein levels. We also performed site-directed mutagenesis to create Iso1 P54A and P105A mutants. In contrast to 3NA mutant, the Pro to Ala (PA) mutants showed minimal change in steady-state levels compared with the wild-type protein (Figure 6C). To assess the influence of Iso1 glycosylation and disruption of proline loops on protein stability, we also performed protein half-life studies. As shown in Figure 6D and quantified in Figure 6E, the Iso1 3NA mutant was highly unstable (t1/2 ~45 min) relative to WT Iso1 (t1/2 >120 min), suggesting the importance of glycosylation in maintaining Iso1 protein stability. The protein half-life of the P105A mutant was comparable with that of the WT protein (Figure 6D and E).
Figure 6.
Asparagine (N) glycosylations of Iso1 are important for protein stability, whereas loops at proline residues impact UBCH5 interaction to modulate functionality. (A) Crystal structure of RNF128 Iso1 (modified from Protein Data Bank 3ICU) showing asparagine (N) and proline (P) residues. (B) Steady-state levels of wild-type and glycosylation-deficient mutants of RNF128 (N48A, N59A, N101A, or triple mutant 3NA) in CpA cells (short exposure [SE] and long exposure [LE]). (C) Similar studies as described earlier showing steady-state levels of P54A and P105A mutants compared with WT and 3NA mutants of Iso1. (D) CpA cells were transfected with either WT, 3NA, or P105A Iso1 mutants. Twenty-four hours after transfection, cells were treated with cycloheximide (50 μg/mL) for the indicated times followed by immunoblotting. (E) To calculate protein half-life, relative band intensities were determined from 3 independent experiments using ImageJ (National Institutes of Health), normalized to the amount of protein at the initial time point and plotted (means ± SD). (F) UBCH5A (WT) protein showed weaker interaction with RNF128 Iso1 P54A and P105A mutants compared with WT Iso1. (G) RNF128 Iso1 (WT, 3NA, P54A, and P105A) proteins were synthesized using Transcription and Translation coupled in vitro to transcription and translation systems. In vitro ubiquitination assay of His-tagged p53 (WT) was performed using the earlier translated E3s along with the recombinant UBCH5A. Subsequently, p53 protein was immunoprecipitated using a specific antibody followed by immunoblotting using indicated antibodies. Fold-change of p53 polyubiquitinated species generated were quantified relative to WT Iso1-V5. CHX, Cycloheximide; DDK, DYKDDDDK tag; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IB, Immunoblotting; IP, Immunoprecipitation.
Proline residues form structural kinks, and these residues, are located proximal to the 2 loops unique to Iso1. Therefore, we rationalized that mutation of P54 and P105 could alter the loop conformation and allow weaker UBCH5A association. As shown in Figure 6F, both the PA mutants showed weaker binding with UBCH5A, supporting our hypothesis. We then performed the in vitro ubiquitination assay of Iso1 PA mutants for comparison with WT protein. The P105A mutant showed increased ubiquitinating ability toward p53 relative to the WT Iso1, however, both the P54A and 3NA mutants showed lower ubiquitin ligase activity similar to WT RNF128 Iso1 (Figure 6G). Previously, we reported that either small interfering RNA (siRNA)-mediated loss of mutant p53 or siRNA-mediated knockdown of RNF128 Iso1, which indirectly promotes mutant p53 down-regulation, can reduce clonogenic survival of mutant p53-driven BE (CpD [(CP-18821) ATCC# CRL-4030 hTERT-immortalized high-grade dysplasia cell line]) cells.2 Glycosylation inhibitors might be tested on impacting the survival of CpD cells, however, instead of a general inhibitor (eg, tunicamycin), a more specific inhibitor may be essential to reduce possible toxicity. We then focused our attention on other factor(s) that may impact RNF128 Iso1 protein stability.
UBCH5A Catalytic Activity Is Important for Maintaining RNF128 Iso1 Protein Stability via Protecting it From β-TrCP1–Mediated Degradation
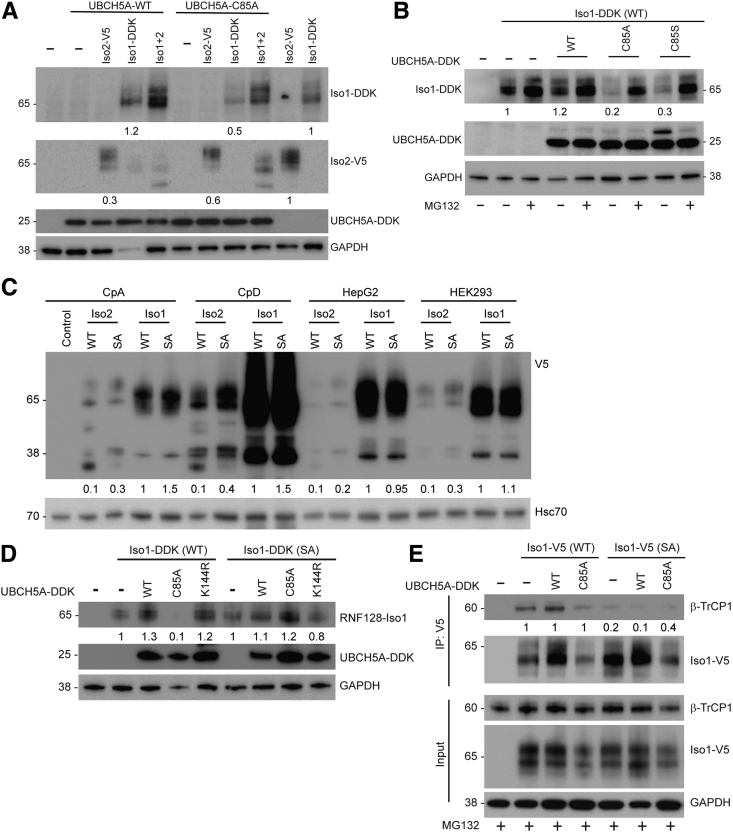
The cysteine located at 85 position (C85) is the catalytic residue for UBCH5A functionality.35 To understand the importance of this residue for its interaction with RNF128 Iso1, we created catalytically dead C85A and C85S mutants. While expressing the UBCH5A C85A mutant in BE cells, we noted a reduction in the steady-state levels of RNF128 Iso1 (Figure 7A). To further confirm and understand the basis for this down-regulation, we performed similar studies using both UBCH5A C85A and C85S catalytic inactive mutants in the presence and absence of the proteasomal inhibitor MG132. As shown in Figure 7B, overexpression of either UBCH5A catalytically dead mutant resulted in significant down-regulation of RNF128 Iso1, which was rescued in the presence of proteasomal inhibitor MG132. Importantly, agents capable of inhibiting UBCH5A activity, thus causing RNF128 Iso1 degradation to impact mutant p53 stability, may have translational value. To identify the ligase responsible for Iso1 degradation in the absence of UBCH5A catalytic activity, we used previously created serine 368/369 to alanine (SA) mutants of RNF128 Iso1 and Iso2, which we had reported as containing a phosphodegron recognized for SCFβ-TrCP1–mediated degradation.2 When we tested these SA mutants in 4 different cell lines, we noted increased steady-state levels for both isoforms (Figure 7C), emphasizing the importance of these serine phosphorylation sites in maintaining RNF128 isoform protein stability. Previously, we identified UBCH5A as the E2 responsible for β-TrCP1 degradation,36 which allowed us to hypothesize that in the absence of UBCH5A catalytic activity, β-TrCP1 may get enriched to promote Iso1 degradation. Consistent with this expectation, the Iso1 SA mutant was resistant to UBCH5A C85A-mediated degradation (Figure 7D). We further show that Iso1-SA mutant protection is owing primarily to the reduced interaction with β-TrCP1 (Figure 7E). When considering these observations together, we conclude that impacting UBCH5A catalytic activity may be a novel approach to target RNF128 Iso1 and reduce mutant p53 accumulation.
Figure 7.
UBCH5A catalytic activity is important in maintaining RNF128 Iso1 stability by protecting it from β-TrCP1–mediated degradation. (A) RNF128 Iso1 and Iso2 steady-state levels were determined in CpA after co-overexpression of either UBCH5A WT or C85A mutants. (B) To determine the effect of UBCH5A catalytic activity on the stability of Iso1, CpA cells overexpressing the indicated proteins were either left untreated or treated with MG132 followed by immunoblotting using the indicated antibodies. (C) Iso1 and Iso2 either WT or β-TrCP1 recognizing phosphodegron-deficient (SA) mutants were overexpressed in multiple cell lines (CpA, CpD, HepG2, and HEK293). Twenty-four hours after transfection, cell lysates were subjected to immunoblotting using the indicated antibodies. (D) Different Iso1 mutants (WT or SA mutant) were co-overexpressed in CpA cells either with WT, C85A, or K144R mutant UBCH5A. Cell lysates were prepared 24 hours after transfection followed by immunoblotting. (E) Interaction of RNF128 Iso1 (WT and SA mutant) with β-TrCP1 in the presence of UBCH5A (WT and C85A mutants). Twenty hours after transfection cells were treated with MG132 for 4 hours and cell lysates were subjected to immunoprecipitation followed by immunoprecipitation as indicated. DDK, DYKDDDDK tag; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Hsc70, Heat shock cognate 71 kDa protein; IP, Immunoprecipitation.
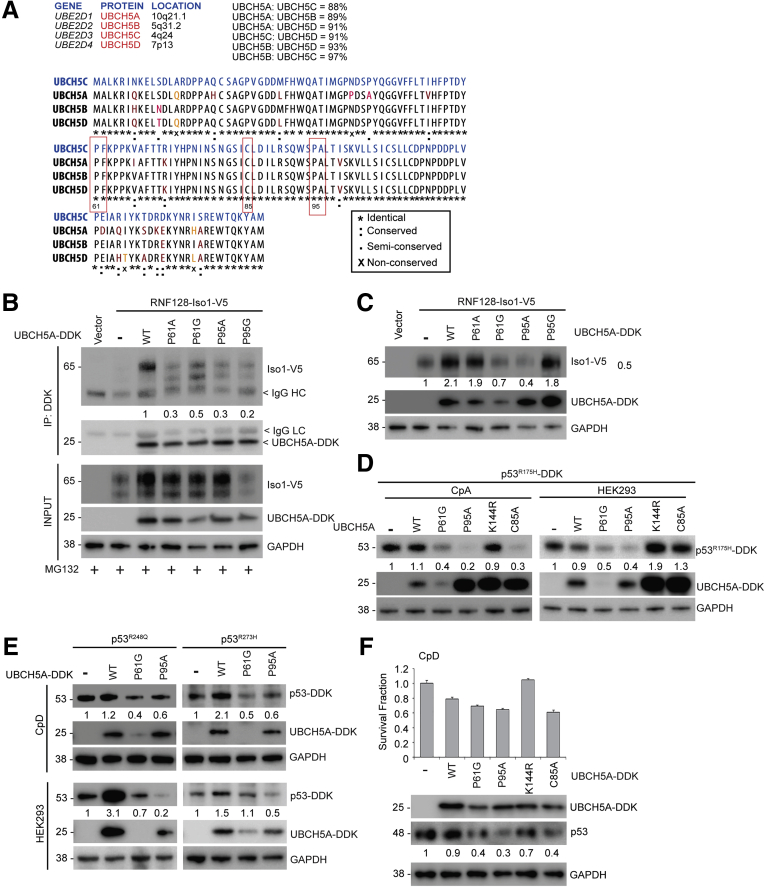
Targeting of RNF128 Iso1 via Altering UBE2D1 Catalytic Activity Destabilizes Mutant p53 and Reduces Clonogenic Survival of CpD Cells
It was reported previously that, in addition to the catalytic cysteine (C85), there are 2 additional sites important for maintaining UBCH5A catalytic activity (61-PF-62 and 95-PA-96), because these residues are known to be key interaction sites for other E3 ligases, including Carboxyl terminus of Hsc70-Interacting Protein (CHIP), another key E3 ligase involved in mutant p53 degradation.37 In an independent study, F62A and A96D mutants of UBCH5A were shown to lose significant catalytic activity.38 These critical residues are conserved among all UBCH5 family members (Figure 8A). To test the importance of these residues, we used a panel of 4 previously created mutants, with 2 for each residue (P61A, P61G, P95A, and P95G),34 to test the effect of each PA mutant’s overexpression on RNF128 Iso1 binding and steady-state levels. In our hands, although P61A/G and P95A/G mutants of UBCH5A interacted weakly with Iso1 (Figure 8B), only P61G and P95A mutants showed effective down-regulation of RNF128 Iso1 (Figure 8C), as seen with the C85A UBCH5A mutant. Consequently, overexpression of UBCH5A P61G and P95A mutants (but not P61A and P95G) efficiently down-regulated a panel of the most commonly observed p53 mutants in BE/EAC39 (R175H, R248Q, and R273H) in CpA (KR-42421) ATCC# CRL-4027 hTERT-immortalized non-dysplastic metaplasia cell line, CpD, and HEK293 cells (Figure 8D and E). In contrast, another UBCH5A mutant (K144R),36 which does not directly impact the E2 catalytic activity, did not down-regulate mutant p53. Consequently, overexpression of P61G, P95A, and C85A UBCH5A mutants trended toward reduced clonogenic survival of mutant p53-driven CpD cells, whereas the K144R mutant did not (Figure 8F). Taken together, we identified that targeting RNF128 Iso1-UBCH5A interaction may compromise E3–E2 catalytic activity to cause mutant p53 degradation, which may be a novel mechanism to cause RNF128 Iso1 degradation, leading to an impact on the survival of the mutant p53-driven BE cells.
Figure 8.
Targeting of RNF128 Iso1 via alteration of UBCH5A catalytic activity to destabilize mutant p53. (A) Upper left panel: Table showing UBE2D family member genes (UBE2D1–4), protein (UBCH5A–D) and their respective chromosomal locations. Upper right panel: Percentage sequence homology between UBCH5 family members. Lower panel: Pairwise amino acid sequence alignments between UBCH5A–D isoforms performed using the CLASTALW multiple sequence alignment tool. Redboxes identify 61PF62, C85. and 95PA96 residues as discussed in the article. (B) The RNF128 Iso1-V5 constructs were co-expressed with the indicated UBCH5A mutants (WT, P61A/G, and P95A/G). Twenty hours after transfection, cells were treated with MG132 as described earlier, followed by immunoprecipitation and immunoblotting as indicated. (C) Similar co-transfection was conducted in CpA cells as described earlier and cell lysates were subjected to immunoblotting with the indicated antibodies. (D) CpA (left panel) and HEK293 (right panel) cells overexpressing P53R175H-DDK mutant were co-transfected with different UBCH5A mutants as indicated and 48 hours after transfection mutant p53 levels were determined using immunoblotting as indicated. (E) As described earlier, p53R248Q and p53R273H mutants showed down-regulation upon overexpression of P61G and P95A mutant UBCH5A in CpD (upper panel) and HEK293 cells (lower panel). (F) UBCH5A (WT, P61G, P95A, K144R, and C85A mutants) were overexpressed in CpD, and 48 hours after transfection cells were plated at clonal density to perform a clonogenic assay. Survival fractions upon UBCH5A overexpression are shown in the top panel and the corresponding cell lysates were subjected to immunoblot analysis using the indicated antibodies. DDK, DYKDDDDK tag; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IP, Immunoprecipitation.
Discussion
Independent studies reported that UBCH5B and UBCH5C have better abilities to degrade p53 relative to UBCH5A.26 In addition, UBCH5C is associated with a favorable prognosis, whereas UBCH5A is a poor prognostic factor for esophageal and lung cancers, respectively.27,28 Mutant p53 is the most frequent mutational event in EAC and is associated with progression of BE to EAC.31 Identifying methods to target the stable mutant p53 protein may provide a means of removing critical precursor cells in BE having the greatest potential to progress to EAC. Our work in BE showed that RNF128 (GRAIL), an E3 ubiquitin ligase, plays a critical role in regulating the stability of the p53 protein.2 RNF128 has 2 isoforms: Iso1 accumulates in the progression of BE to EAC, and Iso2 dramatically decreases in expression. Iso1 stabilizes mutant p53 whereas Iso2 degrades mutant p53. Therefore, the identification of mechanisms that either restore Iso2 expression or reduce the expression or activity of Iso1, might represent novel strategies to target BE cells containing mutant p53. In this study, we characterized the heterogenous cell types found in NDBE and identified RNF128 as highly expressed in BE cells relative to SE cells (Figures 1 and 2). Furthermore, we found that regardless of intestinal vs cardia BE subtypes, RNF128 was expressed in both, and key regulatory E2 family members such as UBE2D1 were low in BE whereas UBE2D3 was highly expressed. We defined potential regulators of RNF128 isoforms at the protein level and examined specific proteins interacting with RNF128 Iso1 and Iso2, identifying several E2 ubiquitin ligases including UBCH5A and UBCH5C. During progression of BE to EAC we saw concurrent loss of RNF128 Iso2 and UBCH5C, both potential critical events for BE progression that allowed Iso1–UBCH5A partner dominance to stabilize mutant p53 (see graphical abstract). Chromosome 4q loss was reported to be a frequent event in EAC,32,33 however, the critical genes remain unclear. Our data suggest that UBE2D3, encoding UBCH5C, may be one of these loss of heterozygosity target genes.
Ubiquitination and the subsequent degradation of mutant p53 requires both E2 and E3 partners, which need to sufficiently, but weakly, interact to transfer the ubiquitin moiety to the substrate molecule.18 Importantly, we found interaction of Iso1 was stronger with both (UBCH5A and UBCH 5C) E2 proteins in marked contrast to Iso2 (Figure 5A). This stability suggests the transfer of ubiquitin to mutant p53 by Iso1 is less efficient and potentially independent of E2 catalytic activity, as indicated by comparing the interaction of Iso1 and Iso2 with either the WT or C85A (CA) mutants of UBCH5A (Figure 5B). As noted here and as we reported previously,2 the 2 RNF128 isoforms consistently are expressed at substantially different levels regardless of the expression vector backbone and under the cytomegalovirus-driven promoter carrying different C-terminal tags (either FLAG or V5). To better understand this difference in isoform expression, we used an in vitro transcription/translation system to synthesize both isoforms. Using this system, comparable quantities of both isoforms were synthesized, suggesting biological factors control exon-mediated, isoform-specific expression of RNF128. Consistent with our previous observation,2 phosphorylation is one of the mechanisms involved in RNF128 protein stability (Figure 7C). Although the site-specific S to A mutation of Iso2 showed increased levels compared with the WT protein, it still was substantially lower when compared with Iso1, suggesting additional regulatory mechanisms control Iso2 protein levels. One caveat of the in vitro transcription/translation system is that the synthesized proteins lack any post-translational modifications including glycosylation, whose effect on RNF128-UBCH5A/UBCH5C interaction remains unknown.
To define the differential binding abilities of the 2 RNF128 isoforms to the E2, we examined several unique aspects of their protein structures. The 2 isoforms have identical C-terminal regions yet differ in the first exon and thus the protein’s N-terminus.2 Inspection of the crystal structure of RNF128 showed 2 unique features of Iso1 including 2 proline residues at positions 54 and 105 (Figure 6). Mutation of the proline to alanine at the 54 and 105 positions resulted in possible disruption of the 2 predicted loops that are putative sites for protein–protein interaction for Iso1. This apparently weakened the interaction between the PA mutants of Iso1 and UBCH5A (Figure 6F), and, importantly for the Iso1 P105A mutant, restoration of p53 ubiquitination ability (Figure 6G). Relative impact of P105 appears to be greater than the P54 site possibly owing to adjacent N101 glycosylation. Expression of the 3NA triple mutant (N48/N59/N101 to A) at these potential glycosylation sites showed minimal improvement on Iso1-mediated p53 polyubiquitination. It may be interesting, however, to create a P105A mutation in a 3NA background to examine the role of these sites on Iso1 ubiquitin ligase activity.
In mutant p53-driven dysplastic BE cells, either the loss of mutant p53 or siRNA-mediated knockdown of RNF128 Iso1 led to reduced clonogenic survival,2 suggesting Iso1 targeting may have translational potential. Because loss of Iso1 glycosylation in 3NA mutant reduced Iso1 protein stability (Figure 6D and E), targeting glycosylation thus may represent a strategy to influence Iso1 abundance. Currently, there are several glycosylation inhibitors undergoing clinical testing,40 presenting an opportunity for future investigation.
We examined 2 sites within UBCH5A critical for its catalytic activity, which when mutated (P61G and P95A) and expressed in BE cells, were both able to reduce Iso1-UBCH5A interaction (Figure 8B) and in turn result in Iso1 down-regulation (Figure 8C). Most importantly, overexpression of these UBCH5A mutants deficient in Iso1 interaction can cause the degradation of commonly observed p53 mutants, including R175H, R248Q, and R273H (Figure 8D and E). Therefore, these studies suggest impacting UBCH5A catalytic activity may be a novel approach to target RNF128 Iso1 for degradation, resulting in the death of mutant p53-containing BE cells, consistent with our previous use of RNF128 Iso1-specific siRNA.2 It was interesting to note that specific amino acid substitution (alanine vs glycine) at the P61 and P95 positions had differential impact on promoting Iso1 down-regulation (Figure 8C). A similar observation on functionality was noted for the enzyme phenylalanine-specific permease in Escherichia coli.41 Because proline residues allow kink formation42 to regulate protein–protein interaction,43 it is plausible that substitution with different amino acids can differentially impact β-TrCP1 recruitment to the complex, which effects RNF128 Iso1 stability differently.
Our study had several limitations. We performed single-cell RNA sequencing in paired patient samples from SE and BE from the 3’ end of the transcript. Because the RNF128 isoforms differ at their 5’ end, our data cannot provide insight into isoform-specific RNF128 alterations in different cell types. In future studies, we plan to experiment with isoform-specific single-cell sequencing and use dysplastic/EAC samples to validate isoform-specific switching of RNF128, UBE2D, and other E2 family members at the single-cell level. We also acknowledge the lack of immunohistochemistry data showing isoform-specific UBCH5 changes during BE progression. However, as shown in Figure 8A, because of the close homology, generation of isoform-specific UBCH5 antibodies may not be feasible. Clonogenic assays (Figure 8F), however, showed evidence for reduced survival of CpD cells upon UBCH5A mutant (P61G, P95A, and C85A) overexpression, and we attribute the minimal differences in colony survival to the transient nature of overexpression. Further experiments are necessary to substantiate and characterize these differences either via establishing stable cell lines overexpressing key variant UBCH5A mutant proteins or by treating dysplastic BE cells carrying mutant p53 with previously reported UBCH5 catalytic inhibitor.44
In conclusion, the increasing incidence of EAC in the United States and other Western countries,45 the high incidence of p53 mutations, and the fact that they arise early in BE progression3 all point to the potential of targeting mutant p53-containing BE cells. We are currently examining small molecules that disrupt RNF128 Iso1–UBCH5A interactions, which have the potential to target mutant p53-containing BE cells.
Methods
Sample Collection
Biopsy specimens of normal esophageal SE and matched NDBE were collected from consented subjects undergoing a scheduled upper endoscopy for BE screening within the University of Michigan Health System between 2017 and 2020. Samples were collected using protocols approved by the University of Michigan Institutional Review Board. Research biopsy specimens were either snap-frozen in liquid nitrogen and stored at −80°C until use or used fresh for single-cell RNAseq processing.
Tissue Processing and Staining
Tissue histology was confirmed on adjacent biopsy specimens using a modified H&E stain. Frozen samples were placed within a mold containing Optimal Cutting Temperature (OCT) cryostat solution. The block was frozen at −80°C and then secured onto the sectioning mount within the cryostat. Sections 10-μm thick were cut at −20°C and placed onto room temperature microscope slides. These slides were submerged in hematoxylin for 1 minute followed by a wash in warm running water for 5 minutes and distilled H2O for an additional 2 minutes. Slides then were dipped in 90% EtOH 10 times and counterstained with 1× Eosin Y for 30 seconds. The tissue was dehydrated in 70% EtOH for 3 minutes × 2 changes, 95% EtOH for 3 minutes × 2 changes, 100% EtOH for 3 minutes × 2 changes, and cleared in Histoclear II (Electron Microscopy Sciences, Hatfield, PA) for 5 minutes × 2 changes. Cover slides were secured using Permount mounting media (Thermo Fisher Scientific, Waltham, MA) before imaging.
Single-Cell RNAseq
Fresh biopsy specimens were manually minced and dissociated using a Neural Tissue Dissociation Kit (130-092-628; Miltenyi Biotec, Auburn, CA) and an abridged manufacturer’s protocol resulting in ice-cold cells at a concentration of 1000 cells/μL. Single-cell droplet (5000–10,000 cells via chromium platform; 10×Genomics, Pleasanton, CA) library preparation (Chromium Next GEM Single-Cell 3ʹ Library Construction Kit v3.1; 10×Genomics) and sequencing (Novaseq platform; Illumina, San Diego, CA) were performed by the University of Michigan Advanced Genomic Core. The 10× chromium at the University of Michigan Advanced Genomics Core facility was used to create single-cell droplets with a target of capturing 5000–10,000 cells. The Chromium Next GEM Single Cell 3’ Library Construction Kit v3.1 prepared single-cell libraries according to the manufacturer’s instructions (10×Genomics). Gene expression matrices were constructed using Ranger (10×Genomics) and the human reference genome (hg19). Previously published pipelines were used for single-cell matrix generation/manipulation,46 cluster identification,47 and cluster visualization,48 with specific parameter details described later.
Tissue Dissociation for Single-Cell RNA Sequencing
A full detailed protocol for tissue dissociation for single-cell RNAseq can be found at www.jasonspencelab.com/protocols. Fresh tissue was placed in a petri dish with ice-cold 1× Hank’s balanced salt solution (HBSS) (with Mg2+, Ca2+). To prevent adhesion of cells, all tubes and pipette tips were prewashed with 1% bovine serum albumin (BSA) in 1× HBSS. The tissue was minced manually using spring–squeeze scissors before being transferred to a 15-mL conical containing 1% BSA in HBSS. Tubes were spun down at 500g for 5 minutes at 10°C, after which excess HBSS was aspirated. Mix 1 from the Neural Tissue Dissociation Kit (130-092-628; Miltenyi) containing dissociation enzymes and reagents was added and incubated at 10°C for 15 minutes. Mix 2 from the Neural Tissue Dissociation Kit was added and the suspension was fluxed through P1000 pipette tips (Gilson Incporporated, Madison, WI), interspersed by 10-minute incubations at 10°C. Flux steps were repeated as needed until cell clumps were no longer visible under a stereo microscope. Cells were filtered through a 1% BSA-coated 70-μm filter using 1× HBSS, spun down at 500g for 5 minutes at 10°C, and resuspended in 500 μL 1× HBSS (with Mg2+, Ca2+). RBC Lysis Buffer (1 mL, 11814389001; Roche Diagnostics Corporation, Indianapolis, IN) was added and tubes were incubated on a rocker at 4°C for 15 minutes. Cells were spun down at 500g for 5 minutes at 10°C, then washed twice in 2 mL 1% BSA, being spun down at 500g for 5 minutes at 10°C each time. A hemocytometer was used to count cells, which then were spun down and resuspended to reach a concentration of 1000 cells/μL and kept on ice.
Single-Cell Library Preparation
The 10× chromium at the University of Michigan Advanced Genomics Core facility was used to create single-cell droplets with a target of capturing 5000–10,000 cells. The Chromium Next GEM Single Cell 3’ Library Construction Kit v3.1 prepared single-cell libraries according to the manufacturer’s instructions (10×Genomics).
Sequencing Data Processing and Cluster Identification
The University of Michigan Advanced Genomic Core Illumina Novaseq performed all single-cell RNA sequencing. Gene expression matrices were constructed from raw data by the 10× Genomic Ranger with hg19. The Single-Cell Analysis for Python (Scanpy) was used for analysis as previously described by Wolf et al.46 Filtering parameters for gene count range, unique molecular identifier (UMI) counts, and mitochondrial transcript fraction were implemented for each data set to verify high-quality input data. For the remainder of processing, all tissue data sets were combined after organ-specific quality filtering had been performed. Highly variable genes were removed, gene expression levels were log normalized, and effects of UMI count and mitochondrial transcript function variations were regressed out via linear regression. Z-transformation then was performed on gene expression values before samples again were separated by organ for downstream analysis. The Uniform Manifold Approximation and Projection (UMAP) algorithm48 was used alongside Louvain algorithm cluster identification within Scanpy, using a resolution of 0.6,47 to perform a graph-based clustering of the top 10–11 principal components. Parameters used for Figure 1 include the following: (a) initial cell count, 19,249; final cell count, 13,332; initial gene count, 32,738; final gene count, 4071. (b) Filtered parameters used were as follows: minimum cells, 0; minimum genes, 500; maximum genes, 7500; maximum counts, 100,000; and max Mitochondria: 0.2. (c) Analysis parameters used were as follows: number of neighbors, 30; number of principal components, 20; spread, 1; minimum distribution: 0.4; and resolution, 1. Parameters for the post hoc extraction of cluster 0 differed in final cell count of 2400 with the number of neighbors of 15, and number of principal components of 11. Analysis of BE biopsy specimens only (n = 2) in Figure 2 includes the following parameters: (a) initial cell count, 7684; final cell count, 4293; initial gene count: 32,738; and final gene count, 4469. (b) Filtered parameters used were as follows: minimum cells, 0; minimum genes, 500; maximum genes, 7500; maximum counts, 100,000; and maximum Mitochondria, 0.2. (c) Analysis parameters used were as follows: number of neighbors, 15; number of principal components, 11; spread, 1; minimum distribution, 0.4; and resolution, 0.4.
Data and Code Availability
All code used for single-cell analysis and data presentation is available via Github at: https://github.com/jason-spence-lab. Submission of raw sequencing data is in process, for inquiries contact the authors. Sequencing data introduced in this study will be deposited at EMBL-EBI ArrayExpress (ArrayExpress: number in progress).
In Silico Identification of Potential E2 Interactors for RNF128 in BE/EAC
To identify potential E2 ubiquitin-conjugating enzyme binding partners for RNF128 in its E3 ligase capacity for p53 ubiquitination in BE/EAC, we used published literature to collate a list of known human E2s, including details pertaining to E3 partnership versatility and history of p53 as a substrate. Using our previously described RNAseq cohort of BE and EAC resection tissues,2 we determined the average BE mRNA expression level of each E2 and expression trends across cataloged BE histology groups (6 NDBE, 20 containing LGD without HGD, 28 containing HGD, and 11 EACs).
Because EAC is characterized by frequent DNA copy number changes,33,49 we noted E2s residing in genomic regions of frequent loss or gain in EAC. We confirmed these copy number trends in the 89 EACs from the TCGA ESCA cohort31 and correlated them with gene expression level for each E2. To estimate BE sample DNA copy number at each E2 locus, we applied a recursive median smoothing algorithm, similar to that used by CaSpER,50 to estimate copy number from RNAseq expression in BE samples. Median expression values (reads per kilobase of exon model per million reads) for each gene from 6 NDBE samples were used as a baseline for log2 expression ratios generated across genome-position ranked (hg38) genes for each individual BE or EAC sample. Only hg38 chromosome-localized genes with expression in all 6 NDBE reference samples were included. Three successive rounds of median smoothing were conducted with increasing window sizes extending 5, 10, and 20 expressed genes on either side of each target locus. The median expression across all X chromosome mapped markers was used to adjust for each sample for ploidy drift. We considered copy number estimates at E2 loci vs E2 expression levels across the 65 individual samples.
Transfection and Protein Analyses
BE cells (CpA and CpD) and hepatocellular carcinoma (HepG2) cells were transfected using Fugene-HD (Thermo Fisher Scientific, Waltham, MA), and human embryonic kidney (HEK293) cells were transfected using calcium phosphate methods as described.2 Immunoprecipitation and immunoblot analyses were performed as earlier standardized.51
Protein Half-Life Studies
RNF128 Iso1 WT and glycosylation-defective (3NA) mutant protein half-lives were calculated after transfection of CpA cells by adding cycloheximide (50 μg/mL) 24 hours after transfection as described.2 Glyceraldehyde-3-phosphate dehydrogenase was used as loading control. The approximate mutant RNF128 protein t1/2 was calculated and compared with the WT protein based on relative band density (arbitrary units) and time (hour) in a linear scale calculated from 3 independent experiments.
Coupled Transcription and Translation and In Vitro Ubiquitination Assay
V5-tagged RNF128 (WT) and (3NA, P54A, and P105A) mutants were in vitro transcribed and translated using the T7 quick-coupled transcription/translation system (Promega Corporation, Madison, WI) according to the manufacturer’s protocol. Protein expression was confirmed using immunoblotting and a V5-specific antibody. The in vitro ubiquitination assay and p53 protein modifications were performed as described.2
Statistical Analysis
Mapped (hg19) and normalized single-cell expression data from SE and BE (Figure 1) or BE only (Figure 2) esophageal biopsy specimens from 2 nonprogressed individuals were grouped into unsupervised fully modular Louvain clusters with UMAP48 (version 0.4 as implemented in the umap-learn python package) using parameters given earlier for each biopsy comparison. Before clustering, cells with the least (<500 genes; dead cells) and most (>7500 genes; doublet cells) genes expressed were removed from analyses, along with cells with high mitochondrial loading (averaged mitochondria to nuclear expression ratio, >0.2) and cells with high UMI counts greater than 10,000. The top 200 most discriminatory genes for each UMAP cluster are shown in Supplementary Table 3 (BE only) and Supplementary Table 2 (squamous and BE). These outputs were generated via the tl.rank_genes_groups function in Scanpy,46 which represents the z-score associated with the P value, and includes the log2-adjusted fold change ratio of average expression for cluster-specific vs all the rest of cells (cells not assigned to the current cluster), P values t test comparison of cell expression in the current cluster vs the rest of cells, and these P values52 were adjusted for multiple testing (P values_adj). The Scanpy function pl.rank_genes_groups_dotplot was used to generate the subclustering dot-plot analysis given in Figure 2A. Figure 2, which focuses on cell clusters 1, 3, 5, and 9, designated as having high BE cell content, based on low squamous cell (KRT4 and KRT15) and high columnar cell (EPCAM, TFF1, TFF2, and TFF3) marker content. Dot-plot expression content is represented with dot circumference as a fraction of positive cells and color intensity as average expression level, as depicted in the key (lower right of Figure 2L).
Standard RNAseq analyses of RNF128 isoforms and the specifically chosen E2s performed using normalized RPMK values,53 as generated previously,2 were graphed in Prism (version 9; GraphPad Software, San Diego, CA), with ANOVA adjusted for 2 × 2 group (NDBE+LGD vs HGD or EAC) comparisons, paired t test (LGD vs patient-matched HGD or EAC), or Pearson correlations used to highlight trends between gene expression and estimated local copy number, or gene expression and RNF128 isoform expression. RNF128 isoform expression was expressed as a log2 ratio between Iso2 and Iso1 levels using previous data2 from our 65-sample BE progression-related cohort.
DNA copy number (capped relative linear copy number or Genomic Identification of Significant Targets in Cancer generated mean copy) and matched RNAseq by expectation maximization54 gene expression data for the EAC portion (cancer type detailed = EAC; n = 87) of the ESCA (esophageal cancer) TCGA cohort31 were extracted from cBioPortal55 for the top E2 conjugation enzyme genes and plotted using Prism to explore correlations between gene dose and copy number, assessed by Pearson correlation.
UBE2D3-specific data (Affymetrix [Santa Clara, CA] U133 array probe set 200668_s_at) for the cohorts of GEO series GSE142029 and GEO series GSE2688630 were extracted using the GEO2R interface,56 in which BE and EAC expression in each cohort was compared using an unequal variances t test. We included normal SE sample groups available for these cohorts in plots for reference, but SE sample groups were excluded from statistical analyses because they were not a feature of validation comparison.
For Pearson comparisons we considered r values with magnitudes of 0.4 (r2 = 0.16) to represent nominal evidence for an association whereby greater than 15% of variance could be attributed to this association. For single-comparison t test–based comparisons we considered P values less than .05 as evidence for an association.
For protein half-life studies, results are presented as means ± SD of at least 3 independent experiments.
All authors had access to the study data and reviewed and approved the final manuscript.
CRediT Authorship Contributions
Paramita Ray (Data curation: Lead; Formal analysis: Lead; Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Supervision: Supporting; Validation: Lead; Writing – original draft: Supporting)
Derek J Nancarrow (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Supporting; Investigation: Supporting; Methodology: Lead; Supervision: Supporting; Validation: Supporting; Visualization: Supporting; Writing – original draft: Equal)
Daysha Ferrer-Torres (Conceptualization: Supporting; Data curation: Equal; Formal analysis: Equal; Methodology: Lead; Validation: Supporting; Visualization: Supporting; Writing – original draft: Supporting)
Zhuwen Wang (Data curation: Equal; Methodology: Equal; Validation: Equal; Visualization: Supporting; Writing – review & editing: Supporting)
May San Martinho (Data curation: Supporting; Methodology: Supporting; Validation: Supporting; Writing – review & editing: Supporting)
Tonaye Hinton (Data curation: Supporting; Methodology: Supporting; Validation: Supporting; Writing – review & editing: Supporting)
Joshua H Wu (Data curation: Equal; Formal analysis: Equal; Investigation: Supporting; Methodology: Equal; Software: Lead; Validation: Supporting; Writing – review & editing: Supporting)
Angeline Wu (Data curation: Supporting; Methodology: Supporting; Validation: Supporting; Writing – review & editing: Supporting)
Danielle K Turgeon (Data curation: Supporting; Resources: Lead; Supervision: Supporting; Writing – review & editing: Supporting)
Max A Hammer (Data curation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Michael K Dame (Data curation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Theodore S Lawrence (Conceptualization: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Project administration: Supporting; Resources: Supporting; Supervision: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Patrick J O’Brien (Conceptualization: Equal; Investigation: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Jason R Spence (Conceptualization: Supporting; Formal analysis: Supporting; Funding acquisition: Lead; Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Resources: Lead; Supervision: Supporting; Writing – review & editing: Supporting)
David G Beer (Conceptualization: Lead; Data curation: Supporting; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Supporting; Project administration: Equal; Resources: Lead; Supervision: Lead; Validation: Supporting; Visualization: Supporting; Writing – original draft: Lead; Writing – review & editing: Supporting)
Dipankar Ray, PhD (Conceptualization: Lead; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Lead; Methodology: Equal; Project administration: Lead; Resources: Lead; Supervision: Lead; Validation: Equal; Visualization: Equal; Writing – original draft: Lead; Writing – review & editing: Lead)
Acknowledgments
The authors thank Steven Kronenberg for graphic assistance.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by National Cancer Institute grants RO1CA215596 (D.R.; D.G.B), U54CA163059 (D.G.B.), P30CA046592 (D.G.B), and F31A200113 (D.F.T), and the John and Carla Klein Family Research Fund (D.G.B.). Also supported by Michigan Institute for Clinical and Health Research (Post-doctoral Translational Science Program) grant UL1TR002240, and by the Training Program in Organogenesis (NIH-National Institute of Child Health and Human Development T32 HD007505) (D.F.T.).
Supplementary Material
References
- 1.Fisher O.M., Lord S.J., Falkenback D., Clemons N.J., Eslick G.D., Lord R.V. The prognostic value of TP53 mutations in oesophageal adenocarcinoma: a systematic review and meta-analysis. Gut. 2017;66:399–410. doi: 10.1136/gutjnl-2015-310888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray D., Ray P., Ferrer-Torres D., Wang Z., Nancarrow D., Yoon H.W., San Martinho M., Hinton T., Owens S., Thomas D., Jiang H., Lawrence T.S., Lin J., Lagisetty K., Chang A.C., Beer D.G. Isoforms of RNF128 regulate the stability of mutant P53 in Barrett's esophageal cells. Gastroenterology. 2020;158:583–597 e1. doi: 10.1053/j.gastro.2019.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stachler M.D., Camarda N.D., Deitrick C., Kim A., Agoston A.T., Odze R.D., Hornick J.L., Nag A., Thorner A.R., Ducar M., Noffsinger A., Lash R.H., Redston M., Carter S.L., Davison J.M., Bass A.J. Detection of mutations in Barrett's esophagus before progression to high-grade dysplasia or adenocarcinoma. Gastroenterology. 2018;155:156–167. doi: 10.1053/j.gastro.2018.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neshat K., Sanchez C.A., Galipeau P.C., Blount P.L., Levine D.S., Joslyn G., Reid B.J. p53 mutations in Barrett's adenocarcinoma and high-grade dysplasia. Gastroenterology. 1994;106:1589–1595. doi: 10.1016/0016-5085(94)90415-4. [DOI] [PubMed] [Google Scholar]
- 5.Weaver J.M.J., Ross-Innes C.S., Shannon N., Lynch A.G., Forshew T., Barbera M., Murtaza M., Ong C.J., Lao-Sirieix P., Dunning M.J., Smith L., Smith M.L., Anderson C.L., Carvalho B., O'Donovan M., Underwood T.J., May A.P., Grehan N., Hardwick R., Davies J., Oloumi A., Aparicio S., Caldas C., Eldridge M.D., Edwards P.A.W., Rosenfeld N., Tavare S., Fitzgerald R.C., OCCAMS Consortium Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet. 2014;46:837–843. doi: 10.1038/ng.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain A.K., Barton M.C. Making sense of ubiquitin ligases that regulate p53. Cancer Biol Ther. 2010;10:665–672. doi: 10.4161/cbt.10.7.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sane S., Rezvani K. Essential roles of E3 ubiquitin ligases in p53 regulation. Int J Mol Sci. 2017;18:442. doi: 10.3390/ijms18020442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y.C., Chan J.Y., Chiu Y.L., Liu S.T., Lozano G., Wang S.L., Ho C.L., Huang S.M. Grail as a molecular determinant for the functions of the tumor suppressor p53 in tumorigenesis. Cell Death Differ. 2013;20:732–743. doi: 10.1038/cdd.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.San Martinho M., Nancarrow D.J., Lawrence T.S., Beer D.G., Ray D. Chaperones and ubiquitin ligases balance mutant p53 protein stability in esophageal and other digestive cancers. Cell Mol Gastroenterol Hepatol. 2021;11:449–464. doi: 10.1016/j.jcmgh.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whiting C.C., Su L.L., Lin J.T., Fathman C.G. GRAIL: a unique mediator of CD4 T-lymphocyte unresponsiveness. FEBS J. 2011;278:47–58. doi: 10.1111/j.1742-4658.2010.07922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh S., Singh A.G., Singh P.P., Murad M.H., Iyer P.G. Statins are associated with reduced risk of esophageal cancer, particularly in patients with Barrett's esophagus: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2013;11:620–629. doi: 10.1016/j.cgh.2012.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siddals K.W., Marshman E., Westwood M., Gibson J.M. Abrogation of insulin-like growth factor-I (IGF-I) and insulin action by mevalonic acid depletion: synergy between protein prenylation and receptor glycosylation pathways. J Biol Chem. 2004;279:38353–38359. doi: 10.1074/jbc.M404838200. [DOI] [PubMed] [Google Scholar]
- 13.Urani C., Melchioretto P., Fabbri M., Bowe G., Maserati E., Gribaldo L. Cadmium impairs p53 activity in HepG2 cells. ISRN Toxicol. 2014;2014:976428. doi: 10.1155/2014/976428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokumoto M., Fujiwara Y., Shimada A., Hasegawa T., Seko Y., Nagase H., Satoh M. Cadmium toxicity is caused by accumulation of p53 through the down-regulation of Ube2d family genes in vitro and in vivo. J Toxicol Sci. 2011;36:191–200. doi: 10.2131/jts.36.191. [DOI] [PubMed] [Google Scholar]
- 15.Lee J.Y., Tokumoto M., Hattori Y., Fujiwara Y., Shimada A., Satoh M. Different regulation of p53 expression by cadmium exposure in kidney, liver, intestine, vasculature, and brain astrocytes. Toxicol Res. 2016;32:73–80. doi: 10.5487/TR.2016.32.1.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markson G., Kiel C., Hyde R., Brown S., Charalabous P., Bremm A., Semple J., Woodsmith J., Duley S., Salehi-Ashtiani K., Vidal M., Komander D., Serrano L., Lehner P., Sanderson C.M. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 2009;19:1905–1911. doi: 10.1101/gr.093963.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michelle C., Vourc'h P., Mignon L., Andres C.R. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J Mol Evol. 2009;68:616–628. doi: 10.1007/s00239-009-9225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart M.D., Ritterhoff T., Klevit R.E., Brzovic P.S. E2 enzymes: more than just middle men. Cell Res. 2016;26:423–440. doi: 10.1038/cr.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Wijk S.J., de Vries S.J., Kemmeren P., Huang A., Boelens R., Bonvin A.M., Timmers H.T. A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol Syst Biol. 2009;5:295. doi: 10.1038/msb.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosekrans S.L., Baan B., Muncan V., van den Brink G.R. Esophageal development and epithelial homeostasis. Am J Physiol Gastrointest Liver Physiol. 2015;309:G216–G228. doi: 10.1152/ajpgi.00088.2015. [DOI] [PubMed] [Google Scholar]
- 21.Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 22.Longman R.J., Douthwaite J., Sylvester P.A., Poulsom R., Corfield A.P., Thomas M.G., Wright N.A. Coordinated localisation of mucins and trefoil peptides in the ulcer associated cell lineage and the gastrointestinal mucosa. Gut. 2000;47:792–800. doi: 10.1136/gut.47.6.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arul G.S., Moorghen M., Myerscough N., Alderson D.A., Spicer R.D., Corfield A.P. Mucin gene expression in Barrett's oesophagus: an in situ hybridisation and immunohistochemical study. Gut. 2000;47:753–761. doi: 10.1136/gut.47.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szachnowicz S., Cecconello I., Ribeiro U., Iriya K., El Ibrahim R., Takeda F.R., Corbett C.E., Vaz Safatle-Ribeiro A. Mucin pattern reflects the origin of the adenocarcinoma in Barrett's esophagus: a retrospective clinical and laboratorial study. World J Surg Oncol. 2009;7:27. doi: 10.1186/1477-7819-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quante M., Graham T.A., Jansen M. Insights into the pathophysiology of esophageal adenocarcinoma. Gastroenterology. 2018;154:406–420. doi: 10.1053/j.gastro.2017.09.046. [DOI] [PubMed] [Google Scholar]
- 26.Saville M.K., Sparks A., Xirodimas D.P., Wardrop J., Stevenson L.F., Bourdon J.C., Woods Y.L., Lane D.P. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 27.Guan G.G., Wang W.B., Lei B.X., Wang Q.L., Wu L., Fu Z.M., Zhou F.X., Zhou Y.F. UBE2D3 is a positive prognostic factor and is negatively correlated with hTERT expression in esophageal cancer. Oncol Lett. 2015;9:1567–1574. doi: 10.3892/ol.2015.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou L., Li Y., Wang Y., Xu D., Cui H., Xu X., Cong Y., Yu C. UBE2D1 RNA expression was an independent unfavorable prognostic indicator in lung adenocarcinoma, but not in lung squamous cell carcinoma. Dis Markers. 2018;2018 doi: 10.1155/2018/4108919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimchi E.T., Posner M.C., Park J.O., Darga T.E., Kocherginsky M., Karrison T., Hart J., Smith K.D., Mezhir J.J., Weichselbaum R.R., Khodarev N.N. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–3154. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q., Ma C., Kemmner W. Wdr66 is a novel marker for risk stratification and involved in epithelial-mesenchymal transition of esophageal squamous cell carcinoma. BMC Cancer. 2013;13:137. doi: 10.1186/1471-2407-13-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network. Analysis Working Group. Asan University; BC Cancer Agency. Brigham and Women’s Hospital. Broad Institute; Brown University. Case Western Reserve University. Dana-Farber Cancer Institute. Duke University. Greater Poland Cancer Centre. Harvard Medical School. Institute for Systems Biology; KU Leuven. Mayo Clinic. Memorial Sloan Kettering Cancer Center. National Cancer Institute. Nationwide Children’s Hospital. Stanford University. University of Alabama. University of Michigan. University of North Carolina. University of Pittsburgh. University of Rochester. University of Southern California. University of Texas MD Anderson Cancer Center. University of Washington. Van Andel Research Institute. Vanderbilt University. Washington University. Genome Sequencing Center. Broad Institute; Washington University in St. Louis. Genome Characterization Centers. BC Cancer Agency. Broad Institute. Harvard Medical School. Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University. University of North Carolina. University of Southern California Epigenome Center. University of Texas MD Anderson Cancer Center. Van Andel Research Institute. Genome Data Analysis Centers. Broad Institute. Brown University. Harvard Medical School. Institute for Systems Biology. Memorial Sloan Kettering Cancer Center. University of California Santa Cruz. University of Texas MD Anderson Cancer Center. Biospecimen Core Resource: International Genomics Consortium. Research Institute at Nationwide Children’s Hospital. Tissue Source Sites: Analytic Biologic Services. Asan Medical Center. Asterand Bioscience. Barretos Cancer Hospital. BioreclamationIVT. Botkin Municipal Clinic. Chonnam National University Medical School. Christiana Care Health System. Cureline; Duke University. Emory University. Erasmus University. Indiana University School of Medicine. Institute of Oncology of Moldova. International Genomics Consortium. Invidumed; Israelitisches Krankenhaus Hamburg. Keimyung University School of Medicine. Memorial Sloan Kettering Cancer Center. National Cancer Center Goyang. Ontario Tumour Bank. Peter MacCallum Cancer Centre. Pusan National University Medical School. Ribeirão Preto Medical School. St. Joseph’s Hospital &Medical Center. St. Petersburg. Academic University. Tayside Tissue Bank. University of Dundee. University of Kansas Medical Center. University of Michigan. University of North Carolina at Chapel Hill. University of Pittsburgh School of Medicine. University of Texas MD Anderson Cancer Center. Disease Working Group. Duke University. Memorial Sloan Kettering Cancer Center. National Cancer Institute. University of Texas MD Anderson Cancer Center. Yonsei University College of Medicine. Data Coordination Center. CSRA Inc. Project Team: National Institutes of Health Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doak S.H., Jenkins G.J., Parry E.M., D'Souza F.R., Griffiths A.P., Toffazal N., Shah V., Baxter J.N., Parry J.M. Chromosome 4 hyperploidy represents an early genetic aberration in premalignant Barrett's oesophagus. Gut. 2003;52:623–628. doi: 10.1136/gut.52.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nancarrow D.J., Handoko H.Y., Smithers B.M., Gotley D.C., Drew P.A., Watson D.I., Clouston A.D., Hayward N.K., Whiteman D.C. Genome-wide copy number analysis in esophageal adenocarcinoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2008;68:4163–4172. doi: 10.1158/0008-5472.CAN-07-6710. [DOI] [PubMed] [Google Scholar]
- 34.Ray P., Raghunathan K., Ahsan A., Allam U.S., Shukla S., Basrur V., Veatch S., Lawrence T.S., Nyati M.K., Ray D. Ubiquitin ligase SMURF2 enhances epidermal growth factor receptor stability and tyrosine-kinase inhibitor resistance. J Biol Chem. 2020;295:12661–12673. doi: 10.1074/jbc.RA120.013519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plechanovova A., Jaffray E.G., Tatham M.H., Naismith J.H., Hay R.T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukla S., Allam U.S., Ahsan A., Chen G., Krishnamurthy P.M., Marsh K., Rumschlag M., Shankar S., Whitehead C., Schipper M., Basrur V., Southworth D.R., Chinnaiyan A.M., Rehemtulla A., Beer D.G., Lawrence T.S., Nyati M.K., Ray D. KRAS protein stability is regulated through SMURF2: UBCH5 complex-mediated beta-TrCP1 degradation. Neoplasia. 2014;16:115–128. doi: 10.1593/neo.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z., Kohli E., Devlin K.I., Bold M., Nix J.C., Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levin I., Eakin C., Blanc M.P., Klevit R.E., Miller S.I., Brzovic P.S. Identification of an unconventional E3 binding surface on the UbcH5 ~ Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc Natl Acad Sci U S A. 2010;107:2848–2853. doi: 10.1073/pnas.0914821107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinho M.S., Nancarrow D.J., Lawrence T.S., Beer D.G., Ray D. Chaperones and ubiquitin ligases balance mutant p53 protein stability in esophageal and other digestive cancers. Cell Mol Gastroenterol Hepatol. 2021;11:449–464. doi: 10.1016/j.jcmgh.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costa A.F., Campos D., Reis C.A., Gomes C. Targeting glycosylation: a new road for cancer drug discovery. Trends Cancer. 2020;6:757–766. doi: 10.1016/j.trecan.2020.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Pi J., Dogovski C., Pittard A.J. Functional consequences of changing proline residues in the phenylalanine-specific permease of Escherichia coli. J Bacteriol. 1998;180:5515–5519. doi: 10.1128/jb.180.21.5515-5519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woolfson D.N., Williams D.H. The influence of proline residues on alpha-helical structure. FEBS Lett. 1990;277:185–188. doi: 10.1016/0014-5793(90)80839-b. [DOI] [PubMed] [Google Scholar]
- 43.Kini R.M., Evans H.J. A hypothetical structural role for proline residues in the flanking segments of protein-protein interaction sites. Biochem Biophys Res Commun. 1995;212:1115–1124. doi: 10.1006/bbrc.1995.2084. [DOI] [PubMed] [Google Scholar]
- 44.Liu L., Hua Y., Wang D., Shan L., Zhang Y., Zhu J., Jin H., Li H., Hu Z., Zhang W. A sesquiterpene lactone from a medicinal herb inhibits proinflammatory activity of TNF-alpha by inhibiting ubiquitin-conjugating enzyme UbcH5. Chem Biol. 2014;21:1341–1350. doi: 10.1016/j.chembiol.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 45.Kroep S., Lansdorp-Vogelaar I., Rubenstein J.H., Lemmens V.E., van Heijningen E.B., Aragones N., van Ballegooijen M., Inadomi J.M. Comparing trends in esophageal adenocarcinoma incidence and lifestyle factors between the United States, Spain, and the Netherlands. Am J Gastroenterol. 2014;109:336–343. doi: 10.1038/ajg.2013.420. quiz 5, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blondel V.D., Guillaume J.L., Hendrickx J.M., de Kerchove C., Lambiotte R. Local leaders in random networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;77 doi: 10.1103/PhysRevE.77.036114. [DOI] [PubMed] [Google Scholar]
- 48.Becht E., McInnes L., Healy J., et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 49.Dulak A.M., Schumacher S.E., van Lieshout J., Imamura Y., Fox C., Shim B., Ramos A.H., Saksena G., Baca S.C., Baselga J., Tabernero J., Barretina J., Enzinger P.C., Corso G., Roviello F., Lin L., Bandla S., Luketich J.D., Pennathur A., Meyerson M., Ogino S., Shivdasani R.A., Beer D.G., Godfrey T.E., Beroukhim R., Bass A.J. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harmanci A., Harmanci A.S., Swaminathan J., Gopalakrishnan V. EpiSAFARI: sensitive detection of valleys in epigenetic signals for enhancing annotations of functional elements. Bioinformatics. 2020;36:1014–1021. doi: 10.1093/bioinformatics/btz702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray D., Ahsan A., Helman A., Chen G., Hegde A., Gurjar S.R., Zhao L., Kiyokawa H., Beer D.G., Lawrence T.S., Nyati M.K. Regulation of EGFR protein stability by the HECT-type ubiquitin ligase SMURF2. Neoplasia. 2011;13:570–578. doi: 10.1593/neo.11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klipper-Aurbach Y., Wasserman M., Braunspiegel-Weintrob N., Borstein D., Peleg S., Assa S., Karp M., Benjamini Y., Hochberg Y., Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 53.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 54.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code used for single-cell analysis and data presentation is available via Github at: https://github.com/jason-spence-lab. Submission of raw sequencing data is in process, for inquiries contact the authors. Sequencing data introduced in this study will be deposited at EMBL-EBI ArrayExpress (ArrayExpress: number in progress).