ABSTRACT
The life cycle of filamentous fungi generally comprises hyphal growth and asexual reproduction. Both growth and propagation processes are critical for invasion growth, spore dissemination, and virulence in fungal pathogens and for the production of secondary metabolites or for biomass accumulation in industrial filamentous fungi. The CCAAT-binding complex (CBC) is a heterotrimeric transcription factor comprising three subunits, HapB, HapC, and HapE, and is highly conserved in fungi. Previous studies revealed that CBC regulates sterol metabolism by repressing several genes in the ergosterol biosynthetic pathway in the human fungal pathogen Aspergillus fumigatus. In the present study, we found dysfunction of CBC caused the abnormal asexual reproduction (conidiation) in submerged liquid culture. CBC suppresses the activation of the brlA gene in the central regulatory pathway for conidiation combined with its upstream regulators fluG, flbD, and flbC by binding to the 5′-CCAAT-3′ motif within conidiation gene promoters, and lack of CBC member HapB results in the upregulation of these genes. Furthermore, when the expression of brlA or flbC is repressed, the submerged conidiation does not happen in the hapB mutant. Interestingly, deletion of HapB leads to enhanced transient cytosolic Ca2+ levels and activates conidiation-positive inducer Ca2+-CrzA modules to enhance submerged conidiation, demonstrating that CrzA works with CBC as a reverse regulator of fungal conidiation. To the best of our knowledge, the finding of this study is the first report for the molecular switch mechanism between vegetative hyphal growth and asexual development regulated by CBC, in concert with Ca2+-CrzA signaling in A. fumigatus.
KEYWORDS: submerged conidiation, CCAAT-binding complex, calcium, CrzA, Aspergillus fumigatus
INTRODUCTION
Aspergillus fumigatus is a common environmental saprobe mold and plays an important role in carbon and nitrogen recycling by decomposing dead organic biomass (1, 2). In addition, A. fumigatus is a leading cause of invasive mold infections in immunocompromised individuals (3–6). In the natural environment or on solid culture plates, A. fumigatus first grows in a vegetative state and is composed of long branched filamentous structures that are able to attach to the substrate upon which the fungus feeds. Under host conditions, a fungal colony is initiated from the germination of a single spore that gives rise to a network of hyphae referred to as invasive hyphal growth, which damages or destroys host organ tissue (6, 7). When vegetative hyphae encounter some unsuitable growth conditions or are exposed to air, aerial hyphae form asexual conidiophores associated with conidia and initiate an asexual reproductive cycle (8–11). A. fumigatus propagates asexually via spores that can be dispersed over large geographical distances in the air, and these spores can germinate to grow under a broad range of environmental conditions (1, 12–14). The genetic mechanisms of asexual reproduction (conidiation) have been extensively studied in the model fungus Aspergillus nidulans, and some have also been studied in A. fumigatus (8, 15, 16). In both species, the activation of brlA encoding a conserved C2H2 zinc finger transcription factor, BrlA (bristle), is a key event in response to the development induction signal (16–19). Previous studies have identified that brlA and its targets, abaA and wetA (BrlA→AbaA→WetA), comprise a central regulatory pathway that controls the spatial and temporal expression of conidiation-specific genes during conidiogenesis (20). In addition to its involvement in this central regulatory pathway, later studies identified that brlA expression could be affected by several upstream developmental activators, fluG, flbA, flbB, flbC, flbD, and flbE, and mutations in these genes result in “fluffy” colonies with downregulated brlA (8, 15, 16, 21–25). Moreover, through combinatorial genetic studies, SfgA, VosA, VeA, VelB, VelC, NsdC, and NsdD, which repress the expression of brlA as negative regulators of conidiation, have been found (26–30). Notably, current knowledge of conidiation-related regulators in Aspergillus was mainly obtained from studies under solid-culture conditions. In comparison, submerged cultured hyphae in A. fumigatus and in all other filamentous fungi undergo constant vegetative growth to form a large number of mycelial pellets, whereas asexual conidiation rarely occurs (27). In addition, during progress of the invasive aspergillosis, germinated spores display high-speed tip growth, and conidiation rarely occurs (17, 31). These phenomena suggest that Aspergillus species have the ability to finely sense the environment and decide when or where to undergo hyphal extension or initiate conidiation. Thus, the transition from vegetative growth to conidiation in Aspergillus species is a crucial process contributing to the long-term survival, propagation, and fitness of fungi in response to various environmental conditions (32, 33). To date, several reports have demonstrated that the loss of each of the velvet regulators VosA, VeA, and VelB; SfgA (the FluG suppressor); and NsdC and NsdD (the sexual reproduction activators) causes abnormal submerged hyperconidiation (26–28, 30, 34), suggesting that these regulators may exert negative influence on asexual conidiation in liquid-submerged culture. However, whether or how these genes work together with a known central regulatory pathway and its upstream developmental activators is still poorly understood.
The CCAAT-binding complex (CBC) is a heterotrimeric transcription factor comprising three subunits (HapB/HapC/HapE) that is involved in the regulation of sterol metabolism by repressing several genes in the ergosterol biosynthetic pathway (35). In the present study, we found a new function for CBC in A. fumigatus as a negative regulator of conidiation under liquid culture conditions. The lack of each member of the heterotrimeric CBC led to abnormal A. fumigatus conidiation in liquid-submerged culture, suggesting that CBC plays an important role in maintaining normal vegetative growth. As a transcription factor, CBC participates in numerous cellular processes, such as iron homeostasis, sterol biosynthesis, oxidative stress response, secondary metabolism, and development, by regulating the expression of related genes. However, the negative regulation of conidiation by CBC in liquid-submerged culture has not yet been reported. Here, we identified that CBC could directly bind to the CCAAT motif within the promoters of brlA, fluG, flbD, and flbC, and lack of CBC member-HapB caused the upregulation of these genes, suggesting that CBC suppresses the activation of the central regulatory pathway combined with its upstream regulators to inhibit conidiation in submerged culture. Notably, the submerged conidiation in the CBC mutants was markedly enhanced by the addition of calcium. In contrast, the calcium-chelating agent EGTA remarkably suppressed the abovementioned submerged conidiation. Calcium, a secondary messenger, plays important roles in regulating fungal growth, development, and reproduction. The transcription factor calcineurin-responsive zinc finger (CrzA) is a central regulator of Ca2+-mediated signals. Through a conditional crzA gene-Tet on/off system, we demonstrated that CrzA, as a positive conidiation inducer, works with CBC to control asexual reproduction in A. fumigatus in submerged culture.
RESULTS
Lack of CBC causes desuppression of A. fumigatus conidiation in liquid-submerged culture.
Generally, conidiation initiation is suppressed in A. fumigatus under liquid shaking culture conditions. As shown in Fig. 1A and B, the parental wild-type (WT) strain displayed constant hyphal vegetative growth to form many colorless mycelial pellets. Interestingly, under the same culture conditions, the CBC mutants (ΔhapB, ΔhapC, and ΔhapE) showed pale-green mycelial pellets presenting conidiophores that popped out, suggesting the occurrence of asexual conidiation in a process referred to as submerged conidiation. Furthermore, the mycelial pellets of the CBC mutants were significantly smaller and exhibited less biomass than those of the WT strain or hapB-complemented strain (hapBC) (Fig. 1A and see Fig. S1A in the supplemental material). Generally, asexual conidiation is accompanied by melanin accumulation on the conidial surface in Aspergillus species (36–38). Next, we extracted the melanin with NaOH lysis buffer from mycelial pellets and found that the ΔhapB, ΔhapC, and ΔhapE mutants truly had highly accumulated melanin contents compared with the WT or the hapBC strain (Fig. 1C). It implies that CBC might be a suppressor of conidiation in liquid culture. However, on solid medium, the A. fumigatus CBC mutants displayed significantly fewer conidia than did the wild-type and the hapBC strains (Fig. S1B to D). These data suggest that CBC may have different functions for asexual development on solid medium than under liquid culture conditions.
FIG 1.
Phenotypes of the CBC mutants in liquid minimal medium (MM). (A and B) A total of 3.5 × 106 spores were grown in liquid MM for the indicated days in a shaker at 37°C. The macromorphology and hyphae/asexual conidiogenous structure of mycelial pellets were observed. Scale bar represents 10 μm. (C) The OD400 value of the supernatant with dissolved melanin represented the relative melanin content. CK indicates the OD400 value of control check which was obtained by measuring the optical density of buffer extracted for melanin at 400 nm. Statistical significance was determined by Student’s t test. *, P < 0.05; **, P < 0.01.
(A) Spores (3.5 × 106) were grown in liquid MM for the indicated days in a shaker at 37°C. The dry weight (biomass) of the resulting mycelial pellets was measured. (B to D) Spores (2 × 105) were inoculated on solid MM at 37°C for the indicated days. Morphology, diameter, and conidial amount of the resulting colonies. Statistical significance was determined by Student’s t test. **, P < 0.01. Download FIG S1, TIF file, 0.9 MB (955.2KB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
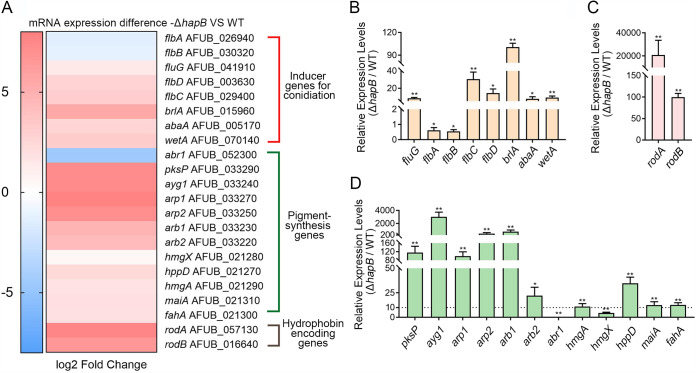
Transcriptome analyses of the ΔhapB mutant reveal altered expression patterns for genes related to conidiation and pigment synthesis.
Given that CBC deficiency induces the abnormal submerged conidiation, we wondered whether CBC affects the expression of conidiation-regulated genes under the liquid culture condition. A transcriptome profiling experiment (RNA-seq) was carried out to identify genes differentially expressed between WT and ΔhapB strains grown in liquid minimal medium (MM). The results showed that 4,592 (46.3%) of the total predicted 9,911 A. fumigatus transcripts were differentially expressed in the transcriptome database, with 2,424 genes shown to be upregulated at least 2-fold in the ΔhapB mutant compared to the WT strain (Data Set S1). Based on the biological processes, Gene Ontology (GO) enrichment analysis was performed. Notably, asexual spore wall assembly and cell cycle arrest belonged to the top 10 biological processes of the upregulated genes (Fig. S2), suggesting the inhibition of hyphal vegetative growth and the activation of asexual conidiation in the ΔhapB mutant. Among these upregulated genes (Fig. 2A), all the key central conidiation regulators (brlA, abaA, and wetA) and their relative upstream genes (fluG, flbC, and flbD) were remarkably upregulated, but flbA and flbB were slightly downregulated, which was further verified by real-time quantitative PCR (RT-qPCR) (Fig. 2B). According to previous reports, these central and upstream regulators of conidiation are mainly responsible for positive regulation during the process of conidiation. In addition, we also analyzed the majority of the reported negative regulators of conidiation at the mRNA level and found that none of the tested reported negative regulators, VosA, VeA, VelB, MpkB, SfgA, NsdC, and NsdD, showed significantly decreased expression. Instead, they displayed slight upregulation or no changes in the ΔhapB mutant compared to the WT strain (Fig. S3A and B). These results suggested that the submerged conidiation caused by the hapB mutant was not due to changed expression of the known negative regulators of conidiation. Notably, the submerged conidiation of the ΔhapB mutant was mainly associated with the upregulation of brlA-related central conidiation regulators and their upstream fluffy genes. Activation of the asexual conidiation pathway induces the synthesis of pigment and conidium-superficial hydrophobin protein (36–40). Correspondingly, we found that most pigment synthesis-related key genes, including dihydroxynaphthalene (DHN)-melanin genes (pksP, ayg1, arp1, arp2, arb1, and arb2) and promelanin genes (hmgA, hppD, maiA, and fahA), and conidium-hydrophobin-encoding genes rodA and rodB, were significantly upregulated in the ΔhapB mutant compared to the WT strain (Fig. 2A, C, and D). RNA-seq showed that the top 10 biological processes of the downregulated genes were mainly mitochondrial electron transport and biotin biosynthesis required for vegetative growth (Fig. S2), corresponding to the phenotype of decreased hyphal biomass in the ΔhapB strain cultured in liquid MM (Fig. S1A). Taken together, our studies suggest that lack of the CBC subunit HapB derepresses conidiation by upregulating brlA-related genes and results in the initiation of conidiation under liquid culture condition.
FIG 2.
A comparison of gene expression related to conidiation and pigment synthesis in RNA-seq and RT-qPCR analyses for the ΔhapB mutant relative to the WT strain. (A) Heat map of RNA-seq data for the 22 selected genes. (B to D) The transcript levels of the indicated genes in WT and ΔhapB strains grown at 37°C for 48 h. Statistical significance was determined by Student’s t test. *, P < 0.05; **, P < 0.01; ns, not significant.
Gene Ontology (GO) term enrichment analysis of differentially expressed genes (fold changes ≥ 2.0) in the RNA-seq database. Download FIG S2, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A and C) Comparison of RNA-seq data in heat map for the previously reported conidiation negative regulator-encoding genes and the selected Ca2+ transporter-related genes between the ΔhapB and WT strains cultured for 48 h in liquid MM. (B and D) The relative transcript levels of the indicated genes were further confirmed by RT-qPCR. Statistical significance was determined by Student’s t test. *, P < 0.05; **, P < 0.01; ns, not significant. Download FIG S3, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The RNA-seq data sets of WT and ΔhapB strains. Download Data Set S1, XLS file, 4.5 MB (4.5MB, xls) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CBC binds to the promoters of genes related to asexual conidiation.
Next, we investigated how CBC affects expression of the conidiation-related genes to regulate conidiation. Since previous studies have shown that CBC can regulate the expression of targets by binding to 5′-CCAAT-3′ within the promoter regions of these target genes (35, 41, 42), we then searched the promoter sequences of the abovementioned genes that are upregulated in the absence of CBC subunit HapB. Interestingly, a motif search analysis revealed that the majority of conidiation-regulatory genes, including brlA, have CCAAT motifs in the promoter region. To further validate whether the A. fumigatus CBC in vitro could bind to the CCAAT motifs within the promoter regions of these genes, electrophoretic mobility shift assays (EMSAs) were carried out in which C-terminally His-tagged HapB, HapC, and HapE proteins were expressed in Escherichia coli (Fig. 3A) and then purified by affinity chromatography. Double-stranded oligomer probes harboring the CCAAT motif were amplified from the promoter regions of selected differentially expressed genes (fluG, flbC, flbD, and brlA) by PCR and labeled with Cy5. The results showed that probes (−4421 and −3175 of brlA, −788 and −335 of fluG, −4340 and −247 of flbD, and −3268 of flbC) mixed with the protein mixture (CBC) composed of HapB, HapC, and HapE (1:1:1, molar ratio) markedly displayed slowed-shift bands compared to the free probe without CBC, suggesting the in vitro binding of CBC to the CCAAT motif-containing promoter fragments of brlA, fluG, flbD, and flbC (Fig. 3B to H). Excess unlabeled probe (cold probe) significantly reduced the binding activity of CBC with the Cy5-labeled probe. The mutant probe without the CCAAT motif showed a clear decrease in binding activity to the CBC, indicating that the interaction between the protein and the DNA specifically occurs via the CCAAT motif. In comparison, CBC was unable to bind to probes at the site of −1587 of brlA and −1027 of flbC (Fig. S6A), suggesting not all probes with CCAAT motifs could directly bind to CBC in vitro. Next, we performed chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) experiments to verify in vivo binding of HapB with DNA fragments by using a green fluorescent protein (GFP)-HapB-expressing strain. The GFP-Trap agarose beads were used to perform ChIP, and then the HapB-bound DNA fragments were amplified by primers flanking the specific CCAAT regions. As a result, ChIP-qPCR data suggest that GFP-HapB was able to pull down the target templates that could be amplified to generate positive PCR products by using primers from the specific promoter regions (Fig. 3I), which cover the previously EMSA-identified CCAAT motifs. These results collectively suggest that the CBC member HapB is capable of binding to promoters of brlA, fluG, flbD, and flbC.
FIG 3.
CBC in vitro (EMSA) and in vivo (ChIP-qPCR) binds to the promoters of genes related to asexual conidiation. (A) The purified HapB, HapC, and HapE proteins are shown by SDS-PAGE and Coomassie blue staining. HapB, HapC, HapE, and their mixture (1:1:1) are shown as labeled, respectively. (B to H) CBC binds to the promoters of target genes in vitro. EMSA of CBC binding to Cy5-labeled promoter fragments of brlA, fluG, flbD, and flbC. Two specific brlA probes (double-stranded DNA) were designed to target the CCAAT motif located at position bp 4421/3175 upstream of the brlA translational start site (−4421/−3175). Similarly, probes for fluG, flbD, and flbC targeting the CCAAT motif located at their own promoters were given. The specificity of EMSA binding was validated by adding specific competitors/cold probes (unlabeled probes) or substituting the Cy5-labeled mutant probe for the original Cy5-labeled probe with the CCAAT motif. (I) In vivo binding of the CBC was confirmed by comparing % recovery of DNA (ChIP-qPCR) from promoter fragments of conidiation-related genes to an unbound region of the AFUB_041590. ChIP-qPCR with IgG was performed as the control. Mean ± SD (n = 3). Unpaired t test. **, P < 0.01; *, P < 0.05; ns, not significant.
(A and B) EMSA for CBC binding to Cy5-labeled promoter fragments of brlA, flbC, and crzA. Probes were designed to target the CCAAT motif located at bp position 1587/1027/1109 (−1587/−1027/−1109) upstream of the brlA/flbC/crzA translational start site. The binding of CBC to the CCAAT motif at −3175/−4340 of brlA/flbD showed the positive binding. (C) A total of 3.5 × 106 spores was grown in liquid MM at 37°C for 96 h. The relative melanin content of mycelial pellets was given. (D) The number of conidia produced by the indicated strains grown in liquid medium. The indicated strains were grown in liquid MM with or without 10 mM CaCl2 for 96 h in a shaker. The culture was filtered by sterile lens-cleaning paper. Then, 500 μl of the filtered supernatant was inoculated onto solid MM. The number of resulting CFU indicates the conidial amount in the inoculated culture. Statistical significance was determined by Student’s t test. *, P < 0.05; **, P < 0.01. Download FIG S6, TIF file, 0.3 MB (310.3KB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
brlA and flbC are required for the submerged conidiation induced by a lack of hapB.
To further explore whether the submerged conidiation of the ΔhapB mutant was related to the expression of central and upstream conidiation genes, brlA and flbC were conditionally expressed under the control of the Tet on-off system in the ΔhapB mutant, yielding ΔhapB Tet-flbC and ΔhapB Tet-brlA, respectively. In this system, the gene was turned on in the presence of doxycycline and was turned off in the absence of doxycycline. The results of the real-time quantitative PCR (RT-qPCR) analysis showed that under repression conditions the mRNA expression of flbC and brlA was significantly reduced to 17% and 10%, respectively, in the ΔhapB Tet-flbC and ΔhapB Tet-brlA mutants, compared to the ΔhapB mutant (Fig. 4A), indicating that flbC and brlA could be significantly decreased by this Tet-off system. Next, we tested the conidiation of related strains grown in liquid medium and found that the submerged conidiation caused by loss of hapB was completely abolished in conditionally repressed flbC or brlA strains (Fig. 4B). Accordingly, repression of flbC or brlA markedly reduced the relative melanin accumulation accompanied by low expression of the melanin synthesis-related gene pksP and hydrophobin-encoding gene rodA (Fig. 4C to E), especially in the ΔhapB Tet-brlA strain, which is consistent with the abovementioned conidiation phenotype. In comparison, the conditional strains under the doxycycline-induced condition showed significant pigmentation accumulation in mycelium pellets and the visible asexual conidiogenous structure (Fig. 4B and C). These results suggest that the submerged conidiation caused by hapB deficiency is dependent on the flbC- and brlA-associated regulatory pathways.
FIG 4.
The conidiation-related genes brlA and flbC were required for asexual reproduction in the hapB mutant in submerged culture. (A and E) The transcript levels of the indicated genes in the ΔhapB, ΔhapB Tet-flbC, and ΔhapB Tet-brlA strains grown in liquid MM with or without 0.6 μM doxycycline (Dox) at 37°C for 48 h in a shaker culture. (B to D) A total of 3.5 × 106 spores were grown in liquid MM with or without 0.6 μM Dox at 37°C for 96 h. Asexual conidiogenous structure, macromorphology, and melanin of mycelial pellets were observed and measured. “(Off)” indicates the condition without Dox treatment. Scale bar represents 10 μm. Statistical significance was determined by Student’s t test. *, P < 0.05; **, P < 0.01; ns, not significant.
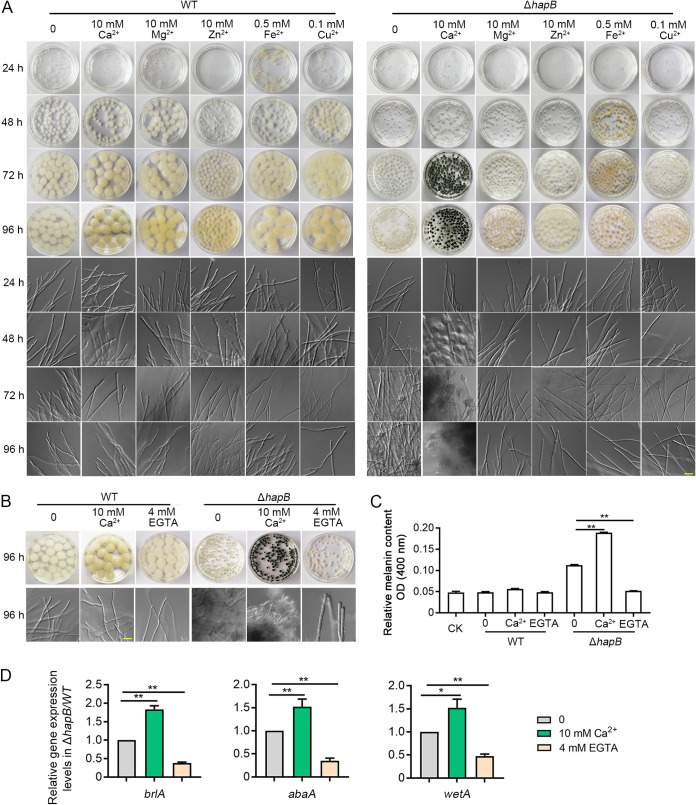
External calcium enhances conidiation caused by the absence of CBC.
To further dissect the molecular mechanism by which CBC negatively regulates conidiation under liquid culture conditions, label-free quantitative proteomics analysis was performed to investigate differentially abundant proteins between WT and ΔhapB strains grown in liquid minimal medium. The results showed that a total of 3,892 proteins were detected and quantified, and proteins with 2-fold change ratios between ΔhapB and WT strains (2.0 ≤ ΔhapB/WT strain ≤ 0.50) were selected for GO annotation and enrichment analysis. The expression levels of 429 and 355 proteins were increased and decreased, respectively, in the ΔhapB mutant compared to the WT strain (Data Set S2). GO enrichment analysis according to molecular function (MF) demonstrated that a large number of ion binding proteins, especially metal ion binding proteins, were enriched in the differentially expressed proteins between the WT and ΔhapB strains (Fig. S4). To further verify whether metal ions could affect submerged conidiation in the CBC mutants, the WT and ΔhapB strains were grown in liquid minimal medium supplemented with different divalent metal ions, including Ca2+, Mg2+, Zn2+, Fe2+, and Cu2+. As shown in Fig. 5A, when the ΔhapB strain was exposed to additional calcium (10 mM) in minimal medium, it displayed much more deep green conidia and conidiophores in mycelial pellets than when grown in MM with other noncalcium divalent metal ions, suggesting that calcium was able to enhance abnormal asexual conidiation caused by a lack of hapB. In contrast, when the 4 mM Ca2+-chelating agent EGTA was added to the medium, the submerged conidiation of the CBC mutants in liquid culture was almost abolished and the mycelial pellets were pale (Fig. 5B and C and Fig. S5A). As shown in Fig. 5D, the mRNA expression of brlA, abaA, and wetA in the ΔhapB strain was significantly upregulated by the addition of Ca2+ and downregulated by supplementation with EGTA. These data collectively indicate that calcium enhances the submerged conidiation of the CBC mutants. Notably, there was no detectable difference for the colony phenotype in CBC mutants on solid medium between the treatment of CaCl2 and EGTA under the tested concentration (Fig. S5B to D).
FIG 5.
Calcium enhanced the submerged conidiation of the CBC mutants. (A to C) A total of 3.5 × 106 spores were grown in liquid MM supplemented with indicated reagents in a shaker at 37°C for the indicated times, and the macromorphology, asexual conidiogenous structure, and melanin content of mycelial pellets were observed and measured. Scale bar represents 10 μm. (D) The relative transcript levels of the indicated genes between the ΔhapB and WT strains grown in liquid MM with/without 10 mM CaCl2 or 4 mM EGTA at 37°C for 48 h. Statistical significance was determined by Student’s t test. *, P < 0.05; **, P < 0.01.
Gene Ontology (GO) term enrichment (top 20) analysis of differentially expressed proteins (fold changes ≥ 2.0) in the proteomics database. Based on molecular function (MF), the top 20 GO terms were identified. Download FIG S4, TIF file, 0.9 MB (935.7KB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) A total of 3.5 × 106 spores was grown in liquid MM with or without 10 mM CaCl2 or 4 mM EGTA in a shaker at 37°C for 4 days, and the macromorphology and asexual conidiogenous structure of mycelial pellets were observed. Scale bar represents 10 μm. (B to D) Spores (2 × 105) were inoculated on solid MM with or without 10 mM CaCl2 or 4 mM EGTA at 37°C for 3 days. Morphology, diameter, and conidial amount of the resulting colonies. Statistical significance was determined by Student’s t test. ns, not significant. Download FIG S5, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The proteomics data sets of WT and ΔhapB strains. Download Data Set S2, XLSX file, 0.3 MB (291.6KB, xlsx) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Lack of HapB causes enhanced transient cytosolic Ca2+ levels and activates the Ca2+-calcineurin-CrzA pathway to enhance submerged conidiation.
We next investigated whether CBC affects calcium homeostasis in A. fumigatus. As shown in Fig. S3C and D, using RT-qPCR, we measured the mRNA expression of the known Ca2+ transporter-encoding genes midA, cchA, figA, vcxA to -E, pmcA to -C, pmrA, srcA, mcuA, and yvcA and found that the majority of these genes in the ΔhapB mutant truly exhibited upregulation to a certain extent compared with those in the WT strain, which is consistent with the RNA-seq data. Of these upregulated genes, the majority of Ca2+ transporters were involved in calcium influx into the cytosol or calcium stores in vacuoles and mitochondria. Therefore, HapB deficiency could affect cellular calcium homeostasis. To further test this hypothesis, we constructed aequorin-containing related strains for real-time monitoring of the dynamics of free cytosolic Ca2+ ([Ca2+]c) in living hyphal cells. As shown in Fig. 6A, the [Ca2+]c amplitude of the hapB mutant strain was significantly increased compared with that of the WT strain, suggesting that the absence of HapB resulted in abnormally higher cellular calcium capacity than that in the parental wild type. It has been reported that cytosolic free Ca2+ can bind to calmodulin, a predominant calcium sensor protein that activates protein phosphatase 2B (calcineurin), which subsequently dephosphorylates and activates the transcription factor CrzA, inducing its nuclear translocation (43–46). We speculated that HapB may have an effect on CrzA translocation and then generated C-terminally GFP-labeled CrzA (CrzA-GFP) strains on the wild-type and hapB mutant backgrounds. As shown in Fig. 6B, CrzA-GFP was located in the cytosol in minimal medium, as expected when CaCl2 was added to medium, and almost all of the CrzA-GFP was translocated into nuclei in the WT strain. However, in the hapB mutant strain, the CrzA-GFP strain showed a constant nuclear localization pattern in either calcium-supplemented medium or minimal medium alone, suggesting that hapB deficiency may lead to constitutive nuclear localization of the calcium-responsive transcription factor CrzA. In addition, we found that deletion of hapB also increased expression of crzA to some extents at the transcriptional level (Fig. 6C). To further test whether HapB could bind to the promoter of CrzA, through a CCAAT motif search analysis, we revealed that the putative crzA promoter has two CCAAT motifs located at position 1,109/207 bp (−1109/−207) upstream of the crzA translational start site. EMSA showed that CBC could in vitro bind to the CCAAT motif at the position −207, but not at the position −1109 (Fig. 6D and Fig. S6B). Furthermore, ChIP-qPCR verified that HapB in vivo bound to the specific region (position: −101 to −230) in the crzA promoter (Fig. 6E). Collectively, HapB not only affects localization of CrzA in A. fumigatus cells but also regulates crzA expression via binding to its promoter.
FIG 6.
Lack of HapB led to enhanced transient cytosolic Ca2+ levels and activated the Ca2+-calcineurin-CrzA pathway. (A) The aequorin-expressing strains were stimulated with 100 mM CaCl2 after growth in MM with or without 2 mM CaCl2. The linear graphs indicated that the real-time [Ca2+]c changes in response to calcium stimuli. [Ca2+]c, free Ca2+ concentration in the cytoplasm. Basal [Ca2+]c was indicated by the resting level prior to extracellular calcium stimulus at the 20-s time point. [Ca2+]c amplitude was indicated by the poststimulatory peak value of [Ca2+]c. Data are the average from at least six experiments. (B) Epifluorescence microscopic images demonstrating the CrzA-GFP distribution under untreated conditions (lower panels) or treatment with CaCl2 (100 mM) for 15 min (upper panels) in the parental wild-type and ΔhapB strains. Hoechst is a nuclear localization signal dye used to visualize the nucleus. Scale bar represents 10 μm. (C and G) The relative transcript levels of the indicated genes in indicated strains grown in liquid MM with/without 10 mM CaCl2 for 48 h. (D) EMSA of CBC binding to the CCAAT motif at the −207 position of crzA promoter. (E) ChIP-qPCR analysis for binding affinity of CBC to the specific promoter fragments of crzA in the GFP-HapB-expressing strain. ChIP-qPCR with IgG was performed as for the control. (F) A total of 3.5 × 106 spores were grown in liquid MM for 36 h, and then the resulting mycelial pellets were transferred into new liquid MM with or without 10 mM CaCl2, and/or 10 μg/ml Dox was added to the medium for 60 h. Asexual conidiogenous structure and macromorphology of mycelial pellets were observed. Scale bar represents 10 μm. Statistical significance was determined by Student’s t test. *, P < 0.05; **, P < 0.01.
We next speculated whether the constant nuclear localization of CrzA contributed to submerged conidiation in the hapB mutant strain under liquid culture condition. To test this hypothesis, we generated a conditionally expressed CrzA strain (ΔhapB Tet-crzA) under the control of the Tet on–off system on the background of hapB deletion. As shown in Fig. 6F and Fig. S6C and D, repressing crzA expression significantly reduced, but did not completely abolish, the submerged conidiation and pigmentation induced by the absence of hapB, in particular when calcium was present. By comparison, the ΔhapB Tet-crzA strain under the doxycycline-induced condition showed similar phenotypes as the ΔhapB strain (Fig. 6F), suggesting that Tet-on state of crzA is functional. Furthermore, turning off crzA in the ΔhapB strain (ΔhapB Tet-crzA) led to decreased expression of conidiation-, pigment-, and hydrophobin-related genes, including fluG, flbC, flbD, brlA, pksP, and rodA (Fig. 6G), which corresponds to the phenotypic difference. These data suggest that calcium-enhanced submerged conidiation of the hapB mutant is primarily dependent on CrzA.
DISCUSSION
The life cycle of filamentous fungi generally consists of hyphal growth and developmental process to generate asexual spores (1, 16, 47). For fungal pathogens, hyphal growth is required for host invasion. Under suboptimal growth conditions, fungi will undergo genetic reprogramming and initiate asexual conidiation, allowing dissemination to new hosts and the beginning of a new infection cycle. Thus, both vegetative hyphal growth and asexual reproduction are critical for invasion, growth, spore dissemination, and virulence in hosts for mycosis (17, 25). In comparison, for industrially important fungi, optimized liquid fermentation processes for biomass production are required (48–51). During this process, the initiation of asexual reproduction is repressed through the suppression of the developmental inductive transcription factor gene brlA, a conserved essential conidiation gene in fungi, and its upstream developmental activation pathway (27). Previous studies have verified that in the model filamentous fungus A. nidulans or A. fumigatus, dysfunction of NsdC, NsdD, MpkB, VosA, SfgA, VeA, or VelB results in the activation of conidiation after a period of vegetative growth under liquid culture conditions (26–28, 30, 34), suggesting that negative regulators exist for suppressing submerged conidiation to maintain constant vegetative growth with increasing mycelial pellets.
In this study, we report a novel negative regulator, the CCAAT-binding complex (CBC), of conidiation in liquid-submerged culture in A. fumigatus. CBC is a heterotrimeric transcription factor comprising three subunits (HapB/HapC/HapE) and is highly conserved from yeast to humans (41, 52–54). In A. fumigatus, we verified that CBC is able to bind to the CCAAT motif within the promoter of brlA, a key regulator of asexual development (conidiation), and its upstream elements fluG, flbC, and flbD to suppress the expression of these genes (8). Lack of CBC subunit HapB causes desuppression of conidiation in conjunction with the action of the calcium transcription factor CrzA required for brlA overexpression in liquid-submerged culture. Furthermore, we found that deletion of hapB resulted in abnormal cellular calcium capacity and that calcium remarkably enhanced the submerged conidiation caused by CBC dysfunction. Notably, hapB deletion leads to constant nuclear localization of CrzA-GFP, and CrzA is required for calcium-enhanced conidiation in the hapB mutant, implying that deletion of the CBC subunit results in calcium-enhanced constitutive activation of CrzA. These data collectively indicate that the transcription factors CrzA and CBC are reverse regulators of key conidiation regulator brlA in response to developmental signals. The findings of this study have explored the molecular switch mechanism between vegetative growth and asexual reproduction in liquid culture. Notably, A. fumigatus CBC mutants showed similar defects as each other in radial growth and conidiation compared to the wild-type strain on solid minimal medium, which is consistent with the previous report on A. nidulans (52).
As a transcription factor, CBC binds to the 5′-CCAAT-3′ motif within the promoters of its target genes to regulate their expression. The cis-acting sequence CCAAT is present in approximately 30% of eukaryotic promoters (55, 56). RNA-seq data analysis revealed that almost half (46.3%) of the A. fumigatus transcripts were differentially expressed at least 2-fold between the wild-type and hapB mutant strains, suggesting that CBC is a potential global transcriptional regulator. Notably, a lack of HapB led to the upregulation of most of the key conidiation regulatory genes, including brlA, which encodes a master regulator of the conidiation pathway. BrlA is a key regulator of conidiation in A. nidulans, A. fumigatus, and other filamentous fungi, and the appropriate expression of conserved brlA is necessary for the transition from vegetative growth to asexual development. In liquid-submerged culture, brlA is expressed at a low level, while a lack of CBC subunit HapB causes abnormally high brlA expression and results in submerged conidiation. Our EMSA data demonstrated that CBC was able to directly bind the CCAAT motif within the promoters of the conidiation inducer genes brlA, fluG, flbC, and flbD. Accordingly, a previous study revealed that CBC-associated ChIP-seq data sets showed that brlA, fluG, flbD, and flbC have at least one HapC-binding peak upstream of their translational start site (53).
Comparison data analyzed in EMSA of this study indicate that some sites/CCAAT motifs (−4421 and −3175 of brlA, −4340 of flbD, and −3268 of flbC) are in close proximity to the in vivo HapC-binding peaks (−4445 and −3192 of brlA, −4327 of flbD, and −3230 of flbC) reported in previous CBC-associated ChIP-seq data sets (53), but others (−1578 of brlA, −788 and −335 of fluG, −247 of flbD, and −1027 of flbC) are relatively far from the nearest HapC-binding peaks (−2603 of brlA, −634 and −592 of fluG, −725 of flbD, and −1548 of flbC). Results displayed that, except for −1578 of brlA and −1027 of flbC, all the other CCAAT motifs can in vitro bind to CBC. These data revealed that the in vitro binding sites of CBC in EMSA are not exactly but mostly matched to sites previously reported for the HapC-binding peak in vivo. Furthermore, through ChIP-qPCR, we tested the in vivo binding of CBC to the CCAAT motifs in relative genes and verified the binding of HapB to the specific promoter regions that harbor the abovementioned CBC-binding CCAAT motifs in EMSA. Therefore, the data collectively demonstrate that CBC is consistently able to in vivo and in vitro bind to CCAAT motifs of indicated genes.
These results indicated that CBC directly regulates both upstream and central conidiation regulators. In line with this evidence, repressing brlA or flbC expression completely abolished the submerged conidiation caused by the hapB deletion (Fig. 4B and C). These results collectively indicated that the abnormal upregulation of brlA and flbC, involved in the upstream and central pathway of conidiation, contributes to submerged conidiation in the CBC mutants. However, based on these data alone, we could not exclude other possible mechanisms whereby submerged conidiation induced by the CBC mutants is due to derepression of other negative regulators of conidiation. In A. nidulans or A. fumigatus, it has been reported that the vosA, veA, velB, sfgA, nsdC, nsdD, and mpkB mutants also displayed asexual conidiation phenotypes similar to those of the CBC mutants in liquid medium (26–28, 30, 34). We then compared the expression differences of the abovementioned genes in the hapB mutant and its parental wild-type strain. There was no significant suppression of their expression in the hapB mutant compared to that in the parental wild-type strain, suggesting that the derepression of brlA and the submerged conidiation in the hapB mutant were unlikely to be attributed to these reported negative regulators of conidiation. Instead, CBC is able to directly and negatively regulate the expression of conidiation-related genes brlA and flbC.
As an intracellular second messenger in eukaryotic cells, the calcium signaling pathway plays important roles in various physiological processes, including asexual conidiation (8, 45, 57). In the present study, the submerged conidiation of CBC mutants was significantly enhanced under calcium-treated conditions and was relatively inhibited by the calcium chelator EGTA, indicating that the calcium signaling pathway is tightly associated with this submerged conidiation. A conserved Ca2+-calcineurin-CrzA pathway in eukaryotic cells has been widely reported to regulate growth and development. Calcium-activated calcineurin can dephosphorylate the transcription factor CrzA, which induces its translocation to the nucleus and then regulates expression of targeted genes (43–46). We found that the hapB deletion caused the enhanced transient cytosolic Ca2+ level which results in the persistent nuclear localization of CrzA. The enhanced transient cytosolic Ca2+ level indicated that lack of CBC would increase cellular calcium. In fact, the RNA-seq data combined with the RT-qPCR results indicated that the majority of calcium influx channels in the plasma membrane and endomembrane system had upregulated expression to some extent in the hapB mutant. It has been reported that enhancement of transient cytosolic Ca2+ levels and persistent nuclear localization of CrzA may affect its transcription activity (43, 45, 58, 59). Thus, these data suggest the hapB deletion may abnormally activate the transcription activity of CrzA. Notably, previously reported results along with ours demonstrated that deleting either cnaA (calcineurin subunit-encoding gene) or crzA leads to reduced brlA expression and conidiation in A. fumigatus. Consistently, when the expression of crzA was conditionally turned off, both the upstream and central conidiation regulators fluG, flbC, flbD, and brlA showed significantly low expression in the hapB mutant, especially under calcium-supplemented culture conditions accompanied by decreased submerged conidiation, demonstrating that Ca2+-calcineurin-CrzA affects conidiation by increasing the expression of conidiation-related regulators. A working model summarizing the findings of this study is shown in Fig. 7. Therefore, the present results demonstrate the molecular mechanism underlying the regulation of asexual reproduction involving a new negative regulator, CBC, in A. fumigatus.
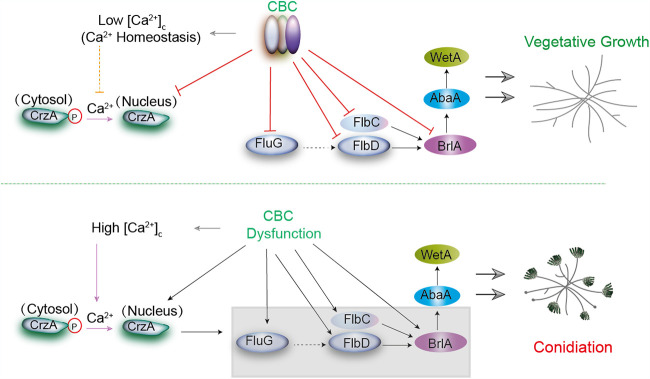
FIG 7.
A working model showing how CBC and CrzA reversely regulate vegetative growth and asexual reproduction in liquid-submerged culture. When the transcription factor CBC is present (top), it represses the brlA-associated central regulatory pathway in combination with its upstream regulators fluG, flbD, and flbC, and crzA (encoding the calcium-responsive transcription factor CrzA). Meanwhile, the cytosolic Ca2+ concentration is at a low level (Ca2+ homeostasis), which represses the dephosphorylation of CrzA and its translocation to the nucleus. In this situation, A. fumigatus undergoes constant vegetative growth in liquid-submerged culture. When CBC is absent, the expressive suppression of the upstream and central regulator in the conidiation pathways and crzA is stopped. Additionally, the absence of CBC also causes enhanced transient cytosolic Ca2+ levels and activates conidiation-positive inducer Ca2+-CrzA modules to further elevate the expression of the indicated genes. As a result, asexual conidiation is activated.
MATERIALS AND METHODS
Strains and culture.
The genotype of each A. fumigatus strain is provided in the supplemental material (Table S1). For conidiation determination, RNA-seq, proteomics, and RT-qPCR, A. fumigatus strains were grown in 100 ml liquid minimal medium (MM) with or without 10 mM CaCl2 at 37°C for the indicated time with shaking at 220 rpm. MM comprises 1% glucose, 1 ml liter−1 trace elements, and 50 ml liter−1 20× salt (60), pH 6.0.
The list of A. fumigatus strains used in this study. Download Table S1, DOCX file, 0.02 MB (17.8KB, docx) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid construction.
The plasmid for the hapB complementation strain was generated as follows. The PCR experiment was performed by using the primers NotI-hapB-F and NotI-hapB-R to amplify the hapB gene from the genomic DNA (gDNA) of A. fumigatus. Then, the hapB gene was subcloned into the NotI site of p-zero-pyr4, generating plasmid p-zero-pyr4-hapB. The resulting plasmid was transformed into the ΔhapB deletion strain, yielding the hapB-complemented strain (hapBC).
Construction of Tet-conditional and GFP-tagged strains.
The MMEJ-CRISPR system was used as described in our previously published papers to induce the Tet-On/Off system/cassette in front of the transcriptional start site of conidiation-related genes and to tag hapB and crzA with GFP at the 5′ and 3′ termini, respectively (60, 61). Briefly, single guide RNA (sgRNA) was synthesized in vitro using the MEGAscript T7 kit. The corresponding repair template was amplified by PCR from plasmid pCH008 (62). Then, the repair template fragments and corresponding sgRNA were cotransformed into ZC03/WT (a Cas9-expressing A. fumigatus recipient strain). The primers and annotations for the repair templates and sgRNA are listed in Table S2. Transformation procedures were performed as previously described (60).
Primers used in this study. Download Table S2, DOCX file, 0.03 MB (29.5KB, docx) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-seq analysis and RT-qPCR.
Fresh A. fumigatus conidia were grown in liquid MM in a rotary shaker at 220 rpm at 37°C for 48 h. For RNA-sequencing (RNA-seq) analysis, mycelial pellets were collected and quickly frozen in liquid nitrogen. After mRNA extraction, purification, and library construction, sequencing was performed by next-generation sequencing (NGS) based on the Illumina sequencing platform. A fold change of ≥2 and a P value of < 0.05 were set as the threshold values for differentially expressed genes. The detailed procedures were performed by Shanghai OE Biotech. Co., Ltd. (China). Each sample was analyzed using three biological repetitions. For RT-qPCR analysis, total RNA was extracted with the UNlQ-10 Column TRIzol total RNA isolation kit (Sangon Biotech, B511361) according to the manufacturer’s directions. The HiScript II Q RT SuperMix for qPCR kit (Vazyme, R223-01) was used to synthesize cDNA. Independent assays were performed with three replicates, and transcript levels were calculated by the comparative threshold cycle (ΔCT) and normalized against the mRNA expression of tubA in A. fumigatus. The 2−ΔΔCT method was used to determine the changes in mRNA expression. All the RT-qPCR primers are given in Table S2.
Comparative label-free quantitative proteomics analysis.
Fresh A. fumigatus conidia were grown in liquid MM in a rotary shaker at 220 rpm at 37°C for 48 h. Quantitative proteomic analysis was performed at Wuhan Genecreate Biological Engineering Co., Ltd., as a commercial service. Samples were first ground to powder in liquid nitrogen and incubated in lysis buffer (7 M urea, 2 M thiourea, 4% SDS, 40 mM Tris-HCl, pH 8.5) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and 2 mM EDTA for 5 min. Then, 10 mM dithiothreitol (DTT) was added to the sample. The suspension was sonicated for 15 min on ice and then centrifuged at 4°C and 13,000 rpm for 20 min. The supernatant was mixed with 4 volumes of precooled acetone at −20°C overnight. After centrifugation, the protein pellets were air dried and resuspended in 8 M urea-100 mM triethylammonium bicarbonate (TEAB) (pH 8.0). Protein samples were reduced with 10 mM DTT at 56°C for 30 min and alkylated with 50 mM iodoacetamide (IAM) at room temperature for 30 min in the dark. After dilution 4 times with 10 mM TEAB, the total protein concentration was measured using the Bradford method. Equal amounts of proteins from each sample were digested with trypsin. After digestion, peptides were desalted using C18 columns, and the desalted peptides were dried with a vacuum concentration meter. The dried peptide powder was redissolved in 20 μl with 0.5 M TEAB for peptide labeling. The dried peptide samples were reconstituted with mobile phase A (2% acetonitrile [ACN], 0.1% formic acid [FA]) and centrifuged at 20,000 × g for 10 min, and the supernatant was taken for injection. Separation was performed on a Thermo UltiMate 3000 ultrahigh-performance liquid chromatograph (UHPLC). The sample was first enriched in a trap column and desalted and then transferred to a tandem self-packed C18 column and separated. The nanoliter liquid phase separation end was directly connected to the mass spectrometer.
The peptides separated by liquid phase chromatography were ionized by a nano-electrospray ionization (nanoESI) source and then passed to a Q-Exactive HF X tandem mass spectrometer for data-dependent acquisition (DDA) mode detection. The resulting tandem mass spectrometry data were processed using the MaxQuant (1.6.10.43) search engine. Tandem mass spectra were searched against the Aspergillus UniProt database concatenated with the reverse decoy database. Proteins with change ratios significantly different from general protein variation (2.0 ≤ ΔhapB/wild type ≤ 0.50) were analyzed for GO terms and by KEGG pathway analysis using the UniProt-GOA database (http://www.ebi.ac.uk/GOA/) and the KEGG database, respectively.
Measurement of the free Ca2+ concentration ([Ca2+]c).
The strains expressing aequorin were cultured for 2 days at 37°C to form fresh spores. Fresh spores were filtered through nylon cloth and washed 10 times in distilled deionized water. One million (106) spores in 100 μl liquid MM with/without 2 mM CaCl2 were inoculated into each well of a 96-well microtiter plate (Thermo Fisher) and incubated at 37°C for 24 h. The subsequent measurement of the free Ca2+ concentration was performed as described previously (63, 64).
Melanin measurement.
Since the melanin could be easily dissolved in alkaline solution (65, 66), we extracted the melanin from mycelial pellets using NaOH lysis buffer. After that, the spectrophotometric method based on the absorption of light at a wavelength (400 nm) for melanin is the most popular method used for the measurement of the relative melanin contents between different samples. In brief, fresh A. fumigatus conidia were grown in liquid MM in a rotary shaker at 220 rpm at 37°C for 96 h. The resulting mycelial pellets were quickly collected and frozen in liquid nitrogen. Fifty milligrams of powder was added to 1 ml of 2 M NaOH. The suspension liquid was incubated at 37°C for 2 weeks and centrifuged at 12,000 × g for 10 min. After diluting twice, the optical density of the supernatant at 400 nm (OD400) was measured by spectrophotometry. The OD400 values were used to compare the relative melanin contents.
Recombinant CBC protein purification and electrophoretic mobility shift assay (EMSA).
To express His-labeled CBC subunits in Escherichia coli, the exons of hapB, hapC, and hapE (three subunits of CBC) were amplified with three pairs of primers, EmsA-hapB-F/EmsA-hapB-R, EmsA-hapC-F/EmsA-hapC-R, and EmsA-hapE-F/EmsA-hapE-R, respectively, and then ligated into the pET30a vector. The resulting vector was subsequently transformed into BL21(DE3) competent cells. The resulting strains were grown in LB medium at 37°C to an OD600 (optical density, measured at 600 nm) between 0.6 and 0.8 and subsequently induced by 0.1 mM isopropyl-β-d-thiogalactoside. Target proteins were purified as previously described (67) using a rapid Ni-nitrilotriacetic acid (NTA) agarose minicolumn. EMSA was performed as previously described with minor modifications (67, 68). The basic reaction mixtures consisted of 6 μl of 5× EMSA binding buffer, 1.5 μl of 1 mg/ml salmon sperm DNA, 60 ng Cy5-labeled probe (double-stranded DNA), 0.5 μg HapB, 0.8 μg HapC, and 0.6 μg HapE. For competitive testing, a 30-fold nonlabeled DNA probe (1,800 ng) as a competitive cold probe was added to the basic reaction mixtures. To confirm the specific binding of CBC to the CCAAT motif, the CCAAT motif within the probe was randomly mutated into a non-CCAAT sequence. The reaction mixtures were incubated at 37°C for 30 min and then separated in a 5% polyacrylamide gel in 0.5× Tris-borate=EDTA buffer. The Cy5-labeled probes were detected with an Odyssey machine.
Fluorescence microscopy.
Fresh conidia of CrzA-GFP-expressing strains in 0.5 ml of liquid MM were grown under different treatments (see figure legends) on sterile glass at 37°C for indicated times. The resulting hyphae were gently washed with phosphate-buffered saline (PBS) buffer three times and then fixed with 4% paraformaldehyde for 1 h. To observe nuclei, the hyphae were stained with Hoechst 33528 at a final concentration of 100 μg/ml for 30 min. For microscopic observation of conidiophore formation, fresh spores were grown in liquid medium at 37°C with shaking at 220 rpm for indicated times, and then the resulting mycelial pellets sandwiched between the microscope slide and coverslips were observed directly by microscopy. The fluorescent images were captured with a Zeiss Axio Imager A1 microscope (Zeiss, Jena, Germany).
ChIP-qPCR analysis.
Spores (3.5 × 106) of the gfp-hapB strain were grown in 100 ml of liquid MM for 48 h at 220 rpm and 37°C. ChIP was carried out using GFP-Trap agarose beads (gta-20; ChromoTek) per manufacturer’s instructions (https://www.chromotek.com/downloads/application-notes/). The IgG agarose beads were used as a control. Immunoprecipitated DNA fragments were reverse cross-linked, treated with RNase A (CWBIO, CW0601), and then purified. ChIP-qPCR was performed as previously described with SYBR Premix Ex Taq II (35). The primers for ChIP-qPCR are listed in Table S2. ChIP-qPCR experiments were run in triplicates.
Data availability.
A processed format of the RNA-seq and proteomics data sets is included in Data Sets S1 and S2, respectively. The raw Illumina sequencing data were uploaded in SRA (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA749483/) at NCBI with accession number PRJNA749483. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD027699) via the iProX partner repository (69) with the data set identifier PXD027699.
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (31861133014), the National Key R & D Program of China (2019YFA0904900), the National Natural Science Foundation of China (31770086), and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions to L.L.
We thank Johannes Wagenera (Ludwig-Maximilians-Universität München) for kindly providing pCH008.
C.Z., Y.R., and L.L. conceived the study; Y.R., C.Z., and Z.C. performed experiments; C.Z., Y.R., and L.L. analyzed and interpreted the data and wrote the manuscript with input from all authors.
We declare that we have no conflict of interests.
Footnotes
Citation Ren Y, Zhang C, Chen Z, Lu L. 2021. The heterotrimeric transcription factor CCAAT-binding complex and Ca2+-CrzA signaling reversely regulate the transition between fungal hyphal growth and asexual reproduction. mBio 12:e03007-21. https://doi.org/10.1128/mBio.03007-21.
Contributor Information
Chi Zhang, Email: bbmczhangchi@163.com.
Ling Lu, Email: linglu@njnu.edu.cn.
Reinhard Fischer, Karlsruhe Institute of Technology (KIT).
REFERENCES
- 1.van de Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latge JP. 2017. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol 15:661–674. doi: 10.1038/nrmicro.2017.90. [DOI] [PubMed] [Google Scholar]
- 2.Osherov N, May GS. 2001. The molecular mechanisms of conidial germination. FEMS Microbiol Lett 199:153–160. doi: 10.1111/j.1574-6968.2001.tb10667.x. [DOI] [PubMed] [Google Scholar]
- 3.Rybak JM, Fortwendel JR, Rogers PD. 2019. Emerging threat of triazole-resistant Aspergillus fumigatus. J Antimicrob Chemother 74:835–842. doi: 10.1093/jac/dky517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toda M, Beer KD, Kuivila KM, Chiller TM, Jackson BR. 2021. Trends in agricultural triazole fungicide use in the United States, 1992–2016 and possible implications for antifungal-resistant fungi in human disease. Environ Health Perspect 129:55001. doi: 10.1289/EHP7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margalit A, Kavanagh K. 2015. The innate immune response to Aspergillus fumigatus at the alveolar surface. FEMS Microbiol Rev 39:670–687. doi: 10.1093/femsre/fuv018. [DOI] [PubMed] [Google Scholar]
- 6.Dagenais TR, Keller NP. 2009. Pathogenesis of Aspergillus fumigatus in Invasive aspergillosis. Clin Microbiol Rev 22:447–465. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latge JP. 2001. The pathobiology of Aspergillus fumigatus. Trends Microbiol 9:382–389. doi: 10.1016/s0966-842x(01)02104-7. [DOI] [PubMed] [Google Scholar]
- 8.Park HS, Yu JH. 2016. Developmental regulators in Aspergillus fumigatus. J Microbiol 54:223–231. doi: 10.1007/s12275-016-5619-5. [DOI] [PubMed] [Google Scholar]
- 9.Cole GT. 1986. Models of cell differentiation in conidial fungi. Microbiol Rev 50:95–132. doi: 10.1128/mr.50.2.95-132.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reese S, Chelius C, Riekhof W, Marten MR, Harris SD. 2021. Micafungin-induced cell wall damage stimulates morphological changes consistent with microcycle conidiation in Aspergillus nidulans. J Fungi (Basel) 7:525. doi: 10.3390/jof7070525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park HS, Yu JH. 2012. Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol 15:669–677. doi: 10.1016/j.mib.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Bandres MV, Modi P, Sharma S. 2021. Aspergillus fumigatus. StatPearls, Treasure Island, FL. [PubMed] [Google Scholar]
- 13.Levetin E. 2004. An atlas of fungal spores. J Allergy Clin Immunol 113:366–368. doi: 10.1016/j.jaci.2003.09.049. [DOI] [PubMed] [Google Scholar]
- 14.Wang F, Sethiya P, Hu X, Guo S, Chen Y, Li A, Tan K, Wong KH. 2021. Transcription in fungal conidia before dormancy produces phenotypically variable conidia that maximize survival in different environments. Nat Microbiol 6:1066–1081. doi: 10.1038/s41564-021-00922-y. [DOI] [PubMed] [Google Scholar]
- 15.Alkhayyat F, Chang Kim S, Yu JH. 2015. Genetic control of asexual development in Aspergillus fumigatus. Adv Appl Microbiol 90:93–107. doi: 10.1016/bs.aambs.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Oiartzabal-Arano E, Perez-de-Nanclares-Arregi E, Espeso EA, Etxebeste O. 2016. Apical control of conidiation in Aspergillus nidulans. Curr Genet 62:371–377. doi: 10.1007/s00294-015-0556-0. [DOI] [PubMed] [Google Scholar]
- 17.Stewart JIP, Fava VM, Kerkaert JD, Subramanian AS, Gravelat FN, Lehoux M, Howell PL, Cramer RA, Sheppard DC. 2020. Reducing Aspergillus fumigatus virulence through targeted dysregulation of the conidiation pathway. mBio 11:e03202-19. doi: 10.1128/mBio.03202-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han S, Adams TH. 2001. Complex control of the developmental regulatory locus brlA in Aspergillus nidulans. Mol Genet Genomics 266:260–270. doi: 10.1007/s004380100552. [DOI] [PubMed] [Google Scholar]
- 19.Twumasi-Boateng K, Yu Y, Chen D, Gravelat FN, Nierman WC, Sheppard DC. 2009. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot Cell 8:104–115. doi: 10.1128/EC.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu MY, Mead ME, Lee MK, Ostrem Loss EM, Kim SC, Rokas A, Yu JH. 2018. Systematic dissection of the evolutionarily conserved WetA developmental regulator across a genus of filamentous fungi. mBio 9:e01130-18. doi: 10.1128/mBio.01130-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etxebeste O, Garzia A, Espeso EA, Ugalde U. 2010. Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol 18:569–576. doi: 10.1016/j.tim.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Wieser J, Adams TH. 1995. flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev 9:491–502. doi: 10.1101/gad.9.4.491. [DOI] [PubMed] [Google Scholar]
- 23.Otamendi A, Perez-de-Nanclares-Arregi E, Oiartzabal-Arano E, Cortese MS, Espeso EA, Etxebeste O. 2019. Developmental regulators FlbE/D orchestrate the polarity site-to-nucleus dynamics of the fungal bZIP transcription factor FlbB. Cell Mol Life Sci 76:4369–4390. doi: 10.1007/s00018-019-03121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otamendi A, Espeso EA, Etxebeste O. 2019. Identification and characterization of Aspergillus nidulans mutants impaired in asexual development under phosphate stress. Cells 8:1520. doi: 10.3390/cells8121520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mah JH, Yu JH. 2006. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot Cell 5:1585–1595. doi: 10.1128/EC.00192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MK, Kwon NJ, Choi JM, Lee IS, Jung S, Yu JH. 2014. NsdD is a key repressor of asexual development in Aspergillus nidulans. Genetics 197:159–173. doi: 10.1534/genetics.114.161430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee MK, Kwon NJ, Lee IS, Jung S, Kim SC, Yu JH. 2016. Negative regulation and developmental competence in Aspergillus. Sci Rep 6:28874. doi: 10.1038/srep28874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo JA, Guan Y, Yu JH. 2003. Suppressor mutations bypass the requirement of fluG for asexual sporulation and sterigmatocystin production in Aspergillus nidulans. Genetics 165:1083–1093. doi: 10.1093/genetics/165.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park HS, Nam TY, Han KH, Kim SC, Yu JH. 2014. VelC positively controls sexual development in Aspergillus nidulans. PLoS One 9:e89883. doi: 10.1371/journal.pone.0089883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alves de Castro P, Valero C, Chiaratto J, Colabardini AC, Pardeshi L, Pereira Silva L, Almeida F, Campos Rocha M, Nascimento Silva R, Malavazi I, Du W, Dyer PS, Brock M, Vieira Loures F, Wong KH, Goldman GH. 2021. Novel biological functions of the NsdC transcription factor in Aspergillus fumigatus. mBio 12:e03102-20. doi: 10.1128/mBio.03102-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tochigi N, Okubo Y, Ando T, Wakayama M, Shinozaki M, Gocho K, Hata Y, Ishiwatari T, Nemoto T, Shibuya K. 2013. Histopathological implications of Aspergillus infection in lung. Mediators Inflamm 2013:809798. doi: 10.1155/2013/809798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyatt TT, Wosten HA, Dijksterhuis J. 2013. Fungal spores for dispersion in space and time. Adv Appl Microbiol 85:43–91. doi: 10.1016/B978-0-12-407672-3.00002-2. [DOI] [PubMed] [Google Scholar]
- 33.Kwon-Chung KJ, Sugui JA. 2013. Aspergillus fumigatus–what makes the species a ubiquitous human fungal pathogen? PLoS Pathog 9:e1003743. doi: 10.1371/journal.ppat.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manfiolli AO, Siqueira FS, dos Reis TF, Van Dijck P, Schrevens S, Hoefgen S, Föge M, Straßburger M, de Assis LJ, Heinekamp T, Rocha MC, Janevska S, Brakhage AA, Malavazi I, Goldman GH, Valiante V. 2019. Mitogen-activated protein kinase cross-talk interaction modulates the production of melanins in Aspergillus fumigatus. mBio 10:e00215-19. doi: 10.1128/mBio.00215-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gsaller F, Hortschansky P, Furukawa T, Carr PD, Rash B, Capilla J, Muller C, Bracher F, Bowyer P, Haas H, Brakhage AA, Bromley MJ. 2016. Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathog 12:e1005775. doi: 10.1371/journal.ppat.1005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lind AL, Lim FY, Soukup AA, Keller NP, Rokas A. 2018. An LaeA- and BrlA-dependent cellular network governs tissue-specific secondary metabolism in the human pathogen Aspergillus fumigatus. mSphere 3:e00050-18. doi: 10.1128/mSphere.00050-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang PK, Cary JW, Lebar MD. 2020. Biosynthesis of conidial and sclerotial pigments in Aspergillus species. Appl Microbiol Biotechnol 104:2277–2286. doi: 10.1007/s00253-020-10347-y. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Cuesta U, Aparicio-Fernandez L, Guruceaga X, Martin-Souto L, Abad-Diaz-de-Cerio A, Antoran A, Buldain I, Hernando FL, Ramirez-Garcia A, Rementeria A. 2020. Melanin and pyomelanin in Aspergillus fumigatus: from its genetics to host interaction. Int Microbiol 23:55–63. doi: 10.1007/s10123-019-00078-0. [DOI] [PubMed] [Google Scholar]
- 39.Schmaler-Ripcke J, Sugareva V, Gebhardt P, Winkler R, Kniemeyer O, Heinekamp T, Brakhage AA. 2009. Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus. Appl Environ Microbiol 75:493–503. doi: 10.1128/AEM.02077-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valsecchi I, Dupres V, Stephen-Victor E, Guijarro JI, Gibbons J, Beau R, Bayry J, Coppee JY, Lafont F, Latge JP, Beauvais A. 2017. Role of hydrophobins in Aspergillus fumigatus. J Fungi (Basel) 4:2. doi: 10.3390/jof4010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hortschansky P, Haas H, Huber EM, Groll M, Brakhage AA. 2017. The CCAAT-binding complex (CBC) in Aspergillus species. Biochim Biophys Acta Gene Regul Mech 1860:560–570. doi: 10.1016/j.bbagrm.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Steidl S, Hynes MJ, Brakhage AA. 2001. The Aspergillus nidulans multimeric CCAAT binding complex AnCF is negatively autoregulated via its hapB subunit gene. J Mol Biol 306:643–653. doi: 10.1006/jmbi.2001.4412. [DOI] [PubMed] [Google Scholar]
- 43.Shwab EK, Juvvadi PR, Waitt G, Soderblom EJ, Barrington BC, Asfaw YG, Moseley MA, Steinbach WJ. 2019. Calcineurin-dependent dephosphorylation of the transcription factor CrzA at specific sites controls conidiation, stress tolerance, and virulence of Aspergillus fumigatus. Mol Microbiol 112:62–80. doi: 10.1111/mmi.14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juvvadi PR, Steinbach WJ. 2015. Calcineurin orchestrates hyphal growth, septation, drug resistance and pathogenesis of Aspergillus fumigatus: where do we go from here? Pathogens 4:883–893. doi: 10.3390/pathogens4040883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soriani FM, Malavazi I, Ferreira MED, Savoldi M, Kress MRV, Goldman MHD, Loss O, Bignell E, Goldman GH. 2008. Functional characterization of the Aspergillus fumigatus CRZ1 homologue, CrzA. Mol Microbiol 67:1274–1291. doi: 10.1111/j.1365-2958.2008.06122.x. [DOI] [PubMed] [Google Scholar]
- 46.Chang PK. 2008. Aspergillus parasiticus crzA, which encodes calcineurin response zinc-finger protein, is required for aflatoxin production under calcium stress. Int J Mol Sci 9:2027–2043. doi: 10.3390/ijms9102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu MY, Mead ME, Lee MK, Neuhaus GF, Adpressa DA, Martien JI, Son YE, Moon H, Amador-Noguez D, Han KH, Rokas A, Loesgen S, Yu JH, Park HS. 2021. Transcriptomic, protein-DNA interaction, and metabolomic studies of VosA, VelB, and WetA in Aspergillus nidulans asexual spores. mBio 12:e03128-20. doi: 10.1128/mBio.03128-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan MI, Shin JH, Kim JD. 2018. The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb Cell Fact 17:36. doi: 10.1186/s12934-018-0879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui J, Zhu N, Mao F, Wu P, Dang Z. 2021. Bioleaching of indium from waste LCD panels by Aspergillus niger: method optimization and mechanism analysis. Sci Total Environ 790:148151. doi: 10.1016/j.scitotenv.2021.148151. [DOI] [PubMed] [Google Scholar]
- 50.Dong L, Lin X, Yu D, Huang L, Wang B, Pan L. 2020. High-level expression of highly active and thermostable trehalase from Myceliophthora thermophila in Aspergillus niger by using the CRISPR/Cas9 tool and its application in ethanol fermentation. J Ind Microbiol Biotechnol 47:133–144. doi: 10.1007/s10295-019-02252-9. [DOI] [PubMed] [Google Scholar]
- 51.Costa C, Menolli RA, Osaku EF, Tramontina R, de Melo RH, do Amaral AE, Duarte PAD, de Carvalho MM, Smiderle FR, Silva J, Mello RG. 2019. Exopolysaccharides from Aspergillus terreus: production, chemical elucidation and immunoactivity. Int J Biol Macromol 139:654–664. doi: 10.1016/j.ijbiomac.2019.08.039. [DOI] [PubMed] [Google Scholar]
- 52.Steidl S, Papagiannopoulos P, Litzka O, Andrianopoulos A, Davis MA, Brakhage AA, Hynes MJ. 1999. AnCF, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol Cell Biol 19:99–106. doi: 10.1128/MCB.19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furukawa T, Scheven MT, Misslinger M, Zhao C, Hoefgen S, Gsaller F, Lau J, Jochl C, Donaldson I, Valiante V, Brakhage AA, Bromley MJ, Haas H, Hortschansky P. 2020. The fungal CCAAT-binding complex and HapX display highly variable but evolutionary conserved synergetic promoter-specific DNA recognition. Nucleic Acids Res 48:3567–3590. doi: 10.1093/nar/gkaa109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuncher A, Sprote P, Gehrke A, Brakhage AA. 2005. The CCAAT-binding complex of eukaryotes: evolution of a second NLS in the HapB subunit of the filamentous fungus Aspergillus nidulans despite functional conservation at the molecular level between yeast, A.nidulans and human. J Mol Biol 352:517–533. doi: 10.1016/j.jmb.2005.06.068. [DOI] [PubMed] [Google Scholar]
- 55.Bucher P. 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol 212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 56.Huber EM, Scharf DH, Hortschansky P, Groll M, Brakhage AA. 2012. DNA minor groove sensing and widening by the CCAAT-binding complex. Structure 20:1757–1768. doi: 10.1016/j.str.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 57.Zeng FS, Menardo F, Xue MF, Zhang XJ, Gong SJ, Yang LJ, Shi WQ, Yu DZ. 2017. Transcriptome analyses shed new insights into primary metabolism and regulation of Blumeria graminis f. sp tritici during conidiation. Front Plant Sci 8:1146. doi: 10.3389/fpls.2017.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manoli MT, Espeso EA. 2019. Modulation of calcineurin activity in Aspergillus nidulans: the roles of high magnesium concentrations and of transcriptional factor CrzA. Mol Microbiol 111:1283–1301. doi: 10.1111/mmi.14221. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, Zhang Y, Zhang C, Wang H, Wei X, Chen P, Lu L. 2020. Mitochondrial dysfunctions trigger the calcium signaling-dependent fungal multidrug resistance. Proc Natl Acad Sci USA 117:1711–1721. doi: 10.1073/pnas.1911560116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang C, Lu L. 2017. Precise and efficient in-frame integration of an exogenous GFP tag in Aspergillus fumigatus by a CRISPR system. Methods Mol Biol 1625:249–258. doi: 10.1007/978-1-4939-7104-6_17. [DOI] [PubMed] [Google Scholar]
- 61.Zhang C, Meng X, Wei X, Lu L. 2016. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet Biol 86:47–57. doi: 10.1016/j.fgb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Helmschrott C, Sasse A, Samantaray S, Krappmann S, Wagener J. 2013. Upgrading fungal gene expression on demand: improved systems for doxycycline-dependent silencing in Aspergillus fumigatus. Appl Environ Microbiol 79:1751–1754. doi: 10.1128/AEM.03626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Zheng Q, Sun C, Song J, Gao L, Zhang S, Munoz A, Read ND, Lu L. 2016. Palmitoylation of the cysteine residue in the DHHC motif of a palmitoyl transferase mediates Ca2+ homeostasis in Aspergillus. PLoS Genet 12:e1005977. doi: 10.1371/journal.pgen.1005977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelson G, Kozlova-Zwinderman O, Collis AJ, Knight MR, Fincham JR, Stanger CP, Renwick A, Hessing JG, Punt PJ, van den Hondel CA, Read ND. 2004. Calcium measurement in living filamentous fungi expressing codon-optimized aequorin. Mol Microbiol 52:1437–1450. doi: 10.1111/j.1365-2958.2004.04066.x. [DOI] [PubMed] [Google Scholar]
- 65.Bashyal BM, Chand R, Kushwaha C, Sen D, Prasad LC, Joshi A. 2010. Association of melanin content with conidiogenesis in Bipolaris Sorokiniana of barley (Hordeum vulgare L.). World J Microbiol Biotechnol 26:309–316. doi: 10.1007/s11274-009-0177-1. [DOI] [Google Scholar]
- 66.Hunt G, Todd C, Cresswell JE, Thody AJ. 1994. Alpha-melanocyte stimulating hormone and its analogue Nle4DPhe7 alpha-MSH affect morphology, tyrosinase activity and melanogenesis in cultured human melanocytes. J Cell Sci 107:205–211. doi: 10.1242/jcs.107.1.205. [DOI] [PubMed] [Google Scholar]
- 67.Huang W, Shang Y, Chen P, Gao Q, Wang C. 2015. MrpacC regulates sporulation, insect cuticle penetration and immune evasion in Metarhizium robertsii. Environ Microbiol 17:994–1008. doi: 10.1111/1462-2920.12451. [DOI] [PubMed] [Google Scholar]
- 68.Long N, Orasch T, Zhang S, Gao L, Xu X, Hortschansky P, Ye J, Zhang F, Xu K, Gsaller F, Straßburger M, Binder U, Heinekamp T, Brakhage AA, Haas H, Lu L. 2018. The Zn2Cys6-type transcription factor LeuB cross-links regulation of leucine biosynthesis and iron acquisition in Aspergillus fumigatus. PLoS Genet 14:e1007762. doi: 10.1371/journal.pgen.1007762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma J, Chen T, Wu S, Yang C, Bai M, Shu K, Li K, Zhang G, Jin Z, He F, Hermjakob H, Zhu Y. 2019. iProX: an integrated proteome resource. Nucleic Acids Res 47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Spores (3.5 × 106) were grown in liquid MM for the indicated days in a shaker at 37°C. The dry weight (biomass) of the resulting mycelial pellets was measured. (B to D) Spores (2 × 105) were inoculated on solid MM at 37°C for the indicated days. Morphology, diameter, and conidial amount of the resulting colonies. Statistical significance was determined by Student’s t test. **, P < 0.01. Download FIG S1, TIF file, 0.9 MB (955.2KB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene Ontology (GO) term enrichment analysis of differentially expressed genes (fold changes ≥ 2.0) in the RNA-seq database. Download FIG S2, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A and C) Comparison of RNA-seq data in heat map for the previously reported conidiation negative regulator-encoding genes and the selected Ca2+ transporter-related genes between the ΔhapB and WT strains cultured for 48 h in liquid MM. (B and D) The relative transcript levels of the indicated genes were further confirmed by RT-qPCR. Statistical significance was determined by Student’s t test. *, P < 0.05; **, P < 0.01; ns, not significant. Download FIG S3, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The RNA-seq data sets of WT and ΔhapB strains. Download Data Set S1, XLS file, 4.5 MB (4.5MB, xls) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A and B) EMSA for CBC binding to Cy5-labeled promoter fragments of brlA, flbC, and crzA. Probes were designed to target the CCAAT motif located at bp position 1587/1027/1109 (−1587/−1027/−1109) upstream of the brlA/flbC/crzA translational start site. The binding of CBC to the CCAAT motif at −3175/−4340 of brlA/flbD showed the positive binding. (C) A total of 3.5 × 106 spores was grown in liquid MM at 37°C for 96 h. The relative melanin content of mycelial pellets was given. (D) The number of conidia produced by the indicated strains grown in liquid medium. The indicated strains were grown in liquid MM with or without 10 mM CaCl2 for 96 h in a shaker. The culture was filtered by sterile lens-cleaning paper. Then, 500 μl of the filtered supernatant was inoculated onto solid MM. The number of resulting CFU indicates the conidial amount in the inoculated culture. Statistical significance was determined by Student’s t test. *, P < 0.05; **, P < 0.01. Download FIG S6, TIF file, 0.3 MB (310.3KB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene Ontology (GO) term enrichment (top 20) analysis of differentially expressed proteins (fold changes ≥ 2.0) in the proteomics database. Based on molecular function (MF), the top 20 GO terms were identified. Download FIG S4, TIF file, 0.9 MB (935.7KB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) A total of 3.5 × 106 spores was grown in liquid MM with or without 10 mM CaCl2 or 4 mM EGTA in a shaker at 37°C for 4 days, and the macromorphology and asexual conidiogenous structure of mycelial pellets were observed. Scale bar represents 10 μm. (B to D) Spores (2 × 105) were inoculated on solid MM with or without 10 mM CaCl2 or 4 mM EGTA at 37°C for 3 days. Morphology, diameter, and conidial amount of the resulting colonies. Statistical significance was determined by Student’s t test. ns, not significant. Download FIG S5, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The proteomics data sets of WT and ΔhapB strains. Download Data Set S2, XLSX file, 0.3 MB (291.6KB, xlsx) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The list of A. fumigatus strains used in this study. Download Table S1, DOCX file, 0.02 MB (17.8KB, docx) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, DOCX file, 0.03 MB (29.5KB, docx) .
Copyright © 2021 Ren et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
A processed format of the RNA-seq and proteomics data sets is included in Data Sets S1 and S2, respectively. The raw Illumina sequencing data were uploaded in SRA (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA749483/) at NCBI with accession number PRJNA749483. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD027699) via the iProX partner repository (69) with the data set identifier PXD027699.