Abstract
Background and Aim
Immune checkpoint inhibitors and their combination with other agents have recently been available in advanced hepatocellular carcinoma (HCC). Hence, a thorough understanding of the tumor microenvironment based on tumor samples is yet to be achieved. This study aimed to explore the tumor microenvironment in advanced HCC in terms of microsatellite instability‐high (MSI‐H) by using tumor samples from advanced HCC patients eligible for systemic therapy.
Methods
MSI‐H was assessed by polymerase chain reaction, and the expression of mismatch repair proteins, PD‐L1, CD8, VEGF, and HLA‐class1 was evaluated by immunohistochemistry. Whole‐exome sequencing was performed for MSI‐H tumor samples.
Results
Of 50 patients, one (2.0%) was confirmed with MSI‐H. In the MSI‐H advanced HCC tumor, a high tumor mutation burden, infiltration of CD8+ lymphocytes, and low expression of VEGF were identified. Although PD‐L1 expression was negative, there was shrinkage of tumor following pembrolizumab. However, another tumor nonresponsive to pembrolizumab was present simultaneously. Checking the Cancer Genome Atlas (TCGA) database, we found a similar case to this patient. The TCGA case had unique gene features of miR‐21 and miR‐155 overexpression and hypermethylation of the MSH2 gene.
Conclusion
We identified a very small number of MSI‐H cases in HCC using one tumor biopsy sample for each patient with advanced HCC. In addition, epigenetic aberrations possibly lead to MSI‐H in HCC patients. Since different HCC clones might coexist in the liver, sampling from multiple tumors should be considered to clarify the true proportion of MSI‐H in HCC and to analyze tumor microenvironments.
Keywords: hepatocellular carcinoma, microsatellite instability, mismatch repair, PD‐L1, tumor microenvironment
We confirmed that microsatellite instability‐high (MSI‐H) in hepatocellular carcinoma (HCC) was an extraordinarily rare case by using tumor samples of advanced HCC patients that were eligible for systemic therapies. Since the existence of heterogeneity in tumor microenvironment of advanced HCC was also implied, sampling from multiple tumors should be considered for analyzing tumor microenvironment including MSI‐H in patients with HCC.
Introduction
Deficient mismatch repair (dMMR) is generally caused by somatic mutations, miRNA‐mediated downregulation, or hypermethylation in MMR protein (MLH1, MSH2, MSH6, and PMS2) genes, which recognize and correct errors in mismatched nucleotides. 1 , 2 , 3 , 4 Errors in MMR lead to the accumulation of mutations in DNA microsatellites, known as microsatellite instability‐high (MSI‐H). MSI‐H tumors have a unique feature, as they express a large number of neoantigens due to their high number of somatic mutations. 5 Therefore, MSI‐H or dMMR tumors are expected to benefit from immunotherapy. Based on this theoretical background, clinical trials of pembrolizumab [anti‐programmed cell death 1 (PD‐1) antibody] have been conducted as a tumor‐agnostic treatment in solid MSI‐H or dMMR tumors. 6 In 2017, pembrolizumab became the first drug to receive tumor‐agnostic approval by the Food and Drug Administration for the treatment of adult and pediatric patients with unresectable or metastatic and MSI‐H or dMMR solid tumors that have progressed following prior treatment and who have no satisfactory alternative treatment options. 7
The incidence of MSI‐H or dMMR is known to vary widely among cancer types, although they are observed in many primary cancers. A latest report has demonstrated that MSI‐H or dMMR are found in approximately 30% of endometrial, 20% of colon or gastric, and less than 5% of most other cancers. 8 In hepatocellular carcinoma (HCC), the incidence is reported to be low (0–2.9%). 9 , 10 , 11 Because of this low incidence, our understanding of HCC with MSI‐H or dMMR has been very limited until now.
Immunotherapy has been developed in advanced HCC in line with other cancers. Initially, clinical trials of immune checkpoint inhibitor (nivolumab and pembrolizumab) monotherapy were conducted in patients with advanced HCC. In early‐phase clinical trials, the modest effectiveness of immune checkpoint inhibitors was established. 12 , 13 However, phase 3 trials failed to demonstrate statistical hypotheses. 14 , 15 In the IMbrave 150 trial, which compared atezolizumab [anti‐programmed cell death ligand 1 (PD‐L1) antibody] plus bevacizumab [a humanized anti‐vascular endothelial growth factor (VEGF) monoclonal antibody] versus sorafenib in advanced HCC patients with no previous history of systemic therapy, atezolizumab plus bevacizumab demonstrated a statistically significant and clinically meaningful improvement in both overall survival (OS) and progression‐free survival (PFS). 16 Nowadays, atezolizumab plus bevacizumab is the standard of care for front‐line systemic therapy in advanced HCC patients.
Exploring the association between the tumor microenvironment and treatment effectiveness by analyzing tumor samples from patients receiving immunotherapy will have essential clinical implications, including regarding mechanisms of resistance and biomarkers. In advanced HCC, to the best of our knowledge, there are only a few reports using specimens from early phase clinical trials of immune checkpoint inhibitors, 12 , 13 , 17 , 18 given the recent development of immunotherapy in this cancer type. Therefore, we aimed to explore the tumor microenvironment of advanced HCC patients focusing on MSI‐H by using tumor samples of patients that were eligible for systemic therapy. In the case of MSI‐H advanced HCC, we obtained further details using comprehensive tumor genome analysis.
Methods
Patient and sample preparation
The present prospective study was designed as a research for MSI‐H in patients with advanced HCC by performing a tumor biopsy directly before the administration of any line of systemic therapy. After obtaining informed consent, we performed the tumor biopsy. Formalin‐fixed, paraffin‐embedded (FFPE) blocks were prepared using biopsy samples, and Hematoxylin‐Eosin staining was performed on fresh 4‐μm sections from each block. We analyzed samples, which were pathologically diagnosed with HCC.
Clinical parameters
Clinical parameters of this study were collected as follows: baseline demographic data (e.g. sex, age, etc.), etiology, Child–Pugh class, alpha‐fetoprotein (AFP), radiological assessment, Eastern Cooperative Oncology Group performance status, and Barcelona Clinic Liver Cancer (BCLC) stage. The neutrophil/lymphocyte ratio (NLR) was defined as neutrophil count divided by lymphocyte count.
MSI‐polymerase chain reaction analysis
MSI analysis was carried out using the MSI (FALCO) Kit (FALCO Biosystems, Japan), which is approved as a companion diagnosis to identify patients suitable for treatment with pembrolizumab. The cumulative number of instable shifts in two or more of the five marker loci defined MSI‐H; in one, MSI‐low; and in zero, microsatellite stable. Details of polymerase chain reaction (PCR) analysis are described in Supplementary Methods.
Analysis of MMR proteins expression
Analysis of MMR protein (including MLH1, MSH2, MSH6, and PMS2) expression was performed by immunohistochemistry (IHC). Nuclei of tumor cells with intact expression of all four MMR proteins were defined as proficient MMR (pMMR), and those with loss of staining or showing a reduced expression of any MMR protein were defined as dMMR. See Supplementary Methods for more information.
Analysis of the tumor microenvironment
The tumor microenvironment was evaluated by PD‐L1, CD8, VEGF, and human leukocyte antigen (HLA)‐class1 IHC analyses. We classified PD‐L1 expression as either negative (<1%) or positive (≥1%) on the basis of the clinical trial assessments of immune checkpoint inhibitors for advanced HCC. 12 , 13 CD8 expression was defined as the mean number of CD8+ lymphocytes per tumor tissue unit in square millimeters. CD8 expression level was classified as low or high using the median value. Based on a previous report on HCC, the VEGF expression level cutoff value was set at 50%. 19 Although there are few reports exploring HLA‐class1 expression based on IHC in patients with HCC, we likewise adopted an HLA‐class1 expression cutoff value of 50% after a discussion with pathologists (Manayu Shiina, Masayuki Ota, and Jun‐ichiro Ikeda). Further instructions can be found in Supplementary Methods.
Whole‐exome sequencing
We performed whole‐exome sequencing of tumor tissues and matched white blood cells in patients who identified as MSI‐H. Oncogenicity was annotated using OncoKB. 20 More details of whole‐exome sequencing are shown in Supplementary Methods.
Data collection and analysis from TCGA‐liver hepatocellular carcinoma
A somatic mutation dataset (ID: mc3_gene_level/LIHC_mc3_gene_level.txt), a gene expression RNA sequencing dataset (ID: TCGA.LIHC.sampleMap/HiSeqV2), an miRNA mature strand expression RNA sequencing dataset (TCGA.LIHC.sampleMap/miRNA_HiSeq_gene), and a DNA methylation dataset (TCGA.LIHC.sampleMap/HumanMethylation450) were downloaded using the UCSC Xena Browser (https://xenabrowser.net/). Subsequently, somatic mutation, MSH2 mRNA expression, MSH6 mRNA expression, miR‐21 expression, miR‐155 expression, and MSH2 DNA methylation in HCC were subjected to secondary analyses.
Tumor mutation burden calculation
Tumor mutation burden (TMB) was defined as the number of nonsynonymous mutations per megabase of coding sequence. In this study, 18 919 genes (32 824 034 bp) were calculated as coding sequences.
Results
Baseline characteristics of the study population
Between March 2019 and April 2020, 58 patients underwent tumor biopsy from intrahepatic lesion (55 patients), bone metastatic lesion (2 patients), or lung metastatic lesion. Based on pathological diagnosis, we excluded eight patients for the reasons mentioned bellow; five patients did not have a definitive pathological diagnosis, and three patients had a pathological diagnosis other than HCC (intrahepatic cholangiocarcinoma, neuroendocrine carcinoma, and gastrointestinal stromal tumor). The baseline characteristics of 50 patients with a pathological diagnosis of HCC are summarized in Table 1. The median age was 71 years old. The majority of the etiology was hepatitis C virus (40.0%), followed by hepatitis B virus (10.0%) and alcohol abuse (10.0%). At the baseline radiological assessments, 26.0 and 36.0% of patients were found to have macrovascular invasion (MVI) and extrahepatic metastasis (EHM), respectively. We observed 27 patients (54.0%) with a previous history of systemic therapy.
Table 1.
Baseline characteristics of 50 advanced hepatocellular carcinoma patients with an indication of systemic therapies
| Demographics/characteristics | |
|---|---|
| Gender, male (n [%]) | 44 (88.0) |
| Age, >71 years (n [%]) | 24 (48.0) |
| HBV positive (n [%]) | 5 (10.0) |
| HCV positive (n [%]) | 20 (40.0) |
| Alcohol abuse (n [%]) | 5 (10.0) |
| Child–Pugh class A (n [%]) | 31 (62.0) |
| MVI (n [%]) | 13 (26.0) |
| EHM (n [%]) | 18 (36.0) |
| BCLC stage C (n [%]) | 25 (50.0) |
| AFP, >400 ng/mL (n [%]) | 15 (30.0) |
| Any pretreatment (n [%]) | 36 (72.0) |
| Treatment history of systemic therapy (n [%]) | 27 (54.0) |
AFP, alpha‐fetoprotein; BCLC, Barcelona clinic liver cancer; EHM, extrahepatic metastasis; HBV, hepatitis B virus; HCV, hepatitis C virus; MVI, macrovascular invasion.
Identification of MSI‐H in advanced HCC patients
In our patient cohort, one patient with MSI‐H was identified using PCR analysis (2.0%). Compared with normal tissue DNA, the tumor DNA showed instable shifts in BAT25 and NR24 (Fig. 1a,b). MMR protein expression was also evaluated in all 50 patients. Compared to a pMMR tumor, the MSI‐H tumor showed reduced expression of MSH2 and MSH6 proteins, making it a dMMR tumor (Fig. 1c,d). The NLR of the patient with the MSI‐H was 3.9, which was higher than for other patients included in this study (median NLR, 3.2).
Figure 1.
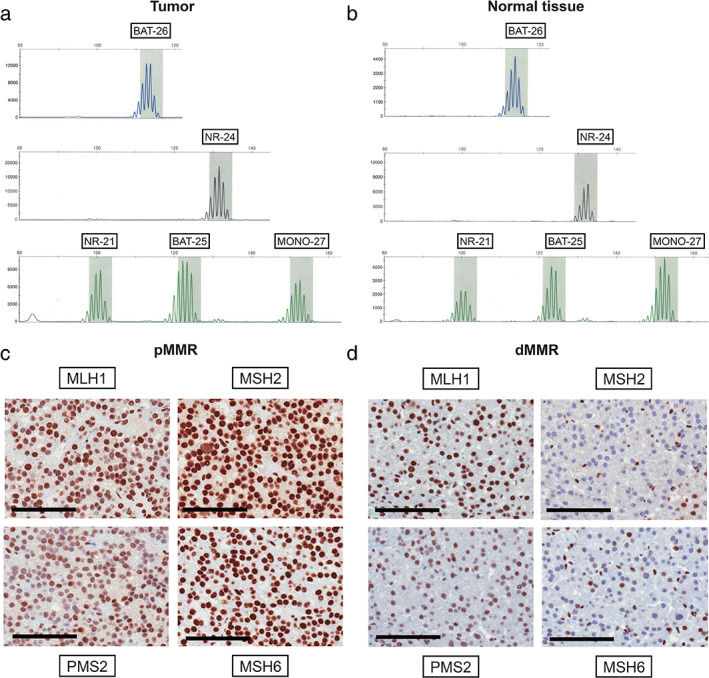
Microsatellite instability‐polymerase chain reaction (MSI‐PCR) analysis and mismatch repair (MMR) protein expression. MSI‐PCR analysis of five mononucleotide repeat markers (BAT‐25, BAT‐26, NR‐21, NR‐24, and MONO‐27). Instable shifts to the left in BAT25 and NR24 were found in the tumor DNA (a) compared with the normal tissue DNA (b). IHC analysis of MMR protein (MLH1, MSH2, MSH6, and PMS2) expression in the proficient MMR (pMMR) tumor. The pMMR tumor showed intact expression of all four MMR proteins (c). IHC analysis of MMR protein expression in the deficient MMR (dMMR) tumor. The dMMR tumor showed a reduced expression of MSH2 and MSH6 proteins (d). Scale bar = 100 μm.
Clinical course of the advanced HCC patient with MSI‐H
The patient we identified as MSI‐H was an 83‐year‐old male who had been treated with local ablation, transcatheter arterial chemoembolization, and sorafenib. We confirmed more than seven nodules in the liver with a maximum diameter of 51 mm on radiological imaging at the time of sorafenib discontinuation. Both MVI and EHM were not found, and the patient was classified as BCLC stage B. He received liver tumor biopsy from tumor #1 (Fig. 2a) immediately after sorafenib failure. The PCR and IHC images of this patient are shown in the previous section. Based on this finding, pembrolizumab was initiated every 3 weeks. He discontinued pembrolizumab 3.3 months (6 cycles) after starting treatment, due to severe ascites. Comparing radiological assessments at the baseline and end of treatment, the tumor diagnosed as MSI‐H on biopsy showed shrinkage (baseline: 51 mm, at the end of treatment: 34 mm). On the other hand, an apparently enlarging tumor was found at the segment 3 in the liver (baseline: 27 mm, at the end of treatment: 46 mm) (Fig. 2b). According to RECIST version 1.1, 21 we determined the best overall response to be stable disease.
Figure 2.
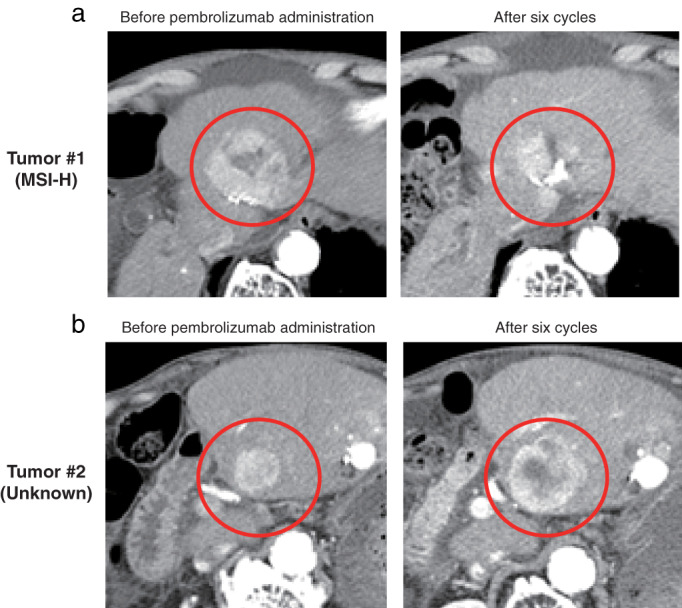
Computed tomography scan images of the microsatellite instability‐high (MSI‐H) advanced hepatocellular carcinoma patient. Tumor #1 was identified as MSI‐H from the biopsy samples. Images comparing tumor #1 status before pembrolizumab administration and after six cycles showed shrinkage (a). Tumor #2 was not examined by MSI‐polymerase chain reaction analysis and it apparently enlarged (b).
Assessment of the tumor microenvironment in the MSI‐H patient according to IHC
We evaluated PD‐L1, CD8, VEGF, and HLA‐class1 to explore the tumor microenvironment of the MSI‐H patient in the advanced HCC cohort. Of 50 patients in the present study, 16 patients (32.0%) classified as PD‐L1 expression level ≥1% (Fig. 3a), although PD‐L1 expression level of the patient with MSI‐H was <1% (Fig. 3b). The median number of CD8+ lymphocytes in all advanced HCC patients was 195/mm2 (Fig. 3c). The number of CD8+ lymphocytes of the MSI‐H tumor was higher than that of the whole study population (median level: 403/mm2) (Fig. 3d). According to the VEGF expression level classification assessed by IHC, 16.0% of advanced HCC patients categorized as high expression group (>50%) (Fig. 3e). We could not observe VEGF expression in the MSI‐H tumor (0%) (Fig. 3f). We also assessed HLA‐class1 expression level by IHC and 56.0% of the whole study population was classified as high expression level (>50%) (Fig. 3g). HLA‐class1 expression level of the patients with MSI‐H was confirmed as lower expression level (≤50%) (Fig. 3h).
Figure 3.
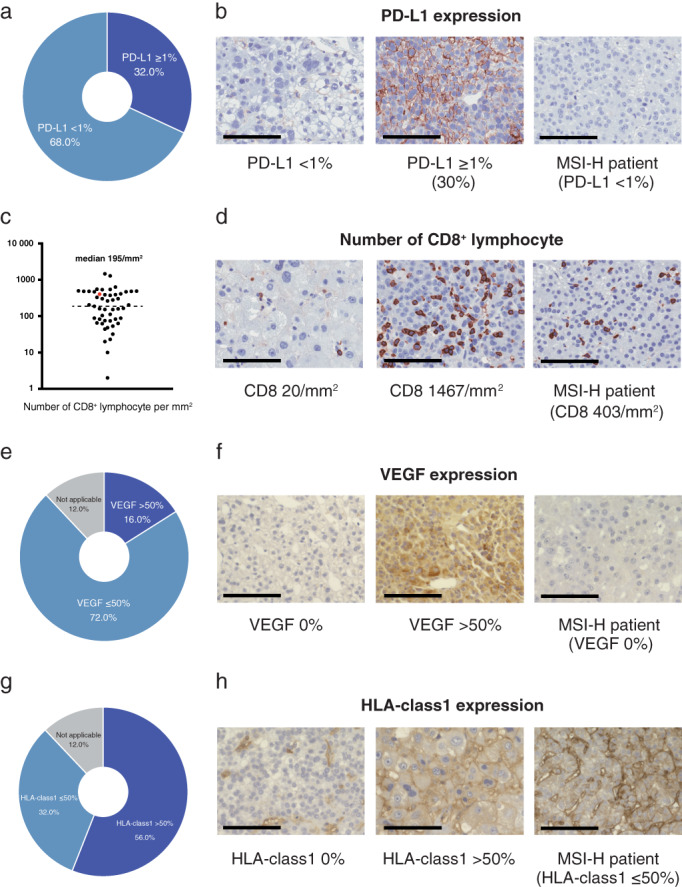
PD‐L1, CD8, VEGF, and HLA‐class1 expression. A total of 32.0% of advanced hepatocellular carcinoma (HCC) patients were classified as PD‐L1 expression level ≥1% (a). PD‐L1 expression level of the patient with microsatellite instability‐high (MSI‐H) was <1% (b). The median number of CD8+ lymphocytes in all advanced HCC patients was 195/mm2 (c). The number of CD8+ lymphocytes of the MSI‐H tumor was 403/mm2 (d). Sixteen percent of advanced HCC patients categorized as high VEGF expression (>50%) (e). VEGF expression levels of the patient with MSI‐H were 0% (f). Among advanced HCC patients, 56.0% categorized as high HLA‐class1 expression (>50%) (g). HLA‐class1 expression levels of the patient with MSI‐H were ≤50% (h). Scale bar = 100 μm.
Comprehensive genome analysis of the MSI‐H tumor patient
Figure 4a indicates the TMB of 363 HCC patients from the TCGA database and the MSI‐H patient from our cohort. The median TMB of the TCGA cohort was 2.2/Mb and that of the MSI‐H patient was extremely higher (15.4/Mb). Single nucleotide variants, insertions and deletions, and copy number variation of oncogenic genes in the MSI‐H tumor are summarized in Tables S1 and S2. Some nonsynonymous mutations and amplifications of oncogenic genes were detected in the MSI‐H tumor. Next, somatic mutations in the MMR genes were examined to explore whether MSI‐H tumors were caused by genetic or epigenetic aberration patterns. In the MSI‐H patient, nonsynonymous mutation was not detected in MSH2 genes (Table S3). Therefore, the hypothesized causes of dMMR in the MSI‐H patient in our cohort were miRNA‐mediated downregulation or hypermethylation in MMR proteins: aberrant epigenetic patterns. 1 , 2 , 3 , 4 Unfortunately, because of the lack of residual specimens, we were unable to perform detailed analyses of the miRNA or the methylation of this MSI‐H patient. Thus, we attempted to explore similar cases in the TCGA database with the assessments of MSH2 mRNA levels. One case identified from the TCGA database had a markedly reduced expression level of MSH2 mRNA without nonsynonymous mutation in the MSH2 gene (Fig. 4b). This case was confirmed to have simultaneous downregulation of both MSH2 and MSH6 mRNA (Fig. 4c). Interestingly, in that case, the correlations between MSH2 mRNA and miR‐21 and between MSH2 mRNA and miR‐155 were markedly inverse (Fig. 4d,e). In addition, we observed hypermethylation of the MSH2 gene in this case with the downregulation of MSH2 mRNA (Fig. 4f).
Figure 4.
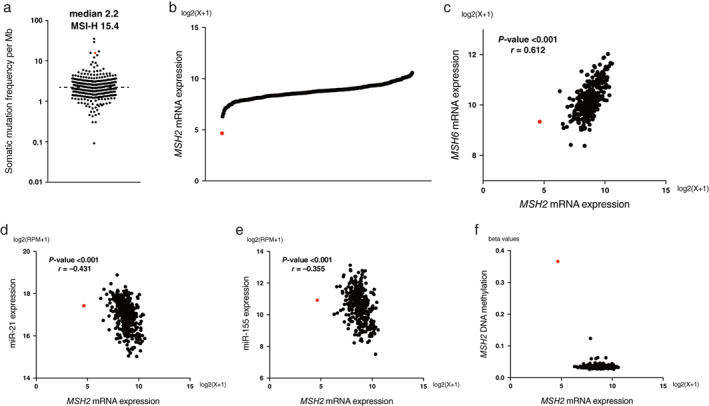
Tumor mutation burden (TMB) and MSH2 epigenetic aberration analyses of hepatocellular carcinoma (HCC) patients from the TCGA database and the microsatellite instability‐high (MSI‐H) patient from our cohort. TMB plots of 363 HCC patients from TCGA database and the MSI‐H patient from our cohort. The median TMB of the TCGA cohort was 2.2/Mb, and that of our patient with MSI‐H was 15.4/Mb (a), MSH2 mRNA expression (b), MSH2 and MSH6 mRNA expression (c), MSH2 mRNA and miR‐21 expression (d), MSH2 mRNA and miR‐155 expression (e), and MSH2 mRNA expression and MSH2 DNA methylation (f) plots of 354 HCC patients from the TCGA database. The patient with a red dot was similar to the MSI‐H patient in our cohort. The correlation was analyzed by Pearson's correlation coefficient. Statistical analyses were performed using the SPSS statistical software version 27 (IBM, Chicago, IL, USA).
Discussion
In the present prospective study, we examined tumor microenvironment of advanced HCC focusing on MSI‐H. In particular, in case of advanced HCC with MSI‐H, we explored the association between its tumor microenvironment and effectiveness of immunotherapy using comprehensive gene analysis. The existence of heterogeneity in tumor microenvironment of advanced HCC was also implied through this study. Furthermore, we investigated the cause of MSI‐H in advanced HCC using the TCGA database. At the threshold of the new era of cancer immunotherapy, we believe that this study might be a clue to elucidate tumor microenvironment in advanced HCC.
We found that the rate of MSI‐H in patients with advanced HCC was 2.0% in the present cohort. In Japanese HCC patients, Kawaoka et al. 22 recently documented the rate of MSI‐H was 2.4%, with 2 of 82 HCC patients who obtained tumor sample from biopsy and resection in 49 and 33 patients, respectively. The rate of MSI in patients with HCC was concordant with that of several previous reports. 9 , 10 , 11 We would like to emphasize that our study obtained tumor samples at the time of advanced HCC when systemic therapies were indicated, while most previous studies included patients that were analyzed using archival samples. As far as the comparison between previous reports and ours, the rate of MSI‐H might not increase even at the time of advanced HCC.
This study also confirmed the clinical outcomes of pembrolizumab in the MSI‐H advanced HCC patient in the present cohort. Although a dramatic response was not achieved in the patient unlike in other reports, 22 , 23 tumor shrinkage was observed in limited nodules, including tumor diagnosed as MSI‐H on biopsy. More interestingly, the patient from our cohort showed discrepancies in the intrahepatic tumor radiological response to pembrolizumab. Radiological findings might suggest that separate clones of the tumor coexist in the patient's liver. HCC is known to often be caused by multicentric carcinogenesis and it is not uncommon that tumors with different clones occur in the liver simultaneously. 24 These features are assumed to affect the tumor microenvironment of HCC. 23 In other cancer types, a combination of dMMR and pMMR cancers was observed in gastric cancer with multiple coexisting tumors. 25 In some cases of multiple intrahepatic tumors in HCC patients, assessment of the tumor microenvironment (including MSI‐H) may need to be conducted for each tumor.
During the present study, we attempted to understand the tumor microenvironment of the MSI‐H advanced HCC patient. This patient was confirmed to have a high TMB, similar to typical MSI‐H patients with solid tumors reported in a previous article. 5 Therefore, the tumor of this patient seemingly released antigens. 26 We also observed infiltration of CD8+ lymphocytes into the tumor in the MSI‐H patient in our study, as well as a low expression of VEGF. However, in this MSI‐H patient, PD‐L1 expression was negative. Although the strong correlations between the expression of PD‐L1 and clinical outcomes of PD‐1/PD‐L1 inhibitors have been well known in several cancers, 27 , 28 the efficacy of PD‐1/PD‐L1 inhibitors have been reported in patients with negative PD‐L1 expression. 29 , 30 , 31 Several latest reports indicated that these mechanisms might be considered to involve the action of PD‐1/PD‐L1 inhibitors on PD‐L1 pathways of dendric cells, macrophages, or NK cells. 32 , 33 , 34 It was demonstrated here and in the results of a clinical trial of PD‐1 inhibitors that the expression rate of PD‐L1 is not high in advanced HCC, approximately 30%. 12 , 13 In addition, the expression of PD‐L1 in advanced HCC has not been shown to be well correlated with the efficacy of PD‐1/PD‐L1 inhibitors and their combination with other agents. 12 , 13 , 17 We consider that exploring the mechanism of the efficacy of PD‐1/PD‐L1 inhibitors and combination therapy with other agents in cases of negative PD‐L1 expression in advanced HCC is interesting research question.
Using the TCGA database, we sought to identify cases that were similar to the MSI‐H patient in our cohort, finding a comparable case with a markedly reduced expression level of MSH2 mRNA without nonsynonymous mutation in the MSH2 gene. The case was also confirmed to have downregulation of both MSH2 and MSH6 mRNAs. Therefore, we considered the possibility that this case from the TCGA database and the MSI‐H patient from our cohort were almost identical. The case from the TCGA database had several unique gene characteristics, including an inverse correlation between the levels of MSH2 mRNA and miR‐21, and MSH2 mRNA and miR‐155 (Fig. 4d,e). Moreover, we found the hypermethylation of the MSH2 gene with downregulation of the MSH2 mRNA in this case (Fig. 4f). Previous studies suggested that the causes of MSH2 epigenetic aberration are miR‐21 or miR‐155 overexpression, or hypermethylation of the MSH2 gene. 4 , 35 , 36 , 37 Surprisingly, the case we identified from the TCGA database exhibited all three of those characteristics. Based on that comparison case and our findings, the dMMR of the MSI‐H patient in our cohort is assumed to be a result of an MSH2 epigenetic aberration caused by miRNA‐mediated downregulation and/or hypermethylation. We consider that the low incidence of MSI‐H in HCC is understandable if only cases with such a unique genetic abnormality lead to MSI‐H.
This study had several limitations but also highlights some directions for future research. First, more than a single nodule should be analyzed when assessing tumor microenvironments in patients with HCC. As much as possible, in the future studies, multiple nodules should be evaluated to take into account tumor heterogeneity in HCC. Second, our inability to analyze the levels of both miRNA expression and MSH2 gene methylation using specimens from the MSI‐H patient led to an insufficient exploration of the mechanism exhibiting dMMR. Although our results may suggest that the dMMR in the MSI‐H patient was the result of miRNA‐mediated downregulation or hypermethylation in the MMR gene instead of gene mutations, a detailed analysis of the mechanism leading to dMMR in HCC was not possible because of the lack of specimens from the MSI‐H patient. Furthermore, assuming that dMMR is an acquired modification in HCC, it would be interesting to reveal the correlation between progression to dMMR and the evolution process of carcinogenesis to advanced cancer in HCC. Recently, Eso et al. 36 demonstrated that MSH2 dysregulation was induced by tumor necrosis factor–α stimulation. They also suggested MSH2 dysregulation as a mechanism of genetic alterations during hepatocarcinogenesis. Based on our results and those of Eso et al., 36 the degradation of MSH2 mRNA caused by an epigenetic aberration may occur at the time of carcinogenesis. A detailed analysis of the association between alternation to dMMR and carcinogenesis will be necessary to understand the whole picture of the tumor microenvironment in HCC. Third, we were unable to clarify whether the effectiveness of pembrolizumab on the MSI‐H patient in our cohort is related to the specific tumor microenvironment according to dMMR. The latest clinical trials indicated that roughly 20% of patients with advanced HCC responded to immune checkpoint inhibitor monotherapy (i.e. nivolumab or pembrolizumab). 12 , 13 Hence, the effect in our MSI‐H patient may not have been related to the tumor microenvironment induced by dMMR. Currently, there is little understanding of the type of tumor microenvironment in which immunotherapy is likely to be effective in HCC. Since atezolizumab plus bevacizumab is widely used in clinical practice and has been positioned as the standard of care in the frontline treatment of advanced HCC, 16 identifying the tumor microenvironment where immunotherapy shows efficacy will be driven by tumor specimens from immunotherapy patients.
In conclusion, using one tumor biopsy sample per case of advanced HCC in patients that were eligible for systemic therapies, we determined that MSI‐H in HCC is extraordinarily rare. Although further verification is essential, epigenetic aberrations caused by downregulation by miRNAs and/or hypermethylation might lead to MSI‐H in patients with HCC. Since different HCC clones might coexist in the liver, collecting samples from multiple tumors should be considered to clarify the true proportion of MSI‐H in HCC and analyze the tumor microenvironment in patients with HCC.
Supporting information
Appendix S1. Supporting information.
Acknowledgments
Authors are grateful to the following people for their contributions to the data management: Satomi Nakamura, Ryoko Arai, and Yuka Iwase.
Shohei Mukai, Hiroaki Kanzaki, and Sadahisa Ogasawara contributed equally to this work.
Declaration of conflict of interest: Sadahisa Ogasawara received honoraria from Bayer, Eisai, Eli Lilly, and Chugai Pharma; and research funding from Bayer, Eisai, Eli Lilly, and AstraZeneca. Yuichi Takiguchi received honoraria from Chugai Pharma, AstraZeneca, and Boeringer Ingerheim; and research funding from Eli Lilly, Ono Pharma, Taiho Pharma, and Chugai Pharma. Naoya Kato received honoraria from Bayer, Eisai, Eli Lilly, and Chugai Pharma; and research funding from Bayer, Eisai, and Eli Lilly. The other authors made no disclosures.
References
- 1. Hendriks YM, de Jong AE, Morreau H et al. Diagnostic approach and management of Lynch syndrome (hereditary nonpolyposis colorectal carcinoma): a guide for clinicians. CA Cancer J. Clin. 2006; 56: 213–25. [DOI] [PubMed] [Google Scholar]
- 2. Armaghany T, Wilson JD, Chu QD, Mills G. Genetic alterations in colorectal cancer. Gastrointest. Cancer Res. 2012; 5: 19–27. [PMC free article] [PubMed] [Google Scholar]
- 3. Volinia S, Calin GA, Liu CG et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U. S. A. 2006; 103: 2257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ligtenberg MJL, Kuiper RP, Chan TL et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009; 41: 112–17. [DOI] [PubMed] [Google Scholar]
- 5. Mardis ER. Neoantigens and genome instability: impact on immunogenomic phenotypes and immunotherapy response. Genome Med. 2019; 11: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Le DT, Uram JN, Wang H et al. PD‐1 blockade in tumors with mismatch‐repair deficiency. N. Engl. J. Med. 2015; 372: 2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prasad V, Kaestner V, Mailankody S. Cancer drugs approved based on biomarkers and not tumor type‐FDA approval of pembrolizumab for mismatch repair‐deficient solid cancers. JAMA Oncol. 2018; 4: 157–8. [DOI] [PubMed] [Google Scholar]
- 8. Le DT, Durham JN, Smith KN et al. Mismatch repair deficiency predicts response of solid tumors to PD‐1 blockade. Science. 2017; 357: 409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonneville R, Krook MA, Kautto EA et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. 2017; 1: PO.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cortes‐Ciriano I, Lee S, Park WY, Kim TM, Park PJ. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 2017; 8: 15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salem ME, Puccini A, Grothey A et al. Landscape of tumor mutation load, mismatch repair deficiency, and PD‐L1 expression in a large patient cohort of gastrointestinal cancers. Mol. Cancer Res. 2018; 16: 805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. El‐Khoueiry AB, Sangro B, Yau T et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open‐label, non‐comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017; 389: 2492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu AX, Finn RS, Edeline J et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE‐224): a non‐randomised, open‐label phase 2 trial. Lancet Oncol. 2018; 19: 940–52. [DOI] [PubMed] [Google Scholar]
- 14. Yau T, Park JW, Finn RS et al. LBA38_PR—CheckMate 459: a randomized, multi‐center phase III study of nivolumab (NIVO) vs sorafenib (SOR) as first‐line treatment in patients (pts) with advanced hepatocellular carcinoma (aHCC). Ann. Oncol. 2019; 30: v874–5. [Google Scholar]
- 15. Finn RS, Ryoo BY, Merle P et al. Pembrolizumab as second‐line therapy in patients with advanced hepatocellular carcinoma in KEYNOTE‐240: a randomized, double‐blind, Phase III trial. J. Clin. Oncol. 2020; 38: 193–202. [DOI] [PubMed] [Google Scholar]
- 16. Finn RS, Qin S, Ikeda M et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 2020; 382: 1894–905. [DOI] [PubMed] [Google Scholar]
- 17. Lee MS, Ryoo BY, Hsu CH. Net al. Atezolizumab with or without bevacizumab in unresectable hepatocellular carcinoma (GO30140): an open‐label, multicentre, phase 1b study. Lancet Oncol. 2020; 21: 808–20. [DOI] [PubMed] [Google Scholar]
- 18. Sangro B, Melero I, Wadhawan S et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab‐treated patients with advanced hepatocellular carcinoma. J. Hepatol. 2020; 73: 1460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamaguchi R, Yano H, Iemura A, Ogasawara S, Haramaki M, Kojiro M. Expression of vascular endothelial growth factor in human hepatocellular carcinoma. Hepatology. 1998; 28: 68–77. [DOI] [PubMed] [Google Scholar]
- 20. Chakravarty D, Gao J, Phillips SM et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017; 1: PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eisenhauer EA, Therasse P, Bogaerts J et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer. 2019; 45: 228–47. [DOI] [PubMed] [Google Scholar]
- 22. Kawaoka T, Ando Y, Yamauchi M et al. Incidence of microsatellite instability‐high hepatocellular carcinoma among Japanese patients and response to pembrolizumab. Hepatol. Res. 2020; 50: 885–8. [DOI] [PubMed] [Google Scholar]
- 23. Ando Y, Yamauchi M, Suehiro Y et al. Complete response to pembrolizumab in advanced hepatocellular carcinoma with microsatellite instability. Clin. J. Gastroenterol. 2020; 13: 867–72. [DOI] [PubMed] [Google Scholar]
- 24. Dong LQ, Peng LH, Ma LJ et al. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J. Hepatol. 2020; 72: 896–908. [DOI] [PubMed] [Google Scholar]
- 25. Takaoka S, Hirotsu Y, Ohyama H et al. Molecular subtype switching in early‐stage gastric cancers with multiple occurrences. J. Gastroenterol. 2019; 54: 674–86. [DOI] [PubMed] [Google Scholar]
- 26. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 2016; 22: 1342–50. [DOI] [PubMed] [Google Scholar]
- 27. Ito K, Miura S, Sakaguchi T et al. The impact of high PD‐L1 expression on the surrogate endpoints and clinical outcomes of anti‐PD‐1/PD‐L1 antibodies in non‐small cell lung cancer. Lung Cancer. 2019; 128: 113–19. [DOI] [PubMed] [Google Scholar]
- 28. Sun L, Zhang L, Yu J et al. Clinical efficacy and safety of anti‐PD‐1/PD‐L1 inhibitors for the treatment of advanced or metastatic cancer: a systematic review and meta‐analysis. Sci. Rep. 2020; 10: 2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herbst RS, Soria JC, Kowanetz M et al. Predictive correlates of response to the anti‐PD‐L1 antibody MPDL3280A in cancer patients. Nature. 2014; 515: 563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Powles T, Eder JP, Fine GD et al. MPDL3280A (anti‐PD‐L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014; 515: 558–62. [DOI] [PubMed] [Google Scholar]
- 31. Shen X, Zhao B. Efficacy of PD‐1 or PD‐L1 inhibitors and PD‐L1 expression status in cancer: meta‐analysis. BMJ. 2018; 362: k3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin H, Wei S, Hurt EM et al. Host expression of PD‐L1 determines efficacy of PD‐L1 pathway blockade‐mediated tumor regression. J. Clin. Invest. 2018; 128: 805–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dong W, Wu X, Ma S et al. The mechanism of anti‐PD‐L1 antibody efficacy against PD‐L1‐negative tumors identifies NK cells expressing PD‐L1 as a cytolytic effector. Cancer Discov. 2019; 9: 1422–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhong R, Zhang Y, Chen D, Cao S, Han B, Zhong H. Single‐cell RNA sequencing reveals cellular and molecular immune profile in a Pembrolizumab‐responsive PD‐L1‐negative lung cancer patient. Cancer Immunol. Immunother. 2021; 70: 2261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Valeri N, Gasparini P, Braconi C et al. MicroRNA‐21 induces resistance to 5‐fluorouracil by down‐regulating human DNA MutS homolog 2 (hMSH2). Proc. Natl. Acad. Sci. U. S. A. 2010; 107: 21098–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eso Y, Takai A, Matsumoto T et al. MSH2 Dysregulation is triggered by proinflammatory cytokine stimulation and is associated with liver cancer development. Cancer Res. 2016; 76: 4383–93. [DOI] [PubMed] [Google Scholar]
- 37. Valeri N, Gasparini P, Fabbri M et al. Modulation of mismatch repair and genomic stability by miR‐155. Proc. Natl. Acad. Sci. U. S. A. 2010; 107: 6982–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting information.


