SUMMARY
To gain insight into the signaling determinants of effector-associated DNA methylation programming among CD8 T cells, we explore the role of interleukin (IL)-12 in the imprinting of IFNg expression during CD8 T cell priming. We observe that anti-CD3/CD28-mediated stimulation of human naive CD8 T cells is not sufficient to induce substantial demethylation of the IFNg promoter. However, anti-CD3/CD28 stimulation in the presence of the inflammatory cytokine, IL-12, results in stable demethylation of the IFNg locus that is commensurate with IFNg expression. IL-12-associated demethylation of the IFNg locus is coupled to cell division through TET2-dependent demethylation in an ex vivo human chimeric antigen receptor T cell model system and an in vivo immunologically competent murine system. Collectively, these data illustrate that IL-12 signaling promotes TET2-mediated effector DNA demethylation programming in CD8 T cells and serve as proof of concept that cytokines can guide induction of epigenetically regulated traits for T cell-based immunotherapies.
In brief
Zebley et al. report that proinflammatory cytokines imprint CD8 T cells with effector-associated DNA methylation programs during T cell priming. Specifically, interleukin-12 promotes division-dependent TET2-mediated demethylation of the IFNg locus during an endogenous CD8 T cell response to an acute infection in mice and during human chimeric antigen receptor T cell expansion.
Graphical Abstract

INTRODUCTION
Adoptive transfer of T cells into patients with cancer has proven efficacious against multiple tumor targets (Garfall et al., 2015; Park et al., 2016, 2018). Despite the potential of this approach, many patients still succumb to their disease, prompting investigation into correlates of clinical response as well as methods to improve upon T cell-based therapies. Interleukin (IL)-12 has been shown to be an essential cytokine for enhancing the anti-tumor ability of T cells in multiple settings including intratumoral administration (van Herpen et al., 2004; Zhao et al., 2011), facilitating successful anti-PD1 immunotherapy (Garris et al., 2018), and improving adoptive cell therapy approaches such as chimeric antigen receptor (CAR) T cell-mediated tumor eradication (Liu et al., 2019; Yeku et al., 2017; Zhang et al., 2015). IL-12-driven tumoricidal activity has been directly linked to an upregulation in interferon gamma (IFNg) expression (Yu et al., 1997). Prior work in murine model systems has established that IL-12 signaling during T cell priming promotes IFNg expression in both effector and memory T cells and is critical for establishing long-lived functional memory CD8 T cells (Curtsinger and Mescher, 2010; Gerner et al., 2013; Xiao et al., 2009). Given the vital role of IL-12 in mediating a successful immune response, a deeper understanding of the molecular mechanisms underlying inflammation-mediated epigenetic (DNA methylation) programming of T cells is needed.
Immunological imprinting of effector programs in CD8 T cells occurs following naive T cell antigen recognition and is tailored to the cytokine signals received from the innate immune system. Antigen-presenting cells (APCs), such as dendritic cells, sense pathogens through Toll-like receptors, which triggers production of proinflammatory cytokines including IL-12 (Trinchieri, 2003; Zundler and Neurath, 2015). While naive CD8 T cells are able to proliferate in response to TCR (signal 1) and CD28 (signal 2), signal 3 provided by either IL-12 or type I IFN is necessary for the development of a functional effector response (Xiao et al., 2009). Importantly, IL-12 or type I IFN signals are not only crucial for the differentiation of naive CD8 T cells into effector cells, but they are also required for the development of memory cells (Xiao et al., 2009). The critical role of IL-12 and type I IFN in memory formation is consistent with previous work in both humans and mice demonstrating that memory CD8 T cells originate from a subset of fate-permissive effector CD8 T cells (Youngblood et al., 2017). These memory CD8 T cells retain effector epigenetic programs that allow them to rapidly recall effector functions upon antigen reencounter (Abdelsamed et al., 2018). The ability of memory CD8 T cells to rapidly elicit effector functions contributes to their overall capacity to provide the host with lifelong protection against previously encountered pathogens.
Persistence of memory CD8 T cells in the absence of their cognate antigen indicates that the cells undergo stable changes to gene regulation that can persist during homeostatic self-renewal (Goldrath et al., 2002). Dividing cells preserve transcriptionally repressive and permissive chromatin states through DNA methylation programs (Youngblood et al., 2012). Propagation of these acquired epigenetic programs preserves a population of long-lived CD8 T cells poised to recall an effector response. Indeed, our previous work shows that the effector DNA methylation programs of human memory CD8 T cells are conserved during antigen-independent self-renewal (Abdelsamed et al., 2017). DNA methylation programming is maintained by DNA methyltransferase 1 (DNMT1), which recognizes hemi-methylated CpG sites in order to re-establish 5-methylcytosine (5mC) on the daughter strand after DNA replication. DNA demethylation is driven by ten-eleven translocation (TET) protein-mediated oxidation of 5mC to 5-hydroxymethlcytosince and can occur passively through decreasing the affinity of DNMT1 binding, resulting in the dilution of 5mC during DNA replication (An et al., 2017). While effector loci have broadly been shown to undergo demethylation during the priming stage of a T cell immune response, the mechanism governing acquisition of a memory T cell’s poised effector potential requires further investigation.
To better understand the molecular mechanisms reinforcing effector-associated epigenetic programs instilled during CD8 T cell differentiation, here we investigate the role of IL-12 in CD8 T cell priming with a focus on IFNg. Using a human CAR model system, we report that IL-12-driven demethylation of the IFNg promotor is mediated by TET2 and occurs following cell division. We extend these results to an endogenous immune response using the well-established lymphocytic choriomeningitis virus (LCMV) murine model of acute viral infection. Our results inform on the signaling events that promote effector-associated DNA demethylation during both human and mouse CD8 T cell effector differentiation and provide clinically relevant mechanistic insight regarding the optimization of adoptive T cell therapies.
RESULTS
IL-12 signaling promotes IFNg locus epigenetic reprogramming during human CD8 T cell effector differentiation
DNA methylation is known to play a crucial role in T cell differentiation. For instance, during effector differentiation, the IFNg locus in CD8 T cells becomes demethylated commensurate with its high level of expression (Kersh et al., 2006). To investigate the role of individual cytokines in effector differentiation-driven epigenetic reprogramming, we isolated naive CD8 T cells from a healthy human donor and cultured them in vitro with anti-CD3/CD28 plus different cytokines known to be crucial for CD8 T cell function, including IL-2, IL-7, IL-12, IL-15, IL-18, and IL-21 (Figure S1). While the majority of cytokines did not impact IFNg protein expression or locus methylation status, naive CD8 T cells cultured in the presence of IL-12 and anti-CD3/CD28 both exhibited an increase in IFNg expression and underwent demethylation of the IFNg promotor, prompting further investigation of IL-12 (Figures 1A and S1). We next proceeded to define the kinetics for IFNg expression and locus demethylation. At the 2-, 7-, and 14-day time points of the in vitro culture, we measured IFNg expression and examined the methylation status of the IFNg locus for naive CD8 T cells stimulated with anti-CD3/CD28 plus IL-12. As early as day 2, the CD8 T cells cultured in the presence of IL-12 exhibited robust IFNg expression as compared to minimal IFNg expression seen in the CD8 T cells stimulated only with anti-CD3/CD28 (Figures 1B–1D). Further, the CD8 T cells stimulated with anti-CD3/CD28 and IL-12 maintained a heightened ability to express IFNg at both the 7- and 14-day time points (Figures 1B–1D). Notably, the ability of these CD8 T cells to express IFNg was coupled to epigenetic changes at the IFNg locus. While unstimulated naive CD8 T cells and naive CD8 T cells stimulated with anti-CD3/CD28 remained methylated, naive CD8 T cells stimulated with anti-CD3/CD28 in the presence of IL-12 underwent demethylation of the IFNg locus by day 7 and the demethylation persisted at day 14 (Figure 1E). These data suggest that the presence of anti-CD3/CD28 signaling alone is not sufficient to fully upregulate IFNg expression or induce complete demethylation of the IFNg locus. The presence of proinflammatory cytokines, specifically IL-12, during CD8 T cell priming is necessary to drive the expression of IFNg and instill the effector-associated epigenetic programs of the IFNg locus.
Figure 1. IL-12 signaling during human CD8 T cell priming promotes IFNg expression and locus demethylation.
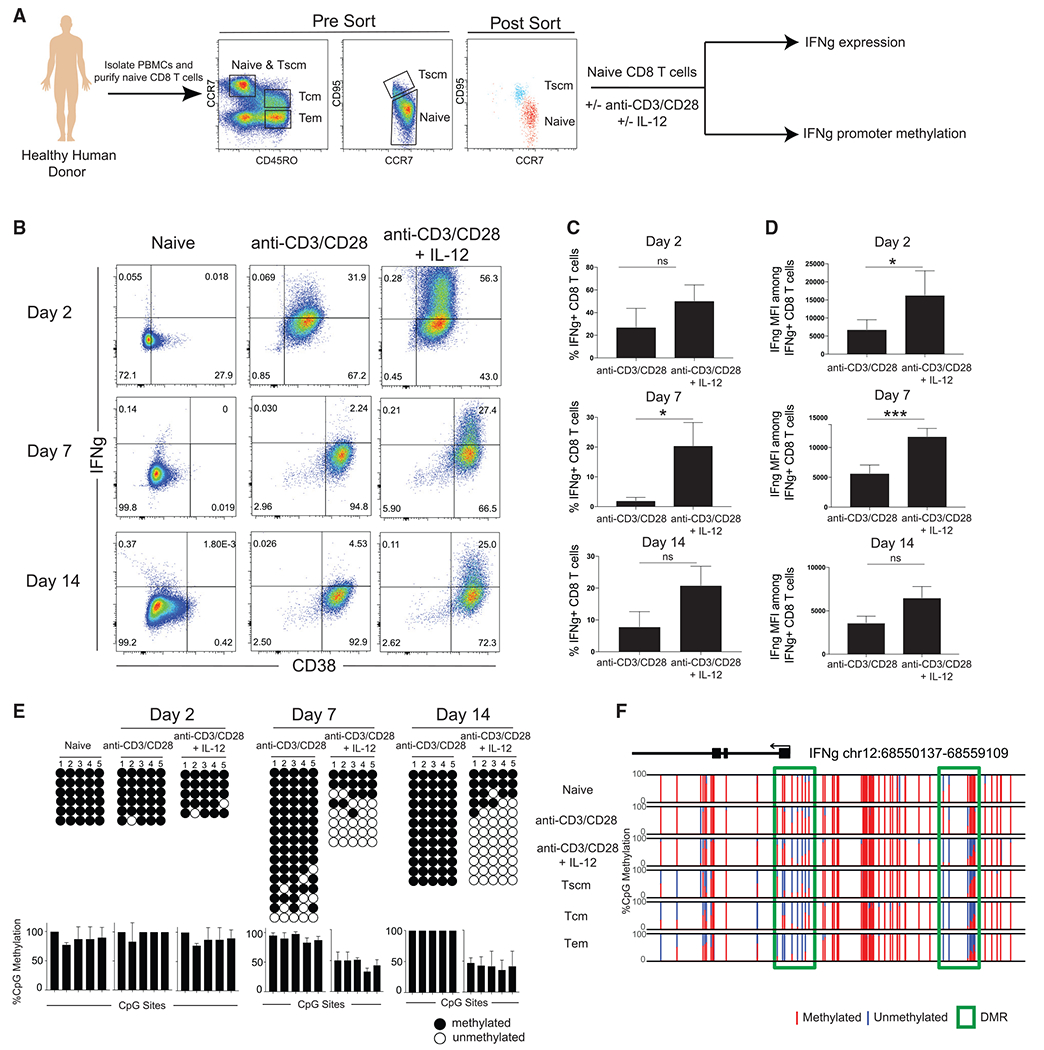
(A) Experimental setup showing isolation of naive CD8 T cells for in vitro stimulation with or without anti-CD3/CD28 and with or without IL-12. PBMC, peripheral blood mononuclear cell.
(B) CD38 and IFNg expression of naive, anti-CD3/CD28-stimulated, and anti-CD3/CD28 + IL-12-stimulated CD8 T cells after 2, 7, and 14 days. n = 2 independent donors.
(C) Summary graph of the percentage of CD8 T cells expressing IFNg. Error bars are defined based on the means ± SEM. *p < 0.05. ns, not significant.
(D) Summary graph of IFNg mean fluorescence intensity (MFI) among IFNg-positive CD8 T cells. Error bars are defined based on the means ± SEM. *p < 0.05 and ***p < 0.001.
(E) Representative IFNg promotor bisulfite sequencing methylation profiles and summary graph of naive, anti-CD3/CD28-stimulated, and anti-CD3/CD28 + IL-12-stimulated CD8 T cells after 2, 7, and 14 days. For all representative bisulfite sequencing analysis, each horizontal line represents a clone and each vertical line represents a different CpG site. Black circles represent methylated CpGs, while white circles represent unmethylated CpGs. n = 2 independent donors.
(F) WGBS nucleotide-resolution methylation profiling of IFNg. Individual CpG sites are represented by vertical lines with red indicating methylation and blue indicating lack of methylation. DMRs are represented by a green box. n = 3 independent donors.
Based on prior published work demonstrating that memory T cells transition through an effector stage of differentiation (Akondy et al., 2017; Youngblood et al., 2017), we proceeded to ask whether IL-12 signaling induced DNA demethylation events were also present in freshly isolated endogenous memory T cells from healthy human donors. Consistent with our loci-specific DNA methylation profiling, genome-wide methylation profiling documented demethylation of the IFNg promotor as well as the 5′ conserved noncoding sequence (CNS) element (Schoenborn et al., 2007) in the presence of anti-CD3/CD28 and IL-12 (Figures 1F and S2A). This specific CNS, which is approximately 5 to 6 kb upstream of the promoter, has previously been shown to directly enhance promoter activity (Schoenborn et al., 2007). Notably, these specific demethylation events were present in memory T cell subsets freshly isolated from healthy human donors (Abdelsamed et al., 2017, 2020). For further comparison, we looked at other effector-associated genes (CCR5, GZMB, TNF) and found similar demethylation events that were enriched in the long-lived endogenous T cell memory populations (Figures 1F and S2B). Given that IL-12 enhances effector differentiation, our methylation data further support a model of memory T cell development whereby the memory CD8 T cells transition through an effector stage of the immune response. To further explore the biological pathways associated with anti-CD3/CD28 and IL-12 signaling, we performed a Gene Ontology (GO) enrichment analysis of the top genes and found regulation of the JAK-STAT cascade as well as IFNg production (Figure S2C). We also performed a HOMER motif analysis for the genome regions that underwent demethylation in the presence of anti-CD3/CD28 and IL-12 (Figure S2D). Notably, the transcription factor consensus sequences were heavily enriched for CpGs, further supporting the role of DNA (de)methylation in regulation of these sites. To further characterize the implementation of IL-12-associated differentially methylated regions (DMRs) among functional memory CD8 T cells, we performed a comparison of naive CD8 T cells to each of the memory subsets (effector memory [Tem], central memory [Tcm], and stem cell memory [Tscm]; Figure S2E). Consistent with IL-12 inducing effector-associated epigenetic programs, we found the most overlap between naive and Tem CD8 T cells, followed by naive and Tcm, and lastly naive and Tscm. Taken together, these data highlight that IL-12 plays a role in establishing effector-associated epigenetic programs in memory CD8 T cells.
Cell division is coupled to epigenetic reprogramming of the IFNg locus during human effector CD8 T cell differentiation
Prior efforts to identify the origin of human memory CD8 T cells have demonstrated that long-lived memory T cells are derived from a subset of T cells that have undergone a proliferative burst during the effector stage of a primary immune response (Akondy et al., 2017). Given that we did not observe notable demethylation of the IFNg locus until after 2 days of stimulation (Figure 1E), we proceeded to evaluate whether cell division plays a role in establishing the observed epigenetic reprogramming. To evaluate the role of cell division in demethylation of the IFNg locus, CFSE-labeled naive CD8 T cells were stimulated in the presence of anti-CD3/CD28 with and without IL-12. After 1 week, the carboxyfluorescein succinimidyl ester (CFSE)-labeled CD8 T cells were assessed for IFNg expression. While both culture conditions resulted in significant cell division after stimulation with anti-CD3/CD28, the majority of IFNg expression was observed in those CD8 T cells that divided in the presence of IL-12 (Figures 2A and 2B). We next used fluorescence-activated cell sorting (FACS) to purify the CD8 T cells into undivided and divided cell populations to determine whether the IFNg expression was correlated with division-dependent changes in epigenetic programs. Bisulfite sequencing revealed that IFNg expression in the divided cell population was coupled to DNA demethylation of the IFNg locus. These results demonstrate that cell division promotes IFNg locus demethylation in CD8 T cells stimulated with anti-CD3/CD28 and IL-12 (Figure 2C). While these results further support the conclusion that IL-12 signaling is critical for inducing IFNg expression, the association with cell division prompted us to further explore the enzymatic mechanism responsible for inducing DNA demethylation.
Figure 2. IL-12-mediated demethylation of the human IFNg promotor occurs after cell division.
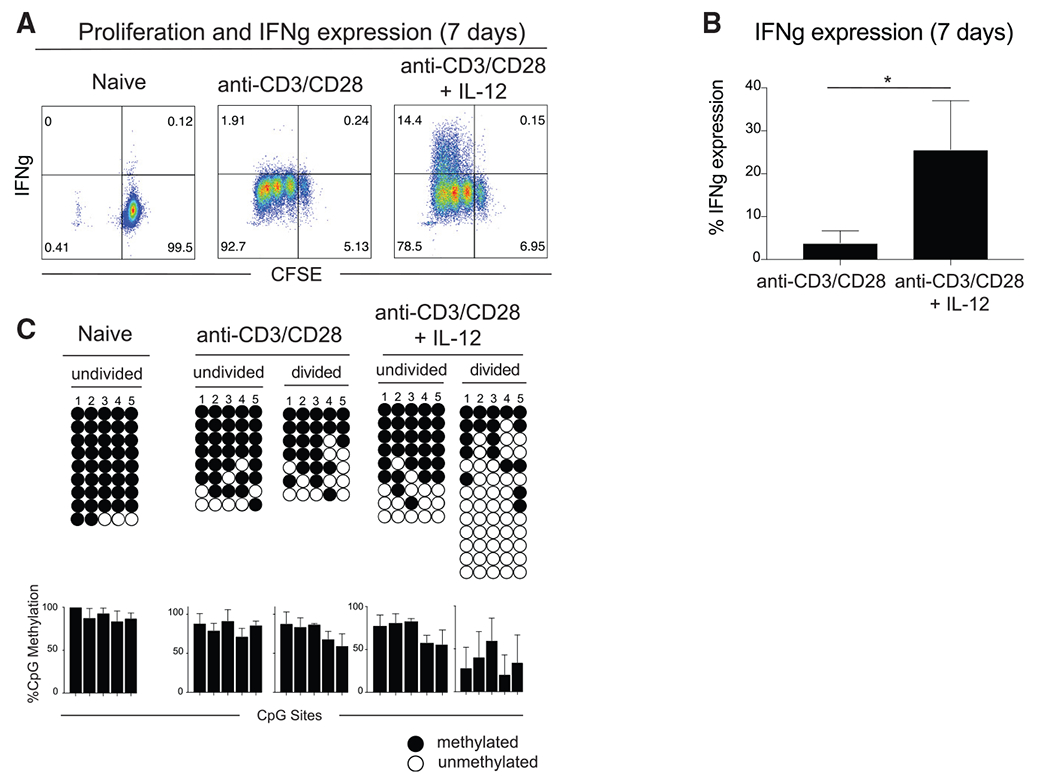
(A) Representative IFNg expression of CFSE-labeled naive, anti-CD3/CD28-stimulated, and anti-CD3/CD28 + IL-12-stimulated CD8 T cells. n = 4 independent donors.
(B) Summary of IFNg expression in CFSE-labeled anti-CD3/CD28-stimulated and anti-CD3/CD28 + IL-12-stimulated CD8 T cells from (A). n = 4 independent donors. Error bars are defined based on the means ± SEM. *p < 0.05.
(C) Representative IFNg promotor bisulfite sequencing methylation profiles and summary graph of naive, anti-CD3/CD28-stimulated, and anti-CD3/CD28 + IL-12-stimulated CD8 T cells in FACS-purified undivided and divided cell populations after 7 days. Divided populations were sorted after three or more CFSE-defined divisions. Error bars are defined based on the means ± SEM. n = 2 independent donors.
IL-12 signaling promotes TET2-driven IFNg locus demethylation during human CAR T cell activation and expansion
Previous studies have shown that the TET enzymes oxidize 5mC to 5-hydroxymethylcytone (5hmC) (An et al., 2017). The maintenance methyltransferase, DNMT1, does not readily recognize 5hmC, which results in a failure to maintain DNA methylation after cell division (An et al., 2017). Given our observation that IFNg demethylation occurs in a division-dependent manner, we used an ex vivo human CAR T cell system to ask whether TET2-mediated 5hmC deposition resulted in DNA demethylation of the IFNg locus. To investigate the role of TET2 in human CD8 T cell effector differentiation, human naive CD8 T cells were FACS purified from a healthy donor, activated in the presence of anti-CD3/CD28 followed by electroporation with Cas9 and gRNA targeting either mCherry (control) or TET2, and subsequently transduced with a lentivirus that expressed the B7-H3-specific CAR. The cells were then expanded in the presence of IL-7 and IL-15 until day 10, at which point they were activated with plate-bound recombinant human B7-H3 with or without IL-12 for 7–14 days prior to DNA methylation profiling (Figure 3A). Loci-specific bisulfite sequencing revealed that DNA demethylation of the IFNg locus was significantly enhanced by IL-12 as seen in the control knockout (KO) but strikingly, IFNg locus demethylation did not occur in the absence of TET2 in conditions either with or without IL-12 (Figure 3B). These findings were confirmed by performing whole-genome methylation profiling on both the control and TET2KO CAR T cells cultured in the presence of IL-12 at 1 and 2 weeks postactivation. Again, we observed IFNg demethylation but only in the presence of TET2. In addition, we looked at the effector-associated genes CCR5, GZMB, and TNF, which had similar TET2-mediated demethylation regions (Figure S3A). In comparing the DMRs between the control and TET2KO CAR T cells, there are 882 DMRs that are methylated after TET2KO compared to only 77 demethylated genes (Figure S3B). GO analysis was performed in order to characterize the biological pathways associated with control and TET2KO CAR T cell DMRs (Figure S3C). Notably, the methylated DMRs were associated with regulation of lymphocyte activation, while the unmethylated DMRs were associated with T cell differentiation and activation. These results document the critical role of TET2 in driving demethylation of the human IFNg locus as well as other effector-associated genes in the setting of inflammatory cytokines.
Figure 3. TET2 drives demethylation of the human IFNg promotor.
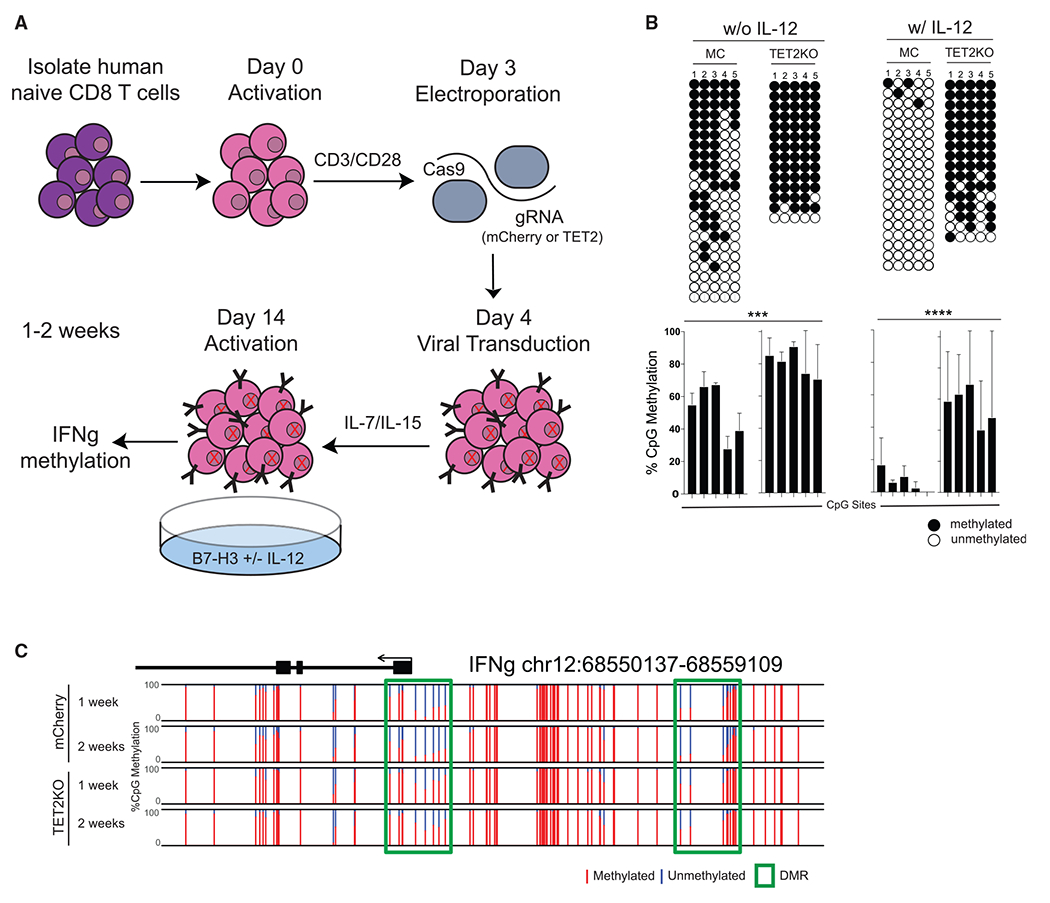
(A) Experimental setup for TET2KO in activated human naive CD8 T cells. Human naive CD8 T cells were FACS purified from a healthy donor, activated in the presence of anti-CD3/CD28 for 48 h, electroporated with cas9 and gRNA targeting either mCherry (control) or TET2, transduced with lentivirus, expanded in the presence of IL-7 and IL-15 until day 14, and activated with plate-bound recombinant B7-H3 with or without IL-12 for 7–14 days prior to DNA methylation profiling.
(B) Representative IFNg promotor bisulfite sequencing methylation profiles and summary graph of control or TET2KO ex vivo expanded CD8 T cells either in the absence or presence of IL-12. Error bars are defined based on the means ± SEM. n = 2 independent donors. ***p < 0.001 and ****p < 0.0001.
(C) WGBS nucleotide-resolution methylation profiling of control or TET2KO ex vivo expanded CD8 T cells in the presence of IL-12 for 1–2 weeks. n = 2 independent donors.
In vivo inflammation during T cell priming promotes TET2-mediated Ifng locus demethylation
We next sought to extend our findings by examining the role of inflammatory cytokines in an endogenous immune response using an immunocompetent model system. Previously, we demonstrated that demethylation of Ifng occurs in memory precursor T cells and persists into the development of all memory T cell subsets (Youngblood et al., 2017)(Figures S4A and S4B). Building on these previous findings, we examined Listeria monocytogenes (LM)-specific effector CD8 T cells to begin dissecting the role of inflammation on DNA methylation of the Ifng locus. LM-specific effector CD8 T cells undergo extensive proliferation during the effector response to acute LM infection, which has been shown to involve various inflammatory cytokines including IL-12 (Nomura et al., 2002). However, unlike acute LCMV-specific effector CD8 T cells, LM-specific effector CD8 T cells are resistant to demethylation of the Ifng 3′ CNS (Figure S4C), a site that has been previously described as a regulatory element for Ifng expression (Schoenborn et al., 2007). Notably, LM-specific effector CD8 T cells do undergo demethylation of the Ifng promotor (Northrop et al., 2006), illustrating inflammatory environment-associated intergenic variability and focusing our efforts on identifying the signaling determinants for demethylation of the Ifng 3′ CNS.
Having identified a dichotomous methylation state among LM-and LCMV-specific effector CD8 T cells, we sought to determine whether the inflammatory milieu of an acute LCMV infection could promote further demethylation of the Ifng locus in LM-specific CD8 T cells. We proceeded to interrogate LM-specific CD8 T cells in the context of single infection with LM or co-infection with LM and acute LCMV. To track an LM-specific CD8 T cell response, we adoptively transferred ova-specific T-cell receptor transgenic OT-1 CD8 T cells into naive C57BL/6 mice prior to either infection with LM-OVA or co-infection with LM-OVA and acute LCMV (Figure 4A). Relative to the OT-1 cells in mice infected only with LM-OVA, the OT-1 CD8 T cells exhibited a more effector-like phenotype in the context of LM-OVA and LCMV co-infection as exemplified by a higher percentage of KLRG1+CD127− cells (Figure S4D). Having observed a difference in the phenotypes, we next FACS purified OT-1 effector CD8 T cells from mice either single infected with LM-OVA or co-infected with LM-OVA and LCMV. The OT-1 effector CD8 T cells remained mostly methylated at the Ifng 3′ CNS after acute LM-OVA infection. Remarkably, the OT-1 CD8 T cells become demethylated in the setting of co-infection with LM-OVA and acute LCMV (Figure 4B), which we believe supports a role for proinflammatory cytokines in providing the signals required to induce epigenetic changes. Demethylation of the Ifng 3′ CNS was maintained in OT-1 memory cells generated during co-infection with LM-OVA and acute LCMV (Figure S4E), demonstrating the stability of DNA methylation programming instilled during effector differentiation. Given that LM infection induces production of IL-12, the differences in Ifng 3′ CNS methylation between LM-OVA and LM-OVA + LCMV-generated OT-1 effector and memory CD8 T cells suggests that demethylation of the Ifng 3′ CNS element is enhanced by additional proinflammatory cytokines in the setting of co-infection.
Figure 4. TET2 promotes inflammatory cytokine-mediated demethylation of murine Ifng during effector CD8 T cell differentiation.

(A) Experimental setup showing adoptive transfer of CD45.1 OT-1 CD8 T cells into WT C57BL/6 CD45.2 mice. One day later, the mice were either infected with LM-OVA or co-infected with LM-OVA and LCMV Armstrong. At the effector (8 days) and memory (>2 months) stages, CD45.1 OT1 CD8 T cells were FACS purified for phenotypic and DNA methylation analysis.
(B) Representative and summary graphs of IFNg CNS bisulfite sequencing methylation profiles of naive CD8 T cells and LM-specific and LCMV-specific effector CD8 T cells at day 8 postinfection. The bottom bar graphs show the percentage of CpG methylation (mean ± SEM) at each individual CpG site of the IFNg locus. n = 3 for LM and n = 2 for LM+LCMV summary graphs.
(C) Experimental setup showing infection of WT C57BL/6 or TET2cKO mice with LCMV Armstrong. At the effector (8 days) stage, LCMV-specific effector CD8 T cells were FACS purified for DNA methylation analysis.
(D) Representative and summary bisulfite sequencing methylation profiles of the Ifng 3′ CNS in LCMV-specific effector CD8 T cells from WT or TET2cKO mice. Error bars are defined based on the means ± SEM.
(E) Experimental setup showing adoptive transfer of Thy1.1+ P14 CD8 T cells into Thy1.2+ WT C57BL/6 or TET2cKO mice. One day later, the mice were infected with LCMV Armstrong. At the effector (8 day) stages, endogenous and adoptively transferred GP33-specific CD8 T cells were phenotypically characterized and FACS purified for DNA methylation analysis.
(F) Representative FACS analysis and summary graph characterizing the terminal effector (KLRG1+ CD127−) and memory precursor (KLRG1− CD127+) phenotype of either endogenous or adoptively transferred LCMV-specific CD8 T cells isolated from WT C57BL/6 or Tet2cKO mice. n = 4 for Tet2cKO and n = 4 for WT C57BL/6 summary graphs. ****p < 0.0001.
(G) Targeted methylation profiling of the IFNg 3′ CNS in naive, endogenous, and WT P14s CD8 Tcells isolated from WT and TET2cKO mice at the effector time point.
After establishing that the cytokine milieu resulting from LCMV infection enables demethylation of the Ifng 3′ CNS, we asked whether Tet2 was playing a role in this inflammatory-mediated process. To answer this question, we infected either wild-type (WT) or TET2cKO mice with acute LCMV and analyzed the LCMV-specific CD8 T cells at the effector time point. Bisulfite sequencing analysis revealed that the Ifng 3′ CNS was primarily demethylated in LCMV-specific CD8 T cells isolated from WT mice but remained mostly methylated in LCMV-specific CD8 T cells isolated from TET2cKO mice (Figures 4C and 4D). To further investigate these differences in Ifng 3′ CNS methylation while controlling for environmental differences between mice, we transferred 5,000 WT Thy1.1 P14s into TET2cKO or C57BL/6 mice and infected the mice with the LCMV Armstrong strain. From the same mouse, the transferred Thy1.1 P14s and endogenous LCMV-specific CD8 T cells were sorted at the effector time point (Figure 4E). Phenotypic analysis revealed that the endogenous LCMV-specific CD8 T cells isolated from the TET2cKO mice had a significantly greater proportion of memory precursors (CD127+ KLRG1−) compared to adoptively transferred P14 cells within the same animal (Figure 4F). Conversely, endogenous LCMV-specific CD8 T cells from WT mice exhibited a similar phenotype to the adoptively transferred P14 cells within the same mouse (Figures 4F and S4F–S4H). These results are consistent with a prior study documenting a role for TET2 in delineating terminal-effector versus memory-precursor differentiation programs during an LCMV T cell-mediated immune response (Carty et al., 2018). We next performed targeted DNA methylation profiling to interrogate the methylation status of the Ifng 3′ CNS. As expected, the endogenous naive population retained methylation at the Ifng locus, while the transferred Thy1.1 P14s underwent greater demethylation at the Ifnfng locus in both the WT and TET2cKO mice (Figure 4G). Importantly, differences in methylation were observed among the endogenous WT and TET2cKO LCMV-specific effector CD8 T cells. While the endogenous LCMV-specific CD8 T cells underwent demethylation of the Ifng locus in the WT mice consistent with the demethylation observed in the transferred Thy1.1 P14s, this was not the case in the TET2cKO mice. The endogenous LCMV-specific CD8 T cells in the TET2cKO mice underwent only partial demethylation of the Ifng locus and remained significantly more methylated than the transferred Thy1.1 P14s isolated from the same mouse (Figure 4E). This result demonstrates that TET2 regulates the demethylation of a cytokine-sensitive CNS element in the Ifng locus during an in vivo CD8 T cell effector response to a viral infection. Collectively, our results provide further mechanistic insight into how memory CD8 T cells acquire effector-associated epigenetic programs during the proliferative burst of an immune response.
DISCUSSION
Here we show that IL-12 acts as a signal 3 cytokine during effector and memory CD8 T cell differentiation to drive TET2-mediated DNA demethylation of the IFNg locus during T cell priming. Collectively, our data broadly demonstrate that communication between the innate and adaptive immune systems via cytokine signaling can lead to stable changes in DNA methylation. While T cells recognize antigenic peptide in the context of the major histocompatibility complex (MHC) and co-stimulation during activation, cytokine exposure introduces variability during the process of T cell differentiation. It has been previously shown that the presence of anti-CD3/CD28 signaling alone is not sufficient to produce a functional effector or memory CD8 T cell (Curtsinger and Mescher, 2010). The data we provide here highlight the importance of inflammatory cytokines in tailoring the epigenetic programs that are acquired during the priming stage of a human immune response that are maintained in long-lived memory T cells. Our in vitro studies characterizing the impact of proinflammatory cytokines on epigenetic programming of early T cell priming during an immune response are further supported by our in vivo analysis showing that the inflammatory milieu of a bystander viral infection can impact the reprogramming of an unrelated antigen-specific T cell undergoing effector differentiation. Our results build on previous work demonstrating that memory T cells arise from fate-permissive effector T cells and subsequently maintain effector-associated DNA methylation programs (Youngblood et al., 2017). Further, these observations are consistent with prior reports that memory T cells divide extensively after infection and are endowed with an effector epigenetic landscape that is maintained by quiescent memory cells (Abdelsamed et al., 2017; Akondy et al., 2017).
Previous work has shown that T-bet requires signal transducer and activator of transcription 4 (Stat4) for complete IL-12-dependent development of murine T helper 1 (Th1) cells (Thieu et al., 2008). Stat4 dimerization regulates chromatin remodeling (Becskei and Grusby, 2007), yet Stat4 is only transiently activated and the mechanisms that dictate Th1 transcriptional regulation in effector and memory CD8 T cells have not been fully elucidated. While IL-12 is known to play a critical role in murine Th1 differentiation (Jacobson et al., 1995), here we show a direct link between signal 3 cytokine signaling and DNA methylation remodeling during human effector and memory CD8 T cell differentiation. Our data suggest that site-specific IFNg demethylation is regulated by TET2 with development of a memory precursor phenotype in TET2-deficient CD8 T cells. These results are consistent with previous studies reporting that loss of TET2 limited effector T cell differentiation (Carty et al., 2018). Our findings provide additional insight into recent reports showing improved clinical efficacy coupled to CAR T cells that had a mutation in TET2 and ultimately enriched for survival of TET2 KO CAR T cells with a central memory phenotype (Fraietta et al., 2018). Here we show that deletion of TET2 during naive to effector differentiation modifies the effector DNA methylation program in CAR T cells.
In order to incorporate mechanisms of memory T cell differentiation into the manufacturing process of CAR T cells, it is important to define when and how memory T cells acquire and maintain their effector potential. Consistent with prior investigations into the origin of human memory T cells, our work here shows that effector-associated epigenetic programs are acquired during the proliferative burst of an effector response and are promoted by signal 3 cytokines present during T cell priming. Future efforts to improve CAR T cell therapeutic approaches are now looking to exploit our understanding of memory T cell identification by incorporating cytokines into conditions for developing CAR T cells. Notably, with the field looking to expand current CAR T cell protocols in treating solid tumors, our work serves as a proof of principle that such engineering approaches may be implemented to enhance the effector potential of long-lived adoptive T cell therapies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ben Youngblood (benjamin.youngblood@stjude.org).
Materials availability
All stable reagents generated in this study are available from the lead contact without restriction.
Data and code availability
Whole genome methylation data have been deposited to GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-Cy7 anti-mouse CD90.1 (clone OX-7) | BD Biosciences | Cat#561401; RRID: AB_10645789 |
| PerCp anti-mouse CD45.1 (clone A20) | BioLegend | Cat#110725; RRID: AB_893347 |
| BV421 anti-mouse CD45.2 (clone 104) | BioLegend | Cat#109831; RRID: AB_10900256 |
| FITC anti-mouse CD8a (clone 53-6.7) | BioLegend | Cat#100705; RRID: AB_312744 |
| PeCy7 anti-mouse CD8a (clone 53-6.7) | BioLegend | Cat#100721; RRID: AB_312760 |
| FITC anti-mouse Klrg1 (clone 2F1) | BioLegend | Cat#138409; RRID: AB_10643998 |
| PE anti-mouse CD127 (clone A7R34) | BioLegend | Cat#135009; RRID: AB_1937252 |
| BV421 anti-mouse/human CD44 (clone IM7) | BioLegend | Cat#103309; RRID: AB_10895752 |
| BV605 anti-mouse CD62L (clone MEL-14) | BioLegend | Cat#104437; RRID: AB_11125577 |
| FITC anti-human CCR7 (clone G043H7) | BioLegend | Cat#353215; RRID: AB_10945291 |
| APCCy7 anti-human CD8 (clone SK1) | BioLegend | Cat#344713; RRID: AB_2044005 |
| PeCy7 anti-human CD95 (clone DX2) | BioLegend | Cat#305621; RRID: AB_2100370 |
| FITC anti-human CD38 (clone HB-7) | BioLegend | Cat#356609; RRID: AB_2561949 |
| Pe anti-human IFNg (clone 4S.B3) | BioLegend | Cat#502508; RRID: AB_315233 |
| APC anti-human CD45RO (clone UCHL1) | BioLegend | Cat#304210; RRID: AB_314426 |
| Alexa Fluor® 647 AffiniPure F(ab’)2 Fragment Goat Anti-Human IgG, F(ab’)2 fragment specific | Jackson ImmunoResearch | Cat#109-606-006; |
| Bacterial and virus strains | ||
| Lymphocytic choriomeningitis virus, Armstrong strain | NA | |
| XL10-Gold Ultracompetent E. Coli | Stratagene | Cat#200314 |
| Listeria monocytogenes | Gift from McGargill lab, St. Jude Children’s Research Hospital | NA |
| Biological samples | ||
| Human PBMCs | St. Jude blood donor bank | NA |
| Chemicals, peptides, and recombinant proteins | ||
| Ghost Dye Violet 510 viability dye | Tonbo Biosciences | Cat#13-0870 |
| BD Cytofix/Cytoperm | BD Biosciences | Cat#554714 |
| LCMV gp33 monomer | Yerkes NIH tetramer core facility | NA |
| LCMV gp33 peptides | Peptide synthesis facility at SJCRH | NA |
| Cas9-NLS, purified protein | MacroLab | NA |
| Protamine Sulfate | Fresenius Kabi | C22905 |
| CD28 Antibody, anti-human | Miltenyi Biotec | 130-093-375 |
| CD3 Antibody, anti-human | Miltenyi Biotec | 130-093-387 |
| Recombinant Human B7-H3 Fc Chimera Protein | R&D systems | 1027-B3 |
| Recombinant human IL-12 p70 | PeproTech | Cat#200-12 |
| Recombinant human IL-7 | PeproTech | Cat#200-07 |
| Recombinant human IL-2 | PeproTech | Cat#200-02 |
| Recombinant human IL-15 | PeproTech | Cat#200-15 |
| Recombinant human IL-21 | PeproTech | Cat#200-21 |
| Recombinant human IL-18 | Medical & biological laboratories | Cat#B001-5 |
| Critical commercial assays | ||
| QIAGEN DNeasy kit | QIAGEN | Cat#69506 |
| EZ DNA methylation kit | Zymo Research | Cat#D5002 |
| EZ DNA methylation-Direct kit | Zymo Research | Cat#50-444-323 |
| pGEM-T Vector cloning kit | Promega | Cat#A3600 |
| JumpStart Taq ReadyMix | Sigma-Aldrich | Cat#P2893 |
| In-Fusion® HD Cloning Kit | TakaraBio | Cat#639650 |
| P3 Primary Cell 4D X Kit S | Lonza | V4XP-3032 |
| Directprep 96 Miniprep kit | QIAGEN | Cat#27361 |
| Taq polymerase-based PCR kit | QIAGEN | Cat#201225 |
| Human CD3/CD28 T cell activator | Stemcell technologies | Cat#10971 |
| EasySep human CD8+ T cell isolation kit | Stemcell | Cat#19053 |
| Deposited data | ||
| WGBS data | This paper | GEO: GSE182968 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | The Jackson Laboratory | Cat#000664 |
| Mouse: CD45.1/1 + | The Jackson Laboratory | Cat#002014 |
| Mouse: B6, Granzyme b-Cre | SJCRH animal house | NA |
| Mouse: Tet2 floxed | The Jackson Laboratory | Cat#017573 |
| Oligonucleotides | ||
| pUC/M13 reverse primer | Promega | Cat#5421 |
| Human IFNg Forward: 5′-GATTTAGAGTAATTTGAAATTTGTGG-3′ | IDT | NA |
| Human IFNg Reverse: 5′-CCTCCTCTAACTACTAATATTTATACC-3′ | IDT | NA |
| Mouse IFNg Forward: 5′-GTTTATTTTTATTGTTGTGGTTGGTAGCTG-3′ | IDT | NA |
| Mouse IFNg Reverse: 5′-CCTTTCTTCTCCAAATTACTTTTAATC-3′ | IDT | NA |
| Mouse Miseq IFNg Forward: 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTATTTTTATTGTTGTGGTTGGTAGCTG-3′ | IDT | NA |
| Mouse Miseq IFNg Reverse: 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCTTTCTTCTCCAAATTACTTTTAATC3′ | IDT | NA |
| TET2 sgRNA:5′-CGAAGCAAGCCTGATGGAACNGG-3′ | Synthego | NA |
| sgRNA mCherry:5′-CAAGUAGUCGGGGAUGTCGG - 3′ | Synthego | NA |
| Recombinant DNA | ||
| Plasmid: pCL45.MND.m276.hCD8aTM.hCD28.z | This paper | NA |
| Software and algorithms | ||
| Prism 6 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo 9.9.5 | FlowJo | NA |
| SnapGene | Insightful Science | snapgene.com |
| Model-based analysis of bisulfite sequencing | https://www.ncbi.nlm.nih.gov/pubmed/26184873/ | |
| Gene Ontology (GO) enrichment analysis | GREAT | http://great.stanford.edu/public/html/index.php |
| Loci-specific bisulfite sequencing | Quma | http://quma.cdb.riken.jp/ |
| Other | ||
| Fortessa flow cytometer | Flow cytometry core facility at SJCRH | NA |
| Illumina NovaSeq | Hartwell Center at SJCRH | NA |
| Illumina HiSeq 4000 | Hartwell Center at SJCRH | NA |
| 4D-Nucleofector | Lonza | Cat#AAF-1002B |
| Ficoll-Paque PLUS | GE Healthcare | |
| BD GolgiPlug | Fisher scientific | Cat#15847968 |
| BD GogliStop | Fisher scientific | Cat#10716676 |
| CFSE | Fisher scientific | Cat#50-591-407 |
| Penicillin-streptomycin (GIBCO) | ThermoFisher | Cat#1514012 |
| Gentamycin (GIBCO) | ThermoFisher | Cat#15750060 |
All original code has been deposited at GEO and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse strains
Tet2 floxed mice were obtained from Jackson laboratories (Catalog #017573) and crossed with granzyme B cre mice (Ghoneim et al., 2017) to generate mice that conditionally delete Tet2 in activated CD8 T cells. Wild-type C57B6 mice were purchased from Jackson laboratories. Female mice age ~6 weeks were used for all experiments. Littermates of the same sex were randomly assigned to experimental groups. Mouse studies were performed in accordance with ISCUC guidelines and adhered to the regulatory standards.
Human samples
This study was conducted with approval from the Institutional Review Board of St. Jude Children’s Research Hospital. Deidentified human peripheral blood mononuclear cells (PBMCs) were collected through the St. Jude Blood Bank from healthy donors, and samples for methylation profiling were collected under IRB protocol XPD15-086.
METHODS DETAILS
Isolation and phenotypic analysis of mouse antigen-specific CD8 T cells
LM specific CD8 T cells were generated by adoptive transfer of ~5000 naive OT-1 anti-CD3/CD28 transgenic CD8 T cells (CD45.1/1+) into C57BL/6 mice (CD45.1/2+). Chimeric mice were subsequently infected with Listeria monocytogenes containing OVA by injection of 1.5×10^4 CFU per mouse. LCMV specific CD8 T cells were generated by adoptive transfer of ~5000 congenically distinct naive P14 CD8 T cells. One day later, we infected the mice with acute lymphocytic choriomeningitis virus (LCMV). Acute LCMV infection was performed by i.p. injection of 2×10^5 PFU Armstrong strain per mouse. Chronic LCMV infections were performed by i.v. injection of 2×10^6 PFU LCMV per mouse using either Clone 13 strain. Antigen specific CD8 T cells were identified by gating on CD44hi CD8+ lymphocytes that were either tetramer+ or congenically labeled. Tetramer was obtained from the NIH Yerkes tetramer core facility. All antigen specific CD8 T cells were phenotypically analyzed by Flow Cytometry after surface staining using monoclonal antibodies for Thy1.1(BD clone OX-7), CD45.1 (Biolegend clone A20), CD45.2 (Biolegend clone 104), CD8 (Biolegend clone 53-6.7), Klrg1 (Biolegend clone 2F1), CD127 (Biolegend clone A7R34), CD44 (Biolegend clone IM7).
Isolation of human naive CD8 T cells from healthy donor blood
PBMCs were purified from platelet apheresis blood unit by density gradient. Briefly, blood was diluted 1:2.5 using sterile Dulbecco’s phosphate-buffered saline (Life Technologies). The diluted blood was then overlayed above Ficoll-Paque PLUS (GE Healthcare) at a final dilution of 1:2.5 (ficoll:diluted blood). The gradient was centrifuged at 400 xg with no brake for 20 minutes at room temperature. The PBMCs interphase layer was collected and washed with 2% fetal bovine serum (FBS)/1mM EDTA PBS buffer and then centrifuged at 400xg for 5 minutes.
Flow cytometric analysis of human naive CD8 T cells
After enrichment of CD8 T cells, naive and memory CD8 T cell subsets were sorted using the following markers, as previously described (Gattinoni et al., 2011; Lugli et al., 2013). Naive CD8 T cells were defined as live CD8+ CCR7+, CD45RO−, CD45RA+, and CD95− cells. CD8 Tem cells were defined as live CD8+, CCR7−, and CD45RO+ cells. TCM cells were defined as live CD8+, CCR7+, and CD45RO+ cells. Tscm cells were defined as live CD8+, CCR7+, CD45RO−, and CD95+ cells. Naive sorted cells were checked for purity (i.e., samples were considered pure if > 90% of the cells had the desired phenotype). The naive CD8 T cells were then cultured ex vivo with or without anti-CD3/CD28 (1:1 ratio) in the presence or absence of the following cytokines: IL-2 (10 ng/mL), IL-7 (5 ng/mL), IL-12 (10ng/mL), IL-15 (5ng/mL), IL-18 (10ng/mL), IL-21 (30ng/mL). At 2, 7, or 14 days, the levels of IFNg expression was examined by intracellular staining after exposure to 4 hours of GolgiStop and GolgiPlug (BD).
Human CD8 T cells were stained with the following antibodies: CCR7 (Biolegend clone G043H7), APCCy7 (Biolegend clone SK1), PeCy7 (Biolegend clone DX2), CD38 (Biolegend clone HB-7), IFNg (Biolegend clone 4S.B3) APC (Biolegend clone UCHL1).
In vitro CD8 T cell ex vivo proliferation
Sorted naive CD8 T cells and memory CD8 T cell subsets were labeled with CFSE (Life Technologies) at a final concentration of 2 μM. CFSE-labeled cells were maintained in culture in RPMI containing 10% FBS, penicillin-streptomycin, and gentamycin. After 7 days of ex vivo culture at 37°C and 5% CO2, undivided and divided cells (third division and higher) were sorted and checked for purity (> 90%). The levels of IFNg protein expression was determined by intracellular staining after exposure to 4 hours of GolgiStop and GolgiPlug (BD).
Genomic methylation analysis
DNA was extracted from the sorted cells by using a DNA-extraction kit (QIAGEN) and then bisulfite treated using an EZ DNA methylation kit (Zymo Research). The bisulfite-modified DNA-sequencing library was generated using the EpiGnome™ kit (Epicenter) per the manufacturer’s instructions. Bisulfite-modified DNA was PCR amplified using the following primers for mouse and human, respectively.
Mouse IFNg Forward: 5′-GTTTATTTTTATTGTTGTGGTTGGTAGCTG-3′
Mouse IFNg Reverse: 5′-CCTTTCTTCTCCAAATTACTTTTAATC-3′
Human IFNg Forward: 5′-GATTTAGAGTAATTTGAA ATTTGTGG-3′
Human IFNg Reverse: 5′-CCTCCTCTAACTACT AATATTTATACC-3′
The PCR amplicon was cloned into a pGEMT easy vector (Promega) and then transformed into XL10-Gold ultracompetent bacteria (Agilent Technologies). Bacterial colonies were selected using a blue/white X-gal selection system after overnight growth, the cloning vector was then purified from individual colonies, and the genomic insert was sequenced. After bisulfite treatment, the methylated CpGs were detected as cytosines in the sequence, and unmethylated CpGs were detected as thymines in the sequence by using QUMA software (Kumaki et al., 2008).
WGBS was performed as described previously. Briefly, bisulfite-modified DNA sequencing libraries were generated using the EpiGenome kit (Epicenter) according to the manufacturer’s instructions. Bisulfite-modified DNA libraries were sequenced using Illumina HiSeq 4000 and NovaSeq 6000 systems (Abdelsamed et al., 2017). Sequencing data were aligned to the HG19 genome using the BSMAP v. 2.74 software (Xi and Li, 2009). Differential analysis of CpG methylation among the datasets was determined with a Bayesian hierarchical model to detect regional methylation differences with at least three CpG sites (Wu et al., 2015).
MiSeq
Naive, endogenous LCMV-specific, and WT P14 LCMV-specific CD8 T cells were FACs purified from either WT B6 or Tet2cKO mice. DNA was isolated from the cells, bisulfite converted, and then PCR amplified using primers from the Integrated DNA Technologies custom oligos tool. Illumina overhang adaptor sequences were added to each respective primer to make the products compatible with Illumina index and sequencing adapters. Amplified samples were analyzed using the MiSeq platform.
Mouse IFNg Forward: 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTATTTTTATTGTTGTGGTTGGTAGCTG-3′
Mouse IFNg Reverse: 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCTTTCTTCTCCAAATTACTTTTAATC3′
Generating human TET2-KO.B7-H3 CAR T cells
Generating B7-H3-CAR lentiviral vectors
Human B7-H3 CAR T cells were generated using the previously described CAR backbone (Nguyen et al., 2020). The vector was modified by removing the insulators from the self-inactivating (SIN) 3′ partially-deleted viral LTRs (Cornetta et al., 2018; McGarrity et al., 2013). Briefly, codon-optimized DNA encoding B7-H3 specific scFv derived from the m276 monoclonal antibody (Haydar et al., 2021; Seaman et al., 2017) was synthesized by GeneArt (Thermo Fisher, Waltham, MA). The scFv was then ligated using InFusion cloning (Takara Bio, Kusatsu, Shiga, Japan) into the backbone encoding a CD8α transmembrane domain, CD28 costimulatory domain, and CD3ζ activation domain. All final constructs were verified by sequencing. High titer lentiviral particles were then generated by the St. Jude Vector Core as described in Bauler et al. (2019).
TET2 knock-out T cells
TET2 and control knock-out T cells were generated using CRISPR-Cas9 technology. At 48 hours post-activation, sorted naive T cells were nucleofected with TET2 or control (mCherry) sgRNAs as Cas9 ribonucleoprotein (RNP) complexes. We used a TET2 sgRNA (5′-CGAAGCAAGCCTGATGGAACNGG-3′, Synthego, Menlo Park, CA) and a control sgRNA targeting mCherry (5′ – CAAGUAGUCGGGGAUGTCGG - 3′, Synthego, Menlo Park, CA). RNPs were pre-complexed at a sgRNA:Cas9 ratio of 4.5:1, prepared by adding 3 μL of 60 μM sgRNA (Synthego, Menio Park, CA) to 1 μL of 40 μM Cas 9 (Macro Lab, University of California, Berkeley), incubated at room temperature for 10 min and then stored at −20°C for later use. Activated T cells were collected by gentle pipetting up and down and then pelleted at 0.5×106 cells per reaction. Cell pellets were resuspended in 17 μL of P3 transfection solution (13.94 μL of Nucleofector Solution with 3.06 μL of supplement, Lonza, Walkersville, MD) and 4 μL of RNP complexes were added for each reaction. Electroporation was then performed by transferring 20 μL of T cell-RNP mixture into the transfection vessel and using the Lonza transfection program EH:115 (4D-Nucleofector, Lonza, Walkersville, MD). Electroporated T cells were then allowed to recover for overnight in RPMI media supplemented with 20% FBS and 1% glutamax and in the presence of IL-7 and IL-15 cytokines at 10ng/mL and 5ng/mL respectively.
Transducing CAR T cells
For generating human CAR T cells, electroporated cells were collected after 24 hours and washed with complete RPMI media (10% FBS with 1% Glutamax). Cells were then plated at 0.5×106 cells/well in 500 μL of complete RPMI media supplemented with IL-7 and IL-15 cytokines. T Cells were transduced by adding the lentiviral particles at a multiplicity of infection of 50, and protamine sulfate at 8ug/ml. Transduced T cells were then expanded until day 10 post-transduction with frequent supplementation with fresh media containing IL-7 and IL-15. CAR detection was performed using anti-Fab specific antibody at day 5 post-transduction (109-606-006, Jackson ImmunoResearch, West Grove, PA).
TET2-KO.B7-H3 CAR T cell activation assay
To evaluate the interaction of signal 3 cytokine signaling and DNA methylation in TET2-KO CAR T cells, transduced cells were activated with plate-bound recombinant human B7-H3 protein. Briefly, non-tissue culture plates were coated overnight at 4°C with recombinant human B7-H3 protein (R&D Systems, Minneapolis, MN) at 10 μg/well in 500 μL of PBS. Wells coated with PBS only served as unstimulated controls. TET2 and control knockout CAR T cells were then washed with complete RPMI media and added to coated wells at 1×106 cells/well in the presence or absence of IL-12 (10ng/mL). Stimulated and control unstimulated CAR T cells were then collected at day 14 post-activation for analysis.
GO annotation
Gene ontology annotation was carried using GREAT web server (http://great.stanford.edu/public/html/index.php). Input regions include top 300 methylated and demethylated DMRs.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details of experiments can be found in the figure legends, including sample sizes and p values. For sample size, n = the number of mice or human samples as specified. For p values, ns = not significant, p < 0.05: *, p < 0.01: **, p < 0.001: ***, p < 0.0001: ****. Unpaired t tests were performed to assess differences. A p value of < 0.05 was considered statistically significant.
Supplementary Material
Highlights.
Bystander inflammation promotes effector DNA methylation programs during T cell priming
The proinflammatory cytokine, IL-12, induces demethylation of IFNg locus CNS elements
IL-12 drives TET2-mediatiated demethylation of the mouse and human IFNg locus
Human memory T cells retain DNA methylation signatures of IL-12 signaling
ACKNOWLEDGMENTS
We thank Drs. Yiping Fan and Jeremy Crawford for bioinformatic analysis of methylation profiling. This work was supported by grants from the National Institutes of Health (R01AI114442 and R01CA237311 to B.Y., R01NS121249 to G.K., and a loan repayment program to C.C.Z.) and the American Lebanese Syrian Associated Charities (ALSAC; to B.Y.). Part of the laboratory studies were performed by the Center for Translational Immunology (CeTI2), which is supported by St. Jude. Some figures were generated with BioRender. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109796.
DECLARATION OF INTERESTS
C.C.Z., G.K., and B.Y. have patents related to epigenetic biomarkers and methods for enhancing CAR T cell function. B.Y. has a consulting agreement with ElevateBio.
REFERENCES
- Abdelsamed HA, Moustaki A, Fan Y, Dogra P, Ghoneim HE, Zebley CC, Triplett BM, Sekaly RP, and Youngblood B (2017). Human memory CD8 T cell effector potential is epigenetically preserved during in vivo homeostasis. J. Exp. Med 214, 1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelsamed HA, Zebley CC, and Youngblood B (2018). Epigenetic maintenance of acquired gene expression programs during memory CD8 T cell homeostasis. Front. Immunol 9, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelsamed HA, Zebley CC, Nguyen H, Rutishauser RL, Fan Y, Ghoneim HE, Crawford JC, Alfei F, Alli S, Ribeiro SP, et al. (2020). Beta cell-specific CD8+ T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes. Nat. Immunol 27, 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. (2017). Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Rao A, and Ko M(2017). TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp. Mol. Med 49, e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauler M, Roberts JK, Wu C-C, Fan B, Ferrara F, Yip BH, Diao S, Kim Y-I, Moore J, Zhou S, et al. (2019). Production of lentiviral vectors using suspension cells grown in serum-free media. Mol. Ther. Methods Clin. Dev 17, 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, and Grusby MJ (2007). Contribution of IL-12R mediated feedback loop to Th1 cell differentiation. FEBS Lett. 581, 5199–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty SA, Gohil M, Banks LB, Cotton RM, Johnson ME, Stelekati E, Wells AD, Wherry EJ, Koretzky GA, and Jordan MS (2018). The loss of TET2 promotes CD8+ T cell memory differentiation. J. Immunol 200, 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornetta K, Duffy L, Turtle CJ, Jensen M, Forman S, Binder-Scholl G, Fry T, Chew A, Maloney DG, and June CH (2018). Absence of replication-competent lentivirus in the clinic: analysis of infused T cell products. Mol. Ther 26, 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger JM, and Mescher MF (2010). Inflammatory cytokines as a third signal for T cell activation. Curr. Opin. Immunol 22, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraietta JA, Nobles CL, Sammons MA, Lundh S, Carty SA, Reich TJ, Cogdill AP, Morrissette JJD, DeNizio JE, Reddy S, et al. (2018). Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfall AL, Maus MV, Hwang WT, Lacey SF, Mahnke YD, Melenhorst JJ, Zheng Z, Vogl DT, Cohen AD, Weiss BM, et al. (2015). Chimeric antigen receptor T cells against CD19 for multiple myeloma. N. Engl. J. Med 373, 1040–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C, Engblom C, Pfirschke C, Siwicki M, Gungabeesoon J, et al. (2018). Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity 49, 1148–1161.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, et al. (2011). A human memory T cell subset with stem cell-like properties. Nat. Med 17, 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner MY, Heltemes-Harris LM, Fife BT, and Mescher MF (2013). Cutting edge: IL-12 and type I IFN differentially program CD8 T cells for programmed death 1 re-expression levels and tumor control. J. Immunol 191, 1011–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, and Youngblood B (2017). De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170, 142–157.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldrath AW, Sivakumar PV, Glaccum M, Kennedy MK, Bevan MJ, Benoist C, Mathis D, and Butz EA (2002). Cytokine requirements for acute and basal homeostatic proliferation of naive and memory CD8+ T cells. J. Exp. Med 195, 1515–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydar D, Houke H, Chiang J, Yi Z, Odé Z, Caldwell K, Zhu X, Mercer KS, Stripay JL, Shaw TI, et al. (2021). Cell-surface antigen profiling of pediatric brain tumors: B7-H3 is consistently expressed and can be targeted via local or systemic CAR T-cell delivery. Neuro-oncol. 23, 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson NG, Szabo SJ, Weber-Nordt RM, Zhong Z, Schreiber RD, Darnell JE Jr., and Murphy KM (1995). Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J. Exp. Med 181, 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersh EN, Fitzpatrick DR, Murali-Krishna K, Shires J, Speck SH, Boss JM, and Ahmed R (2006). Rapid demethylation of the IFN-gamma gene occurs in memory but not naive CD8 T cells. J. Immunol 176, 4083–4093. [DOI] [PubMed] [Google Scholar]
- Kumaki Y, Oda M, and Okano M (2008). QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 36, W170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Di S, Shi B, Zhang H, Wang Y, Wu X, Luo H, Wang H, Li Z, and Jiang H (2019). Armored inducible expression of IL-12 enhances antitumor activity of glypican-3-targeted chimeric antigen receptor-engineered T cells in hepatocellular carcinoma. J. Immunol 203, 198–207. [DOI] [PubMed] [Google Scholar]
- Lugli E, Gattinoni L, Roberto A, Mavilio D, Price DA, Restifo NP, and Roederer M (2013). Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat. Protoc 8, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarrity GJ, Hoyah G, Winemiller A, Andre K, Stein D, Blick G, Greenberg RN, Kinder C, Zolopa A, Binder-Scholl G, et al. (2013). Patient monitoring and follow-up in lentiviral clinical trials. J. Gene Med 15, 78–82. [DOI] [PubMed] [Google Scholar]
- Nguyen P, Okeke E, Clay M, Haydar D, Justice J, O’Reilly C, Pruett-Miller S, Papizan J, Moore J, Zhou S, et al. (2020). Route of 41BB/41BBL costimulation determines effector function of B7-H3-CAR.CD28ζ T cells. Mol. Ther. Oncolytics 18, 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kawamura I, Tsuchiya K, Kohda C, Baba H, Ito Y, Kimoto T, Watanabe I, and Mitsuyama M (2002). Essential role of interleukin-12 (IL-12) and IL-18 for gamma interferon production induced by listeriolysin O in mouse spleen cells. Infect. Immun 70, 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop JK, Thomas RM, Wells AD, and Shen H (2006). Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J. Immunol 177, 1062–1069. [DOI] [PubMed] [Google Scholar]
- Park JH, Geyer MB, and Brentjens RJ (2016). CD19-targeted CAR T-cell therapeutics for hematologic malignancies: interpreting clinical outcomes to date. Blood 127, 3312–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Riviére I, Gonen M, Wang X, Sénéchal B, Curran KJ, Sauter C, Wang Y, Santomasso B, Mead E, et al. (2018). Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med 378, 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, and Wilson CB (2007). Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat. Immunol 8, 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman S, Zhu Z, Saha S, Zhang XM, Yang MY, Hilton MB, Morris K, Szot C, Morris H, Swing DA, et al. (2017). Eradication of tumors through simultaneous ablation of CD276/B7-H3-positive tumor cells and tumor vasculature. Cancer Cell 31, 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, and Kaplan MH (2008). Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity 29, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G (2003). Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol 3, 133–146. [DOI] [PubMed] [Google Scholar]
- van Herpen CM, Looman M, Zonneveld M, Scharenborg N, de Wilde PC, van de Locht L, Merkx MA, Adema GJ, and de Mulder PH (2004). Intratumoral administration of recombinant human interleukin 12 in head and neck squamous cell carcinoma patients elicits a T-helper 1 profile in the locoregional lymph nodes. Clin. Cancer Res 10, 2626–2635. [DOI] [PubMed] [Google Scholar]
- Wu H, Xu T, Feng H, Chen L, Li B, Yao B, Qin Z, Jin P, and Conneely KN (2015). Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res. 43, e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, and Li W (2009). BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Casey KA, Jameson SC, Curtsinger JM, and Mescher MF (2009). Programming for CD8 T cell memory development requires IL-12 or type I IFN. J. Immunol 182, 2786–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeku OO, Purdon TJ, Koneru M, Spriggs D, and Brentjens RJ (2017). Armored CAR T cells enhance antitumor efficacy and overcome the tumor microenvironment. Sci. Rep 7, 10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Wherry EJ, and Ahmed R (2012). Acquired transcriptional programming in functional and exhausted virus-specific CD8 T cells. Curr. Opin. HIV AIDS 7, 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, et al. (2017). Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552, 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WG, Ogawa M, Mu J, Umehara K, Tsujimura T, Fujiwara H, and Hamaoka T (1997). IL-12-induced tumor regression correlates with in situ activity of IFN-gamma produced by tumor-infiltrating cells and its secondary induction of anti-tumor pathways. J. Leukoc. Biol 62, 450–457. [DOI] [PubMed] [Google Scholar]
- Zhang L, Morgan RA, Beane JD, Zheng Z, Dudley ME, Kassim SH, Nahvi AV, Ngo LT, Sherry RM, Phan GQ, et al. (2015). Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin. Cancer Res 21, 2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Bose A, Komita H, Taylor JL, Kawabe M, Chi N, Spokas L, Lowe DB, Goldbach C, Alber S, et al. (2011). Intratumoral IL-12 gene therapy results in the crosspriming of Tc1 cells reactive against tumor-associated stromal antigens. Mol. Ther 19, 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zundler S, and Neurath MF (2015). Interleukin-12: Functional activities and implications for disease. Cytokine Growth Factor Rev. 26, 559–568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome methylation data have been deposited to GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-Cy7 anti-mouse CD90.1 (clone OX-7) | BD Biosciences | Cat#561401; RRID: AB_10645789 |
| PerCp anti-mouse CD45.1 (clone A20) | BioLegend | Cat#110725; RRID: AB_893347 |
| BV421 anti-mouse CD45.2 (clone 104) | BioLegend | Cat#109831; RRID: AB_10900256 |
| FITC anti-mouse CD8a (clone 53-6.7) | BioLegend | Cat#100705; RRID: AB_312744 |
| PeCy7 anti-mouse CD8a (clone 53-6.7) | BioLegend | Cat#100721; RRID: AB_312760 |
| FITC anti-mouse Klrg1 (clone 2F1) | BioLegend | Cat#138409; RRID: AB_10643998 |
| PE anti-mouse CD127 (clone A7R34) | BioLegend | Cat#135009; RRID: AB_1937252 |
| BV421 anti-mouse/human CD44 (clone IM7) | BioLegend | Cat#103309; RRID: AB_10895752 |
| BV605 anti-mouse CD62L (clone MEL-14) | BioLegend | Cat#104437; RRID: AB_11125577 |
| FITC anti-human CCR7 (clone G043H7) | BioLegend | Cat#353215; RRID: AB_10945291 |
| APCCy7 anti-human CD8 (clone SK1) | BioLegend | Cat#344713; RRID: AB_2044005 |
| PeCy7 anti-human CD95 (clone DX2) | BioLegend | Cat#305621; RRID: AB_2100370 |
| FITC anti-human CD38 (clone HB-7) | BioLegend | Cat#356609; RRID: AB_2561949 |
| Pe anti-human IFNg (clone 4S.B3) | BioLegend | Cat#502508; RRID: AB_315233 |
| APC anti-human CD45RO (clone UCHL1) | BioLegend | Cat#304210; RRID: AB_314426 |
| Alexa Fluor® 647 AffiniPure F(ab’)2 Fragment Goat Anti-Human IgG, F(ab’)2 fragment specific | Jackson ImmunoResearch | Cat#109-606-006; |
| Bacterial and virus strains | ||
| Lymphocytic choriomeningitis virus, Armstrong strain | NA | |
| XL10-Gold Ultracompetent E. Coli | Stratagene | Cat#200314 |
| Listeria monocytogenes | Gift from McGargill lab, St. Jude Children’s Research Hospital | NA |
| Biological samples | ||
| Human PBMCs | St. Jude blood donor bank | NA |
| Chemicals, peptides, and recombinant proteins | ||
| Ghost Dye Violet 510 viability dye | Tonbo Biosciences | Cat#13-0870 |
| BD Cytofix/Cytoperm | BD Biosciences | Cat#554714 |
| LCMV gp33 monomer | Yerkes NIH tetramer core facility | NA |
| LCMV gp33 peptides | Peptide synthesis facility at SJCRH | NA |
| Cas9-NLS, purified protein | MacroLab | NA |
| Protamine Sulfate | Fresenius Kabi | C22905 |
| CD28 Antibody, anti-human | Miltenyi Biotec | 130-093-375 |
| CD3 Antibody, anti-human | Miltenyi Biotec | 130-093-387 |
| Recombinant Human B7-H3 Fc Chimera Protein | R&D systems | 1027-B3 |
| Recombinant human IL-12 p70 | PeproTech | Cat#200-12 |
| Recombinant human IL-7 | PeproTech | Cat#200-07 |
| Recombinant human IL-2 | PeproTech | Cat#200-02 |
| Recombinant human IL-15 | PeproTech | Cat#200-15 |
| Recombinant human IL-21 | PeproTech | Cat#200-21 |
| Recombinant human IL-18 | Medical & biological laboratories | Cat#B001-5 |
| Critical commercial assays | ||
| QIAGEN DNeasy kit | QIAGEN | Cat#69506 |
| EZ DNA methylation kit | Zymo Research | Cat#D5002 |
| EZ DNA methylation-Direct kit | Zymo Research | Cat#50-444-323 |
| pGEM-T Vector cloning kit | Promega | Cat#A3600 |
| JumpStart Taq ReadyMix | Sigma-Aldrich | Cat#P2893 |
| In-Fusion® HD Cloning Kit | TakaraBio | Cat#639650 |
| P3 Primary Cell 4D X Kit S | Lonza | V4XP-3032 |
| Directprep 96 Miniprep kit | QIAGEN | Cat#27361 |
| Taq polymerase-based PCR kit | QIAGEN | Cat#201225 |
| Human CD3/CD28 T cell activator | Stemcell technologies | Cat#10971 |
| EasySep human CD8+ T cell isolation kit | Stemcell | Cat#19053 |
| Deposited data | ||
| WGBS data | This paper | GEO: GSE182968 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | The Jackson Laboratory | Cat#000664 |
| Mouse: CD45.1/1 + | The Jackson Laboratory | Cat#002014 |
| Mouse: B6, Granzyme b-Cre | SJCRH animal house | NA |
| Mouse: Tet2 floxed | The Jackson Laboratory | Cat#017573 |
| Oligonucleotides | ||
| pUC/M13 reverse primer | Promega | Cat#5421 |
| Human IFNg Forward: 5′-GATTTAGAGTAATTTGAAATTTGTGG-3′ | IDT | NA |
| Human IFNg Reverse: 5′-CCTCCTCTAACTACTAATATTTATACC-3′ | IDT | NA |
| Mouse IFNg Forward: 5′-GTTTATTTTTATTGTTGTGGTTGGTAGCTG-3′ | IDT | NA |
| Mouse IFNg Reverse: 5′-CCTTTCTTCTCCAAATTACTTTTAATC-3′ | IDT | NA |
| Mouse Miseq IFNg Forward: 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTATTTTTATTGTTGTGGTTGGTAGCTG-3′ | IDT | NA |
| Mouse Miseq IFNg Reverse: 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCTTTCTTCTCCAAATTACTTTTAATC3′ | IDT | NA |
| TET2 sgRNA:5′-CGAAGCAAGCCTGATGGAACNGG-3′ | Synthego | NA |
| sgRNA mCherry:5′-CAAGUAGUCGGGGAUGTCGG - 3′ | Synthego | NA |
| Recombinant DNA | ||
| Plasmid: pCL45.MND.m276.hCD8aTM.hCD28.z | This paper | NA |
| Software and algorithms | ||
| Prism 6 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo 9.9.5 | FlowJo | NA |
| SnapGene | Insightful Science | snapgene.com |
| Model-based analysis of bisulfite sequencing | https://www.ncbi.nlm.nih.gov/pubmed/26184873/ | |
| Gene Ontology (GO) enrichment analysis | GREAT | http://great.stanford.edu/public/html/index.php |
| Loci-specific bisulfite sequencing | Quma | http://quma.cdb.riken.jp/ |
| Other | ||
| Fortessa flow cytometer | Flow cytometry core facility at SJCRH | NA |
| Illumina NovaSeq | Hartwell Center at SJCRH | NA |
| Illumina HiSeq 4000 | Hartwell Center at SJCRH | NA |
| 4D-Nucleofector | Lonza | Cat#AAF-1002B |
| Ficoll-Paque PLUS | GE Healthcare | |
| BD GolgiPlug | Fisher scientific | Cat#15847968 |
| BD GogliStop | Fisher scientific | Cat#10716676 |
| CFSE | Fisher scientific | Cat#50-591-407 |
| Penicillin-streptomycin (GIBCO) | ThermoFisher | Cat#1514012 |
| Gentamycin (GIBCO) | ThermoFisher | Cat#15750060 |
All original code has been deposited at GEO and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.


