Abstract
Oral squamous cell carcinoma (OSCC) is a common type of malignant tumor worldwide. Claudin-7 (CLDN7) has been reported to exhibit low expression in tissues of patients with OSCC; however, the underlying mechanisms of CLDN7 remain to be elucidated. The present study aimed to investigate the effects of CLDN7 on the progression of OSCC and identify its potential regulatory mechanisms. CLDN7 and interferon regulatory factor-2 (IRF2) expression in several OSCC cell lines were detected using reverse transcription-quantitative PCR (RT-qPCR) and western blotting. Following CLDN7 overexpression, cell proliferation, invasion and migration were determined using a Cell Counting Kit-8, colony formation, Transwell and wound healing assays, respectively. The potential binding sites of IRF2 on the CLDN7 promoter were analyzed using the PROMO and JASPAR databases, which were verified via chromatin immunoprecipitation and RT-qPCR assays. The effects of IRF2 and CLDN7 on the biological functions of OSCC cells were examined by transfection with short hairpin RNA (shRNA) against CLDN7 (sh-CLDN7), or IRF2 and CLDN7 overexpression plasmids. The results revealed that CLDN7 and IRF2 expression were significantly downregulated in OSCC cell lines, and CLDN7 overexpression reduced the proliferation, invasion and migration of OSCC cells. Additionally, IRF2 was confirmed to combine with the CLDN7 promoter. CLDN7 silencing reversed the inhibitory effects of IRF2 overexpression on the proliferation, invasion and migration of OSCC cells. Taken together, these findings demonstrated that IRF2-induced CLDN7 upregulation suppressed the proliferation, invasion and migration of OSCC cells, suggesting the possibility of CLDN7 and IRF2 as novel targets for the treatment of OSCC.
Keywords: interferon regulatory factor-2, Claudin-7, oral squamous cell carcinoma, proliferation, invasion, migration
Introduction
Oral squamous cell carcinoma (OSCC), a type of head and neck squamous cell carcinoma, is one of the most common malignant tumors diagnosed worldwide with high morbidity and mortality rates (1). According to the 2018 global cancer statistics, OSCC accounted for ~3% of new cases and 1.9% of deaths of all malignant cancers (2). Over the past two decades, although a multidisciplinary approach, including chemotherapy, targeted therapy and surgical therapy, has been applied for the treatment of OSCC, the overall 5-year survival rate of patients with OSCC has remained <50% (3,4). Hence, it is of great importance to widen the current understanding of the mechanisms underlying OSCC progression and discover effective biomarkers and therapeutic strategies for improving OSCC treatment and prognosis.
Claudin-7 (CLDN7), one of the 24 members of the Claudin family, is involved in regulating tight junction assembly, intercellular exchange and cell polarity maintenance (5). An increasing number of studies have confirmed that the abnormal expression of CLDN7 can disrupt the integrity of tight junctions, thereby resulting in the loss of cell polarity, and inducing abnormal proliferation, invasion and metastasis (6,7). Accumulating evidence has shown that CLDN7 is abnormally expressed in multiple types of cancer tissues, such as ovarian carcinoma, salivary adenoid cystic carcinoma and colorectal cancer (6,8,9), indicating that alterations in CLDN7 expression may be closely related to the occurrence of these tumors. Emerging evidence supports the notion that low expression of CLDN7 serves as a crucial feature of OSCC, and loss of CLDN7 is considered as a negative prognostic factor for invasion and metastasis in OSCC (10). However, the exact regulatory mechanisms of CLDN7 in OSCC remains to be elucidated. Interferon regulatory factor-2 (IRF2) was predicted as an upstream regulator of CLDN7 expression, as determined using the PROMO database, and the highest bound fraction between IRF2 and CLDN7 was confirmed using the JASPAR database. The roles of IRF2 on carcinogenesis are controversial. Initially, it was shown that IRF-2 served as an oncogenic protein, and the expression levels of IRF-2 were also found to be upregulated in esophageal and pancreatic cancer (11,12). The downregulation of IRF2 is responsible for the anti-tumorigenic effects in prostate cancer cells overexpressing microRNA-221(13). In contrast, IRF2 served as a tumor suppressor and its inactivation led to impaired P53 function (14). IRF2 is reported to inhibit the invasion and migration of gastric cancer cells by inactivating MMP-1 expression (15). However, the function of IRF2 in OSCC remains unknown, to the best of our knowledge.
The present study aimed to investigate the effects of CLDN7 on proliferation, invasion and migration of OSCC cells, and identify whether IRF2 could transcriptionally activate CLDN7 to affect the progression of OSCC.
Materials and methods
Cell culture
SCC-9, CAL-27, SCC-15 and SCC090 human OSCC cell lines were purchased from the American Type Culture Collection. The human oral keratinocyte HOK cell line was provided by Shanghai Haoyuan Biotech Co., Ltd. These cell lines were maintained in DMEM supplemented with 10% FBS (HyClone; Cytiva) at 37˚C in a humidified incubator with 5% CO2.
Cell transfection
CAL-27 cells in the logarithmic growth phase were collected and plated into 6-well plates at a density of 1x106 cells/well at 37˚C. When cells reached 85% confluence, cells were transfected with 2 µg CLDN7 plasmid (pc-CLDN7), 2 µg IRF2 plasmid (pc-IRF2) or 2 µg empty vector plasmid (pcDNA3.1) using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C according to the standard protocol (16). The aforementioned plasmids were purchased from Shanghai GenePharma Co., Ltd. Additionally, 2 µg short hairpin RNA (shRNA) targeting CLDN7 (sh-CLDN7) was constructed in a U6/GFP/Neo plasmid (GenePharma, Shanghai, China) and transfected into CAL-27 cells using Lipofectamine® 2000 reagent at 37˚C. Western blot analysis was employed to evaluate the transfection efficiency 48 h post-transfection.
Cell viability assay
Cell Counting Kit-8 (CCK-8; Shanghai Yeasen Biotechnology Co., Ltd.) assays were utilized to assess cell viability, according to the manufacturer's instructions. CAL-27 cells were inoculated into 96-well plates and cultured at 37˚C overnight. At the indicated time points (24, 48 or 72 h), 10 µl CCK-8 solution was added to each well after 2 h of culture at 37˚C. The absorbance was determined at 450 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
Cells in the logarithmic growth phase were seeded into a 6-well culture plate (500 cells/well). Cells were cultured at 37˚C for 2 weeks until colony formation became visible to the naked eye. Then, cells were fixed with 4% paraformaldehyde for 30 min at room temperature, followed by staining with 0.5% crystal violet solution (Sigma-Aldrich; Merck KGaA) for 5 min at room temperature. The number of colony cells with a minimum of 50 cells was counted under a light microscope (magnification, x200).
Transwell invasion assay
A Matrigel-based assay in 24-well 8-µm pore Transwell chambers (Costar; Corning, Inc.) was utilized for the evaluation of cell invasion. A total of 2.5x105 CAL-27 cells were inoculated in serum-free medium in the upper chamber. DMEM supplemented with 10% FBS was added to the lower compartment as a chemoattractant. After 24 h of incubation, cells remaining on the top of the Matrigel were gently removed with a cotton swab. Cells which had invaded to the lower surface of the membrane were fixed in 4% paraformaldehyde for 20 min at 37˚C and stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for 30 min at 37˚C. Images were captured using an inverted fluorescence microscope (magnification, x200; Olympus Corporation). The number of cells which had invaded were counted using ImageJ version 1.52r (National Institutes of Health).
Wound healing assay
CAL-27 cells were seeded into 6-well plates (5x104 cells/well) and cultured to 80% confluence. The cells were serum-starved overnight at 37˚C prior to the experiment. Then, cells were scratched with a sterile plastic micropipette tip to generate a wound in the monolayer. After 24 h of incubation, the wound was imaged under an inverted microscope (Olympus Corporation) and the separation distance was analyzed using ImageJ.
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract the total RNA from CAL-27 cells, according to the manufacturer's instructions. Then, RNA was reverse transcribed into cDNA using a RT kit according to the manufacturer's protocol (Beijing Transgen Biotech Co., Ltd.). qPCR was performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) on an ABI 7500 Real-Time PCR Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The RT-qPCR conditions were as follows: Pre-denaturation at 95˚C for 2 min; followed by 35 cycles of 95˚C for 35 sec, 58˚C for 45 sec and 72˚C for 30 sec; and 72˚C for 5 min. The sequences of the primers were: CLDN7 forward, 5'-AAAGTGAAGAAGGCCCGTATA-3' and reverse, 5'-TAATGTTGGTAGGGATCAAAGG-3'; IRF2 forward, 5'-TCACTAGTGTTATTACATCCTTGTGGCAC-3' and reverse, 5'-GAACTAGTGAAGTCATGCAAAACGCTCA-3'; GAPDH forward, 5'-ACAACTTTGGTATCGTGGAAGG-3' and reverse, 5'-GCCATCACGCCACAGTTTC-3'. Analysis of gene expression was performed using the 2-IICq method (17). GAPDH was used as an internal reference gene.
Chromatin immunoprecipitation (ChIP) assay
The binding of IRF2 to the CLDN7 promoter was examined using a ChIP assay with EZ-ChIPTM Chromatin Immunoprecipitation Kit (Millipore), according to the manufacturer's protocol. After fixing with formaldehyde (1%) for 10 min at room temperature to produce cross-linked protein and DNA, cultured CAL-27 cells were incubated with 1X Buffer B for 10 min at room temperature and incubated with ice-cold 1X Buffer C for a further 10 min. Cells were subsequently incubated with the 1X Buffer D/PI mix and chromatin fragments were obtained using sonication (5 times for 10 sec each) on ice for 10 min. IRF2 antibody (cat. no. sc-374327; Santa Cruz Biotechnology, Inc.) was utilized to generate immunoprecipitants, whereas an IgG antibody (cat. no. sc-69786; Santa Cruz Biotechnology, CA, USA) was used as the blank control group to exclude the influence of other factors. The recuperated DNA fragments were evaluated via qPCR. The relative levels of CLDN7 promoter was normalized according to the average level of the IgG group.
Luciferase reporter assay
Luciferase reporter plasmids (Promega Corporation) were constructed using wild-type and mutant 3' untranslated regions of the CLDN7 promoter. Firefly luciferase activity represented the primary reporter used to monitor the binding of proteins to cloned target sequences. Renilla luciferase was regarded as a control reporter for normalization. The luciferase reporter plasmids and regulating factors were co-transfected into CAL-27 cells using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the luciferase activities were quantified using the Dual-Luciferase Reporter assay system (Promega Corporation).
Western blot analysis
For immunoblotting, total cellular extracts were prepared using RIPA lysis buffer containing protease inhibitor cocktail (Beyotime Institute of Biotechnology). Protein concentration was determined using a bicinchoninic acid kit (Beyotime Institute of Biotechnology). From each sample, 40 µg protein was added and separated via SDS-PAGE on a 10% gel, and separated proteins were subsequently transferred to a nitrocellulose blotting membrane (Cytiva). Non-specific binding was blocked by 5% skimmed milk for 1.5 h at room temperature and then the membranes were incubated overnight at 4˚C with specific primary antibodies. The following day, the membranes were incubated with HRP-conjugated goat anti-rabbit secondary antibody (cat. no. 7074S; Cell Signaling Technology, Inc.) for 1 h at room temperature. Signals were visualized using the Odyssey Western Blot Analysis system (LI-COR Biosciences). Anti-CLDN7 (cat. no. ab265583) antibody was provided by Abcam, anti-IRF2 (cat. no. sc-374327) and anti-GAPDH (cat. no. sc-47724) antibodies were purchased from Santa Cruz Biotechnology, Inc. The relative intensity of each band was semi-quantified using ImageJ. GAPDH was used as the loading control.
Statistical analysis
All experiments were repeated independently three times. Data are presented as the mean ± standard deviation. Comparisons between two groups were performed using a Student's t-test. A one-way ANOVA followed by a Tukey's post-hoc test was used to assess differences between multiple groups. All analyses were performed using GraphPad Prism version 6.0 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Bioinformatics
PROMO (alggen.lsi.upc.es/; version, 3.0.2) was utilized to predict potential transcription factors of CLDN7, and JASPAR (jaspar.genereg.net/; version, 2020) was used to predict the potential binding sites between IRF2 and the CLDN7 promoter.
Results
CLDN7 expression is significantly downregulated in OSCC cell lines
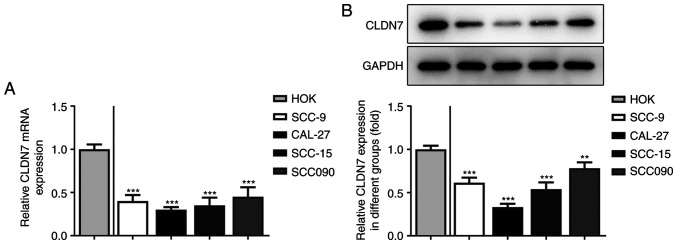
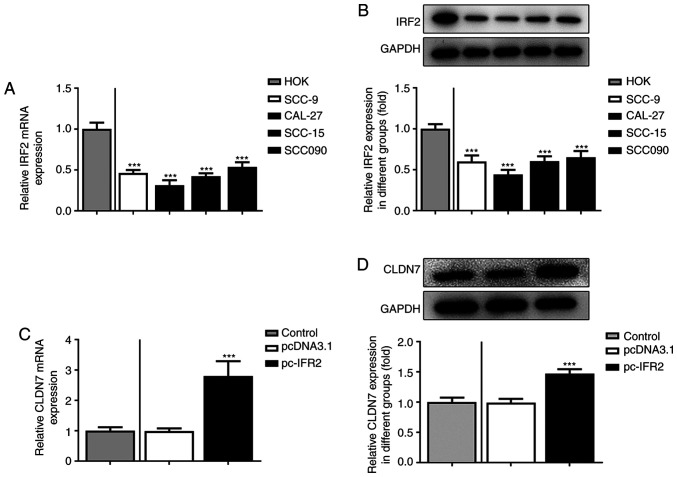
Firstly, the expression of CLDN7 in several OSCC cell lines (SCC-9, CAL-27, SCC-15 and SCC090) was detected. As shown in Fig. 1A and B, the expression levels of CLDN7 at the mRNA and protein level were notably reduced in the OSCC cell lines compared with the human oral keratinocyte HOK cell line. Lowest CLDN7 expression was observed in the CAL-27 cells, therefore, this cell line was used in subsequent experiments.
Figure 1.
CLDN7 expression is notably downregulated in OSCC cell lines. The expression of CLDN7 in several OSCC cells (SCC-9, CAL-27, SCC-15 and SCC090) and a human oral keratinocyte cell line HOK was examined via (A) reverse transcription-quantitative PCR and (B) western blotting. **P<0.01, ***P<0.001 vs. HOK. CLDN7, Claudin-7; OSCC, oral squamous cell carcinoma.
CLDN7 overexpression inhibits the proliferation, invasion and migration of OSCC cells
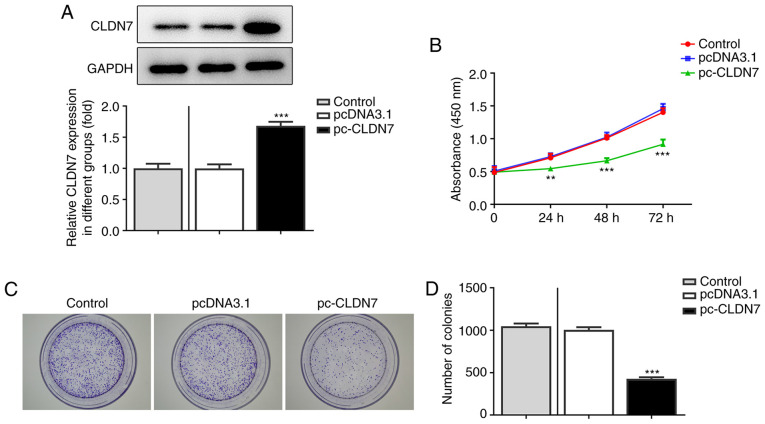
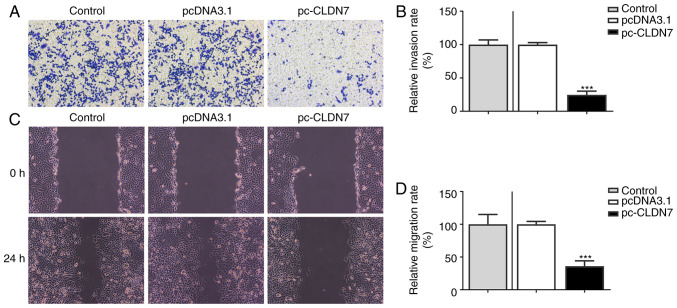
To study the effects of CLDN7 on the progression of OSCC, CLDN7 was overexpressed by transfection with a CLDN7 plasmid. As shown in Fig. 2A, CLDN7 expression was notably upregulated after transfection compared with the empty vector group. Next, the proliferative ability of CAL-27 cells was evaluated using CCK-8 and colony formation assays. It was found that CLDN7 overexpression markedly suppressed cell viability compared with the pcDNA3.1 group (Fig. 2B). Consistently, a significant decrease in the number of colonies was observed after CLDN7 overexpression compared with the vector control group (Fig. 2C and D). Subsequently, Transwell and wound healing assays were used to evaluate cell invasion and migration. As shown in Fig. 3A-D, CLDN7 overexpression notably inhibited the invasion and migration of CAL-27 cells. These data showed that overexpression of CLDN7 suppressed the proliferation, invasion and migration of OSCC cells.
Figure 2.
CLDN7 overexpression suppresses the proliferation of oral squamous cell carcinoma cells. (A) CLDN7 expression in CAL-27 cells was determined via western blotting after transfection with a CLDN7 overexpression plasmid. (B) Cell viability was detected using a Cell Counting Kit-8 assay. (C and D) A colony formation assay was performed to determine the proliferative abilities of CAL-27 cells. **P<0.01, ***P<0.001 vs. pcDNA3.1. CLDN7, Claudin-7.
Figure 3.
CLDN7 overexpression inhibits the invasion and migration of oral squamous cell carcinoma cells. (A and B) The invasive ability of CAL-27 cells was measured using a Transwell assay following CLDN7 overexpression. (C and D) The cell migratory ability of CAL-27 cells was detected using a wound healing assay. ***P<0.001 vs. pcDNA3.1. CLDN7, Claudin-7.
IRF2 regulates CLDN7 expression by directly binding to the CLDN7 promoter
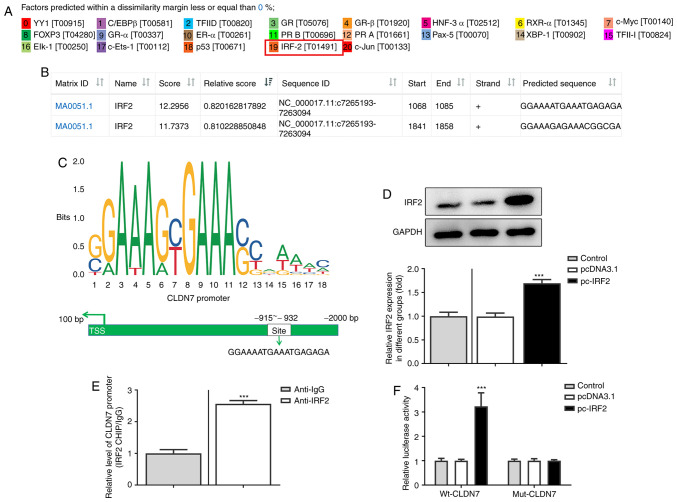
To further explore the molecular mechanism by which CLDN7 regulated proliferation, invasion and migration of OSCC cells, the potential transcription factor binding sites in the promoter region of CLDN7 were predicted using PROMO and JASPAR databases. IRF2 was predicted as a potential transcription factor upstream of CLDN7, and the highest bound fraction between the IRF2 and CLDN7 promoter was confirmed using the JASPAR database (Fig. 4A and B). As presented in Fig. 4C, one possible binding site of IRF2 was identified in the CLDN7 promoter (site-932 to -915). Next, IRF2 was overexpressed by transfection (Fig. 4D). In subsequent ChIP assays performed with extracts from CAL-27 cells, notable enrichment of the CLDN7 promoter sequence was obtained through immunoprecipitation with an anti-IRF2 antibody, but not with the control IgG antibody (Fig. 4E). Moreover, the luciferase activity of Wt-CLDN7 was activated by IRF2 overexpression (Fig. 4F). These observations showed that IRF2 promoted CLDN7 transcription through directly binding to the CLDN7 promoter region.
Figure 4.
CLDN7 expression is induced by the transcription factor IRF2. Potential transcription factor binding with the promoter of CLDN7 was predicated by (A) PROMO and (B) JASPAR. (C) IRF2 sequence and binding sites (Site 1 and 2) to CLDN7 promoter region were predicted using the JASPAR database. (D) IRF2 expression was examined via western blot analysis after transfection with IRF2 plasmid. ***P<0.001 vs. pcDNA3.1. (E) The direct binding of IRF2 to CLDN7 promoter in CAL-27 cells was confirmed via a chromatin immunoprecipitation assay. ***P<0.001 vs. anti-IgG. (F) The binding between IRF2 and the promoter region was examined using a luciferase reporter assay. ***P<0.001 vs. pcDNA3.1. CLDN7, Claudin-7; IRF2, interferon regulatory factor-2.
IRF2 overexpression promotes the expression of CLDN7 in OSCC cells
Subsequently, the expression of IRF2 in several OSCC cell lines was assessed using RT-qPCR and western blotting. As shown in Fig. 5A and B, IRF2 mRNA and protein expression levels were significantly downregulated in OSCC cell lines compared with the HOK cells. Moreover, IRF2 was overexpressed by transfection with the IRF2 plasmid in CAL-27 cells, and the increase in IRF2 expression is shown in Fig. 5C. Subsequently, CLDN7 expression was markedly enhanced following IRF2 overexpression (Fig. 5D). Overall, these data suggested that IRF2 overexpression promoted the expression of CLDN7 in CAL-27 cells.
Figure 5.
IRF2 overexpression upregulates the expression of CLDN7 in OSCC cells. (A) IRF2 expression in several OSCC cells (SCC-9, CAL-27, SCC-15 and SCC090) and the human oral keratinocyte cell line HOK was detected via (A) RT-qPCR and (B) western blotting. ***P<0.001 vs. HOK. (C) IRF2 expression was evaluated using RT-qPCR following transfection with IRF2 plasmid in CAL-27 cells. ***P<0.001 vs. pcDNA3.1. (D) Western blotting was used to assess the expression of CLDN7 following IRF2 overexpression. ***P<0.001 vs. pcDNA3.1. CLDN7, Claudin-7; IRF2, interferon regulatory factor-2; RT-qPCR, reverse transcription-quantitative PCR; OSCC, oral squamous cell carcinoma.
CLDN7 knockdown reverses the inhibitory effects of IRF2 overexpression on the proliferation, invasion and migration of CAL-27 cells
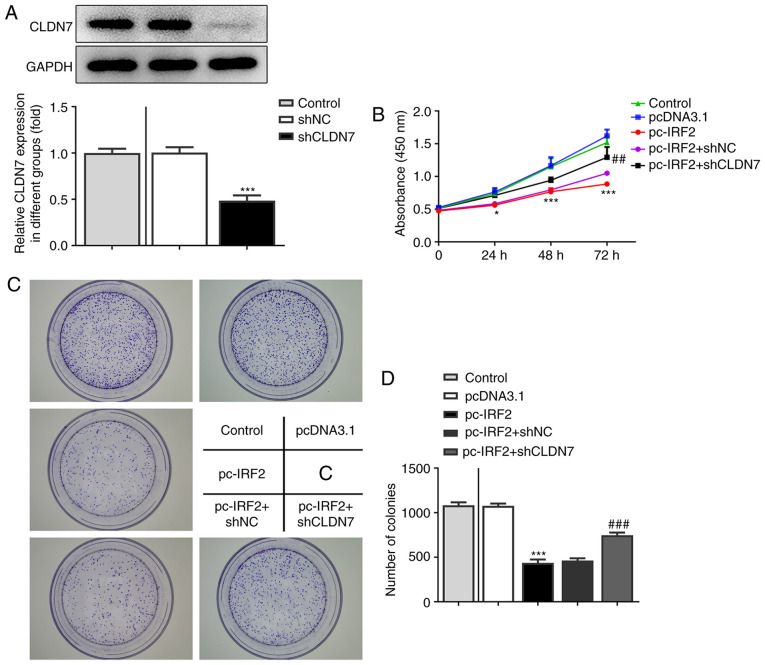
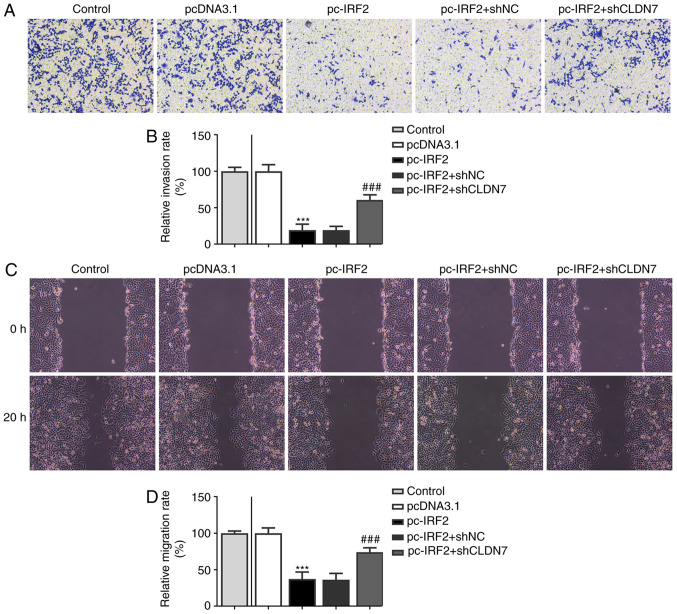
To further confirm the regulation of CLDN7 and IRF2, function-related rescue experiments were performed. Firstly, CLDN7 expression was silenced by transfection with shCLDN7, which is presented in Fig. 6A. As shown in Fig. 6B-D, the proliferative ability of CAL-27 cells was significantly inhibited following IRF2 overexpression, whereas this inhibitory effect was alleviated following the knockdown of CLDN7 expression. Consistently, notably suppressed invasion (Fig. 7A and B) and migration (Fig. 7C and D) of CAL-27 cells was observed in the pc-IRF2 group compared with the pcDNA3.1 group, which was restored after co-transfection with pc-IRF2 and shCLDN7 into CAL-27 cells.
Figure 6.
CLDN7 knockdown reverses the inhibitory effects of IRF2 overexpression on cell proliferation in OSCC cells. (A) Knockdown efficiency of CLDN7 was demonstrated using western blotting. (B) A Cell Counting Kit-8 assay was used to evaluate cell viability in the Control group, pcDNA3.1 group, pc-IRF2 group, pc-IRF2 + shNC group and pc-IRF2 + shCLDN7 group. (C and D) Colony formation assays were used to assess the number of cell colonies in the Control group, pcDNA3.1 group, pc-IRF2 group, pc-IRF2 + shNC group and pc-IRF2 + shCLDN7 group. *P<0.05, ***P<0.001 vs. pcDNA3.1; ##P<0.01, ###P<0.001 vs. pc-IRF2 + shNC. CLDN7, Claudin-7; IRF2, interferon regulatory factor-2; OSCC, oral squamous cell carcinoma; sh, short hairpin RNA; NC, negative control.
Figure 7.
CLDN7 knockdown reverses the inhibitory effects of IRF2 overexpression on cell migration and invasion in OSCC cells. (A and B) Invasive ability of CAL-27 cells in the Control group, pcDNA3.1 group, pc-IRF2 group, pc-IRF2 + shNC group and pc-IRF2 + shCLDN7 group was measured using a Transwell assay. (C and D) Migration rates of CAL-27 cells in the Control group, pcDNA3.1 group, pc-IRF2 group, pc-IRF2 + shNC group and pc-IRF2 + shCLDN7 group was detected using a wound healing assay. ***P<0.001 vs. pcDNA3.1; ###P<0.001 vs. pc-IRF2 + shNC. CLDN7, Claudin-7; IRF2, interferon regulatory factor-2; OSCC, oral squamous cell carcinoma; sh, short hairpin RNA; NC, negative control.
Discussion
OSCC is one of the most aggressive neoplasms amongst head and neck malignant tumors, and is associated with a poor prognosis (18). Improving our understanding of the underlying mechanisms of the development and progression of OSCC is critical for identifying effective targets with therapeutic potential to improve the survival rates of patients with OSCC. The primary aim of the present study was to investigate the role of CLDN7 in the progression of OSCC. It The results showed that CLDN7 expression was significantly decreased in OSCC cells, and CLDN7 overexpression notably inhibited the proliferation, invasion and migration of OSCC cells, which was directly induced by the IRF2 transcription factor.
Proliferation and metastasis are both hallmarks of the malignant biological behavior of OSCC, and the inhibition of these processes is a crucial factor to improve biomedical treatment worldwide (19). CLDN7, a member of the Claudin family, is reported to be a tumor-related gene. At present, it is known that CLDN7 plays either tumor promoting or tumor suppressing roles based on the specific type of tumor. A growing body of literature has shown that the loss of CLDN7 is closely associated with a poor prognosis in nasopharyngeal cancer, ovarian carcinoma and esophageal squamous cell carcinoma (8,20,21). CLDN7 silencing induces invasion and metastasis in colorectal cancer via the promotion of epithelial-mesenchymal transition (6). Dysregulation of CLDN7 results in loss of E-Cadherin expression and the enhanced invasion of esophageal squamous cell carcinoma cells (22). Conversely, CLDN7 is notably elevated in cervical and gastric cancer (23,24). CLDN7 suppresses proliferation and metastasis in salivary adenoid cystic carcinoma (9). Of note, loss of CLDN7 expression may be associated with the pathogenesis of OSCC and this loss is associated with a poor prognosis (25). Loss of CLDN7 is considered a negative prognostic factor for invasion and metastasis in OSCC (10). However, the exact regulatory mechanisms by which CLDN7 is regulated in OSCC remains unknown. The current study revealed that CLDN7 was downregulated in OSCC cell lines, and CLDN7 overexpression markedly inhibited the proliferation, invasion and migration of OSCC cells, suggesting the potential tumor suppressor role of CLDN7 in OSCC.
According to PROMO and JASPAR database analyses, IRF2 could potentially bind to the promoter region of CLDN7. IRF2, one of the members of the IRF family of transcription factors, can transcriptionally induce multiple direct target genes, which has been identified to participate in the regulation of immune responses and immune cell development (26-28). An increasing number of studies have confirmed that IRF2 is closely related to the physiological and pathological processes of various types of cancer (13,29). Existing reports have shown that IRF-2 inhibits the invasion and migration of gastric cancer through downregulation of MMP-1 expression (15). MicroRNA-664 suppresses the progression of cutaneous squamous cell carcinoma via inhibiting IRF2 expression (30). IRF2 serves an inhibitory effect on the invasion and migration of osteosarcoma, where it is regulated by microRNA-18a-5p (31). Of note, IRF2 has been reported to bind to inositol polyphosphate 4-phosphatase type II (INPP4B) promoter and increase INPP4B expression, thereby participating in the development of acute myeloid leukemia (32). In the present study, significantly downregulated levels of IRF2 expression were observed in OSCC cells, and overexpression of IRF2 significantly suppressed the proliferation, invasion and migration of OSCC cells. Furthermore, the binding effect of IRF2 to the promoter of CLDN7 was confirmed via ChIP and luciferase reporter assays. The rescue experiments demonstrated that CLDN7 silencing blocked the inhibitory effects of IRF2 overexpression on the proliferation, invasion and migration of OSCC cells.
In summary, the present study found that both CLDN7 and IRF2 expression levels were downregulated in OSCC cells. IRF2 was determined to have a direct regulatory effect on the transcription of CLDN7, and the biological functions of CLDN7 in OSCC cells were closely regulated by the transcriptional factor IRF2. These findings provided evidence for a novel regulatory mechanism involving CLDN7 and IRF2 in OSCC, thus potentially identifying novel targets for therapy. However, the use of only one OSCC cell line to clarify the effects of CLDN7 and IRF2 in OSCC is a potential limitation of the present study. Subsequent experiments will incorporate more typical OSCC cell lines to support this conclusion.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XL and WY designed the experiments. XL analyzed the experimental data and wrote the manuscript. WY helped to correct the manuscript. XL carried out the experiments. XL and WY confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sasahira T, Kirita T. Hallmarks of cancer-related newly prognostic factors of oral squamous cell carcinoma. Int J Mol Sci. 2018;19(2413) doi: 10.3390/ijms19082413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Sequeira I, Neves JF, Carrero D, Peng Q, Palasz N, Liakath-Ali K, Lord GM, Morgan PR, Lombardi G, Watt FM. Immunomodulatory role of Keratin 76 in oral and gastric cancer. Nat Commun. 2018;9(3437) doi: 10.1038/s41467-018-05872-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li YY, Tao YW, Gao S, Li P, Zheng JM, Zhang SE, Liang J, Zhang Y. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine. 2018;36:209–220. doi: 10.1016/j.ebiom.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao D, Xu T, Qi X, Ning W, Ren S, Ru Z, Ji K, Ma Y, Yu T, Li Y, et al. CLAUDIN7 modulates trophectoderm barrier function to maintain blastocyst development in pigs. Theriogenology. 2020;158:346–357. doi: 10.1016/j.theriogenology.2020.09.038. [DOI] [PubMed] [Google Scholar]
- 6.Wang K, Li T, Xu C, Ding Y, Li W, Ding L. Claudin-7 downregulation induces metastasis and invasion in colorectal cancer via the promotion of epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2019;508:797–804. doi: 10.1016/j.bbrc.2018.10.049. [DOI] [PubMed] [Google Scholar]
- 7.Poon CE, Madawala RJ, Day ML, Murphy CR. Claudin 7 is reduced in uterine epithelial cells during early pregnancy in the rat. Histochem Cell Biol. 2013;139:583–593. doi: 10.1007/s00418-012-1052-y. [DOI] [PubMed] [Google Scholar]
- 8.Romani C, Zizioli V, Silvestri M, Ardighieri L, Bugatti M, Corsini M, Todeschini P, Marchini S, D'Incalci M, Zanotti L, et al. Low expression of Claudin-7 as potential predictor of distant metastases in high-grade serous ovarian carcinoma patients. Front Oncol. 2020;10(1287) doi: 10.3389/fonc.2020.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji H, Ding X, Zhang W, Zheng Y, Du H, Zheng Y, Song H, Li M, Jiang Y, Xie J, et al. Claudin-7 inhibits proliferation and metastasis in salivary adenoid cystic carcinoma through Wnt/β-catenin signaling. Cell Transplant. 2020;29(963689720943583) doi: 10.1177/0963689720943583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshizawa K, Nozaki S, Kato A, Hirai M, Yanase M, Yoshimoto T, Kimura I, Sugiura S, Okamune A, Kitahara H, et al. Loss of claudin-7 is a negative prognostic factor for invasion and metastasis in oral squamous cell carcinoma. Oncol Rep. 2013;29:445–450. doi: 10.3892/or.2012.2161. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Liu DP, Chen PP, Koeffler HP, Tong XJ, Xie D. Involvement of IFN regulatory factor (IRF)-1 and IRF-2 in the formation and progression of human esophageal cancers. Cancer Res. 2007;67:2535–2543. doi: 10.1158/0008-5472.CAN-06-3530. [DOI] [PubMed] [Google Scholar]
- 12.Cui L, Deng Y, Rong Y, Lou W, Mao Z, Feng Y, Xie D, Jin D. IRF-2 is over-expressed in pancreatic cancer and promotes the growth of pancreatic cancer cells. Tumour Biol. 2012;33:247–255. doi: 10.1007/s13277-011-0273-3. [DOI] [PubMed] [Google Scholar]
- 13.Kneitz B, Krebs M, Kalogirou C, Schubert M, Joniau S, van Poppel H, Lerut E, Kneitz S, Scholz CJ, Ströbel P, et al. Survival in patients with high-risk prostate cancer is predicted by miR-221, which regulates proliferation, apoptosis, and invasion of prostate cancer cells by inhibiting IRF2 and SOCS3. Cancer Res. 2014;74:2591–2603. doi: 10.1158/0008-5472.CAN-13-1606. [DOI] [PubMed] [Google Scholar]
- 14.Pettersson S, Kelleher M, Pion E, Wallace M, Ball KL. Role of Mdm2 acid domain interactions in recognition and ubiquitination of the transcription factor IRF-2. Biochem J. 2009;418:575–585. doi: 10.1042/BJ20082087. [DOI] [PubMed] [Google Scholar]
- 15.Chen YJ, Liang L, Li J, Wu H, Dong L, Liu TT, Shen XZ. IRF-2 inhibits gastric cancer invasion and migration by down-regulating MMP-1. Dig Dis Sci. 2020;65:168–177. doi: 10.1007/s10620-019-05739-8. [DOI] [PubMed] [Google Scholar]
- 16.Tian JB, Cao L, Dong GL. Long noncoding RNA DDX11-AS1 induced by YY1 accelerates colorectal cancer progression through targeting miR-873/CLDN7 axis. Eur Rev Med Pharmacol Sci. 2019;23:5714–5729. doi: 10.26355/eurrev_201907_18309. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Binmadi NO, Basile JR. Perineural invasion in oral squamous cell carcinoma: A discussion of significance and review of the literature. Oral Oncol. 2011;47:1005–1010. doi: 10.1016/j.oraloncology.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Takahara T, Kasamatsu A, Yamatoji M, Iyoda M, Kasama H, Saito T, Takeuchi S, Endo-Sakamoto Y, Shiiba M, Tanzawa H, Uzawa K. SIPA1 promotes invasion and migration in human oral squamous cell carcinoma by ITGB1 and MMP7. Exp Cell Res. 2017;352:357–363. doi: 10.1016/j.yexcr.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 20.Suren D, Yildirim M, Kaya V, Elal R, Selcuk OT, Osma U, Yildiz M, Gunduz S, Sezer C. Expression patterns of claudins 1, 4, and 7 and their prognostic significance in nasopharyngeal carcinoma. J BUON. 2015;20:212–217. [PubMed] [Google Scholar]
- 21.Usami Y, Chiba H, Nakayama F, Ueda J, Matsuda Y, Sawada N, Komori T, Ito A, Yokozaki H. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol. 2006;37:569–577. doi: 10.1016/j.humpath.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 22.Lioni M, Brafford P, Andl C, Rustgi A, El-Deiry W, Herlyn M, Smalley KS. Dysregulation of claudin-7 leads to loss of E-cadherin expression and the increased invasion of esophageal squamous cell carcinoma cells. Am J Pathol. 2007;170:709–721. doi: 10.2353/ajpath.2007.060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Lin Y, Bao QF, Zheng YT, Lan L. MiR-1193 inhibits the malignancy of cervical cancer cells by targeting claudin 7 (CLDN7) OncoTargets Ther. 2020;13:4349–4358. doi: 10.2147/OTT.S247115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Z, Shi J, Song Y, Zhao J, Sun J, Chen X, Gao P, Wang Z. Claudin-7 (CLDN7) is overexpressed in gastric cancer and promotes gastric cancer cell proliferation, invasion and maintains mesenchymal state. Neoplasma. 2018;65:349–359. doi: 10.4149/neo_2018_170320N200. [DOI] [PubMed] [Google Scholar]
- 25.Lourenço SV, Coutinho-Camillo CM, Buim ME, de Carvalho AC, Lessa RC, Pereira CM, Vettore AL, Carvalho AL, Fregnani JH, Kowalski LP, Soares FA. Claudin-7 down-regulation is an important feature in oral squamous cell carcinoma. Histopathology. 2010;57:689–698. doi: 10.1111/j.1365-2559.2010.03685.x. [DOI] [PubMed] [Google Scholar]
- 26.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 27.Sun L, Jiang Z, Acosta-Rodriguez VA, Berger M, Du X, Choi JH, Wang J, Wang KW, Kilaru GK, Mohawk JA, et al. HCFC2 is needed for IRF1- and IRF2-dependent Tlr3 transcription and for survival during viral infections. J Exp Med. 2017;214:3263–3277. doi: 10.1084/jem.20161630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao GN, Jiang DS, Li H. Interferon regulatory factors: At the crossroads of immunity, metabolism, and disease. Biochim Biophys Acta. 2015;1852:365–378. doi: 10.1016/j.bbadis.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 29.Sakai T, Mashima H, Yamada Y, Goto T, Sato W, Dohmen T, Kamada K, Yoshioka M, Uchinami H, Yamamoto Y, Ohnishi H. The roles of interferon regulatory factors 1 and 2 in the progression of human pancreatic cancer. Pancreas. 2014;43:909–916. doi: 10.1097/MPA.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Zhou C, Zhang C, Xie X, Zhou Z, Zhou M, Chen L, Ding Z. MicroRNA-664 functions as an oncogene in cutaneous squamous cell carcinomas (cSCC) via suppressing interferon regulatory factor 2. J Dermatol Sci. 2019;94:330–338. doi: 10.1016/j.jdermsci.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Lu C, Peng K, Guo H, Ren X, Hu S, Cai Y, Han Y, Ma L, Xu P. miR-18a-5p promotes cell invasion and migration of osteosarcoma by directly targeting IRF2. Oncol Lett. 2018;16:3150–3156. doi: 10.3892/ol.2018.9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F, Zhu J, Li J, Zhu F, Zhang P. IRF2-INPP4B axis participates in the development of acute myeloid leukemia by regulating cell growth and survival. Gene. 2017;627:9–14. doi: 10.1016/j.gene.2017.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.