Abstract
The basic helix-loop-helix-PAS transcription factor (CLOCK, Circadian Locomoter Output Cycles Protein Kaput) was discovered in 1994 as a circadian clock. Soon after its discovery, circadian clock, Aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL, also call BMAL1), was shown to regulate adiposity and body weight by controlling on the brain hypothalamic suprachiasmatic nucleus (SCN). Farther, circadian-clock genes were determined to exert several of lipid metabolic and diabetes effects, overall indicating that CLOCK and BMAL1 act as a central master circadian clock. Master circadian clock acts through the neurons and hormones, with expression in the intestine, liver, kidney, lunge, heart, SCN of brain and other various cells types of the organization. Among circadian-clock genes numerous metabolic syndrome are the most important in the regulation of food intake (via regulation of circadian-clock genes or clock-controlled genes in peripheral tissue), which lead to a variation in plasma Phospholipids and tissue Phospholipids. Circadian-clock genes affects the regulation of transporters and proteins included in the regulation of Phospholipid -metabolism. These genes have recently received increasing recognition because a pharmacological target of circadian-clock genes may be of therapeutic worth to make better resistance against insulin, diabetes, obesity, metabolism syndrome, atherosclerosis and brain diseases. In this book chapter, we focus on the regulation of circadian clock and summarize its phospholipid effect as well as discuss the chemical, physiology and molecular value of circadian clock pathway regulation for the treatment of plasma lipids and atherosclerosis.
Keywords: Circadian clock, Phospholipids metabolism, lipid metabolism, atherosclerosis, Diet-induced obesity
1. Introduction
Sleep disorders are now a major health threat to our life, may result in more than 22 million Americans suffering from sleep disorder annually [1–15]. At least 38,000 people die from heart disease directly complicated by a sleep disorder [1–15]. A sleep disorder now affects almost every ethnicity and cultural society, setting an enormous load on the modern healthcare system in the United States and worldwide. From the numerous complications associated with sleep disorder are major metabolic syndrome, atherosclerosis, hypertension, dyslipidemia, obesity, diabetes mellitus, cardiovascular diseases, several cancers, and certain types of brain diseases such as Alzheimer’s disease [1–15]. Emphasizing the consequences of sufficient lipid buffering, atherosclerosis represents, to date, the very high common cause of lipid-related diseases [16–20]. According to sleep being a major risk factor for development of atherosclerosis, ample sleep achieved by either dieting, lifestyle, pharmacology, changes in circadian rhythms, proinflammatory responses and metabolic effects improving sleep quality and pattern, have shown, in numerous pre-clinical studies, many promising effects. For example, the ATP binding cassette subfamily G member 5/8 (ABCG5/8), N-terminal Niemann-Pick C1 (NPC1) intracellular cholesterol transporter 1 (NPC1L1) and Microsomal triglyceride transfer protein (MTTP) inhibitor are adequate to indicate significant improvements in systemic lipid metabolism and atherosclerosis-linked comorbidities [4;21–29]. Further emphasizing, the direct relationship between atherosclerosis and lipid regulation, plasma cholesterols reduced by ABCG5/8, NPC1L1 and MTP inhibitors, which are regulated by circadian-clock genes, most often result in whole resolution of atherosclerosis, an opinion that encouraged the American Heart Association and National Institutes of Health to recommend such inhibitors under assured conditions for the treatment of atherosclerosis. Since the correlation between food intake and Phospholipid regulation is highly confirmed by several basic research studies, inhibitors to inhibit food intake intuitively appear promising to improve Phospholipid metabolism [30–34]. Under this reason, remarkable examples of such strategies are the administration of MTP inhibitor, which not only reduces plasma cholesterol through their MTTP inhibition but also decreases Phospholipid metabolism through their ability to reduce lipid absorption via circadian rhythm regulation of food intake [35;36]. We have shown that the circadian-clock genes can regulate plasma triglycerides and cholesterol, and regulate cholesterol and triglyceride absorption and metabolism [28;29;37–40]. A prominent example of circadian-clock gene regulation, is the circadian-clock with a mutant CLOCK gene, which improve body fat mass and body weight through regulation of intestinal lipid absorption and adipose lipid metabolism [38;40–43]. However, whether circadian-clock genes regulate Phospholipid metabolism is not commonly known.
Phospholipids are polar, ionic compounds composed of an alcohol that is attached by a phosphodiester bridge to either diacylglycerol or sphingosine [35]. There are two classes of Phospholipids: those that have glycerol (from glucose) as a backbone are called glycerophospholipids and those that have a sphingosine (from serine and palmitate) are called sphingophospholipids. Most Phospholipids are synthesized in the smooth endoplasmic reticulum [35]. From there, they are transported to the Golgi apparatus and then to membranes of organelles or the plasma membrane of organelles [44]. They could also be secreted from the cell by exocytosis. Phosphatidylcholine (also called PC) and Phosphatidylethanolamine (also called PE) are the most abundant Phospholipids in most eukaryotic cells [35;45]. The primary route of their synthesis uses choline and ethanolamine obtained either from food intake or from the turnover of the body’s Phospholipids [44]. Sphingomyelin is one of the principal structural lipids of the membranes of nerve tissues. It is synthesized from ceramide (an acyl sphingosine) and phosphatidyl choline. Sphingomyelin is also hydrolyzed into ceramide and phosphoryl choline [46]. Ceramide is further degraded to sphingosine and free fatty acid (FA) [46]. In this book chapter, we summarized the regulation of circadian-clock genes with a special focus on their role to control Phospholipid metabolism. A key central field will thereby be the topic of whether disordering the circadian-clock genes will regulate transcription factors, and will the function of a protein pathway be of chronotherapeutic value to progress Phospholipid metabolism?
2. Origins of the mammalian clock
Several biological, physiological, and behavioral activities show characteristic recurrence with 24-h intervals related to sunrise and sunset. Light entrains the central clock present in two lateral SCN in the hypothalamus via the retinohypothalamic tract. The master circadian clock arises from auto regulatory transcriptional, translational, and posttranslational feedback loops of few transcription factors encoded by “clock” genes, including circadian locomotor output cycles kaput (CLOCK), brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1 (BMAL1), neuronal PAS containing protein 2 (NPAS2), period genes (Period1/2/3, PER1/2/3), and cryptochrome genes (CRY1/2) [24;47–50]. The BMAL1:CLOCK and BMAL1:NPAS2 heterodimers bind to cis-acting E-box sequences present in the promoter regions of PER1/2/3 and CRY1/2 and enhances their expression, constituting a positive feed-forward loop. Unlike CLOCK and BMAL1, PER1-3 and CRY1-2 protein can dimerize and translocate to the nucleus, then dimerize the PER1–3:CRY1–2 complex, inhibiting the activity of CLOCK:BMAL1 or NPAS2-BMAL1. In the center of the hypothalamus, circadian-clock genes are localized in the SCN and express neuronally and hormonally [16;25;51]. Zhang et al has reported that the liver had the most circadian genes, then kidney as the 2nd, whereas the hypothalamus had the fewest (Figure 1) [52]. SCN is responsible for controlling circadian rhythms, these circadian-clock protein’s neuronal and hormonal activities regulate different body functions in the 24- hour cycle, such as body temperature, wakeup/sleep, and food intake. To activate CLOCK:BMAL1 or NPAS2:BMAL1 there are 450 unique protein modifications [52]. Clock genes also need multiple post-translational modifications, including phosphorylation, ubiquitination, acetylation and SUMOylation to regulate various physiological functions [53–55]. This post-translational modification of BMAL1 is regulated by ubiquitin-specific protease 2 (USP2) [54;55]. USP2 is essential to deubiquitinating PER1, CRY1 and CRY2 in vivo [54–57]. This mechanism was demonstrated by the absence of deubiquitinated Per1, Bmal1, Cry1 and Cry2 in mice deficient in USP2 [55].
Figure 1. Number of circadian clock genes detected in each organ.
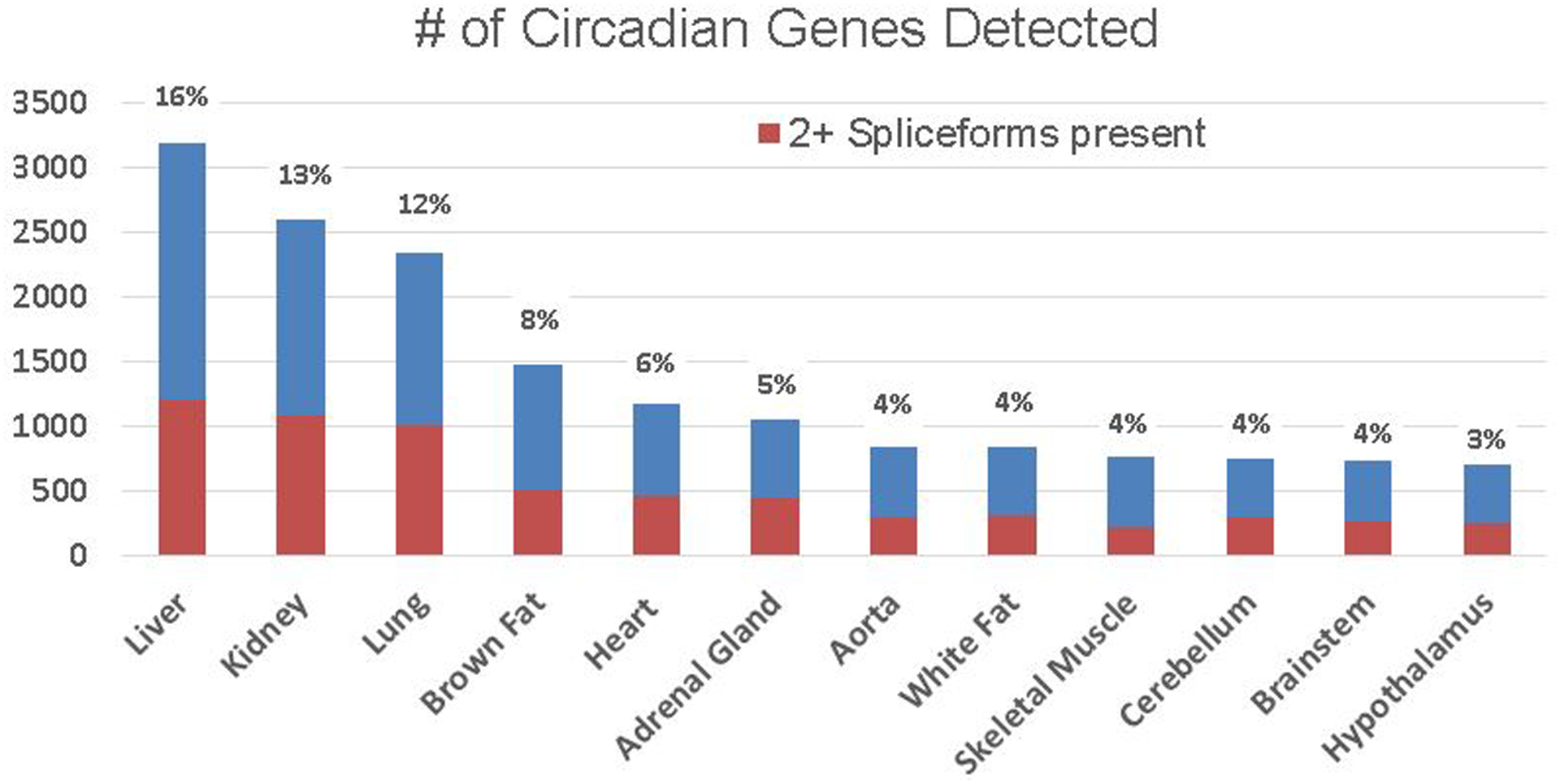
Circadian expression of protein-coding genes in different tissue. Blue marks indicate the number of genes with at least one spliceform detected by RNA-seq. Orange marks indicate the number of genes with at least two spliceforms detected by RNA-seq. Blue numbers to the top of each bar states the percentage of protein-coding genes with rhythmic expression in each organ of Zhang et al. publication. Figure modified according to Zhang et al publication in PNAS52.
BMAL1:CLOCK is a heterodimer formed via CLOCK with 361 amino acids and BMAL1 with 387 amino acids. CLOCK and BMAL1 have the same one, basic, helix-loop-helix (bHLH) binding of protein to DNA via recognized E-box sites, through hydrogen bonding, between serine residues and DNA. As we know, E-box sites are about 20 base pairs upstream of genes with a major 5’-CACGTG-3” canonical motif. CLOCK and BMAL1, or NPAS2 (a paralog of clock) can not only recruit transcription factors to the E-box site, but also can upregulate transcription of the target genes such as, nuclear receptor subfamily 1, group D, member 1 (NR1D1, Rev-erbα), D-Box Binding PAR BZIP transcription factor (DBP), peroxisome proliferator-activated receptor (PPAR) alpha (PPARα), small heterodimer partner (SHP), GATA binding protein 4 (GATA4), and paired box protein 4 (PAX4) [16;24;28;29;40;47–50;58] (Figure 2). These genes can also, through recruitment of histone acetyl transferases, de-condense the nucleosome into heterochromatin allowing transcriptional machinery access to the DNA, such as mutant CLOCK, which down- regulates upstream transcription factor2 (USF2). In addition, USF1 serves as a suppressor of the circadian clock mutant, revealing the nature of the DNA-binding of the Clock:Bmal1 complex in mice [59]. This data also suggests that USF1 and USF2 are an important modulator of molecular and behavioral circadian rhythms in mammals. In addition, it is possible that CLOCK regulates USF2 through the histone acetyl transferase pathway. However, more experiments are required to understand this mechanism.
Figure 2. Clock and clock-collected genes regulate metabolic function.
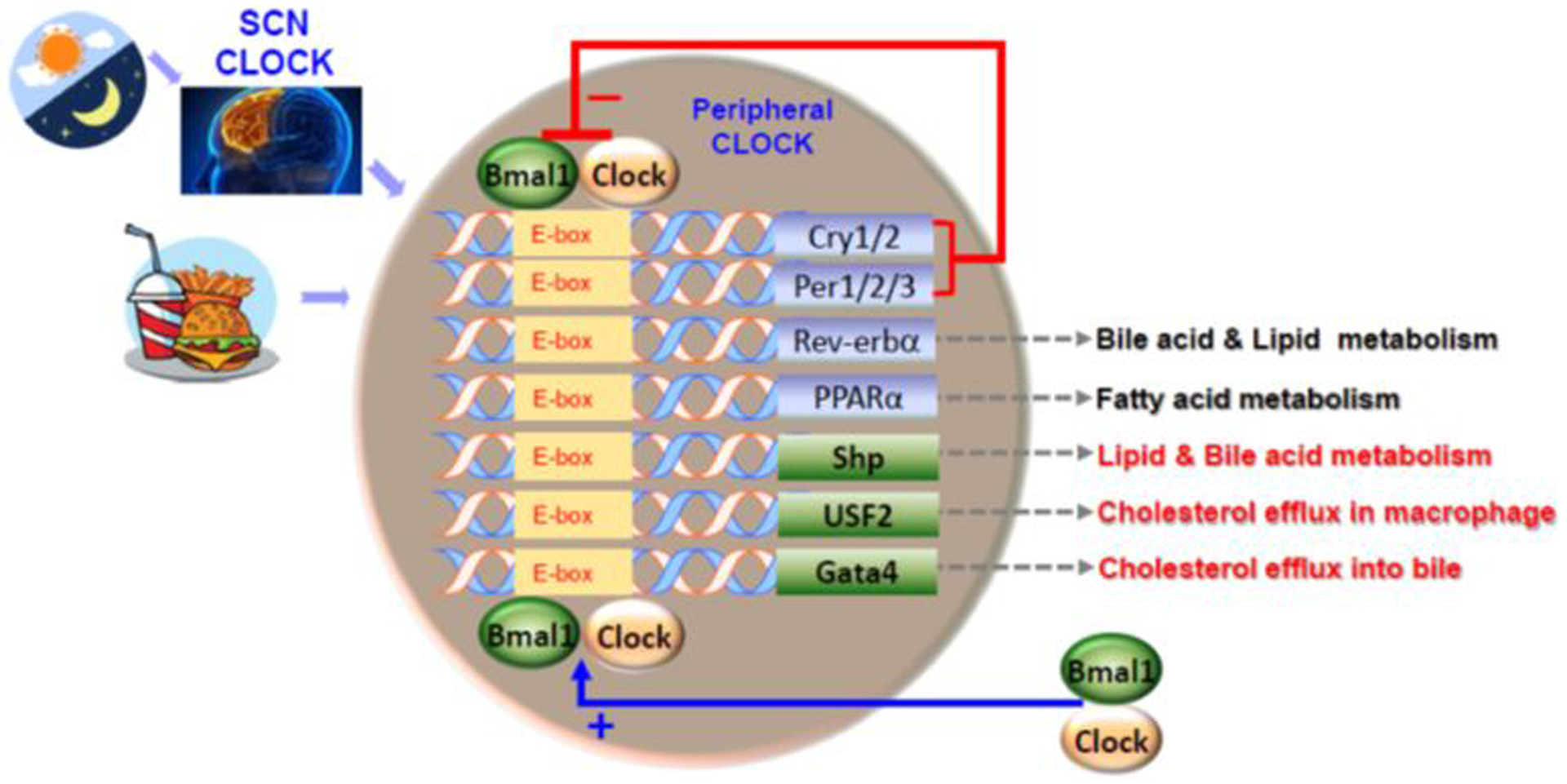
Both light- and food-entrained oscillators appear to affect the expression of circadian-clock genes and clock-collected genes in the peripheral tissue. In SCN and peripheral, Clock :Bmal1 heterodimerize to activate transcription of circadian target genes including the genes of Per1/2/3 and Cry1/2. Per1/2/3 and Cry1/2 interact and inhibit Bmal1 and Clock. We have shown that Clock and Bmal1 regulate several transcription factors such as Shp, Usf2 and Gata4 regulating the expression of several genes involved in lipid metabolism as well as other pathways that affect metabolism.
The reported levels of CLOCK and BMAL protein do not show dramatic circadian oscillations in mammalian brains, however, reflecting the species-related phosphorylation in circadian clock protein show clear circadian oscillations with time-dependent, post-translational regulation [53]. The degradation of CLOCK and BMAL1 is more important for transcription activation of clock-controlled genes through E-boxes in their promoters [60]. For example, estrogen receptors are regulated by CLOCK [61]. CLOCK:BMAL1 proteins can bioaccumulate by proteasome inhibitor MG132 by preventing their protein degradation. MG132 is an inhibitor decreasing E-Box-mediated transcription by interfering with CLOCK:BMAL1 regulation cycles in humans. Whereas in rodents, CLOCK19 protein is hypo-phosphorylated to a higher extent than those of wild-type CLOCK [62]. In vitro studies have also shown several enzymes (such as Casein kinase I/II, Glycogen synthase kinase 3 beta (GSK-3β) and Cyclin-dependent kinase 5) are responsible for CLOCK:BMAL1 degradation [63–65] [66;66;67]. For example, GSK-3β-catalyzed phosphorylation can phosphorylate Ser431 of CLOCK dependent site Ser427 and Thr21 of BMAL1 dependent site with Ser17, to induce higher activity of CLOCK and BMAL1 under unstable conditions. Similarly, protein kinase CR can phosphorylate and stabilize BMAL1 by eliminating BMAL1 polyubiquitination [66;67]. Regulation of CLOCK:BMAL1 phosphorylation affects transcription through alterations in DNA binding [68]. In addition, CLOCK:BMAL1 activity is affected not only by phosphorylation, but also by ubiquitination to induce its transactivation and degradation [69]. For example, SUMOylation and O-GlcNAcylation induce ubiquitination of BMAL1 at Lys259 and at Ser418, respectively, to increase BMAL1 transactivation and degradation [69]. In addition, HECT-type E3 ligase can promote ubiquitination of BMAL1 and CLOCK [66;70;71]. Moreover, there are several studies showing that Sirtuin 1 binds to CLOCK and BMAL1 and deacetylates BMAL1 at lys537 [71], thus preventing CRY1 recruitment and restarting the transactivation of the clock gene. Furthermore, HAT activity required site motif A of CLOCK and acetylation site of BMAL1 are required to rescue the cellular clock-controlled gene rhythm [72;73]. Histone modifications were essential for normal clock function [71;72;74–77]. CLOCK:BMAL1 heterodimers shuttle between the nucleus and the cytosol, thus suggesting that the dimer-protein modulation are involved in several post-translation and transcription levels.
3. Physiological functions of Circadian clock
While major studies indicate that most metabolic functions of circadian clock require transcription and post-translation levels, there is gain experience indicating that circadian clock genes have physiologically related functions on a body metabolism, potentially through several pathways that have yet to be identified. Circadian-clock genes respond to external stimuli and the one prominent effect of the circadian-clock gene is its ability to diurnal control food intake. We have shown that circadian-clock genes and lipid transport proteins are expressed in the small intestinal enterocytes and respond to food entrainment in wild-type mice [38]. Dominant-negative Clock mutant protein mice (ClockΔ19/Δ19 or Clkmt/mt) disrupt the circadian expression and food entrainment of the clock genes [38;41]. In addition, the absorption of lipids was high in Clock mutant mice [38;40]. Our data also suggests that Clock plays an important role in light and food entrainment of intestinal function. To understand the mechanism of Clock genes regulating lipid absorption and metabolism, we studied the role of Clock gene in the diurnal regulation of plasma triglyceride-rich apolipoprotein B-lipoprotein and MTTP. Clock mutant mice showed sustained hypertriglyceridemia and high MTTP expression. We found that CLOCK-knockdown-activated MTTP promoter and reduced SHP, in the Human liver cell line Huh7 cells, CLOCK temporally interacts with the E-box site and increases SHP expression, whereas SHP reduces MTTP expression by differentially interacting with Hepatocyte nuclear factor 4 alpha and the liver receptor homolog−1 [40]. In Clockmt/mt mice, however, the binding of Clock to SHP promoter did not show cyclic change and SHP mRNA levels were relatively low and did not change [40]. This data shows that a decreased interaction of SHP with these transcription factors is associated with increased MTTP expression. Therefore, SHP is a clock-controlled gene that transmits information from Clock to MTTP. Additionally, we showed, for the first time, that ClockΔ19/Δ19 mutant protein enhances plasma cholesterol and atherosclerosis in the low density lipoprotein receptor knockout (Ldlr−/−) and apolipoprotein E knockout (ApoE−/−) atherosclerosis animal models [29]. In addition, Clock mutant protein affects macrophage function. Macrophages from ClockΔ19/Δ19 mice took up more oxidized lipids and were defective in cholesterol efflux. Molecular studies showed that Clock regulates ATP Binding Cassette Subfamily A Member 1 expression and cholesterol efflux in macrophages via Usf2 [29]. In addition, we recently showed that global Bmal1-deficient mice or hepatic-specific Bmal1 knockout mice also have an impaired cholesterol metabolism, display hepatic cholesterol efflux into bile, develop atherosclerosis when fed with an atherogenic diet and potentiate the development of atherosclerotic lesions in the Ldlr−/− and ApoE−/− atherosclerosis animal models [28]. Liver-specific inactivation of Bmal1 led to elevated plasma low density lipoprotein (LDL) and /very low-density lipoprotein (VLDL) cholesterol levels as a consequence of the disruption of the Pcsk9/Ldl receptor regulatory axis [22;28;78].
Phosphatidylcholine is one of Phospholipid that occupies 70% of VLDL Phospholipids. Phosphatidylcholine biosynthesis is known to be required for VLDL secretion [78]. This has also shown that diurnal variation of VLDL concentration is linked to the clock-controlled production of Phosphatidylcholine. Furthermore, Ma et al. have identified two distinct groups exhibiting rhythmic and non-rhythmic patterns of gene expression during light-dark cycles, according to database of the circadian regulation of lipid-associated Genome-Wide Association Studies (GWAS) candidate genes in mouse liver [79]. Liver-specific Bmal1 knockout mice increased plasma Ldl/Vldl cholesterol levels through disordered Pcsk9/Ldl receptor expression [79].
In line with this idea, circadian-clock genes affect food intake, body weight, plasma glucose and lipids, has protective effects on the adipose tissue, heart, liver, intestine and affects Phospholipid metabolism via several pathways [4;21;80–82]. In other words, the circadian-clock may through several mechanisms control Phosphatidylcholine (is one Phospholipids that make up 50% of total cellular Phospholipid biosynthesis), as the Phosphatidylcholine phenotype can be copied by different circadian-clock gene mutations [83]. Wild-type mice in normal light and dark cycles, display a rhythmic accumulation of hepatic phosphatidylcholine with a peak at Zeitgeber time (ZT) 22–0. Bmal1-deficient (Bmal1−/−) mice show elevated phosphatidylcholine levels in the liver associated with an atherogenic lipoprotein profile [78]. To investigate whether the circadian variation of Phosphatidylcholine levels is the result of a circadian regulation of Phosphatidylcholine biosynthesis, Grechez-Cassiau et al. found that Choline Kinase alpha (Chkα) gene is a clock-controlled gene in the liver [78]. Chkα gene expression is regulated by the Rev-erbα and RAR related Orphan Receptor A (Rorα) nuclear receptors [78]. Thus, hepatic phosphatidylcholine is regulated by the circadian-clock gene through a Bmal1-Rev-erbα-Chkα axis and suggests that an intact circadian timing system is important for the temporal coordination of Phospholipid metabolism. The Rev-erbα subtype appears to be a key circadian regulator of Phosphatidylcholine metabolism in the liver through the rhythmic transcriptional repression of the Chkα gene. Thus, a likely mechanism by which hepatic Phosphatidylcholine levels are increased in the Bmal1−/− mice is that Chkα up regulates by the high of total Choline Kinase activity [78]. In addition, there is a low Rev-erbα gene expression level in the Per1/Per2 double knockout mice [84]. Twenty-four out of twenty-seven Phosphatidylcholine species were arrhythmic by the lipidomic profiling, although 16% of lipid metabolites were still oscillating in liver [84]. These studies suggest that a genetic disruption of the circadian clock system compromises Phosphatidylcholine homeostasis.
4. Preclinical studies on diurnal rhythm in Phospholipid metabolism
Minami et al. found oscillatory peaks of phospholipids was detected by Liquid chromatography–mass spectrometry (among these time-indicating metabolites) [85]. Fourteen oscillatory peaks were identified as various types of lysophosphatidylcholines with different unsaturated FAs [85]. As mentioned above, in mammalian cells, Phosphatidylcholine is one of all Phospholipid, constituting 50% of total cellular Phospholipids [86], Phosphatidylcholine is also the main circulating Phospholipid in plasma, where it is critical for the assembly and secretion of lipoproteins by the liver. Hepatica Phospholipids enter in bile-salt mediated micelle formation in the intestinal lumen, which facilitates the absorption of lipid-soluble nutrients from the diet [87]. Several studies have shown that serum Phosphatidylcholine is shown to be subjected to temporal control that could be correlated with rest-activity cycles and feeding [84;88]. Phosphatidylcholine plays an important role in mammalian cell signaling [89] as well as in oncogenic signaling pathways [78;89–91]. Numerous studies have evaluated the circadian-clock genes effect on Phospholipid metabolism. Diurnal rhythm of retinal–Phospholipid–synthetic enzyme has been shown in the retina of rats [92]. Retinal Phospholipid synthetic enzymes showed daily variations, in retinal ganglion cells (RGCs) of chicken when in constant darkness. [32P]Phospholipid display circadian oscillations both in, in vivo chicken kept in constant light, and in cultures of immunopurified embryonic RGCs [92]. Several distinct enzymes, lysophospholipid acyltransferases, phosphatidate phosphohydrolase, and diacylglycerol lipase, in the pathway of Phospholipid biosynthesis and degradation have shown diurnal variation [93]. These activities of these enzymes are high during the subjective day and low at night, as were the metabolic changes observed in the in vivo labeling of Phospholipid in cultures of purified embryonic RGCs [93;94]. In addition, glycerophospholipid synthesis has also shown diurnal rhythm in retinal inner nuclear layer cells [93;94]. Biosynthesis of Phospholipid has shown the circadian cycle by serum shock in cultured quiescent NIH3T3 cells, this cycle is abolished by knock–down Per1 gene, suggesting that the biosynthesis of Phospholipid circadian cycle in cultured fibroblasts depends on the endogenous circadian clock [94;95;95–97]. Ruggiero et al. showed that the diurnal rhythm of phospholipid phosphatidylserine demarcation of photoreceptor outer segments tip, is not intrinsic to rod photoreceptors but requires activities of the retinal pigment epithelium as well [98]. In line with the circadian cycle of Phospholipid or Phospholipid biosynthesis in vivo and in vitro, levels of serum Phospholipids such as phosphatidylcholines (18:0/18:1) or 1-stearoy1-2-oleoyl-sn-glycero-3-phosphocholine, are typically regulated in mice lacking circadian clock-collected gene PPAR gamma (PPARδ) activity [98]. Serum Phosphatidylcholine (18:0/18:1) can reduce postprandial lipid levels and Phosphatidylcholine can increase FA utilization through muscle PPAR alpha (PPARα) [98]. When mice were fed with a high fat diet, the rhythm of Phosphatidylcholine (18:0/18:1) was diminished. Phosphatidylcholine (1:0/18:1) administration in db/db mice (a model for diabetic dyslipidemia) can improve metabolic homeostasis, suggesting that alterations in diurnal hepatic PPARδ-Phosphatidylcholine (18:0/18:1) signaling affects metabolic disorders, including obesity [99]. Obesity can alter circadian rhythms in multiple tissues. Diet induced obesity altered the rhythm pattern of serum Phosphatidylcholine [99;100]. As a Phospholipid outcome, ceramide, a class of sphingolipids, Jang et al. showed that the ceramide concentration in WT mice showed a strong peak at Zeitgeber Time 9 (ZT9; 9 h after lights-on time) and ZT21 but no rhythmicity in ceramide expression was seen in Per1/Per2 double KO mice [101]. To understand the mechanism of diurnal rhythm of ceramide, they also measure several gene expressions including via sphingomyelinase (SMase), or by ceramide synthase (CerS)-mediated synthesis, both are important for sphingomyelin hydrolysis to ceramide. Jang et al. found that CerS2 expression levels showed a biphasic pattern of expression in WT mice but no rhythmicity in Per1/Per2 double KO mice [102]. While the neutral SMase (nSMase) and acidic SMase (aSMase) mRNA in WT mice were expressed in a circadian variation, the correlation between the expression levels of these SMases with times of day was weak in Per1/Per2 double KO mice [102]. Collectively, this study suggests that both SMases and CerS2 mRNA expression are regulated by the presence of mPer1/mPer2 circadian-clock genes in vivo, and imply that ceramide may play a vital role in circadian rhythms and physiology [102]. However, the molecular mechanism of circadian-clock genes regulating phospholipid metabolism are still unclear and limited.
5. Clinical studies on circadian clocks role in Phospholipid metabolism
Animal research shows a clear involvement of membrane-derived Phospholipid in circadian rhythms. Additionally, 7–20% of metabolites in human blood have been observed showing circadian variation [85;103–106]. Under a series of preclinical studies, the existence of both daily change and seasonal variations, affect the composition of Phospholipids in human cell membranes [12;107;108]. Over 1 year, in 20 healthy subjects, Ruf et al. found that 11 of 13 Phospholipids’ FAs content showed significant daily rhythms and were largely synchronous among subjects [108]. This data is supported by several other studies, overall indicating that human physiology is still dominated by geophysical sunrise and sunset, resulting in a strong daily cycle [107;109]. However, seasonal rhythms are less well defined. FA’s derived from Phospholipids also play a role as precursors of prostaglandins, thromboxanes, and leukotrienes. A much more likely candidate for such a function of rhythmicity is the interaction between membrane FA’s and transmembrane proteins. It is a possible explanation for rhythmic alterations of membrane composition [108]. In particular, a link between sleep deprivation and Phosphatidylcholine is also showed by the result that both the circadian system and plasma lipids display a reciprocal correlation over the day with a subset of Phosphatidylcholine and triglyceride species in plasma being high when sleep deprived in twenty total subjects of young-aged-healthy-ethnic Chinese males [110].
Epidemiological studies comfort the relationship between the circadian system and the regulation of diurnal rhythm of Phospholipids. A marked circadian variation was recorded in plasma total-cholesterol, high-density-lipoprotein–cholesterol, Phospholipid, and total lipid concentration in healthy Indians of different age groups of 162 total subjects [111]. Plasma Phospholipid concentrations were characterized by a circadian rhythm in all age groups. Females had numerically higher values than males. However, the rhythm peak was significantly changed by age, reaching a maximum in middle adulthood and decreasing in the older age group [111;112]. This suggests that the diurnal rhythm of plasma Phospholipids is associated with age, gender, diet and smoking and affects circulating plasma lipid components in healthy Indians. In addition, a 24-hr time series of plasma metabolites has been simultaneously assessed in Type 2 Diabetes, compared with an age and weight-matched control group during a controlled daily routine [113]. Similarly, a total of 100 of 663 metabolites, representing all metabolite categories, showed diurnal rhythmic concentrations that exceeded the bonferroni threshold, showing the peak times of all Phospholipids were clustered during the afternoon-midnight [114;115].
We previously showed that peptide-like drugs H+-peptide cotransporter 1, Pept1, showing diurnal rhythm, could influence the pharmacokinetics of peptide-like drugs [116–118]. Drugs statins, a HMG-CoA reductase inhibitor that is in clinical evaluation for the treatment of Type 2 Diabetes and atherosclerosis, shows beneficial effects on plasma lipids [119–121]. Interestingly, statins was recommended to be administered in the evening [119;121]. However opinions differ on the best time to take statins. Simvastatin was reportedly better in the evening too, but, simvastatin taken in the evening was not better than when it was taken in the morning by a different study group [122–124]. It remains in clinical evaluation for treatment. Lipidomics can be used to examine differences in circadian responses to medications that target lipid pathways, such as statins, and to better characterize the mode of action of such drugs. So far, there is collecting preclinical and clinical studies overall suggesting a beneficial effect of chronotherapeutics. Beyond circadian-clock’s direct Phospholipid role, it has to be noticed that food intake and body weight change because of circadian clock pathway regulation might provide a particular potential for secondy improvement of Phospholipid treatment.
6. Conclusion
The circadian clock system has, over the last twenty years, been researched act involved in a number of metabolic functions that go well over their primary classification as a regulator affecting wakeup/sleep and food intake. Along with circadian clocks regulation in Phospholipid metabolism; various studies evaluated the therapeutic effect of Phospholipid modulation. The circadian clock correlating to Phospholipids might offer a potential treatment for atherosclerosis and obesity associated with pathological atherosclerosis. Circadian clock altering of molecular time will be of chronotherapeutic value to reduce metabolic disorders, impaired immune function, and accelerated aging, and to improve Phospholipid metabolism and cardiovascular diseases. Importantly, while disordered circadian-clock genes and sleep disorder is known to affect more than 50 million U.S. residents (https://www.ncbi.nlm.nih.gov/books/NBK19961/), it is possible that other physiology function of circadian clock are yet to be understood.
Acknowledgement:
This work was supported in part by NIH National Heart, Lung, and Blood Institute Grant R56 HL137912-01 and American Heart Association Grant-In-Aid 16GRNT30960027 to X. Pan.
Abbreviations:
- ABCA-1
ATP-binding cassette transporter 1
- ABCG5/8
ATP binding cassette subfamily G member 5/8
- ApoE−/−
Apolipoprotein E knockout
- aSMase
Acidic SMase
- BMAL1
Aryl hydrocarbon receptor nuclear translocator-like protein 1
- CerS
Ceramide synthase
- Chkα
Choline Kinase alpha
- CLOCK
Circadian Locomoter Output Cycles Protein Kaput
- ClockΔ19/Δ19 or Clkmt/mt
Dominant-negative Clock mutant protein mice
- CRY1/2
cryptochrome 1,2
- FA
Fatty Acid
- GSK-3β
Glycogen synthase kinase 3 beta
- HAT
Histone acetyltransferase
- HMG-CoA reductase
3-hydroxy-3-methyl-glutaryl-CoA reductase
- Ldlr−/−
Low density lipoprotein receptor knockout
- LDL
Low-density lipoprotein
- MTTP
Microsomal triglyceride transfer protein
- NPAS2
Neuronal PAS containing protein 2
- NPC1L1
NPC1 intracellular cholesterol transporter 1
- nSMase
Neutral SMase
- PC
Phosphatidylcholine
- PC
Phosphatidylcholine
- PCSK9
Proprotein convertase subtilisin/kexin type 9
- PE
Phosphatidylethanolamine
- Period1/2/3
Period genes 1, 2, 3
- PPARδ
Peroxisome proliferator-activated receptor delta
- Rev-erbα
Nuclear receptor subfamily 1, group D, member 1
- RGCs
Retinal ganglion cells
- Rorα
RAR related Orphan Receptor A
- SCN
Suprachiasmatic nucleus
- SHP
Small heterodimer partner
- SIRT1
Sirtuin 1
- SMase
Sphingomyelinase
- UBE3A
HECT-type E3 ligase
- USF2
Upstream Transcription Factor 2
- VLDL
Very low-density lipoprotein
Reference List
- 1.Fernando S, Biggs SN, Horne RSC, Vollenhoven B, Lolatgis N, Hope N et al. The impact of melatonin on the sleep patterns of women undergoing IVF: a double blind RCT. Hum Reprod Open 2017; 2017:hox027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grajewski B, Whelan EA, Lawson CC, Hein MJ, Waters MA, Anderson JL et al. Miscarriage among flight attendants. Epidemiology 2015; 26:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grajewski B, Whelan EA, Nguyen MM, Kwan L, Cole RJ. Sleep Disturbance in Female Flight Attendants and Teachers. Aerosp Med Hum Perform 2016; 87:638–645. [DOI] [PubMed] [Google Scholar]
- 4.Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest 2011; 121:2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson CL, Redline S, Emmons KM. Sleep as a potential fundamental contributor to disparities in cardiovascular health. Annu Rev Public Health 2015; 36:417–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TW, Jeong JH, Hong SC. The impact of sleep and circadian disturbance on hormones and metabolism. Int J Endocrinol 2015; 2015:591729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knutson KL, Lauderdale DS. Sleep duration and overweight in adolescents: self-reported sleep hours versus time diaries. Pediatrics 2007; 119:e1056–e1062. [DOI] [PubMed] [Google Scholar]
- 8.Labyak S, Lava S, Turek F, Zee P. Effects of shiftwork on sleep and menstrual function in nurses. Health Care Women Int 2002; 23:703–714. [DOI] [PubMed] [Google Scholar]
- 9.Mahoney MM. Shift work, jet lag, and female reproduction. Int J Endocrinol 2010; 2010:813764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maung SC, El SA, Chapman C, Cohen D, Cukor D. Sleep disorders and chronic kidney disease. World J Nephrol 2016; 5:224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNeil J, Doucet E, Chaput JP. Inadequate sleep as a contributor to obesity and type 2 diabetes. Can J Diabetes 2013; 37:103–108. [DOI] [PubMed] [Google Scholar]
- 12.Potter GD, Skene DJ, Arendt J, Cade JE, Grant PJ, Hardie LJ. Circadian Rhythm and Sleep Disruption: Causes, Metabolic Consequences, and Countermeasures. Endocr Rev 2016; 37:584–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiegel K, Knutson K, Leproult R, Tasali E, Van CE. Sleep loss: a novel risk factor for insulin resistance and Type 2 diabetes. J Appl Physiol (1985) 2005; 99:2008–2019. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan V, Singh J, Pandi-Perumal SR, Brown GM, Spence DW, Cardinali DP. Jet lag, circadian rhythm sleep disturbances, and depression: the role of melatonin and its analogs. Adv Ther 2010; 27:796–813. [DOI] [PubMed] [Google Scholar]
- 15.Ting L, Malhotra A. Disorders of sleep: an overview. Prim Care 2005; 32:305–18, v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain MM, Pan X. Clock genes, intestinal transport and plasma lipid homeostasis. Trends Endocrinol Metab 2009; 20:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain MM, Fatma S, Pan X, Iqbal J. Intestinal lipoprotein assembly. Curr Opin Lipidol 2005; 16:281–285. [DOI] [PubMed] [Google Scholar]
- 18.Hussain MM, Rava P, Pan X, Dai K, Dougan SK, Iqbal J et al. Microsomal triglyceride transfer protein in plasma and cellular lipid metabolism. Curr Opin Lipidol 2008; 19:277–284. [DOI] [PubMed] [Google Scholar]
- 19.Bobulescu IA. Renal lipid metabolism and lipotoxicity. Curr Opin Nephrol Hypertens 2010; 19:393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rui L. Brown and Beige Adipose Tissues in Health and Disease. Compr Physiol 2017; 7:1281–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science 2016; 354:994–999. [DOI] [PubMed] [Google Scholar]
- 22.Gooley JJ. Circadian regulation of lipid metabolism. Proc Nutr Soc 2016; 75:440–450. [DOI] [PubMed] [Google Scholar]
- 23.Hussain MM, Pan X. Circadian Regulation of Macronutrient Absorption. J Biol Rhythms 2015; 30:459–469. [DOI] [PubMed] [Google Scholar]
- 24.Hussain MM, Pan X. Circadian regulators of intestinal lipid absorption. J Lipid Res 2015; 56:761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain MM, Pan X. Clock regulation of dietary lipid absorption. Curr Opin Clin Nutr Metab Care 2012; 15:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molusky MM, Hsieh J, Lee SX, Ramakrishnan R, Tascau L, Haeusler RA et al. Metformin and AMP Kinase Activation Increase Expression of the Sterol Transporters ABCG5/8 (ATP-Binding Cassette Transporter G5/G8) With Potential Antiatherogenic Consequences. Arterioscler Thromb Vasc Biol 2018; 38:1493–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douris N, Kojima S, Pan X, Lerch-Gaggl AF, Duong SQ, Hussain MM et al. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr Biol 2011; 21:1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan X, Bradfield CA, Hussain MM. Global and hepatocyte-specific ablation of Bmal1 induces hyperlipidaemia and enhances atherosclerosis. Nat Commun 2016; 7:13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X, Jiang XC, Hussain MM. Impaired cholesterol metabolism and enhanced atherosclerosis in clock mutant mice. Circulation 2013; 128:1758–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zambo V, Simon-Szabo L, Szelenyi P, Kereszturi E, Banhegyi G, Csala M. Lipotoxicity in the liver. World J Hepatol 2013; 5:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiedeman AM, Barr SI, Green TJ, Xu Z, Innis SM, Kitts DD. Dietary Choline Intake: Current State of Knowledge Across the Life Cycle. Nutrients 2018; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lordan R, Tsoupras A, Zabetakis I. Phospholipids of Animal and Marine Origin: Structure, Function, and Anti-Inflammatory Properties. Molecules 2017; 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parchem K, Bartoszek A. Phospholipids and products of their hydrolysis as dietary preventive factors for civilization diseases. Postepy Hig Med Dosw (Online) 2016; 70:1343–1361. [DOI] [PubMed] [Google Scholar]
- 34.Palamiuc L, Ravi A, Emerling BM. Phosphoinositides in autophagy: current roles and future insights. FEBS J 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang XC. Phospholipid transfer protein: its impact on lipoprotein homeostasis and atherosclerosis. J Lipid Res 2018; 59:764–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Park TS, Li Y, Pan X, Iqbal J, Lu D et al. Serine palmitoyltransferase (SPT) deficient mice absorb less cholesterol. Biochim Biophys Acta 2009; 1791:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan X, Munshi MK, Iqbal J, Queiroz J, Sirwi AA, Shah S et al. Circadian regulation of intestinal lipid absorption by apolipoprotein AIV involves forkhead transcription factors A2 and O1 and microsomal triglyceride transfer protein. J Biol Chem 2013; 288:20464–20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res 2009; 50:1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan X, Hussain MM. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J Biol Chem 2007; 282:24707–24719. [DOI] [PubMed] [Google Scholar]
- 40.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab 2010; 12:174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005; 308:1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998; 280:1564–1569. [DOI] [PubMed] [Google Scholar]
- 43.Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem J 2005; 386:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent C Eukaryotic phospholipid biosynthesis. Annu Rev Biochem 1995; 64:315–343. [DOI] [PubMed] [Google Scholar]
- 45.Jiang XC, Goldberg IJ, Park TS. Sphingolipids and cardiovascular diseases: lipoprotein metabolism, atherosclerosis and cardiomyopathy. Adv Exp Med Biol 2011; 721:19–39. [DOI] [PubMed] [Google Scholar]
- 46.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol 2008; 9:162–176. [DOI] [PubMed] [Google Scholar]
- 47.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 2011; 13:125–137. [DOI] [PubMed] [Google Scholar]
- 48.Brown SA. Circadian Metabolism: From Mechanisms to Metabolomics and Medicine. Trends Endocrinol Metab 2016; 27:415–426. [DOI] [PubMed] [Google Scholar]
- 49.Eckel-Mahan K, Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol Rev 2013; 93:107–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi JS. Molecular components of the circadian clock in mammals. Diabetes Obes Metab 2015; 17 Suppl 1:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi JS, Shimomura K, Kumar V. Searching for genes underlying behavior: lessons from circadian rhythms. Science 2008; 322:909–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 2014; 111:16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirano A, Fu YH, Ptacek LJ. The intricate dance of post-translational modifications in the rhythm of life. Nat Struct Mol Biol 2016; 23:1053–1060. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, Duguay D, Bedard N, Rachalski A, Baquiran G, Na CH et al. Regulation of behavioral circadian rhythms and clock protein PER1 by the deubiquitinating enzyme USP2. Biol Open 2012; 1:789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y, Duguay D, Fahrenkrug J, Cermakian N, Wing SS. USP2 regulates the intracellular localization of PER1 and circadian gene expression. J Biol Rhythms 2014; 29:243–256. [DOI] [PubMed] [Google Scholar]
- 56.Scoma HD, Humby M, Yadav G, Zhang Q, Fogerty J, Besharse JC. The de-ubiquitinylating enzyme, USP2, is associated with the circadian clockwork and regulates its sensitivity to light. PLoS One 2011; 6:e25382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong X, Buelow K, Guha A, Rausch R, Yin L. USP2a protein deubiquitinates and stabilizes the circadian protein CRY1 in response to inflammatory signals. J Biol Chem 2012; 287:25280–25291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sussman W, Stevenson M, Mowdawalla C, Mota S, Ragolia L, Pan X. BMAL1 controls glucose uptake through paired-homeodomain transcription factor 4 in differentiated Caco-2 cells. Am J Physiol Cell Physiol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimomura K, Kumar V, Koike N, Kim TK, Chong J, Buhr ED et al. Usf1, a suppressor of the circadian Clock mutant, reveals the nature of the DNA-binding of the CLOCK:BMAL1 complex in mice. Elife 2013; 2:e00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stratmann M, Suter DM, Molina N, Naef F, Schibler U. Circadian Dbp transcription relies on highly dynamic BMAL1-CLOCK interaction with E boxes and requires the proteasome. Mol Cell 2012; 48:277–287. [DOI] [PubMed] [Google Scholar]
- 61.Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell 2003; 11:695–707. [DOI] [PubMed] [Google Scholar]
- 62.Yoshitane H, Takao T, Satomi Y, Du NH, Okano T, Fukada Y. Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol Cell Biol 2009; 29:3675–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spengler ML, Kuropatwinski KK, Schumer M, Antoch MP. A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle 2009; 8:4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kwak Y, Jeong J, Lee S, Park YU, Lee SA, Han DH et al. Cyclin-dependent kinase 5 (Cdk5) regulates the function of CLOCK protein by direct phosphorylation. J Biol Chem 2013; 288:36878–36889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone-Corsi P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS One 2010; 5:e8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamaru T, Hattori M, Honda K, Nakahata Y, Sassone-Corsi P, van der Horst GT et al. CRY Drives Cyclic CK2-Mediated BMAL1 Phosphorylation to Control the Mammalian Circadian Clock. PLoS Biol 2015; 13:e1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamaru T, Hirayama J, Isojima Y, Nagai K, Norioka S, Takamatsu K et al. CK2alpha phosphorylates BMAL1 to regulate the mammalian clock. Nat Struct Mol Biol 2009; 16:446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J Biol Chem 2002; 277:17248–17254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J, Lee Y, Lee MJ, Park E, Kang SH, Chung CH et al. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol 2008; 28:6056–6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gossan NC, Zhang F, Guo B, Jin D, Yoshitane H, Yao A et al. The E3 ubiquitin ligase UBE3A is an integral component of the molecular circadian clock through regulating the BMAL1 transcription factor. Nucleic Acids Res 2014; 42:5765–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008; 134:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 2006; 125:497–508. [DOI] [PubMed] [Google Scholar]
- 73.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 2007; 450:1086–1090. [DOI] [PubMed] [Google Scholar]
- 74.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012; 338:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol 2010; 17:1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 2011; 333:1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duong HA, Weitz CJ. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol 2014; 21:126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grechez-Cassiau A, Feillet C, Guerin S, Delaunay F. The hepatic circadian clock regulates the choline kinase alpha gene through the BMAL1-REV-ERBalpha axis. Chronobiol Int 2015; 32:774–784. [DOI] [PubMed] [Google Scholar]
- 79.Ma D, Liu T, Chang L, Rui C, Xiao Y, Li S et al. The Liver Clock Controls Cholesterol Homeostasis through Trib1 Protein-mediated Regulation of PCSK9/Low Density Lipoprotein Receptor (LDLR) Axis. J Biol Chem 2015; 290:31003–31012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marcheva B, Ramsey KM, Peek CB, Affinati A, Maury E, Bass J. Circadian clocks and metabolism. Handb Exp Pharmacol 2013;127–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maury E, Hong HK, Bass J. Circadian disruption in the pathogenesis of metabolic syndrome. Diabetes Metab 2014; 40:338–346. [DOI] [PubMed] [Google Scholar]
- 82.Peek CB, Ramsey KM, Marcheva B, Bass J. Nutrient sensing and the circadian clock. Trends Endocrinol Metab 2012; 23:312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 2009; 106:21453–21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, Kraut-Cohen J et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab 2014; 19:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A 2009; 106:9890–9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yao ZM, Vance DE. Reduction in VLDL, but not HDL, in plasma of rats deficient in choline. Biochem Cell Biol 1990; 68:552–558. [DOI] [PubMed] [Google Scholar]
- 87.Li Z, Vance DE. Phosphatidylcholine and choline homeostasis. J Lipid Res 2008; 49:1187–1194. [DOI] [PubMed] [Google Scholar]
- 88.Gorne LD, costa-Rodriguez VA, Pasquare SJ, Salvador GA, Giusto NM, Guido ME. The mouse liver displays daily rhythms in the metabolism of phospholipids and in the activity of lipid synthesizing enzymes. Chronobiol Int 2015; 32:11–26. [DOI] [PubMed] [Google Scholar]
- 89.van MG, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 2008; 9:112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer 2011; 11:835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramirez de MA, Gallego-Ortega D, Sarmentero-Estrada J, Lagares D, Gomez Del PT, Bandres E et al. Choline kinase as a link connecting phospholipid metabolism and cell cycle regulation: implications in cancer therapy. Int J Biochem Cell Biol 2008; 40:1753–1763. [DOI] [PubMed] [Google Scholar]
- 92.Ikemoto A, Fukuma A, Fujii Y, Okuyama H. Diurnal rhythms of retinal phospholipid synthetic enzymes are retained but their activities are decreased in rats under alpha-linolenic acid deficiency. Arch Biochem Biophys 2000; 383:108–113. [DOI] [PubMed] [Google Scholar]
- 93.Garbarino-Pico E, Carpentieri AR, Castagnet PI, Pasquare SJ, Giusto NM, Caputto BL et al. Synthesis of retinal ganglion cell phospholipids is under control of an endogenous circadian clock: daily variations in phospholipid-synthesizing enzyme activities. J Neurosci Res 2004; 76:642–652. [DOI] [PubMed] [Google Scholar]
- 94.Garbarino-Pico E, Valdez DJ, Contin MA, Pasquare SJ, Castagnet PI, Giusto NM et al. Rhythms of glycerophospholipid synthesis in retinal inner nuclear layer cells. Neurochem Int 2005; 47:260–270. [DOI] [PubMed] [Google Scholar]
- 95.costa-Rodriguez VA, Marquez S, Salvador GA, Pasquare SJ, Gorne LD, Garbarino-Pico E et al. Daily rhythms of glycerophospholipid synthesis in fibroblast cultures involve differential enzyme contributions. J Lipid Res 2013; 54:1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Arriba Zerpa GA, Guido ME, Bussolino DF, Pasquare SJ, Castagnet PI, Giusto NM et al. Light exposure activates retina ganglion cell lysophosphatidic acid acyl transferase and phosphatidic acid phosphatase by a c-fos-dependent mechanism. J Neurochem 1999; 73:1228–1235. [DOI] [PubMed] [Google Scholar]
- 97.Marquez S, Crespo P, Carlini V, Garbarino-Pico E, Baler R, Caputto BL et al. The metabolism of phospholipids oscillates rhythmically in cultures of fibroblasts and is regulated by the clock protein PERIOD 1. FASEB J 2004; 18:519–521. [DOI] [PubMed] [Google Scholar]
- 98.Ruggiero L, Connor MP, Chen J, Langen R, Finnemann SC. Diurnal, localized exposure of phosphatidylserine by rod outer segment tips in wild-type but not Itgb5−/− or Mfge8−/− mouse retina. Proc Natl Acad Sci U S A 2012; 109:8145–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu S, Brown JD, Stanya KJ, Homan E, Leidl M, Inouye K et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature 2013; 502:550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 2007; 6:414–421. [DOI] [PubMed] [Google Scholar]
- 101.Guan D, Xiong Y, Borck PC, Jang C, Doulias PT, Papazyan R et al. Diet-Induced Circadian Enhancer Remodeling Synchronizes Opposing Hepatic Lipid Metabolic Processes. Cell 2018; 174:831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jang YS, Kang YJ, Kim TJ, Bae K. Temporal expression profiles of ceramide and ceramide-related genes in wild-type and mPer1/mPer2 double knockout mice. Mol Biol Rep 2012; 39:4215–4221. [DOI] [PubMed] [Google Scholar]
- 103.Gooley JJ, Chua EC. Diurnal regulation of lipid metabolism and applications of circadian lipidomics. J Genet Genomics 2014; 41:231–250. [DOI] [PubMed] [Google Scholar]
- 104.Pan X, Hussain MM. Gut triglyceride production. Biochim Biophys Acta 2012; 1821:727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A 2012; 109:2625–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumar JP, Challet E, Kalsbeek A. Circadian rhythms in glucose and lipid metabolism in nocturnal and diurnal mammals. Mol Cell Endocrinol 2015; 418 Pt 1:74–88. [DOI] [PubMed] [Google Scholar]
- 107.Navara KJ, Nelson RJ. The dark side of light at night: physiological, epidemiological, and ecological consequences. J Pineal Res 2007; 43:215–224. [DOI] [PubMed] [Google Scholar]
- 108.Ruf T, Arnold W. Daily and Seasonal Rhythms in Human Mucosa Phospholipid Fatty Acid Composition. J Biol Rhythms 2015; 30:331–341. [DOI] [PubMed] [Google Scholar]
- 109.Nelson RJ. Seasonal immune function and sickness responses. Trends Immunol 2004; 25:187–192. [DOI] [PubMed] [Google Scholar]
- 110.Chua EC, Shui G, Cazenave-Gassiot A, Wenk MR, Gooley JJ. Changes in Plasma Lipids during Exposure to Total Sleep Deprivation. Sleep 2015; 38:1683–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singh R, Sharma S, Singh RK, Mahdi AA, Singh RK, Lee GC et al. Effect of gender, age, diet and smoking status on chronomics of circulating plasma lipid components in healthy Indians. Clin Chim Acta 2016; 459:10–18. [DOI] [PubMed] [Google Scholar]
- 112.Singh R, Sharma S, Singh RK, Cornelissen G. Circadian Time Structure of Circulating Plasma Lipid Components in Healthy Indians of Different Age Groups. Indian J Clin Biochem 2016; 31:215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Isherwood CM, van d, V, Johnston JD, Skene DJ. Twenty-four-hour rhythmicity of circulating metabolites: effect of body mass and type 2 diabetes. FASEB J 2017; 31:5557–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gu F, Klerman EB, Kim S, Moore S, Yu K, Albert PS et al. Diurnal variation of metabolites in three individual participants. Chronobiol Int 2019; 36:332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gu F, Xu S, Devesa SS, Zhang F, Klerman EB, Graubard BI et al. Longitude Position in a Time Zone and Cancer Risk in the United States. Cancer Epidemiol Biomarkers Prev 2017; 26:1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pan X, Terada T, Okuda M, Inui K. Altered diurnal rhythm of intestinal peptide transporter by fasting and its effects on the pharmacokinetics of ceftibuten. J Pharmacol Exp Ther 2003; 307:626–632. [DOI] [PubMed] [Google Scholar]
- 117.Pan X, Terada T, Irie M, Saito H, Inui K. Diurnal rhythm of H+-peptide cotransporter in rat small intestine. Am J Physiol Gastrointest Liver Physiol 2002; 283:G57–G64. [DOI] [PubMed] [Google Scholar]
- 118.Pan X, Terada T, Okuda M, Inui K. The diurnal rhythm of the intestinal transporters SGLT1 and PEPT1 is regulated by the feeding conditions in rats. J Nutr 2004; 134:2211–2215. [DOI] [PubMed] [Google Scholar]
- 119.Miettinen TA. Diurnal variation of LDL and HDL cholesterol. Ann Clin Res 1980; 12:295–298. [PubMed] [Google Scholar]
- 120.Saito Y, Yoshida S, Nakaya N, Hata Y, Goto Y. Comparison between morning and evening doses of simvastatin in hyperlipidemic subjects. A double-blind comparative study. Arterioscler Thromb 1991; 11:816–826. [DOI] [PubMed] [Google Scholar]
- 121.De GA, Mallozzi MA, Fabbian F, Portaluppi F, Manfredini R. Circadian rhythms and medical diseases: does it matter when drugs are taken? Eur J Intern Med 2013; 24:698–706. [DOI] [PubMed] [Google Scholar]
- 122.Cilla DD Jr., Gibson DM, Whitfield LR, Sedman AJ. Pharmacodynamic effects and pharmacokinetics of atorvastatin after administration to normocholesterolemic subjects in the morning and evening. J Clin Pharmacol 1996; 36:604–609. [DOI] [PubMed] [Google Scholar]
- 123.Wallace A, Chinn D, Rubin G. Taking simvastatin in the morning compared with in the evening: randomised controlled trial. BMJ 2003; 327:788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wright DF, Pavan K,V, Al-Sallami HS, Duffull SB. The influence of dosing time, variable compliance and circadian low-density lipoprotein production on the effect of simvastatin: simulations from a pharmacokinetic-pharmacodynamic model. Basic Clin Pharmacol Toxicol 2011; 109:494–498. [DOI] [PubMed] [Google Scholar]


