Abstract
Latent transforming growth factor β (TGF-β) binding protein 2 (LTBP-2) is an integral component of elastin-containing microfibrils. We studied the expression of LTBP-2 in the developing mouse and rat by in situ hybridization, using tropoelastin expression as a marker of tissues participating in elastic fiber formation. LTBP-2 colocalized with tropoelastin within the perichondrium, lung, dermis, large arterial vessels, epicardium, pericardium, and heart valves at various stages of rodent embryonic development. Both LTBP-2 and tropoelastin expression were seen throughout the lung parenchyma and within the cortex of the spleen in the young adult mouse. In the testes, LTBP-2 expression was seen within lumenal cells of the epididymis in the absence of tropoelastin. Collectively, these results imply that LTBP-2 plays a structural role within elastic fibers in most cases. To investigate its importance in development, mice with a targeted disruption of the Ltbp2 gene were generated. Ltbp2−/− mice die between embryonic day 3.5 (E3.5) and E6.5. LTBP-2 expression was not detected by in situ hybridization in E6.5 embryos but was detected in E3.5 blastocysts by reverse transcription-PCR. These results are not consistent with the phenotypes of TGF-β knockout mice or mice with knockouts of other elastic fiber proteins, implying that LTBP-2 performs a yet undiscovered function in early development, perhaps in implantation.
The elastic resilience and structural integrity of the lungs, skin, and large blood vessels of vertebrate animals are imparted by elastic fibers. These fibers consist of the protein elastin and a network of 10- to 12-nm microfibrils, which are composed of a number of glycoproteins (1, 33). In elastic fibers, microfibrils are located around the periphery of the amorphous elastin component of the fiber, as has been shown at the electron microscopic level (4, 10). Microfibrils are also present in tissues devoid of elastin, such as the ocular ciliary zonules and the periodontal ligament. In these structures, the microfibrils are nearly indistinguishable from microfibrils found in elastic tissue. In the skin, microfibrils extend from the epidermis into the dermis. Superficially, these fibers are not associated with elastin but become associated with elastin as they traverse the dermis.
The largest microfibrillar proteins identified thus far are the fibrillins. Two fibrillins have been isolated and demonstrated to be integral structural components of elastic fibers (35, 42). Both are 350-kDa molecules rich in six-cysteine epidermal growth factor-like repeats, which bind calcium through hydroxylation of asparagine and aspartic acid residues. Both also contain unique eight-cysteine repeats which are also found in the latent transforming growth factor β (TGF-β) binding proteins (12). Marfan syndrome is an autosomal dominant disorder linked to the fibrillin 1 gene on chromosome 15 (3, 9, 15, 18). This disease is characterized by skeletal, ocular, and cardiovascular abnormalities. Another disorder, congenital contractural arachnodactyly, is linked to the fibrillin 2 gene on chromosome 5 (15).
The latent TGF-β binding proteins (LTBPs) share a high degree of homology with the fibrillins. Currently, there are four members of the LTBP family. All have several copies of six-cysteine epidermal growth factor-like repeats as well as a more limited number of eight-cysteine repeats unique to this family of proteins and the fibrillins. While all of the LTBPs appear to be constituents of the extracellular matrix, LTBP-1 and -2 may be unique in their localization to elastin-containing microfibrils. The major portion of LTBP-2 in elastic tissues is strongly bound to microfibrils, as 6 M guanidine and dithiothreitol are required to extract it (7). The requirement of guanidine for extraction suggests that LTBP-2 is integrally associated with the elastic fiber matrix, and the requirement of dithiothreitol suggests that the covalent attachment occurs at least in part through disulfide bonding. Taken together with its high sequence similarity with the fibrillins, which are known structural components of elastic fibers, it is likely that LTBP-2 performs a structural role within elastic fibers.
The TGF-βs are a family of multifunctional polypeptides which affect the growth, differentiation, adhesion, migration, and matrix synthesizing capacity of a variety of cell types (17, 19, 39). Most cell types that produce TGF-β secrete this protein in an inactive form (21). TGF-β noncovalently associated with its propeptide (also called the latency associated peptide [LAP]) is referred to as the small latent complex. In order for TGF-β to exert cellular effects, it must dissociate from the LAP to bind its receptor. TGF-β can also form a large latent complex when a molecule of LTBP-1 covalently bound to the LAP of the small latent complex via a disulfide bond is secreted (8, 12, 20, 34). Incorporation of the large latent complex into the extracellular matrix requires cross-linking by transglutaminase (13, 24), while release of this complex from the matrix is protease dependent (40, 41). Activation of complexed TGF-β appears to involve the action of cell-surface-associated plasmin, which cleaves the LAP and disrupts the noncovalent interaction between the mature TGF-β and the LAP (6, 14, 25). While this paradigm has developed exclusively with LTBP-1, it has been suggested that human (23) and murine (5) LTBP-2 can form a large latent complex with TGF-β in cotransfection experiments.
The purpose of this study was to determine the temporal and spatial pattern of LTBP-2 expression in the mouse and rat in order to gain insight into the likely function of LTBP-2 in development and to determine the consequences of eliminating LTBP-2 expression in mice by targeted disruption of the gene. LTBP-2 expression was localized previously in embryonic day 16.5 (E16.5) mouse embryos (5), but expression at other embryonic time points as well as in adult tissues was not examined. In this study, we demonstrate coexpression of LTBP-2 with tropoelastin in elastogenic tissues, suggestive of a structural role for LTBP-2 in these tissues. However, inactivation of the gene was found to result in lethality early in development, suggesting that LTBP-2 plays a role unrelated to elastic fiber homeostasis or TGF-β regulation, perhaps as a structural component of elastin-free microfibrils during implantation.
MATERIALS AND METHODS
In situ hybridization.
Embryos were harvested from timed pregnant Swiss Webster mice (Taconic). Tissues were also harvested from 5-week-old Swiss Webster mice. Embryos or adult tissues were fixed for 1 to 3 days in 10% buffered formalin (DeRuscio and Associates, St. Louis, Mo.). Tissues were processed by shaking 2 times for 15 min each time in phosphate-buffered saline, followed by gentle agitation in 30% ethanol for 1 h, 50% ethanol for 1 h, and 70% ethanol overnight prior to processing. All solutions were treated with diethylpyrocarbonate (0.1%). A fragment of the mouse LTBP-2 cDNA (nucleotides 777 to 1140) was generated by PCR using the primers 5′GCAATTAACCCTCACTAAAGGCCACCATCACCACCTCCATC3′ and 5′CGTAATACGACTCACTATAGGAAGCCAGACTTGGGGTCA3′, in which the binding sites for T7 or T3 RNA polymerases were incorporated into the primers. Sense (T3) and antisense (T7) probes were generated by using 35S-UTP and 0.1 μg of template DNA with the Riboprobe system (Promega, Madison, Wis.). Synthesis of an in situ hybridization probe for rat tropoelastin was done as described previously (29). In situ hybridization was carried out as described previously (31), with exposure times ranging from 2 to 12 weeks.
Northern hybridization.
The LTBP-2 template DNA used for in situ hybridization was labeled with [32P]dCTP using the Redi-Prime system (Amersham Pharmacia Biotech, Piscataway, N.J.). A multiple-tissue Northern blot including eight adult mouse tissue sources was purchased from Clontech (Palo Alto, Calif.). Northern hybridization was done according to the manufacturer's instructions.
Construction of the LTBP-2 targeting vector.
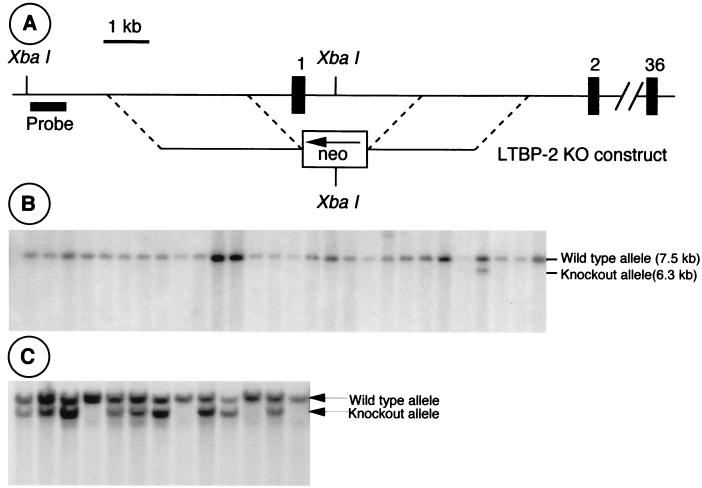
A mouse 129 genomic library in a bacteriophage P1 vector (Genome Systems, St. Louis, Mo.) was screened by PCR using primers 1F (5′TGATGGGGACAAGTCATGCCC3′) and 1R (5′ATGGCTTCTCCGAGTCTGGAC3′), which were predicted and subsequently shown to be located in exon 1. Four clones were identified, one of which was digested with ApaI or KpnI. Fragments were subcloned into pBluescript SK(−) (Stratagene, La Jolla, Calif.), and subclones containing exon 1 were identified by colony hybridization. Overlapping 6.0-kb ApaI and 5.2-kb KpnI subclones were characterized by restriction mapping, PCR, and DNA sequencing. A targeting construct was generated in pBluescript SK(−) that included 3.5 kb of 5′ homology and 2.7 kb of 3′ homology. The 5′ homology consisted of an EcoRI fragment from the ApaI subclone that encompassed part of the Ltbp2 promoter, ending ∼800 bp 5′ to exon 1. The 3′ homology consisted of an ApaI fragment from the KpnI subclone and encompassed part of intron 1, beginning about 1 kb downstream of exon 1. A cassette containing the neomycin phosphotransferase cDNA driven by the phosphoglycerate kinase promoter (PGK-neo) was used to replace the intervening region, which includes exon 1. The targeting construct was linearized with NotI prior to electroporation of embryonic stem cells.
Generation of Ltbp2 heterozygous mice.
The linearized targeting construct was used to electroporate clone RW4 129/SvJ embryonic stem cells. G418-resistant clones were analyzed by Southern blot analysis of XbaI-digested genomic DNA, probed with a 1-kb BamHI fragment located 5′ to the targeting construct. Two heterozygous clones representing homologous recombinants were identified out of 350 that were screened. Heterozygous ES cells were injected into C57BL/6J blastocysts, which were transferred into the uteri of pseudopregnant females. Chimeric males were mated initially to C57BL/6J females, and agouti offspring were screened by Southern blotting of tail genomic DNA for germline transmission of the targeted Ltbp2tm1Ship allele (i.e., Ltbp2−/− allele). Once productive chimeric males were identified, these mice were bred to 129/SvJ females, and heterozygous offspring on a pure 129/SvJ background were identified by Southern blotting.
Identification of Ltbp2−/− embryos.
Embryos at E12 or older embryos from heterozygous matings were genotyped by Southern blotting. Embryos between E6.5 and E11 were genotyped by PCR. In the case of E6.5 embryos, embryos were removed from the decidua by microdissection. The ectoplacental cone was removed and the embryos were digested for 2 h in tail buffer (50 mM Tris [pH 8], 25 mM EDTA, 100 mM NaCl, 1% Triton X-100) containing 2 mg of proteinase K per ml at 55°C in a volume of 20 μl. After boiling for 10 min, 2 μl of the cleared lysate was used for PCR with the primers 5′AGAAGCAGTTCATCTGGGTC3′ and 5′CTCCTTCCTCGTCTATGCTC3′ located 1.2 kb 5′ and 1.2 kb 3′ to exon 1, respectively. Klentaq LA polymerase (Sigma, St. Louis, Mo.) was used with an annealing temperature of 58°C for 35 cycles.
Genotyping of blastocyst-stage embryos was done with two rounds of PCR. Blastocysts were collected by flushing the uteri with phosphate-buffered saline (PBS) and treated briefly with acidic Tyrode buffer (11) to remove the zona pellucida. Blastocysts were incubated with 200 μg of proteinase K per ml in Klentaq LA PCR buffer containing all PCR components except Klentaq LA for 2 h at 55°C. Samples were heated for 10 min at 85°C, and 1 μl of Klentaq LA was added along with mineral oil. The first round of PCR was for 40 cycles at an annealing temperature of 58°C with the primers described above used to genotype later embryos. A 4-μl volume of this reaction mixture was used in a second PCR which incorporated two sets of primers, one set which amplifies exon 1 of Ltbp2 which is only present in the normal allele and one set which amplifies part of the neomycin cassette only present in the targeted allele. The exon 1 primers 1F and 1R (described above) amplify a 289-bp product, while the neo primers (5′ATGATTGAACAAGATGGATTGCAC3′ and 5′TTCGTCCAGATCATCCTGATCGAC3′) amplify a 500-bp product. The second round of PCR was for 35 cycles at an annealing temperature of 58°C.
Detection of LTBP-2 expression by reverse transcription (RT)-PCR.
Wild-type blastocysts were isolated at E3.5 by flushing the uterine horns of timed-pregnant mice with PBS. Blastocysts were treated briefly in acidic Tyrode buffer to remove the zona pellucida and transferred to PBS. Groups of five blastocysts were collected in a minimal volume of PBS (∼10 μl), and diethylpyrocarbonate-treated water was added to bring the final volume to 25 μl. A single tube was subjected to three freeze-thaw cycles, boiled for 2 min, and centrifuged for 5 min. The supernatant was divided into two tubes. Reverse transcription was carried out on one sample with random hexamers using the Superscript preamplification system following the manufacturer's instructions (Life Technologies, Grand Island, N.Y.). The other sample was mock treated with all components except reverse transcriptase. Following cDNA synthesis, PCR was done on both samples for 45 cycles (94°C for 1 min, 55°C for 2 min, and 72°C for 1 min) using the primers 1F (see above) and 2R (5′GTTTGATACAGTGGTTGGTGC3′), located in exons 1 and 2, respectively. Specific products were visualized by Southern blotting, with a cDNA fragment encompassing exons 1 and 2 used as the probe.
RESULTS
Expression of LTBP-2 in mid- to late-gestation embryos.
Because LTBP-2 may play at least two important roles in development, as both a structural component of the elastic fiber and a regulator of TGF-β activity, we anticipated that its expression would reflect these functions. To gain insight into the function(s) of this protein, we investigated LTBP-2 expression in the developing mouse and rat by in situ hybridization. Expression of LTBP-2 was not detected at E10.5 (data not shown). Expression of LTBP-2 was first detected at E13.5 in perichondrial regions surrounding the developing vertebrae and ribs (Fig. 1). Tropoelastin is also expressed by the same cells, as well as in other perichondrial regions which do not appear to express LTBP-2. This may reflect differences in microfibrillar composition in different elastic fibers. Some tropoelastin-positive regions which did not express LTBP-2 were also negative for fibrillin 1 expression (not shown). LTBP-2 expression was also seen in other perichondrial regions at this stage, such as in the developing limb buds (data not shown). Sense-strand probes for both LTBP-2 and tropoelastin showed no specific hybridization in any tissues investigated (data not shown).
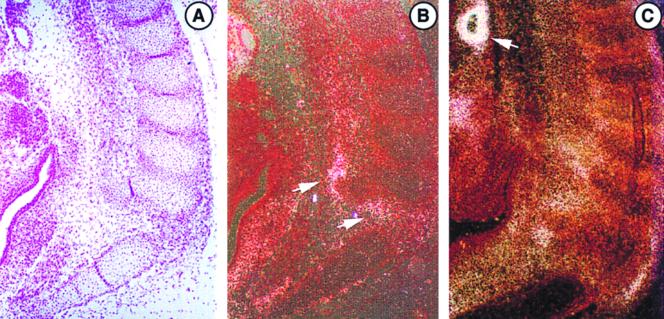
FIG. 1.
Expression of LTBP-2 and tropoelastin mRNAs in E13.5 mouse embryos. In situ hybridization was done to localize expression of each. (A) Bright-field staining of developing ribs. (B) Section hybridized to a LTBP-2 antisense probe showing perichondrial expression (arrows). (C) Section hybridized to a tropoelastin antisense probe, showing perichondrial expression similar to LTBP-2 as well as arterial expression (arrow). Sense probes showed no specific hybridization (not shown). Magnification, ×90.
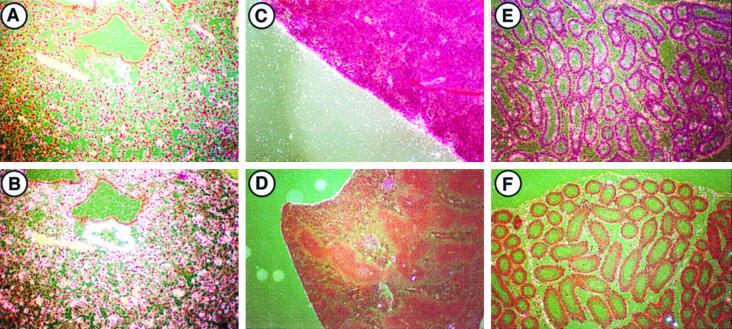
At E15.5, LTBP-2 and tropoelastin are both expressed in the snout, tongue, lungs, and dermis (Fig. 2). While tropoelastin expression is observed throughout perichondrial areas in the snout, LTBP-2 expression is more limited, consistent with the distribution seen in the perichondrium at E13.5. However, areas that express LTBP-2 also appear to express elastin, suggesting a structural role for LTBP-2 in the developing snout. In the lungs and the dermis at E15.5, LTBP-2 and tropoelastin are expressed only by a few cell layers directly underlying the epithelial basement membrane (lining airways in the case of the lung). In both the lungs and the skin, the majority of mesenchymal cells do not express either LTBP-2 or tropoelastin to an appreciable extent. Both tropoelastin (Fig. 2D) and LTBP-2 (data not shown) are expressed within the large blood vessels as well. At E17.5, the most prominent tissue coexpressing both LTBP-2 and tropoelastin is large arterial vessels (Fig. 3). Both mRNAs are expressed primarily in the medial layer of the aorta and other arterial vessels composed largely of smooth muscle cells. Fibrillin 2 has also been shown to be expressed within the aorta of the developing human embryo (42). Coexpression of LTBP-2 and tropoelastin in a number of elastogenic tissues within developing embryos, together with the identification of LTBP-2 as a structural component of elastic microfibrils in developing bovine tissues (7), strongly suggests that LTBP-2 plays a similar structural role within elastogenic tissues of the developing mouse.
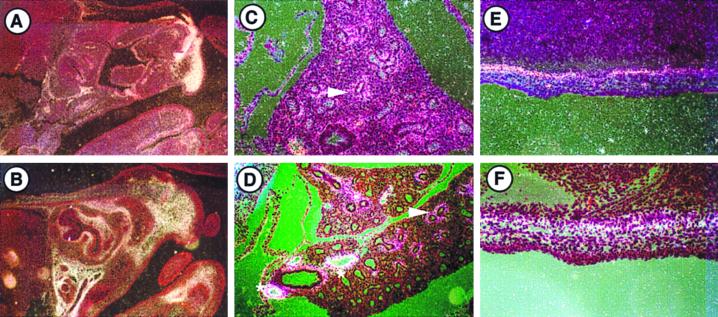
FIG. 2.
Expression of LTBP-2 and tropoelastin mRNAs in E15.5 mouse embryos. In situ hybridization was done to localize expression of LTBP-2 in the snout (A), lung (C), and dermis (E) and expression of tropoelastin in the snout (B), lung (D), and dermis (F). Perichondrial expression of both mRNAs is seen throughout the snout. Expression in the lung is observable in a limited number of cell layers of the subepithelial mesenchyme of developing airways (arrows), and intense tropoelastin expression in arterial vessels (asterisks) is evident. Both signals are also seen in the dermis. Sense probes showed no specific hybridization (not shown). Magnification for panels A and B, ×36. Magnification for panels C through F, ×180.
FIG. 3.
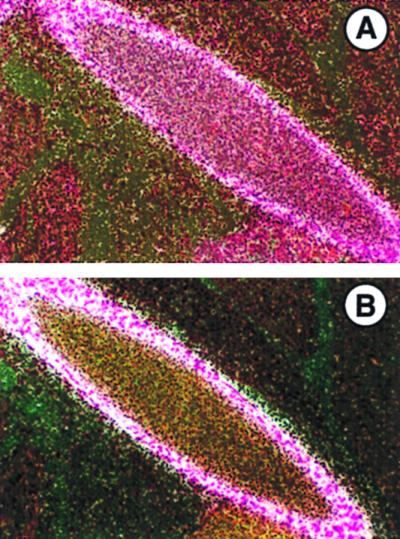
Expression of LTBP-2 (A) and tropoelastin (B) mRNAs in E17.5 mouse embryos. In situ hybridization was done to localize expression of each. Expression of both mRNAs is seen by medial cells of the descending aorta. Sense probes showed no specific hybridization (not shown). Magnification, ×200.
LTBP-2 expression was also examined in the developing rat. At E14 in the developing rat thorax (Fig. 4A), LTBP-2 expression is seen in the pericardium, epicardium, heart valves, large vessels, and lung. Expression in the pericardium and epicardium is consistent with a structural elastic role. As in the mouse at E15.5, expression in the lung is limited to a few mesenchymal cell layers underlying the airway epithelium, suggesting a role for LTBP-2 in lung growth or branching morphogenesis. This pattern is even more pronounced at E18 in the developing rat lung for LTBP-2 as well as for tropoelastin (Fig. 4B and C).
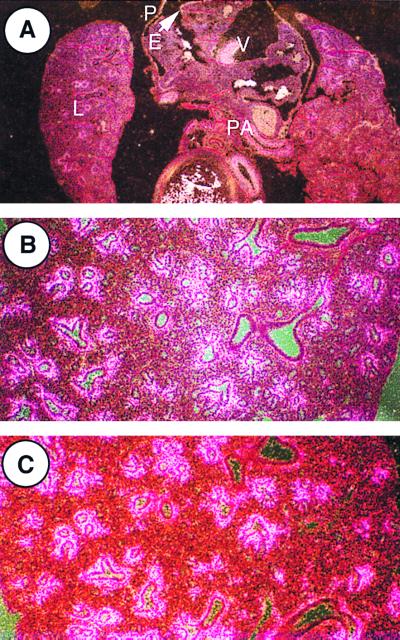
FIG. 4.
Expression of LTBP-2 and tropoelastin mRNAs in the developing rat thorax. In situ hybridization was done to localize expression of each. (A) Expression of LTBP-2 in the E14 rat thorax. Specific hybridization is seen in the lungs (L), the pericardium (P), the epicardium (E), the heart valves (V), and the pulmonary artery (PA), as well as in other large vessels. The pattern of expression is identical to that seen with tropoelastin probes (data not shown). Magnification, ×36. (B) Expression of LTBP-2 in the E18 rat lung. Expression in the lung is observable in a limited number of cell layers of the subepithelial mesenchyme of developing mid-sized airways, with no or minimal corresponding expression seen in the smallest and largest airways. Magnification, ×90. (C) Expression of tropoelastin in the E18 rat lung. The pattern of expression is identical to that of LTBP-2. Sense probes showed no specific hybridization (data not shown). Magnification, ×90.
Expression of LTBP-2 in young adult mice.
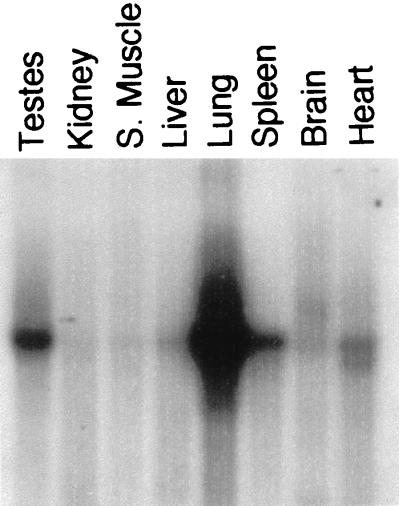
In order to guide in situ hybridization studies of LTBP-2 expression in adult mouse tissues, a multiple-tissue Northern blot prepared with poly(A)+ RNA from a variety of adult mouse tissues (Clontech) was probed for LTBP-2 (Fig. 5). LTBP-2 is most abundantly expressed in the adult lung. In fact, densitometric scanning indicates that the level of expression in lung is at least 10-fold higher than that in any other tissue examined. Significant levels of LTBP-2 mRNA are observed in the testes, spleen, and heart as well. LTBP-2 expression was examined in several tissues of 5-week-old mice by in situ hybridization. Both LTBP-2 and tropoelastin were expressed throughout the mesenchyme of the lungs of 5-week-old mice (Fig. 6A and B). Mice at 5 weeks of age are still growing, and it is therefore not surprising that one might still find elastin expression. Elastin synthesis in other mammals declines after birth, with little or no elastin synthesized during adulthood under physiologic conditions (26). The pattern of both tropoelastin and LTBP-2 expression in the 5-week-old lung is markedly different from that of the late-gestation lung. Expression of each is much more ubiquitous throughout the mesenchyme in the young adult mice. Expression is still abundant within large vessels, although much more so for tropoelastin than LTBP-2. Both are also expressed by cells underlying the fibrous capsule of the spleen (Fig. 6C and D).
FIG. 5.
Northern blot analysis of LTBP-2 expression in adult mouse tissues. A multiple tissue Northern blot including eight adult mouse tissue sources (Clontech) was hybridized to a LTBP-2 cDNA probe. A 7.5-kb mRNA is most abundantly detected in the lung, spleen, and testes.
FIG. 6.
Expression of LTBP-2 and tropoelastin mRNAs in 5-week-old mouse tissues. In situ hybridization was done to localize expression of LTBP-2 in the lung (A) and spleen (C) and expression of tropoelastin in the lung (B) and spleen (D). Coexpression of LTBP-2 and tropoelastin is seen throughout the mesenchyme and vasculature of the lung and the capsule of the spleen. In the testes, LTBP-2 (E) but not tropoelastin (F) is expressed by lumenal cells of the epididymis. Sense probes showed no specific hybridization (data not shown). Magnification for panels A, B, E, and F, ×90. Magnification for panels C and D, ×36.
The expression patterns of LTBP-2 and tropoelastin are clearly distinct in the testes (Fig. 6E and F). In the epididymis, tropoelastin expression is weak and is observable only in cells lining the outside of the spermatic ducts. In contrast, intense LTBP-2 expression is observed in cells within the lumen of the epididymis. This is the first clear example of LTBP-2 expression where the function of LTBP-2 is likely distinct from its role as a structural component of elastic fibers. However, neither TGF-β1, -β2, nor -β3 appears to be coexpressed with LTBP-2 in the testes by in situ hybridization (data not shown). TGF-β expression within the adult mouse testes has been reported (38). The potential function of LTBP-2 in the testes is therefore unclear. LTBP-2 signal was not detected in the heart by in situ hybridization (data not shown). Expression of LTBP-2 in the liver, brain, ovary, oviduct, uterus, and kidney was also undetectable (data not shown).
Ltbp2−/− mice.
In order to directly examine the role of LTBP-2 in mouse development, a targeting construct was made to eliminate its expression in mice (Fig. 7A). Exon 1, as well, the proximal regions of the promoter and intron 1, was replaced in the targeting vector with a PGK-neo cassette. The targeting vector was introduced into 129/SvJ embryonic stem cells by electroporation. DNA from G418-resistant ES clones was analyzed by Southern blotting of XbaI-digested genomic DNA, using a probe located 5′ to the targeting construct (Fig. 7B and C). Two homologous recombinants were identified out of 350 screened and were used to create chimeric mice. Chimeric males were mated to C57BL/6J and 129/SvJ females to generate heterozygotes on mixed and pure genetic backgrounds, respectively. Heterozygous mice appear phenotypically normal in all respects. Additionally, the architecture of elastic tissues such as the lung and aorta appears normal in heterozygotes by Verhoeff Van Gieson staining, which stains elastic fibers (data not shown). Over 400 live offspring of heterozygous matings were screened by Southern blotting. No live Ltbp2−/− mice were identified, and the ratio of heterozygous to normal mice was approximately 2:1, indicating that the knockout resulted in an embryonic lethal phenotype (Fig. 7).
FIG. 7.
Generation of Ltbp2−/− mice. (A) The LTBP-2 targeting construct. The 5′ and 3′ homologies were derived from the promoter and intron 1, respectively. The intervening region including exon 1, which contains the ATG, is replaced with a PGK-neo cassette. (B) Southern blot of ES cell DNA screened as described for panel A, showing one targeted clone. (C) Live progeny of heterozygous matings were genotyped by Southern blotting. No Ltbp2−/− mice lived to birth.
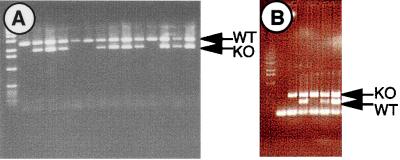
To determine the point at which Ltbp2−/− embryos die, timed matings of heterozygous mice were carried out, and at E10.5 through E19.5, progeny were genotyped by Southern blotting. Again, no Ltbp2−/− embryos were identified in ∼20 litters examined. Younger postimplantation embryos were genotyped by PCR. At E6.5, no Ltbp2−/− embryos were identified among six litters examined (Fig. 8A). These were the youngest postimplantation embryos that we could isolate. However, knockout embryos were identified among E3.5 preimplantation blastocysts (Fig. 8B), implying that LTBP-2 may play an essential developmental role during implantation (E4.5), much earlier than we had initially detected its expression.
FIG. 8.
Genotyping of early postimplantation and preimplantation embryos. (A) E6.5 early-postimplantation embryos from heterozygous matings were genotyped by PCR. No Ltbp2−/− embryos were identified at this stage. (B) E3.5 preimplantation blastocysts, in which knockout embryos were identified, were genotyped by PCR in an assay similar to that used for panel A. The lowest band in all lanes corresponds to unincorporated PCR primers. The first lane following the molecular weight standards is a mock PCR in the absence of target DNA. KO, knockout; WT, wild type.
LTBP-2 expression by preimplantation and early postimplantation embryos.
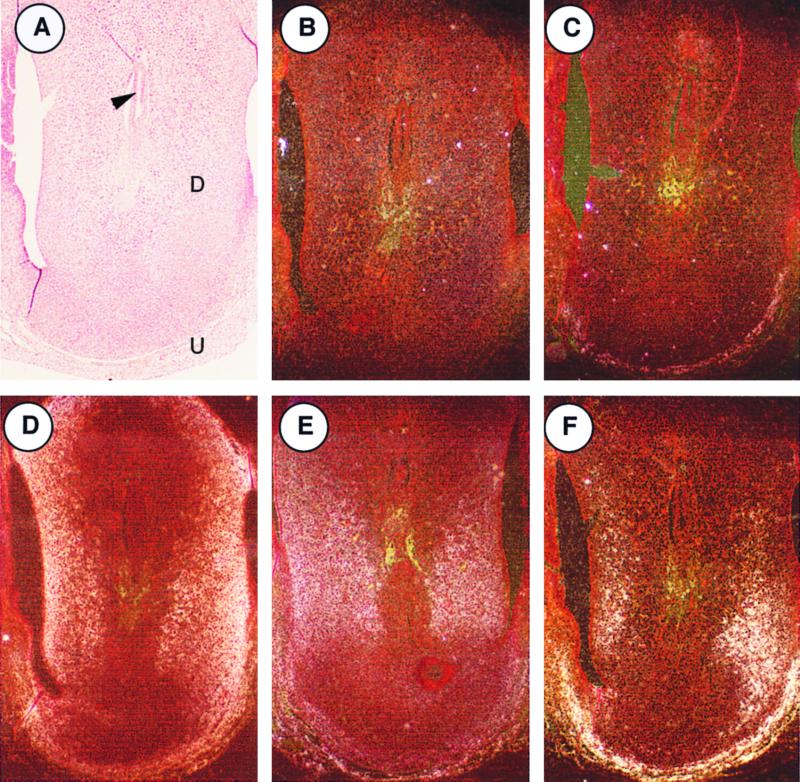
In situ hybridization was done to determine whether LTBP-2 is expressed by E6.5 postimplantation embryos (Fig. 9). LTBP-2 does not appear to be expressed to an appreciable extent in the embryo itself at E6.5. However, much of the decidual stroma, which is maternally derived, as well as the uterine muscle is weakly positive for LTBP-2 expression by in situ hybridization. In situ hybridization for a number of other elastic fiber mRNAs including MAGP-2, fibrillin 1, and tropoelastin revealed similar patterns of expression of these components. Expression of LTBP-2 by preimplantation blastocysts (E3.5) was investigated by RT-PCR (Fig. 10). Primers that span exons 1 and 2 were used so that PCR products arising from genomic DNA (7 kb) would be much larger than the mRNA-derived product (0.5 kb) and undetectable under the conditions used. A specific band of the predicted size hybridizing to an LTBP-2 probe was detected only in the presence of reverse transcriptase. The particular cell type within the blastocyst responsible for this expression is unknown, as in situ hybridization experiments on blastocysts were not successful. However, LTBP-2 expression was not detected by Northern blotting of embryonic stem cell RNA, suggesting that trophectoderm cells express LTBP-2.
FIG. 9.
Expression of LTBP-2 mRNA at E6.5. LTBP-2 expression by E6.5 embryos was investigated by in situ hybridization. The bright-field image (A) and a section probed with a sense probe (B) are shown. Serial sections were probed for LTBP-2 (C) and other elastic fiber components, including MAGP-2 (D), fibrillin 1 (E), and tropoelastin (F). A signal for all of the elastic fiber components, including LTBP-2, is seen within the maternally derived decidual stroma (D) and the uterine muscle (U) but not within the embryo itself (arrow).
FIG. 10.
Expression of LTBP-2 by preimplantation embryos. LTBP-2 expression by E3.5 preimplantation blastocysts was investigated by RT-PCR. Blastocysts were subjected to reverse transcriptase (+RT) or mock reactions lacking reverse transcriptase (−RT) prior to PCR, as described in Materials and Methods. Southern blotting and hybridization with an LTBP-2 cDNA probe identified a band of the predicted size (473 bp) which was present only in samples treated with reverse transcriptase.
DISCUSSION
We have shown that LTBP-2 expression in the developing mouse largely parallels that of tropoelastin. Both are expressed at E13.5 in the perichondrium surrounding the developing vertebrae; at E15.5 in the snout, lungs, and dermis; and at E17.5 primarily in the aorta and other large vessels. Both are also coexpressed in the developing rat lung, pericardium, epicardium, and heart valves. In the young adult mouse, both are expressed in the capsule of the spleen and ubiquitously throughout the mesenchyme of the lung. In all of these cases, coexpression suggests a structural role for LTBP-2 in these tissues. Additionally, there are places where tropoelastin is expressed in the absence of LTBP-2, such as some perichondrial regions throughout the head of the developing mouse. This is not completely surprising considering the distribution of fibrillins 1 and 2 in the mouse. It is apparent that elastic microfibrils vary in composition between tissues, and our data suggests that not all elastic fibers include LTBP-2. The basis for these differences between tissues is not well understood and is a topic of investigation in several laboratories.
The only tissue where LTBP-2 was found to be expressed in the absence of tropoelastin expression was the testes of the young adult mouse, where it is expressed by cells lining the lumen of the epididymis. This pattern of expression suggests that LTBP-2 performs an alternative function in the testes, perhaps one involving the regulation of TGF-β activity. It is unlikely that LTBP-2 plays a structural role within elastin-free microfibrils within the testes, largely because expression occurs within lumenal cells and because fibrillin 1 is not expressed to any appreciable extent here either (data not shown). TGF-β is known to be expressed within the adult testes (38). However, neither TGF-β1, -β2, nor -β3 is coexpressed with LTBP-2 in the testes by in situ hybridization (data not shown). In fact, there is disagreement on the issue of whether LTBP-2 can bind TGF-β. While LTBP-2 was initially shown to have the capability to bind TGF-β in transfection studies (5, 23), extensive mutagenesis studies in transfected cells have suggested that TGF-β binding to LTBP-2 is very weak when compared to its binding to other LTBP-2s (J. Keski-Oja, personal communication). The functional role of LTBP-2 in the testes remains unclear.
Because we could not detect LTBP-2 expression at E10.5, it was surprising that Ltbp2−/− embryos died prior to E6.5. The basis for the early embryonic lethality of the Ltbp2 knockout remains an enigma, considering what is known about knockouts of other elastic fiber components. Elastin knockout mice die shortly after birth from either vascular or pulmonary complications (16). Mice homozygous for targeted disruptions of the microfibril-associated glycoprotein 1 (R. P. Mecham, unpublished observations) and fibrillin 1 (27, 28) genes live into adulthood. The finding that mice lacking elastin can survive past birth suggests that a lethal phenotype associated with a microfibrillar protein such as LTBP-2 is not due to a function related to elastic fibers. However, others have suggested that microfibrils may have elastic properties of their own in the absence of elastin, perhaps thus explaining why mice lacking a microfibrillar component expressed very early in development might have a phenotype markedly different from that of the elastin knockout mouse. Microfibrils are known to exist devoid of associated elastin in some tissues such as the ciliary zonules of the eye and the periodontal ligament. It is therefore possible that LTBP-2 performs an essential structural function in elastin-free microfibrils, but the presence or absence of LTBP-2 has not been reported within elastin-free microfibrils. In any case, it is clear that LTBP-2 performs an essential function that cannot be compensated for by other members of the LTBP family.
Several findings suggest that the severity of the Ltbp2 knockout phenotype is not likely related to regulation of TGF-β activity. The most compelling is the observation in transfection studies that LTBP-2 binds TGF-β very poorly if at all in comparison to the other LTBPs (J. Keski-Oja, personal communication), raising questions about the physiologic significance of this interaction should it occur in vivo. However, if LTBP-2 retains any ability to bind TGF-β in the preimplantation embryo, its potential effect on TGF-β activity is not entirely predictable. LTBP-1 has been shown to facilitate the secretion of TGF-β (22). If this were also true for LTBP-2, one might predict that mice lacking LTBP-2 would have a phenotype similar to one or more of the TGF-β knockouts. The phenotypes of the TGF-β1, -β2, and -β3 knockouts are not similar to that of the Ltbp2 knockout. Although TGF-β1 is expressed in the preimplantation embryo (32), TGF-β knockout mice survive past this stage of development. A percentage of TGF-β1 knockout mice die in utero at ∼E10.5 from a defect in yolk sac vasculogenesis (2), while most survive past birth and die within the first month from infections (37). The TGF-β2 (36) and TGF-β3 (30) knockouts die in the perinatal period. Alternatively, LTBP-1 has been shown to mediate localization of TGF-β to the extracellular matrix, and activation of TGF-β in this scenario requires a proteolytic cleavage event which would release it from the matrix, enabling binding to cell-surface receptors. If this were the case for LTBP-2, then loss of LTBP-2 expression would perhaps result in aberrant localization and/or activation of TGF-β. While this is a formal possibility, we feel that the relative inability of LTBP-2 to bind TGF-β in transfection assays makes this unlikely.
The window of time in which Ltbp2−/− embryos die, E3.5 to E6.5, coincides with implantation of the embryo into the uterine wall. The inconsistency of the Ltbp2 knockout phenotype with that of other elastic fiber knockouts suggests that LTBP-2 may play a structural role related to elastin-free microfibrils at this point in development or that it perhaps has a signaling function. Alternatively, LTBP-2 may have an as-yet-undefined critical function between E3.5 and E6.5. The lack of LTBP-2 expression detected at E6.5 (Fig. 9) and E5.5 (data not shown) suggests that this function occurs between E3.5 and E5.5. RT-PCR on blastocysts for other microfibrillar components should help delineate whether these genes are expressed during implantation (E4.5), which has not been previously determined.
ACKNOWLEDGMENTS
This work was funded by NIH HL60647 (J.M.S.), NIH AR41474 (R.P.M. and J.R.), HL 29594 (morphology core), the Alan A. and Edith L. Wolff Charitable Trust (J.M.S.), and the Parker B. Francis Foundation (J.M.S.).
We are grateful to Zena Werb, Babette Heyer, and Julie Rinkenberger for assistance with embryological techniques and to Gail Griffin for assistance with in situ hybridization.
REFERENCES
- 1.Cleary E G. The microfibrillar component of the elastic fibers. Morphology and biochemistry. In: Uitto J, Perejda A J, editors. Connective tissue disease. Molecular pathology of the extracellular matrix. New York, N.Y: Marcel Dekker, Inc.; 1987. pp. 55–81. [Google Scholar]
- 2.Dickson M C, Martin J S, Cousins F M, Kulkarni A B, Karlsson S, Ackhurst R J. Defective hematopoiesis and vasculogenesis in transforming growth factor-beta 1 knockout mice. Development. 1995;121:1845–1854. doi: 10.1242/dev.121.6.1845. [DOI] [PubMed] [Google Scholar]
- 3.Dietz H C, Cutting G R, Pyeritz R E, Maslen C L, Sakai L Y, Corson G M, Puffenberger E G, Hamosh A, Nanthakumar E J, Curristan S M, Stetten G, Meyers D A, Francomano C A. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 4.Fahrenbach W H, Sandberg L B, Cleary E G. Ultrastructural studies on early elastogenesis. Anat Rec. 1966;155:563–576. [Google Scholar]
- 5.Fang J, Li X, Smiley E, Francke U, Mecham R P, Bonadio J. Mouse latent TGF-β binding protein-2: molecular cloning and developmental expression. Biochim Biophys Acta. 1997;1354:219–230. doi: 10.1016/s0167-4781(97)00104-8. [DOI] [PubMed] [Google Scholar]
- 6.Flaumenhaft R, Abe M, Sato Y, Miyazono K, Harpel J, Heldin C-H, Rifkin D B. Role of latent transforming growth factor-β binding protein in the activation of latent transforming growth factor-β by co-cultures of endothelial and smooth muscle cells. J Cell Biol. 1993;120:995–1002. doi: 10.1083/jcb.120.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson M A, Hatzinikolas G, Davis E C, Baker E, Sutherland G R, Mecham R P. Bovine latent transforming growth factor beta 1-binding protein 2: molecular cloning, identification of tissue isoforms, and immunolocalization to elastin-associated microfibrils. Mol Cell Biol. 1995;15:6932–6942. doi: 10.1128/mcb.15.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gleizes P-E, Beauis R C, Mazzieri R, Shen B, Rifkin D B. Identification and characterization of an eight-cysteine repeat of the latent transforming growth factor-β binding protein-1 that mediates binding to the latent transforming growth factor-β1. J Biol Chem. 1996;271:29891–29896. doi: 10.1074/jbc.271.47.29891. [DOI] [PubMed] [Google Scholar]
- 9.Godfrey M, Menashe V, Weleber R G, Koler R D, Bigley R H, Lovrien E, Zonana J, Hollister D W. Cosegregation of elastin-associated microfibrillar abnormalities with the Marfan phenotype in families. Am J Hum Genet. 1990;46:652–660. [PMC free article] [PubMed] [Google Scholar]
- 10.Greenlee T K J, Ross R, Hartman J L. The fine structure of elastic fibers. J Cell Biol. 1966;30:59–71. doi: 10.1083/jcb.30.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Press; 1994. [Google Scholar]
- 12.Kanzaki T, Olofsson A, Moren A, Wernstedt C, Hellman U, Miyazono K, Claesson-Welsh L, Heldin C-H. TGF-β1 binding protein: a component of the large latent complex of TGF-β1 with multiple repeat sequences. Cell. 1990;61:1051–1061. doi: 10.1016/0092-8674(90)90069-q. [DOI] [PubMed] [Google Scholar]
- 13.Kojima S, Kiyomitsu N, Rifkin D B. Requirement of transglutaminase in the activation of transforming growth factor-β in bovine endothelial cells. J Cell Biol. 1993;121:439–448. doi: 10.1083/jcb.121.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kojima S, Rifkin D B. Mechanism of retinoid-induced activation of latent transforming growth factor-β in bovine endothelial cells. J Cell Physiol. 1993;155:323–332. doi: 10.1002/jcp.1041550213. [DOI] [PubMed] [Google Scholar]
- 15.Lee B, Godfrey M, Vitale E, Hori H, Mattei M, Sarfarazi M, Tsipouras P, Ramirez F, Hollister D W. Linkage of Marfan syndrome and a phenotypically related disorder to two different fibrillin genes. Nature. 1991;352:330–334. doi: 10.1038/352330a0. [DOI] [PubMed] [Google Scholar]
- 16.Li D Y, Brooke B, Davis E C, Mecham R P, Sorensen L K, Boak B B, Eichwald E, Keating M T. Elastin is an essential determinant of arterial morphogenesis. Nature. 1998;393:276–280. doi: 10.1038/30522. [DOI] [PubMed] [Google Scholar]
- 17.Lyons R M, Moses H L. Transforming growth factor-β and the regulation of cell proliferation. Eur J Biochem. 1990;187:467–473. doi: 10.1111/j.1432-1033.1990.tb15327.x. [DOI] [PubMed] [Google Scholar]
- 18.Maslen C L, Corson G M, Maddox B K, Glanville R W, Sakai L Y. Partial sequence of a candidate gene for the Marfan syndrome. Nature. 1991;352:334–337. doi: 10.1038/352334a0. [DOI] [PubMed] [Google Scholar]
- 19.Massague J. The transforming growth factor-β family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 20.Miyazono K, Hellman U, Wernstedt C, Heldin C. Latent high molecular weight complex of transforming growth factor β1. J Biol Chem. 1988;263:6407–6415. [PubMed] [Google Scholar]
- 21.Miyazono K, Ichijo H, Heldin C-H. Transforming growth factor-beta: latent forms, binding proteins, and receptors. Growth Factors. 1993;8:11–22. doi: 10.3109/08977199309029130. [DOI] [PubMed] [Google Scholar]
- 22.Miyazono K, Oloffson A, Colosetti P, Heldin C-H. A role of the latent TGF-β1 binding protein in the assembly and secretion of TGF-β1. EMBO J. 1991;10:1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moren A, Olofsson A, Stenman G, Sahlin P, Kanzaki T, Claesson-Welsh L, ten Dijke P, Miyazono K, Heldin C H. Identification and characterization of LTBP-2, a novel latent transforming growth factor-beta-binding protein. J Biol Chem. 1994;269:32469–32478. [PubMed] [Google Scholar]
- 24.Nunes I, Gleizes P-E, Metz C N, Rifkin D B. Latent transforming growth factor-β binding protein domains involved in activation and transglutaminase-dependent cross-linking of latent transforming growth factor-β. J Cell Biol. 1997;136:1151–1163. doi: 10.1083/jcb.136.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunes I, Shapiro R L, Rifkin D B. Characterization of latent transforming growth factor-β activation by murine peritoneal macrophages. J Immunol. 1995;155:1450–1459. [PubMed] [Google Scholar]
- 26.Parks W C, Pierce R A, Lee K A, Mecham R P. Elastin. In: Kleinman H K, editor. Advances in molecular and cell biology. Vol. 6. Greenwich, Conn: JAI Press, Inc.; 1993. pp. 133–182. [Google Scholar]
- 27.Pereira L, Andrikopoulos K, Tian J, Lee S Y, Keene D R, Ono R, Reinhardt D P, Sakai L Y, Biery N J, Bunton T, Dietz H C, Ramirez F. Targeting of the gene encoding fibrillin-1 recapitulates the vascular aspect of Marfan syndrome. Nat Genet. 1997;17:218–222. doi: 10.1038/ng1097-218. [DOI] [PubMed] [Google Scholar]
- 28.Pereira L, Lee S Y, Gayraud B, Andrikopoulos K, Shapiro S D, Bunton T, Biery N J, Dietz H C, Sakai L Y, Ramirez F. Pathogenetic sequence for aneurysm revealed in mice underexpressing fibrillin-1. Proc Natl Acad Sci USA. 1999;96:3819–3823. doi: 10.1073/pnas.96.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierce R A, Deak S B, Stolle C A, Boyd C D. Heterogeneity of rat tropoelastin mRNA revealed by cDNA cloning. Biochemistry. 1990;29:9677–9683. doi: 10.1021/bi00493a024. [DOI] [PubMed] [Google Scholar]
- 30.Proetzel G, Pawlowski S A, Wiles M V, Yin M, Boivin G P, Howles P N, Ding J, Ferguson M W, Doetschman T. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prosser I W, Stenmark K R, Suthar M, Crouch E C, Mecham R P, Parks W C. Regional heterogeneity of elastin and collagen gene expression in intralobar arteries in response to hypoxic pulmonary hypertension as demonstrated by in situ hybridization. Am J Pathol. 1989;135:1073–1088. [PMC free article] [PubMed] [Google Scholar]
- 32.Rappolee D A, Brenner C A, Schultz R, Mark D, Werb Z. Developmental expression of PDGF, TGF-alpha, and TGF-beta genes in preimplantation mouse embryos. Science. 1988;241:1823–1825. doi: 10.1126/science.3175624. [DOI] [PubMed] [Google Scholar]
- 33.Rosenbloom J, Abrams W B, Mecham R. Extracellular matrix 4: the elastic fiber. FASEB J. 1993;7:1208–1218. [PubMed] [Google Scholar]
- 34.Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-β with an 8-cysteine repeat of its binding protein LTBP-1. EMBO J. 1996;15:245–253. [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai L Y, Keene D R, Engvall E. Fibrillin, a new 350-kD glycoprotein, is a component of extracellular microfibrils. J Cell Biol. 1986;103:2499–2509. doi: 10.1083/jcb.103.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanford L P, Ormsby I, Gittenberger-de Groot A C, Sariola H, Friedman R, Boivin G P, Cardell E L, Doetschman T. TGF-beta 2 knockout mice have multiple developmental defects that are non-overlapping with other TGF-beta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shull M M, Ormsby I, Kier A B, Pawlowski S, Diebold R J, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, Annuziata N, Doetschman T. Targeted disruption of the mouse transforming growth factor-beta-1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skinner M K, Moses H L. Transforming growth factor gene expression and action in the seminiferous tubule: peritubular cell-Sertoli cell interactions. Mol Endocrinol. 1989;3:625–634. doi: 10.1210/mend-3-4-625. [DOI] [PubMed] [Google Scholar]
- 39.Sporn M B, Roberts A B. Transforming growth factor-β: recent progress and new challenges. J Cell Biol. 1992;119:1017–1021. doi: 10.1083/jcb.119.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taipale J, Lohi J, Saharinen J, Kovanen P T, Keski-Oja J. Human mast cell chymase and leukocyte elastase release latent transforming growth factor-β1 from the extracellular matrix of cultured human epithelial and endothelial cells. J Biol Chem. 1995;270:4689–4696. doi: 10.1074/jbc.270.9.4689. [DOI] [PubMed] [Google Scholar]
- 41.Taipale J, Koli K, Keski-Oja J. Release of transforming growth factor-β1 from the pericellular matrix of cultured fibroblasts and fibrosarcoma cells by plasmin and thrombin. J Biol Chem. 1992;267:25378–25384. [PubMed] [Google Scholar]
- 42.Zhang H, Apfelroth S D, Hu W, Davis E C, Sanguineti C, Bonadio J, Mecham R P, Ramirez F. Structure and expression of fibrillin-2, a novel microfibrillar component preferentially located in elastic matrices. J Cell Biol. 1994;124:855–863. doi: 10.1083/jcb.124.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]