Abstract
Mammalian SET domain-containing proteins define a distinctive class of chromatin-associated factors that are targets for growth control signals and oncogenic activation. SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9, contains both SET and chromo domains, signature motifs for proteins that contribute to epigenetic control of gene expression through effects on the regional organization of chromatin structure. In this report we demonstrate that SUV39H1 represses transcription in a transient transcriptional assay when tethered to DNA through the GAL4 DNA binding domain. Under these conditions, SUV39H1 displays features of a long-range repressor capable of acting over several kilobases to silence basal promoters. A possible role in chromatin-mediated gene silencing is supported by the localization of exogenously expressed SUV39H1 to nuclear bodies with morphologic features suggestive of heterochromatin in interphase cells. In addition, we show that SUV39H1 is phosphorylated specifically at the G1/S cell cycle transition and when forcibly expressed suppresses cell growth. Growth suppression as well as the ability of SUV39H1 to form nuclear bodies and silence transcription are antagonized by the oncogenic antiphosphatase Sbf1 that when hyperexpressed interacts with the SET domain and stabilizes the phosphorylated form of SUV39H1. These studies suggest a phosphorylation-dependent mechanism for regulating the chromatin organizing activity of a mammalian su(var) protein and implicate the SET domain as a gatekeeper motif that integrates upstream signaling pathways to epigenetic regulation and growth control.
The formation and propagation of higher-order chromatin states are dynamic processes that establish distinct domains that are either permissive or restrictive for transcription. The functions of such domains have been implicated in the epigenetic control of developmental gene expression in Drosophila (38), proper sister chromatid segregation during meiosis (11), and telomeric and centromeric silencing in yeast (22, 35). The molecular mechanisms that regulate these and other properties of higher-order chromatin are essentially unknown.
Genetic analyses in Drosophila and yeast have identified several genes that participate in the formation of euchromatin or heterochromatin states. Some of these encode proteins that contribute to either an enhancement [E(var)] or suppression [Su(var)] of position effect variegation (PEV) (42). PEV is a gene silencing mechanism that results from the spreading of heterochromatin, thus implicating E(var) and Su(var) proteins in the formation of euchromatic and heterochromatic domains, respectively. Several E(var) and Su(var) proteins share distinctive motifs that are important for their ability to organize chromatin domains. The Su(var)3-9 protein and its mammalian ortholog SUV39H1 are unique in being the only characterized PEV modifiers that share two of these consensus motifs, the chromo and SET domains (1, 50). Chromo domains are 40-amino-acid modular motifs that are implicated in protein self-association (8) and the assembly of site-specific multimeric complexes on chromatin (39, 46). SET domains are 130-amino-acid motifs named for three proteins in which they were originally identified: Su(var)3-9, Enhancer-of-zeste, and Trithorax (25). Enhancer-of-zeste and Trithorax are members of the Polycomb group (PcG) and Trithorax group (TrG) proteins, respectively, that antagonistically maintain Hox gene expression profiles once they have been established during Drosophila development (15, 51). These and other SET domain proteins have been shown to either physically or indirectly associate with chromatin (1, 7, 40). In yeast, mutations in the SET domains of CLR4 and SET1 disrupt centromeric silencing in Schizosaccharomyces pombe and telomeric silencing in Saccharomyces cerevisiae, respectively (22, 35). Although found in over 30 proteins from human, Drosophila, Caenorhabditis elegans and yeast, the molecular functions for SET domains are not known. However, their presence in both PcG and TrG proteins suggests that they may serve a regulatory role in the formation of silent or active chromatin states (25).
Several lines of evidence suggest that mammalian SET domain proteins are targets for growth control signals and oncogenic mutations. Enx-1, a human homolog of Enhancer-of-zeste, interacts with Vav, a signaling protein originally identified as the product of a retrovirally transduced oncogene (19). A human homolog of Drosophila Trithorax, MLL, is encoded by a proto-oncogene that is frequently mutated by chromosomal translocations in human leukemias (12, 16, 48). The SET domain of MLL, which is deleted in oncogenic forms of the protein (53), mediates interactions with INI1, a component of the mammalian hSWI/SNF chromatin remodeling complex (43). INI1 is targeted by inactivating mutations in malignant rhabdoid tumors (52), raising the possibility that loss of hSWI/SNF function or disrupted interaction with SET domain proteins may constitute alternate pathways to oncogenesis (24).
Another protein reported to interact with SET domains is Sbf1, which displays features of a so-called antiphosphatase (21). Sbf1 is similar to dual-specificity phosphatases of the myotubularin family but lacks several crucial residues in the catalytic pocket which render it catalytically inactive as a phosphatase. The pocket is sufficiently preserved, however, to bind phosphorylated synthetic substrates (9), suggesting a possible role as a protective factor that competes with functional phosphatases for substrate interaction (55). Mutated forms of Sbf1 are highly oncogenic, and a conserved motif in Sbf1 that mediates interactions with SET domains in vitro is necessary and sufficient for oncogenic activity (9, 10). These results implicate SET domains as critical transducers of growth control signals and suggest that SET domain proteins are important effectors of growth as well as differentiation programs. Several studies have suggested that phosphorylation influences the activity or effects of E(var) and Su(var) proteins on higher-order chromatin. Notably, heterochromatin binding by the heterochromatin protein 1 [Su(var)2-5] is regulated by phosphorylation (57). Another dominant suppressor of variegation [Su(var)3-6] is itself a type I protein phosphatase (3).
This study was conducted to characterize phosphorylation-dependent growth control pathways that impinge on SET domain proteins. Using a truncated oncogenic form of Sbf1 as a molecular probe, we identified SUV39H1, the mammalian ortholog of Su(var)3-9, as an endogenous SET domain protein that is differentially phosphorylated in the presence of the Sbf1 oncoprotein. Our data demonstrate that SUV39H1 forms large discrete nuclear bodies and has growth-suppressive and transcriptional repressive properties that are modulated by the oncogenic form of Sbf1. In addition, we show that upon mitogenic activation, SUV39H1 is phosphorylated specifically at the G1/S cell cycle transition and that its phosphorylation is enhanced by coexpressed oncogenic Sbf1. Taken together, these data define a SET domain-dependent phosphorylation mechanism for regulating the contributions of a Su(var) protein to cellular growth control.
MATERIALS AND METHODS
DNA constructs.
All DNA constructions were produced by PCR and standard cloning techniques. A SUV39H1 cDNA encoding the complete open reading frame (amino acids 1 to 412) was procured from the IMAGE consortium (clone 23658) and used as template for PCR to create a minimal construct containing the SUV39H1 coding region flanked by EcoRI sites at both ends. The C-terminal deletion mutant SUV39H1ΔC (amino acids 1 to 195) was generated by truncation of the open reading frame at an internal SmaI site (bp 585). SUV39H1ΔSET (amino acids 1 to 286) was generated by inserting a stop codon at an internal BglII site. The expression construct (FLAG)SUV39H1 for immunolocalization studies was made by insertion of SUV39H1 into pYDF30 in frame with the N-terminal FLAG epitope. For protein-protein interaction studies, SUV39H1 and SUV39H1ΔC were tagged at their N termini with epitopes from the hemagglutinin antigen (HA) to generate the constructs (HA2)SUV39H1 and (HA2)SUV39H1ΔC. SUV39H1SET (provided by T. Jenuwein) contains the SET domain downstream of an engineered nuclear localization sequence and localizes to the nucleus (30a). To construct GAL4 DNA binding domain (DBD) fusion proteins, SUV39H1 was cloned in frame with amino acids 1 to 147 of GAL4 in the pM3 vector (provided by R. Baer) (44). Reporter constructs for transient transcriptional assays contained a firefly luciferase gene with (pLUC/GAL4) or without (pLUC) four GAL4 sites upstream of the myelomonocytic growth factor promoter (provided by R. Eisenman) (2). Reporter constructs containing five tandem GAL4 sites at variable distances upstream of the simian virus 40 (SV40) promoter (provided by J. Milbrandt) have been described previously (47).
Retroviral vectors containing an internal ribosome entry site (IRES) were used for coexpression studies. SUV39H1-IRES-EGFP was generated by cloning SUV39H1 into the LZRSpBMN-IRES-EGFP vector (provided by G. Nolan), which expresses the enhanced green fluorescent protein (EGFP) from the IRES element. Retroviral constructs for coexpression of SUV39H1 with Sbf1 or Sbf1HCS were generated by first cloning Sbf1 or Sbf1HCS into the retroviral vector MSCVneoEB (Clontech). A DNA fragment containing SUV39H1 linked to an IRES element (SUV39H1-IRES) was then inserted upstream to generate SUV39H1-IRES-Sbf1 and SUV39H1-IRES-Sbf1HCS, respectively. SUV39H1ΔSET-IRES-Sbf1 and SUV39H1ΔSET-IRES-Sbf1HCS were constructed in a similar manner.
Generation of anti-SUV39H1 MAbs.
Maltose binding protein-SUV39H1 and glutathione S-transferase–SUV39H1 fusion proteins were expressed in Escherichia coli and purified using maltose (New England Biolabs) or glutathione (Sigma)-agarose, respectively. BALB/c mice were immunized against the purified maltose binding protein-SUV39H1 fusion protein in adjuvant by repeated subcutaneous injections. Splenocytes from immune mice were fused with the fusion partner SP2/0 (American Type Culture Collection) using established procedures (17). Monoclonal antibodies (MAbs) were purified as previously described (32). The MAb used for these studies was isotyped as immunoglobulin G1-kappa (IgG1-kappa) and recognized an epitope in the first 195 N-terminal amino acids of SUV39H1.
Transcriptional assays.
DNA constructs were transfected into COS7 cells by either calcium phosphate coprecipitation (6) or the Effectene reagent (Qiagen). Transfections were internally controlled by cotransfection of pCMV-lacZ (0.5 μg/well), which expresses β-galactosidase under control of the cytomegalovirus promoter. Two days after transfection, luciferase assays were performed using commercially prepared reagents (Promega). Light emission was measured using a luminometer (Analytical Luminescence Laboratory), and values were normalized based on the β-galactosidase levels. Data points represent the average normalized activity in lysates prepared from two identically transfected samples.
Cell cycle and growth inhibition assays.
HeLa S3 cells were growth arrested by serum starvation in Dulbecco modified Eagle medium (DMEM) containing 0.2% fetal bovine serum (FBS) for a period of 48 h. Cells were stimulated to reenter the cell cycle by addition of DMEM containing 10% FBS. Thirty minutes prior to harvest, half of the culture was incubated with 50 mM BrdU (bromodeoxyuridine) and subsequently used for quantitation of BrdU incorporation, which was detected and visualized as recommended by the supplier (Boehringer Mannheim). The remaining half of the culture was harvested in sodium dodecyl sulfate (SDS) lysis buffer (2% SDS, 50 mM Tris [pH 6.8], 10% glycerol) and lysate proteins (50 μg) were subjected to SDS–10% polyacrylamide gel electrophoresis (PAGE) and Western blot analysis. The effects of SUV39H1 on cell cycle kinetics were measured in NIH 3T3 cells stably transduced with retroviral constructs. Logarithmically growing NIH 3T3 cells were incubated with 50 mM BrdU (Boehringer Mannheim) for 3 h.
Protein phosphorylation analysis.
Logarithmically growing HeLa S3 cells were washed once in phosphate-buffered saline in (PBS) and incubated for 20 min in phosphate-free DMEM (GIBCO-BRL) supplemented with 10% dialyzed FBS. [32P]orthophosphate (0.5 mCi/ml) was then added, and the cells were incubated for an additional 3 h. Labeled cells were washed once in PBS and lysed in buffer A (20 mM HEPES pH 7.9, 10 mM KCl, 1 mM EDTA, 1 mM dithrothreitol, 1 mM phenylmethylsulfonyl fluoride) supplemented with 40 mM NaF and 1 mM NaVO4. The lysed cells were centrifuged for 5 min at 5,000 × g at 4°C, and the pellet was resuspended in radioimmunoprecipitation assay buffer containing 400 mM NaCl. The nuclear fraction was centrifuged for 20 min at 14,000 × g at 4°C, and the supernatant was taken for immunoprecipitation analysis with an anti-SUV39H1 MAb.
Immunoprecipitation and protein analysis.
COS7 cells were harvested 2 days after transfection, washed once with PBS, resuspended in buffer A, and then lysed in buffer A containing 0.2% NP-40 and 400 mM NaCl by agitation at 4°C for 20 min. Cell debris was removed by centrifugation at 14,000 × g for 20 min, and the supernatant was incubated on ice for 3 h with an anti-Sbf1 (10) or anti-SUV39H1 MAb (5 μg/ml). Immune complexes were precipitated using protein G-agarose beads (Boehringer Mannheim) for 3 h at 4°C. The agarose beads were pelleted, washed five times in immunoprecipitation wash buffer (250 mM NaCl, 20 mM HEPES [pH 7.9], 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) and resuspended in 2× SDS sample buffer (4% SDS, 10% 2-mercaptoethanol, 100 mM Tris [pH 6.8], 20% glycerol). Eluted proteins were boiled, separated by SDS-PAGE, transferred to nitrocellulose (Bio-Rad), and subjected to Western blot analysis using an MAbs specific for Sbf1, SUV39H1, or the HA epitope (Boehringer Mannheim). Immune complexes were detected using a secondary horseradish peroxidase-conjugated goat anti-mouse or anti-rat antibody (Jackson ImmunoResearch) and visualized by chemiluminescence (Amersham).
Immunofluorescence and immunoelectron microscopy.
The subcellular localization of SUV39H1 was detected by indirect immunofluorescence microscopy. COS7 cells that had been transfected 48 h previously were fixed in PBS–4% paraformaldehyde for 15 min. Preparations were then blocked in PBS–5% normal goat serum for 30 min followed by incubation with the primary anti-FLAG MAb (M5; Sigma) at a dilution of 1:500. Immune complexes containing epitope-tagged SUV39H1 were visualized with a fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody. Cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole) (Boehringer Mannheim) and mounted onto slides.
For immunoelectron microscopy, pelleted COS7 cells were fixed with freshly prepared 2% paraformaldehyde–0.5% glutaraldehyde for 50 min at 25°C. Fixed cells were washed in several changes of PBS and dehydrated through a series of ethanol washes. The pellet was infiltrated with absolute ethanol-LR White (1:1) followed by pure LR White (Electron Microscopy Sciences, Fort Washington, Pa.) before polymerization in gelatin capsules at 48°C. Silver sections were placed onto gold grids, blocked in Tris-buffered saline–5% bovine serum albumin–0.5% normal goat serum for 1 h, and then incubated with mouse anti-FLAG antibody overnight at 4°C. Grids were washed several times and incubated with 10 nM colloidal gold-conjugated goat anti-mouse secondary antibody (Amersham Corp., Arlington Heights, Ill.) 1:20 for 3 h at 25°C. The treated grids were washed in Tris-buffered saline followed by filtered deionized water and then air dried. The sections were lightly counterstained with uranyl acetate and lead citrate. Electron micrographs were taken on a Hitachi EM300 (Nissei Sangyo America, Ltd., Mountain View, Calif.).
RESULTS
Sbf1 physically associates with the SET domain of SUV39H1.
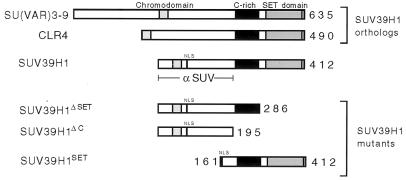
Oncogenic Sbf1 interacts in vitro with the SET domains of several proteins and, when forcibly expressed in vivo, alters the phosphorylation profile of several cellular proteins (9). In this phosphorylation screen, one protein of approximate 45 kDa was identified as a potential target of Sbf1 based on its high degree of differential phosphorylation (9). The only known SET domain protein of this size is SUV39H1, a recently reported mammalian ortholog of Drosophila Su(var)3-9 that also displays extensive similarity with S. pombe CLR4 (1). All three proteins share highly conserved C-terminal SET domains as well as N-terminal chromo domains and internal cysteine-rich regions (Fig. 1).
FIG. 1.
Conservation and expression of SUV39H1, a mammalian ortholog of Su(var)3-9. Schematic depictions of the predicted protein compositions for human SUV39H1 and the orthologous Drosophila Su(var)3-9 and S. pombe CLR4 indicate the conserved chromo domains (light stipple), cysteine-rich regions (black box), SET domains (heavy stipple), and putative nuclear localization sequence (NLS). The portion of SUV39H1 used as immunogen for production of MAbs (α SUV) is shown below. SUV39H1SET is an N-terminal deletion mutant that contains an engineered N-terminal NLS. SUV39H1ΔSET and SUV39H1ΔC are C-terminal deletion mutants used in this study.
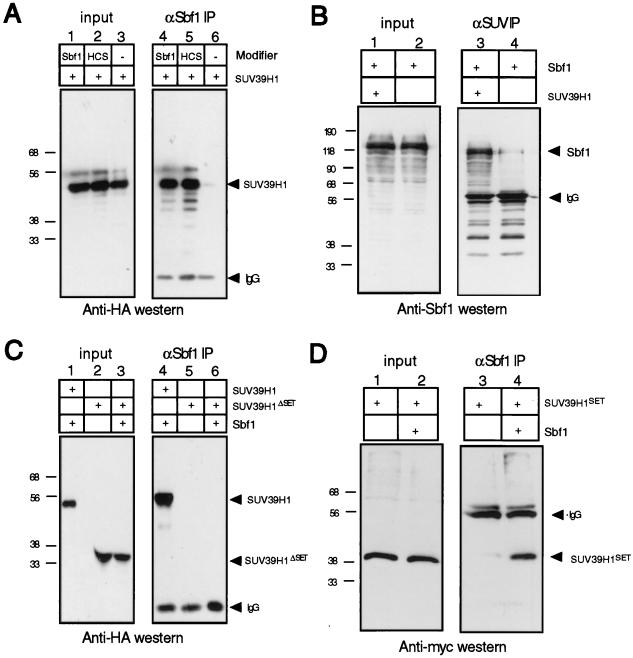
To specifically demonstrate the association of SUV39H1 with Sbf1, we performed coprecipitation analyses in cells transiently expressing full-length SUV39H1 and the oncogenic form of Sbf1. Extracts of 293t cells expressing HA-tagged SUV39H1 with or without cotransfected Sbf1 were subjected to immunoprecipitation analysis with an anti-Sbf1 MAb. Western blot analysis of the immunoprecipitates using an anti-HA MAb showed that SUV39H1 was precipitated in the presence but not absence of cotransfected Sbf1 (Fig. 2A, compare lanes 4 and 6). In a complementary coimmunoprecipitation assay, Sbf1 was more highly precipitated in the presence but not the absence of cotransfected SUV39H1 (Fig. 2B). A small amount of coprecipitating Sbf1 in the latter may be explained by the presence of endogenous SUV39H1 in the 293 cell line.
FIG. 2.
SUV39H1 displays SET domain-dependent physical association with the anti-phosphatase Sbf1. (A) 293t cells were cotransfected with expression constructs encoding HA-tagged SUV39H1, Sbf1, and Sbf1HCS as indicated above the gel lanes. Whole cell extracts prepared 48 h after transfection were subjected to immunoprecipitation (IP) using an anti-Sbf1 MAb. Detection of coprecipitating SUV39H1 by Western blot analysis using an anti-HA antibody demonstrated that it was capable of associating with both Sbf1 and Sbf1HCS. (B) Lysates of 293t cells transfected with constructs expressing the proteins indicated above the gel lanes were subjected to immunoprecipitation using an anti-SUV39H1 antibody. Coprecipitating Sbf1 was detected by Western blot analysis using an anti-Sbf1 MAb. (C and D) Lysates of 293t cells transfected with tagged constructs expressing the proteins indicated above the gel lanes were subjected to immunoprecipitation using an anti-Sbf1 MAb. Coprecipitating SUV39H1 proteins were detected by Western blot analysis with an anti-HA or anti-Myc antibody. The anti-rat secondary antibody (A and C) cross-reacted with mouse IgG heavy chain used in the immunoprecipitations. The amount of lysate in each input lane (input) is equivalent to 2% of the amount applied to beads (IP). Protein migrations are indicated by arrows; sizes are indicated in kilodaltons.
SUV39H1 was also coprecipitated from 293t cells cotransfected with Sbf1HCS, a nontransforming mutant of Sbf1 (Fig. 2A, lane 5). Unlike Sbf1, Sbf1HCS dephosphorylates synthetic phosphotyrosine- and phosphoserine-containing substrates due to several amino acid substitutions engineered into its phosphatase catalytic pocket (9). Since both proteins associate with SUV39H1, the interaction does not appear to result from trapping of SUV39H1 by the nonfunctional phosphatase pocket of Sbf1. This is consistent with previous observations (9) that a defined motif (SID [SET interaction domain]) in Sbf1 mediates in vitro interactions with SET domains. Furthermore, our data indicate that association with SUV39H1 is not exclusively a property of oncogenic forms of Sbf1.
To determine whether the SET domain of SUV39H1 was necessary for interaction with Sbf1, mutants that contained or lacked the SET domain (SUV39H1SET and SUV39H1ΔSET, respectively) were tested in the coprecipitation assay. While SUV39H1SET was precipitated in the presence of coexpressed Sbf1, SUV39H1ΔSET was not (Fig. 2C and D). Taken together, our results demonstrate that the SET domain of SUV39H1 is necessary to mediate SUV39H1/Sbf1 interaction in vivo.
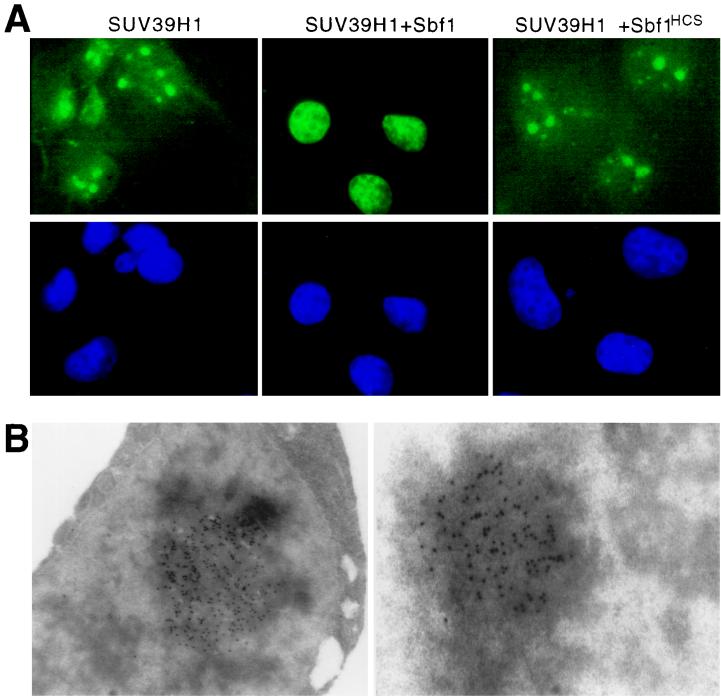
SUV39H1 localizes within distinct nuclear bodies that are dispersed by oncogenic Sbf1.
SUV39H1, like Su(var)3-9, enhances PEV when forcibly expressed in Drosophila and, similar to its yeast ortholog CLR4, localizes to the centromeric regions of metaphase chromosomes (1). Therefore, we tested the possible effects of SUV39H1-Sbf1 association on formation or spreading of heterochromatin under conditions of hyperexpression in mammalian cells. COS cells transfected with FLAG-tagged SUV39H1 were examined by indirect immunofluorescence microscopy to evaluate the subcellular localization of SUV39H1. Under these conditions SUV39H1 formed large, distinct nuclear bodies in interphase cells (Fig. 3A). Immunoelectron microscopy showed that these structures were electron dense and amorphous (Fig. 3B). The nuclear distribution of SUV39H1, however, was dramatically different in cells cotransfected with Sbf1. In cells expressing both proteins, immunofluorescence analysis revealed a more diffuse nuclear distribution for SUV39H1 that was not concentrated into large nuclear bodies (Fig. 3A). This effect was specific for the phosphatase-inactive form of Sbf1 since cotransfected Sbf1HCS did not disrupt the ability of SUV39H1 to form nuclear bodies (Fig. 3A) in spite of its ability to associate and coprecipitate with SUV39H1 (Fig. 2A). Modulation of this phenomenon specifically by Sbf1 but not Sbf1HCS suggested a possible phosphorylation-dependent mechanism for regulating the ability of SUV39H1 to organize higher-order chromatin.
FIG. 3.
SUV39H1 forms nuclear bodies in vivo that are dispersed by Sbf1. (A) COS7 cells were examined by immunofluorescence 48 h after cotransfection of constructs expressing FLAG-tagged SUV39H1 in the presence or absence of a 10-fold excess of expression constructs for Sbf1 or Sbf1HCS. Green fluorescence corresponds to FLAG-tagged SUV39H1 staining which was revealed using primary anti-FLAG and secondary fluorescein isothiocyanate-conjugated antibodies. DAPI staining is shown in blue. Expression of transfected Sbf1 and Sbf1HCS was comparable as detected by Western blot analysis (data not shown). Magnification, ×630. (B) 293t cells were analyzed by immunoelectron microscopy 48 h after transfection with a construct expressing FLAG-tagged SUV39H1. Immune complexes were visualized using a primary antibody directed against the FLAG epitope tag and a secondary goat anti-mouse IgG conjugated with colloidal gold. Magnification, ×42,300.
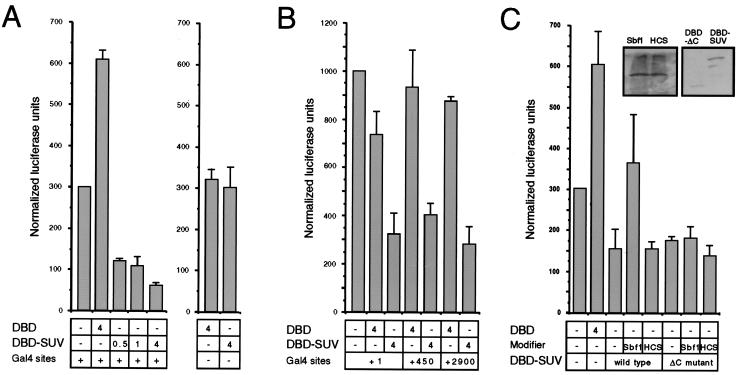
SUV39H1 represses transcription when tethered to DNA.
Previous studies have demonstrated that chromo domain-containing proteins, including Su(var)3-9 and its orthologs, localize to regions of chromatin that are transcriptionally silenced (1, 22, 39). Our own subnuclear localization of SUV39H1 to electron-dense nuclear foci supports the notion that SUV39H1 may associate with transcriptionally inactive chromatin. To test this hypothesis, we evaluated its ability to silence transcription of a reporter gene under control of the monomyelocytic growth factor promoter (2) in transfected COS cells. As a fusion protein containing the GAL4 DBD, SUV39H1 repressed transcription only when tethered to DNA through upstream GAL4 DNA binding sites (Fig. 4A). The level of observed repression was directly dependent on the amount of input SUV39H1-GAL4 expression plasmid. At highest concentrations, the repressive effect was approximately 10-fold compared to the GAL4 DBD alone, whose ability to activate transcription due to a cryptic activation domain has been previously reported (2, 28). Repression was also observed (Fig. 4B) using a reporter gene under control of the SV40 early promoter (47). Comparable levels of transcriptional repression were observed regardless of the distance (0 to 2,900 bp) SUV39H1 was tethered upstream from the promoter (Fig. 4B). The repressive properties of SUV39H1 localized to its N-terminal half since a C-terminal deletion mutant (SUV39H1ΔC [Fig. 1A]) displayed no loss of repressive potential (Fig. 4C). Therefore, SUV39H1 represses transcription when tethered to DNA, and its ability to do so appears promoter and distance independent consistent with chromatin-mediated silencing as opposed to promoter interference (5).
FIG. 4.
SUV39H1 displays transcriptional repressor properties that are modulated by Sbf1. (A) Expression constructs coding for the GAL4 DBD itself or a GAL4-SUV39H1 fusion protein (DBD-SUV) were cotransfected into COS7 cells in combination with a luciferase reporter gene under control of the myelomonocytic growth factor promoter. The amount (micrograms) of each construct present in the transfections is indicated below the histograms. Transcriptional activation is expressed as normalized luciferase units that have been corrected for β-galactosidase expression from an internal control lacZ construct in each transfection. The data represent the means from at least three independent experiments. Transcriptional repression observed for GAL4-SUV was dependent on the presence of GAL4 binding sites in the reporter gene and not observed if SUV39H1 was untethered to the GAL4 DBD (not shown). (B) Transcriptional assays were conducted as described for panel A except that the luciferase reporter gene constructs contained a minimal SV40 promoter separated by variable distances (indicated below histograms) from upstream GAL4 DNA binding sites. (C) Transcriptional assays were performed as described for panel A with the addition of expression constructs encoding Sbf1 (amino acids 700 to 1931) or Sbf1HCS (as indicated below the histograms) at fivefold excess concentration compared to cotransfected SUV39H1 constructs. Repression of the myelomonocytic growth factor promoter by GAL4-SUV was partially alleviated by coexpressed Sbf1 but not Sbf1HCS. Repression was also observed by GAL4-SUVΔC but was not relieved by coexpressed Sbf1. Western blots demonstrating comparable expression levels of transfected Sbf1 and Sbf1HCS as well as GAL4-SUV and GAL4-SUVΔC are shown as insets.
Physical interaction with Sbf1 modulates transcriptional repression by SUV39H1.
We next evaluated whether the transcriptional effects of SUV39H1 were influenced by heterologous interactions with Sbf1 proteins. SUV39H1-GAL4 chimeras were expressed alone or together with Sbf1 or Sbf1HCS in transfected COS cells. While coexpression of Sbf1HCS had no effect on repression by SUV39H1, coexpressed Sbf1 substantially increased reporter gene expression above the repressed levels observed for SUV39H1 alone (Fig. 4C). Thus, Sbf1 partially canceled the repressive effect of SUV39H1 on transcription. However, Sbf1 was unable to reverse repression mediated by SUV39H1ΔC (Fig. 4C), demonstrating a dependence on the SET domain of SUV39H1. These data suggest that the ability of Sbf1 to cancel transcriptional repression by SUV39H1 is critically dependent on physical interactions with the SET domain of SUV39H1. However, derepression appears to require more than association of Sbf1 and SUV39H1 since Sbf1HCS, which also associates with SUV39H1, was unable to similarly neutralize the transcriptional effects of SUV39H1.
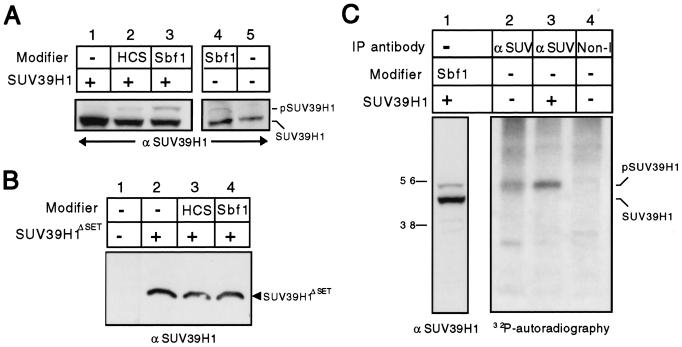
Sbf1 modulates the phosphorylation state of SUV39H1.
The foregoing data indicate that both Sbf1 and Sbf1HCS physically interact with SUV39H1, but only the oncogenic form of Sbf1 modulates its nuclear localization and effects on transcription. Since Sbf1 and Sbf1HCS biochemically differ only in the catalytic properties of their phosphatase pockets, we tested whether Sbf1 may impact SUV39H1 functions by affecting its phosphorylation state. To this end, SUV39H1 was efficiently expressed in cells using retroviral vectors that also coexpressed Sbf1 or Sbf1HCS by means of an IRES element. Proteins from whole cell extracts were subjected to Western blot analysis using an anti-SUV39H1 MAb. This revealed that a small fraction of SUV39H1 was shifted to a slower-migrating form that was substantially more abundant in cells coexpressing Sbf1 but not Sbf1HCS (Fig. 5A, lanes 1 to 3). Expression of exogenous Sbf1 had similar effects on the migration of endogenous SUV39H1, resulting in a 2- to 5-kDa shift in its apparent size (Fig. 5A, lanes 4 versus 5). No shift, however, was detected when SUV39H1ΔSET was coexpressed with Sbf1 (Fig. 5B), indicating that the SET domain was necessary for this modification.
FIG. 5.
SUV39H1 undergoes SET-dependent phosphorylation that is enhanced by Sbf1. (A) Bosc cells were transduced with retroviral vectors coexpressing Sbf1 or Sbf1HCS (from an IRES element) with SUV39H1. Cells were harvested 2 days after transduction, and equal amounts of whole cell lysate used for Western blotting. Shifted (pSUV39H1) and nonshifted (SUV39H1) forms of SUV39H1 were detected using an anti-SUV39H1 antibody. Similar shifts in the migration of exogenous (lane 3) or endogenous (lane 4) SUV39H1 were induced by forced expression of Sbf1. Expression of transfected Sbf1 and Sbf1HCS was comparable as detected by Western blot analysis (data not shown). (B) Analyses similar to those in panel A, substituting SUV39H1ΔSET for SUV39H1, showed no shifted migration of SUV39H1ΔSET following coexpression with Sbf1. (C) HeLa cells transfected with control or SUV39H1-expressing vectors were metabolically labeled with [32P]orthophosphate. Equal amounts of nuclear extracts were immunoprecipitated (IP) using anti-SUV39H1 or anti-Pbx1 (nonimmune) antibodies. Precipitated proteins were fractionated by SDS-PAGE and subjected to autoradiography. In parallel on the same gel, lysate from cells cotransfected with Sbf1 and SUV39H1 was analyzed by Western blotting to determine the migration of shifted (pSUV39H1) and nonshifted (SUV39H1) forms of SUV39H1.
To determine whether the observed shift in migration may be due to phosphorylation, SUV39H1 was immunoprecipitated from HeLa cells that had been metabolically labeled with [32P]orthophosphate. A major phosphoprotein of approximately 50 kDa was detected in the anti-SUV precipitate but not the nonimmune precipitate (Fig. 5C, lanes 2 versus 4). This phosphoprotein was present at elevated levels in cells expressing exogenous SUV39H1 (Fig. 5C, lane 3) and displayed a migration identical to the shifted form of SUV39H1 detected by Western blotting (Fig. 5C, lane 1). No phosphorylated band was observed at a position corresponding to the more abundant unshifted form of SUV39H1 (45 kDa) indicating that most of the protein under these conditions was unphosphorylated. Taken together, these results demonstrate that a fraction of cellular SUV39H1 is phosphorylated and the relative amount is enhanced by coexpressed Sbf1.
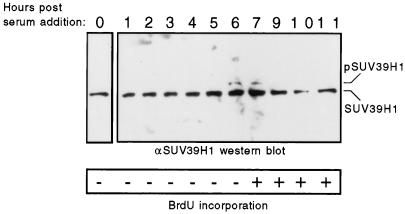
SUV39H1 is transiently phosphorylated at the cell cycle G1/S transition.
The properties of several chromatin-associated proteins are differentially regulated by cell cycle-specific phosphorylation (14, 18, 33). Therefore, we examined whether the minor fraction of SUV39H1 that was phosphorylated in cycling HeLa cells may correlate with a specific phase of the cell cycle. HeLa cells were growth arrested by serum deprivation and then induced by the addition of serum to synchronously reenter the cell cycle. As a surrogate marker of phosphorylation, the relative migration of endogenous SUV39H1 was determined by Western blot analysis of whole cell extracts taken at hourly time points following serum stimulation. Arrested cells and those within 5 h of stimulation showed a single protein band corresponding to the unshifted form of SUV39H1 (Fig. 6). However, at 6 and 7 h, a minor fraction of shifted SUV39H1 was detected in addition to the predominant unshifted form. This correlated with entry into S phase as determined by BrdU incorporation (Fig. 6). The shifted form of SUV39H1 was no longer evident at 9 or more h following serum addition. These observations suggested that SUV39H1 was specifically phosphorylated during the cell cycle at the transition from G1 to S phase.
FIG. 6.
SUV39H1 is phosphorylated at the transition from G1 to S phase of the cell cycle. HeLa cells were growth arrested by serum starvation for 48 h in tissue culture medium. Cells were then stimulated to synchronously reenter the cell cycle by addition of serum-rich medium. Protein lysates were prepared from nonstimulated cells (0) and at hourly time points (indicated above the gel lanes) following serum stimulation. Endogenous SUV39H1 proteins were detected by Western blotting using an anti-SUV39H1 MAb. Migrations of hypo- and hyperphosphorylated SUV39H1 proteins are indicated. The entry of cells into S phase was determined by measuring BrdU incorporation (indicated by + or − below the panel) in parallel cultures following 30-min BrdU pulse-labeling.
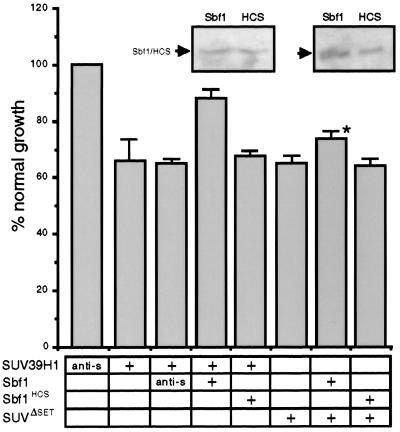
SUV39H1 has growth-inhibitory effects that are partially reversed by Sbf1.
Given the differential phosphorylation of SUV39H1 at G1/S transition, we examined the potential effects of its forced expression on cell cycle progression. NIH 3T3 cells were transduced with retroviral vectors coexpressing SUV39H1 and GFP. The relative growth rates of cells stably expressing SUV39H1 plus GFP or GFP alone were determined by measuring BrdU incorporation. Cells transduced with SUV39H1 showed a 37% decrease in growth rate compared to cells expressing GFP alone (Fig. 7), indicating growth-inhibitory effects that did not completely arrest the cells. To evaluate whether Sbf1 proteins could override SUV39H1-induced growth inhibition, they were coexpressed with SUV39H1 using IRES-containing retroviruses. Constructs were confirmed to be expressing SUV39H1 and/or Sbf1 in transduced cells by Western blotting. Coexpression of oncogenic Sbf1 reversed the growth inhibition of SUV39H1 to 88% of normal, whereas Sbf1HCS had no effect (Fig. 7). SUV39H1ΔSET, which lacks a SET domain, also impaired the growth of NIH 3T3 cells, but in contrast to SUV39H1 its growth-inhibitory effects were not significantly reversed by Sbf1 (Fig. 7). These data indicate that high levels of SUV39H1 partially inhibit cells from entry into S phase, implicating SUV39H1 in growth regulation. Furthermore, this property of SUV39H1 is modulated by Sbf1 in a SET domain-dependent mechanism, suggesting that SUV39H1 may be a downstream target for the oncogenic Sbf1.
FIG. 7.
SUV39H1 has growth-inhibitory properties that are reversed by Sbf1. NIH 3T3 cells were transduced with retroviral stocks expressing SUV39H1 alone or in combination with GFP, Sbf1 (amino acids 1091 to 1861), or Sbf1HCS (indicated below histogram), using an IRES element. SUV39H1 and Sbf1 protein expression in transduced cells was confirmed by Western blotting using anti-SUV39H1 and Sbf1 antibodies. Growth rates were determined by measuring BrdU incorporation in equal numbers of transduced NIH 3T3 cells that were plated 24 h previously. Cells staining positively for BrdU incorporation were counted as a fraction of cells that expressed GFP (growth fraction) or total cells. The growth fraction of cells infected with GFP alone was arbitrarily set at 100%, and percent growth rate was calculated accordingly. Western blots showing expression levels of exogenous Sbf1 and Sbf1HCS are shown as insets above their corresponding panels. Presented data represent the means and standard deviations from three separate experiments. anti-s, cDNA insert in reverse orientation; ∗, growth fraction was not significantly different from SUVΔSET alone (P > 0.05).
DISCUSSION
In this report we provide evidence that the chromatin-organizing activity of a mammalian su(var) protein is regulated through a phosphorylation-dependent mechanism that impinges on its SET domain. Previous studies have shown that SUV39H1 is a structural and functional ortholog of Drosophila Su(var)3-9, a suppressor of PEV (1). Our studies extend these earlier observations by delineating novel roles for this mammalian SET domain protein in transcriptional silencing and cell cycle control. Furthermore, we demonstrate that endogenous SUV39H1 is specifically phosphorylated during G1/S transition of the cell cycle following mitogenic activation and its forced expression antagonizes cellular growth. In each cellular assay of SUV39H1 function, its activity was negatively regulated by the antiphosphatase Sbf1, an oncoprotein that interacts with the SET domain and stabilizes the phosphorylated form of SUV39H1. The SET domain-dependent modulation of SUV39H1 by Sbf1 establishes a phosphorylation-dependent mechanism for regulating the contributions of a su(var) protein in gene silencing and cellular growth control.
Functional analysis of SUV39H1 as a growth and transcriptional regulatory protein.
Using a transient transcriptional assay, we demonstrated that SUV39H1 represses transcription when tethered to DNA through a heterologous DBD. Under these conditions, SUV39H1 displayed features of a long-range repressor capable of acting over several kilobases to silence basal promoters. These properties are characteristic of multiprotein repressor complexes that induce long-lived alterations in chromatin, as opposed to short-range repressors that inhibit or quench activators or components of the basal transcription machinery (5, 37). Consistent with this possible mechanism, SUV39H1 associates with M31 (HP1β), the only other characterized mammalian su(var) homolog (1), and their cosedimentation supports participation in a su(var) complex distinct from two previously described mammalian PcG complexes (45, 46). When SUV39H1 is forcibly expressed under our experimental conditions, it accumulates in distinct nuclear bodies that are large and electron dense, with ultrastructural features suggestive of heterochromatin. These findings as well as the subnuclear localization of endogenous SUV39H1 to heterochromatic regions suggest a role for SUV39H1 in chromatin-mediated gene silencing (1) analogous to the role of Su(var)3-9 in PEV. Indeed, chromatin-dependent gene regulation by SUV39H1 is evident by its ability to increase repression of the pericentromeric white marker gene in transgenic flies (1). Thus, SUV39H1 shares properties with other chromo domain proteins that have been shown to form multimeric complexes capable of transcriptional repression (4, 26).
The ability of SUV39H1 to repress transcription in a transient assay was used to evaluate the functional role of its SET domain, in addition to testing the effects of a SET-interacting protein on SUV39H1 function. The repressor property of SUV39H1 localized to its amino-terminal half which also contains a chromo domain, a modular motif that self-associates and assembles into multimeric complexes on chromatin (31, 39, 46). Notably, transcriptional repression by SUV39H1 did not require its SET domain. This is consistent with previous proposals for a function other than merely repression or activation (36) based on the presence of this highly conserved motif in protein components of both positive and negative regulatory complexes. However, the SET domain was required for cancellation of SUV39H1-mediated repression by Sbf1. These observations are most consistent with a model in which the effector activities of SUV39H1 may be modulated by heterologous interactions that impinge on the SET motif.
Our data also demonstrate that SUV39H1 has features of a growth suppressor protein since its forced expression significantly reduced the growth of NIH 3T3 cells in culture. Although su(var) proteins have not been previously implicated in growth control pathways, transcriptional repression by other multicomponent chromatin modifying complexes containing PcG and TrG proteins has been linked with cell cycle control and senescence. The tumor suppressors p16 and p19Arf, products of the ink4a gene, are critical downstream targets for Bmi-1, an oncoprotein and ortholog of Drosophila Posterior-sex-combs (PSC), a PcG protein (23). Bmi-1 and PSC are components of multimember complexes containing several other PcG proteins and the purified Drosophila complex inhibits the ability of SWI/SNF to remodel nucleosomal arrays in vitro (45). Hbrm/BRG1, a component of the hSWI/SNF complex and an ortholog of Drosophila TrG protein brahma, cooperates with the retinoblastoma protein to inhibit transcription of E2F1 promoters by remodeling chromatin and causes growth arrest when forcibly expressed in mammalian cells (13, 30, 49). These studies provide a paradigm for conceptualizing the possible involvement of SUV39H1 in transcriptional repression of growth control genes through the formation of higher-order chromatin domains, in addition to its likely role in centromere structure and function.
The growth-suppressing effects of SUV39H1, similar to its transcriptional repression, required the SET domain for modulation by Sbf1. Signaling pathways that impinge on chromatin remodeling complexes and regulate growth arrest are complex and not completely defined. However, acetylation (27, 29, 30), phosphorylation (14, 20, 33, 54), and phosphoinositol binding (56) have been shown to affect the ability of several chromatin regulators to form higher-order chromatin domains. Our studies demonstrate that a fraction of total cellular SUV39H1 is specifically phosphorylated during the cell cycle at the G1/S transition, an important checkpoint for entry into S phase (41). The SET domain of SUV39H1 is required for its phosphorylation and the presence of several conserved S/P and T/P sites suggest that it may be a target for cyclin–cyclin-dependent kinase recognition (34). Transient phosphorylation of SUV39H1 at this critical transition point may cancel its repressive transcriptional effects on genes that promote S-phase entry. This would correlate with the ability of Sbf1, which stabilizes the phosphorylated form of SUV39H1, to partially cancel its growth-suppressive effects as well as its ability to repress transcription and form heterochromatin.
Association of SUV39H1 with Sbf1 establishes a novel oncogenic signaling pathway.
In previous studies we demonstrated that the oncogenic activity of Sbf1, in fibroblasts and lymphoid progenitors, required a conserved domain (SID) that mediates interactions with SET domains in vitro (9, 10). Furthermore, the SID was not only necessary but also sufficient for oncogenic activity. However, restoration of phosphatase activity to Sbf1 (Sbf1HCS) completely abrogated its oncogenic effects. These studies suggested a model in which neoplastic transformation induced by Sbf1 (or the SID) resulted from antagonism of endogenous phosphatases and consequent impaired dephosphorylation of critical subordinate proteins. Sbf1 and STYX are the only proteins identified to date that contain naturally occurring inactivating mutations in their catalytic pockets that abrogate their capacity to function as phosphatases (55). Their ability to bind but not dephosphorylate synthetic phosphopeptides suggests that they may function as protective factors to prevent dephosphorylation of substrates (21). Our identification of SUV39H1 as an in vivo binding partner for Sbf1 allowed an evaluation of its hypothesized function as a protective factor. Coexpression of SUV39H1 and oncogenic Sbf1 in cells led to an enhancement of the phosphorylated state of SUV39H1. Sbf1HCS, containing partially restored in vitro phosphatase activity (9), interacted with SUV39H1 but was unable to enhance its phosphorylation. Thus, stabilization of the phosphorylated state of exogenous as well as endogenous SUV39H1 by Sbf1 provides strong support for its proposed role as a phosphorylation-protective factor.
Our studies raise the possibility that the oncogenic effects of Sbf1 may be mediated in part through direct inhibition of the growth-suppressive actions of SUV39H1. The physiological consequences of Sbf1-SUV39H1 interactions are illustrated by the exclusive ability of the oncogenic form of Sbf1 to block the transcriptional repressive properties of SUV39H1, modulate its subnuclear localization, and partially override its growth-suppressive properties. The inability of nononcogenic Sbf1HCS to similarly modulate SUV39H1 function in these assays suggests that part of the mechanism by which Sbf1 transforms cells may involve enhancement of SUV39H1 phosphorylation and subsequent cancellation of its growth-suppressive properties. Since the growth-inhibitory effects of SUV39H1 are modest, we presume that other SET domain proteins serve as targets for the oncogenic Sbf1. More detailed mutational analysis of Sbf1 is required to further characterize the mechanisms for its oncogenic activation and its impact on the effector properties of SUV39H1 and other SET domain proteins. We must also qualify our conclusions regarding the modulation of SUV39H1 by Sbf1, as they are mostly based on assays in which Sbf1 is hyperexpressed. However, our observations that oncogenic Sbf1 modulates SUV39H1 activity in a phosphorylation-dependent manner serves as a useful model for how SET domains may function as gatekeeper motifs to integrate upstream phosphorylation signals with chromatin-dependent gene expression and growth control.
ACKNOWLEDGMENTS
This work was supported by grant CA55029 from the National Institutes of Health. R.F. was supported by training grant 5T32GM07365 from the National Institute of General Medical Sciences.
We acknowledge T. Jenuwein, R. Eisenmann, R. Baer, and J. Milbrandt for providing DNA clones. We thank Peter Nagy for helpful discussion, Bich-Tien Rouse for antibody preparation, Thomas Jenuwein for sharing of unpublished data, and Phil Verzola for photographic assistance.
REFERENCES
- 1.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A S, Reuter G, Jenuwein T. Functional mammalian homologues of the drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baksa K, Morawietz H, Dombradi V, Axton M, Taubert H, Szabo G, Torok I, Udvardy A, Gyurkovics H, Szoor B. Mutations in the protein phosphatase I gene at 87B can differentially affect suppression of position-effect variegation and mitosis in Drosophila melanogaster. Genetics. 1993;135:117–125. doi: 10.1093/genetics/135.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunker C A, Kingston R E. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol Cell Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai H N, Arnosti D N, Levine M. Long-range repression in Drosophila embryo. Proc Natl Acad Sci USA. 1996;93:9309–9314. doi: 10.1073/pnas.93.18.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinwalla V, Jane E P, Harte P J. The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with Polycomb at many sites. EMBO J. 1995;14:2056–2065. doi: 10.1002/j.1460-2075.1995.tb07197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowell I G, Austin C A. Self-association of chromo domain peptides. Biochim Biophys Acta. 1997;1337:198–206. doi: 10.1016/s0167-4838(96)00165-3. [DOI] [PubMed] [Google Scholar]
- 9.Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary M L. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 10.De Vivo I, Cui X, Domen J, Cleary M L. Growth stimulation of primary B cell precursors by the anti-phosphatase Sbf1. Proc Natl Acad Sci USA. 1998;95:9471–9476. doi: 10.1073/pnas.95.16.9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dernburg A F, Sedat J W, Hawley R S. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 12.Djabali M, Selleri L, Parry L, Bower M, Young B D, Evans G A. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 13.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 14.Fujita M, Yamada C, Tsurumi T, Hanaoka F, Matsuzawa K, Inagaki M. Cell cycle- and chromatin binding state-dependent phosphorylation of human MCM heterohexameric complexes. A role for cdc2 kinase. J Biol Chem. 1998;273:17095–17101. doi: 10.1074/jbc.273.27.17095. [DOI] [PubMed] [Google Scholar]
- 15.Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 16.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 18.Herrera R E, Chen F, Weinberg R A. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc Natl Acad Sci USA. 1996;93:11510–11515. doi: 10.1073/pnas.93.21.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobert O, Jallal B, Ullrich A. Interaction of Vav with ENX-1, a putative transcriptional regulator of homeobox gene expression. Mol Cell Biol. 1996;16:3066–3073. doi: 10.1128/mcb.16.6.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang D W, Fanti L, Pak D T, Botchan M R, Pimpinelli S, Kellum R. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol. 1998;142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter T. Anti-phosphatases take the stage. Nat Genet. 1998;18:303–305. doi: 10.1038/ng0498-303. [DOI] [PubMed] [Google Scholar]
- 22.Ivanova A V, Bonaduce M, Ivanov S V, Klar A J. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs J J L, Kieboom K, Marino S, DePinho R A, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 24.Jacobsen S, Pillus L. Modifying chromatin and concepts of cancer. Curr Opin Genet Dev. 1999;9:175–184. doi: 10.1016/S0959-437X(99)80027-6. [DOI] [PubMed] [Google Scholar]
- 25.Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehming N, Le Saux A, Schuller J, Ptashne M. Chromatin components as part of a putative transcriptional repressing complex. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y S, Carey M F, Ptashne M, Green M R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 29.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 30.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 30a.Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol. 2000;20:3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messmer S, Franke A, Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992;6:1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- 32.Miyamoto A, Cui X, Naumovski L, Cleary M L. Helix-loop-helix proteins LYL1 and E2a form heterodimeric complexes with distinctive DNA-binding properties in hematolymphoid cells. Mol Cell Biol. 1996;16:2394–2401. doi: 10.1128/mcb.16.5.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muchardt C, Reyes J C, Bourachot B, Leguoy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 34.Nigg E A. Targets of cyclin-dependent protein kinases. Curr Opin Cell Biol. 1993;5:187–193. doi: 10.1016/0955-0674(93)90101-u. [DOI] [PubMed] [Google Scholar]
- 35.Nislow C, Ray E, Pillus L. SET1, a yeast member of the Trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2431. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlando V, Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr Opin Genet Dev. 1995;5:174–179. doi: 10.1016/0959-437x(95)80005-0. [DOI] [PubMed] [Google Scholar]
- 37.Paro R, Strutt H, Cavalli G. Heritable chromatin states induced by the Polycomb and trithorax group genes. Novartis Found Symp. 1998;214:51–61. doi: 10.1002/9780470515501.ch4. [DOI] [PubMed] [Google Scholar]
- 38.Pirrotta V. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 1997;13:314–318. doi: 10.1016/s0168-9525(97)01178-5. [DOI] [PubMed] [Google Scholar]
- 39.Platero J S, Hartnett T, Eissenberg J C. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rastelli L, Chan C S, Pirrotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed S I. Control of the G1/S transition. Cancer Surv. 1997;29:7–23. [PubMed] [Google Scholar]
- 42.Reuter G, Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- 43.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce C M, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 45.Shao Z, Raible F, Mollaaghababa R, Guyon J R, Wu C T, Bender W, Kingston R E. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 46.Strutt H, Paro R. The polycomb group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol Cell Biol. 1997;17:6773–6783. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swirnoff A H, Apel E D, Svaren J, Sevetson B R, Zimonjic D B, Popescu N C, Milbrandt J. Nab1, a corepressor of NGFI-A (Egr-1), contains an active transcriptional repression domain. Mol Cell Biol. 1998;18:512–524. doi: 10.1128/mcb.18.1.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tkachuk D C, Kohler S, Cleary M L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 49.Trouche D, LeChaloney C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tschiersch B, Hoffman A, Krauss V, Dorn R, Korge G, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Lohuizen M, Tijms M, Voncken J W, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Versteege I, Sevenet N, Lange J, Rousseau-Merck M F, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 53.Waring P, Cleary M L. Disruption of a homolog of trithorax by 11q23 translocations: leukemogenic and transcriptional implications. Curr Top Microbiol Immunol. 1997;220:1–23. doi: 10.1007/978-3-642-60479-9_1. [DOI] [PubMed] [Google Scholar]
- 54.Wei Y, Yu L, Bowen J, Gorovsky M A, Allis C D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 55.Wishart M J, Dixon J E. Gathering STYX: phosphatase-like form predicts functions for unique protein-interaction domains. Trends Biochem Sci. 1998;23:301–306. doi: 10.1016/s0968-0004(98)01241-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhao K, Wang W, Rando O J, Xue Y, Swiderek K, Kuo A, Crabtree G R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 57.Zhao T, Eissenberg J C. Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J Biol Chem. 1999;274:15095–15100. doi: 10.1074/jbc.274.21.15095. [DOI] [PubMed] [Google Scholar]