Abstract
Introduction
Steroid-induced osteonecrosis of the femoral head (ONFH) is a disease of the bone. Metabolism and genetic factors are generally considered to play an important role. The purpose of this study was to investigate the relationship between single-nucleotide polymorphisms (SNPs) in MIR17HG and MIR155HG and the risk of steroid-induced ONFH in the population of northern China.
Methods
A total of 199 steroid-induced ONFH patients and 506 healthy controls were recruited for the study. Four SNPs of MIR17HG and seven SNPs of MIR155HG were genotyped by Sequenom MassARRAY. ORs and 95% CIs were used to evaluate the relationship between these SNPs and steroid-induced ONFH.
Results
In the codominant model, patients with the MIR17HG SNPs (rs7318578) AA genotype had an increased risk of steroid-induced ONFH (OR = 1.79, p = 0.039); in the recessive model, patients with the MIR17HG SNP (rs7318578) AA genotype had an increased risk of steroid-induced ONFH (OR = 1.78, p = 0.032). Stratified analysis showed that a MIR17HG SNP (rs7318578) and the MIR155HG SNPs (rs77218221, rs11911469, rs34904192 and rs4143370) were closely related to different unornamented phenotypes of steroid-induced ONFH. Analysis of the clinical indicators revealed significant differences in high-density lipoprotein (HDL-C) levels between the ONFH group and the control group (p = 0.005). In the MIR17HG SNP (rs75267932), patients with different genotypes had different levels of triglyceride (TG). The MIR155HG SNPs (rs77699734, rs1893650, and rs34904192) showed differences in triglyceride (TG), high-density lipoprotein (HDL-C) and low-density lipoprotein (LDL-C) levels in patients with different genotypes.
Conclusion
Our results confirm that MIR17HG and MIR155HG gene mutations are associated with steroid-induced ONFH susceptibility in the population of northern China, providing new evidence for the early detection and prevention of ONFH.
Keywords: Steroid-induced osteonecrosis of the femoral head, MIR17HG, MIR155HG, Single-nucleotide polymorphism, Case–control study
Introduction
Osteonecrosis of the femoral head (ONFH), also known as avascularity necrosis of the femoral head, occurs mostly in young adults aged 30–50 and is a refractory disease in the Department of Orthopedics [23]. Due to hidden early symptoms, most patients with osteonecrosis develop to the middle and late stages when they are diagnosed with osteonecrosis, and there are varying degrees of femoral head collapse or loss of hip joint function, which seriously affect the quality of life of patients [30, 32, 36]. ONFH has different causes including steroids, alcohol, trauma, and idiopathic origins. Among them, steroid-induced ONFH accounts for the highest proportion and has become the main cause of femoral head necrosis [3]. A national epidemiological study on ONFH in Japan showed that steroid-induced ONFH accounted for 51% of the total cases [5]. Steroid-induced ONFH is a complex and multifactorial disease. A variety of internal and external factors jointly promote intramural microvascular lesions; thrombosis leads to insufficient blood supply and oxygen supply to the femoral head, and finally leads to bone cell death. However, the exact pathogenesis is still unclear. Clinical observations have found that not all patients treated with glucocorticoids have experienced femoral head necrosis, suggesting that there may be other risk factors or individual differences in the occurrence of glucocorticoid-induced ONFH. Some studies have shown that the difference between individuals may be related to gene polymorphism [18, 32].
Osteoblasts (OBs) and osteoclasts (OCs) are the main cells regulating bone homeostasis. Both cell types play a key role in bone tissue reconstruction, bone system integrity, mineral homeostasis in bone tissue, and bone metabolism balance [1, 13]. In the ONFH study, one of the pathogeneses was due to the imbalance of the OB/OC ratio and activity in bone tissue, which disrupted the dynamic balance between bone resorption and bone formation [21, 34].
MicroRNAs (miRNAs) are small, single-stranded, endogenous noncoding RNAs with a length of 21–25 nucleotides. They play biological roles by inhibiting the expression of target proteins at the posttranscriptional level [1, 29, 38]. According to the conformation analysis, more than one third of human genes are regulated by miRNAs, which indicates that miRNAs play an important role in regulating gene expression [10, 11]. MiR-17 and miR-155 are transcripts of MIR17HG and MIR155HG, respectively. MiR-17 and miR-155 are both multifunctional miRNAs, that play a consequential role in the occurrence, development, and prognosis of tumors [28, 38]. They also have important regulatory effects on bone metabolism. MiR-17-92 is located on chromosome 13q31.3 of the host gene MIR17HG (also known as c13orf2 or mirage). Its expression is downregulated with the differentiation of OBs, and its expression is the lowest in mature OBs. These results indicate that miR-17-92 regulates the proliferation, differentiation, and apoptosis of normal OBs [37]. In the study of OCs, it was found that miR-155 inhibits osteoclast differentiation by mediating IFN-b [33].
Single-nucleotide polymorphisms (SNPs) are the most common type of genetic variation. At present, there is no study on the relationship between MIR17HG and MIR155HG SNPs and steroid-induced ONFH. In this study, we conducted a case–control study in a Chinese Han population to analyze the relationship between 11 SNPs and the risk of steroid-induced ONFH. Our results can be used as molecular markers for the diagnosis of osteonecrosis and provide a new theoretical basis for the early detection and prevention of osteonecrosis.
Materials and methods
Study participants
From 2018 to 2020, a total of 199 blood samples from steroid-induced ONFH patients were collected at Zhengzhou’s Traditional Chinese Medicine Hospital trauma center. The diagnosis of steroid-induced ONFH was defined as prednisolone with an average routine daily dose of 16.6 mg or a maximum daily common dose of 80 mg for at least 1 year [35]. All patients underwent anterior, posterior, and frog X-ray examinations. The diagnosis of ONFH was confirmed by MRI. All patients with steroid-induced ONFH had no history of chemotherapy or radiotherapy.
At the same time, 506 unrelated healthy people were recruited from the Zhengzhou Traditional Chinese Medicine Hospital as a control. All the subjects were of Han nationality and lived in Zhengzhou and its surrounding areas. Steroid-induced ONFH was excluded as follows: traumatic dislocation of the hip or other hip diseases, more than 400 ml of alcohol per week or having a clearly familial hereditary disease. In general, subjects with chronic diseases and diseases involving the brain, heart, liver, lungs, and other vital organs were excluded from the study.
The Ethical Committee of Zhengzhou Traditional Chinese Medicine Traumatology Hospital agreed to the study, and all participants signed written informed consent forms.
Genotyping
Five-milliliter blood samples were collected in EDTA test tubes, centrifuged at 2000 rpm for 10 min, and stored at − 80 °C for future experiments. Genomic DNA was extracted by GoldMag extraction (GoldMag, China) and stored at 20 °C. The multichannel SNP quality extension detection method was developed using Agena MassARRAY Assay Design 4.0 software. The Agena MassARRAY system was used for SNP genotyping. Agena Bioscience Typer 4.0 software was used for data management and analysis. In addition, approximately 10% of the total samples were randomly selected for repeat genotyping, and the reproducibility was 100%.
Statistical analysis
SPSS 20.0 software (SPSS, Chicago, IL, USA) was used for statistical analysis of the data. A chi-squared test was used to analyze whether the genotype distribution of the control group accorded with Hardy Weinberg’s equilibrium (HWE). In this study, p values less than 0.05 were statistically significant. We used a chi-squared test to calculate the distribution differences of alleles, genotypes, and haplotypes. We analyzed the relationship between genotypes of MIR17HG and MIR155HG polymorphisms and the risk of steroid-induced ONFH using different genetic models including codominant, dominant, recessive, and log-additive models; and calculated the odds ratios (ORs) and 95% confidence intervals (CIs) of logistic regression adjusted for age and sex. Then, stratified analysis was conducted according to age, gender, hip joint disease, and course of disease. Finally, linkage disequilibrium (LD) and haplotype structure were estimated by half software (version 4.2).
Analysis of clinical indicators
Blood samples of the subjects were further analyzed. The Roche Cobas 8000-701 biochemical analyzer was used to detect blood lipid indexes in the body, and six biochemical indexes (TC, TG, HDL, LDL, ApoA1, ApoB). Two-tailed T test was used for quantitative variables, and chi-squared test was used for categorical variables. The expression of clinical indexes between the case group and the control group as well as different genotypes were analyzed. All p values in this study were two sided, and p value of less than 0.05 was the cutoff value for statistical significance.
KEGG analysis of downstream genes regulated by MIR155HG
We searched out the downstream genes regulated by MIR155HG in (http://www.bio-bigdata.net/LncACTdb/index.html) and (http://mirtarbase.cuhk.edu.cn/) databases, using R software. KEGG analysis was performed on these genes.
Result
Information on steroid-induced ONFH in patients and healthy individuals is shown in Table 1. A total of 199 patients (116 males and 83 females) were enrolled, with an average age of 41.21 ± 12.90 years; 506 healthy persons (423 males and 83 females) with an average age of 42.58 ± 13.15 years were included. Basic features included gender, age, hip lesions (unilateral or bilateral), and course ( > 25 months or ≤ 25 months). Steroid-induced ONFH patients were matched with the age of the control group (p = 0.212).
Table 1.
Characteristics of the individuals in controls and steroid-induced ONFH patients
| Variables | Cases (n = 199) | Controls (n = 506) | p |
|---|---|---|---|
| N (%) | N (%) | ||
| Age, years | |||
| Mean ± SD | 41.21 ± 12.90 | 42.58 ± 13.15 | 0.212a |
| ≤ 45 | 129 (65%) | 275 (54%) | |
| > 45 | 70 (35%) | 231 (46%) | |
| Gender | < 0.001b | ||
| Male | 116 (58%) | 423 (84%) | |
| Female | 83 (42%) | 83 (16%) | |
| Hip lesions | |||
| Unilateral | 142 (71%) | ||
| Bilateral | 55 (28%) | ||
| Course, months | |||
| > 25 | 67 (34%) | ||
| ≤ 25 | 132 (66%) |
P < 0.05 indicates statistical significance.
aIndependent sample t test.
bTwo-sided chi-squared test.
The basic information of 11 SNPs in MIR17HG and MIR155HG, including gene, chromosome, location, allele, MAF, etc., is shown in Table 2. All these SNPs were in Hardy Weinberg’s equilibrium in the control group. The Χ2 test was used to calculate the difference in the allele frequency distribution between the case and the control group. We did not find any loci that affect the genetic susceptibility of steroid-induced ONFH (Table 2).
Table 2.
Basic information of candidate SNPs in this study
| SNP | Gene | Chromosome | Position | Alleles | MAF | HWE | ORs | 95% CI | pb | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A/B | Case | Control | pa | ||||||||
| rs75267932 | MIR17HG | 13 | 91351812 | A/G | 0.114 | 0.124 | 0.411 | 0.90 | 0.63 | 1.30 | 0.575 |
| rs7336610 | MIR17HG | 13 | 91352883 | C/T | 0.528 | 0.481 | 0.129 | 1.21 | 0.95 | 1.52 | 0.116 |
| rs7318578 | MIR17HG | 13 | 91353215 | A/C | 0.327 | 0.292 | 0.389 | 1.18 | 0.92 | 1.51 | 0.206 |
| rs17735387 | MIR17HG | 13 | 91353800 | A/G | 0.191 | 0.176 | 0.358 | 1.1 | 0.81 | 1.48 | 0.537 |
| rs4143370 | MIR155HG | 21 | 25564661 | C/G | 0.136 | 0.146 | 0.597 | 0.92 | 0.66 | 1.28 | 0.610 |
| rs77218221 | MIR155HG | 21 | 25565063 | C/T | 0.058 | 0.046 | 1.000 | 1.26 | 0.75 | 2.10 | 0.377 |
| rs12482371 | MIR155HG | 21 | 25566041 | C/T | 0.332 | 0.323 | 0.104 | 1.04 | 0.81 | 1.33 | 0.758 |
| rs77699734 | MIR155HG | 21 | 25566995 | C/G | 0.090 | 0.103 | 0.636 | 0.87 | 0.58 | 1.29 | 0.486 |
| rs11911469 | MIR155HG | 21 | 25567971 | A/C | 0.138 | 0.130 | 0.545 | 1.12 | 0.80 | 1.57 | 0.522 |
| rs1893650 | MIR155HG | 21 | 25568503 | C/T | 0.196 | 0.202 | 0.491 | 0.97 | 0.72 | 1.29 | 0.813 |
| rs34904192 | MIR155HG | 21 | 25569623 | A/G | 0.226 | 0.251 | 0.288 | 0.87 | 0.66 | 1.15 | 0.328 |
SNP, single-nucleotide polymorphism; HWE, Hardy–Weinberg’s equilibrium; OR, odds ratio; 95% CI, 95% confidence interval; MAF, minor allele frequency
ap values were calculated by exact test
bp values were calculated by Pearson’s chi-squared test
We further evaluated the association between SNPs and the risk of steroid-induced ONFH in four genetic models (codominant, dominant, recessionary, and additive) by logistic regression analysis adjusted for gender and age. Table 3 shows that SNP rs7318578 was associated with an increased risk of steroid-induced ONFH in the age, gender-adjusted codominant model (OR = 1.01, 95% CI: 0.70–1.46, p = 0.039) and recessionary model (OR = 1.78, 95% CI: 1.05–3.00, p = 0.032) (Table 3).
Table 3.
Genotypic model analysis of relationship between SNPs and steroid-induced ONFH
| SNP | Model | Genotype | Group = control | Group = case | OR (95% CI) | p |
|---|---|---|---|---|---|---|
| rs7318578 | Codominant | C/C | 47 (9.3%) | 27 (13.6%) | 1 | 0.039* |
| C/A | 200 (39.5%) | 76 (38.2%) | 1.01 (0.70–1.46) | |||
| A/A | 256 (50.6%) | 96 (48.2%) | 1.79 (1.03–3.09) | |||
| Dominant | C/C | 47 (9.3%) | 27 (13.6%) | 1 | 0.427 | |
| C/A–A/A | 456 (90.1%) | 172 (86.4%) | 1.15 (0.82–1.62) | |||
| Recessive | C/C–C/A | 247 (48.8%) | 103 (51.8%) | 1 | 0.032* | |
| A/A | 256 (50.6%) | 96 (48.2%) | 1.78 (1.05–3.00) | |||
| Log-additive | - | - | - | 1.23 (0.95–1.58) | 0.115 |
*p < 0.05 indicates statistical significance.
The data were stratified according to age, gender, unilateral or bilateral hip joint disease, and course of disease (> 25 months or ≤ 25 months). We further used the Χ2 test to calculate the difference in allele frequency distribution between the case and the control group in different subgroups, and the results are shown in Table 4. Above age 45, rs77218221 in the MIR155HG polymorphism was associated with an increased risk of steroid-induced ONFH (OR = 2.03, 95% CI: 1.02–4.04, p = 0.041). In males, rs11911469 in the MIR155HG polymorphism was associated with an increased risk of steroid-induced ONFH (OR = 1.53, 95% CI: 1.01–2.30, p = 0.0396), whereas rs34904192 reduced the risk of steroid-induced ONFH (OR = 0.67, 95% CI: 0.47–0.96, p = 0.0297). In women, rs11911469 in the MIR155HG polymorphism reduced the risk of steroid-induced ONFH (OR = 0.50, 95% CI: 0.26–0.94, p = 0.0289), and rs4143370 was associated with an increased risk of steroid-induced ONFH (OR = 1.86, 95% CI: 1.04–3.32, p = 0.035). In the course of disease, rs77218221 (OR = 2.73,95% CI: 1.16–6.40, p = 0.017) and rs34904192 (OR = 1.72, 95% CI: 1.06–2.78, p = 0.027) in the MIR155HG polymorphism were associated with an increased risk of steroid-induced ONFH (Table 4). The effects of allele and SNP genotypes in different subgroups on steroid-induced ONFH risk were further evaluated, as shown in Table 5. Stratified analysis by age showed that rs77218221 was associated with an increased risk of steroid-induced ONFH in the group older than 45 years of age (OR = 2.75, 95% CI: 1.24–6.08, p = 0.013), in the dominant model (OR = 2.65, 95% CI: 1.20–5.82, p = 0.016), and in the additive model (OR = 2.39, 95% CI: 1.12–5.08, p = 0.023). Rs7318578 was associated with risk reduction of steroid-induced ONFH in patients senior than 45 years of age in the codominant model (OR = 0.82, 95% CI: 0.43–1.55, p = 0.042) and the recessive model (OR = 0.83, 95% CI: 1.61–6.92, p = 0.022). In the male group, rs34904192 reduced the risk of steroid-induced ONFH in the codominant model (OR = 0.61, 95% CI: 0.39–0.95, p = 0.030), the dominant model (OR = 0.61, 95% CI: 0.40–0.94, p = 0.024), and the additive model (OR = 0.67, 95% CI: 0.47–0.98, p = 0.037). The codominant model of rs11911469 (OR = 0.47, 95% CI: 0.23–0.94, p = 0.034), the dominant model (OR = 0.45, 95% CI: 0.22–0.91, p = 0.026), and the additive model (OR = 0.45, 95% CI: 0.23–0.88, p = 0.021) were associated with reduced risk of steroid-induced ONFH in the female group. In the course group, rs77218221 was associated with an increased risk of steroid-induced ONFH in the dominant model (OR = 2.64, 95% CI: 1.06–6.55, p = 0.037), additive model (OR = 2.64, 95% CI: 1.06–6.55, p = 0.038) and rs34904192 under the codominant model (OR = 1.22, 95% CI: 0.63–2.35, p = 0.043) (Table 5).
Table 4.
The subgroup information of the MIR17HG gene and MIR155HG gene
| Subgroup | SNP | Alleles | MAF | HWE-pa | ORs | 95% CI | pb | ||
|---|---|---|---|---|---|---|---|---|---|
| A/B | Case | Control | |||||||
| Age,> 45 | rs77218221 | C/T | 0.1 | 0.052 | 0.467 | 2.03 | 1.02 | 4.04 | 0.041 |
| Gender, male | rs11911469 | A/C | 0.164 | 0.113 | 1.000 | 1.53 | 1.02 | 2.3 | 0.040 |
| rs34904192 | A/G | 0.19 | 0.259 | 0.312 | 0.67 | 0.47 | 0.96 | 0.030 | |
| Gender, female | rs11911469 | A/C | 0.1 | 0.19 | 0.282 | 0.50 | 0.26 | 0.94 | 0.029 |
| Course, months | rs4143370 | C/G | 0.187 | 0.11 | 0.186 | 1.86 | 1.04 | 3.32 | 0.035 |
| Case, > 25 | rs77218221 | C/T | 0.097 | 0.038 | 1.000 | 2.73 | 1.16 | 6.40 | 0.017 |
| Control, ≤ 25 | rs34904192 | A/G | 0.291 | 0.193 | 0.783 | 1.72 | 1.06 | 2.78 | 0.027 |
SNP, single-nucleotide polymorphism; HWE, Hardy–Weinberg’s equilibrium; OR, odds ratio; 95% CI, 95% confidence interval; MAF, minor allele frequency
ap values were calculated by exact test
bp values were calculated by Pearson’s chi-squared test
Table 5.
The relationship between MIR17HG and MIR155HG gene polymorphism and steroid-induced ONFH subgroup analysis
| Subgroup analysis | SNP | Model | Genotype | Control | Case | OR (95% CI) | p |
|---|---|---|---|---|---|---|---|
| Age, > 45 | rs77218221 | Codominant | C/C | 1 (0.4%) | 0 | 1 | 0.013* |
| C/T | 22 (9.5%) | 14 (20.0%) | 2.75 (1.24–6.08) | ||||
| T/T | 208 (90.0%) | 56 (80.0%) | - | ||||
| Dominant | C/C | 1 (0.4%) | 0 | 1 | 0.016* | ||
| C/T–T/T | 230 (99.6%) | 70 (100.0%) | 2.65 (1.20–5.82) | ||||
| Recessive | C/C–C/T | 23 (10.0%) | 14 (20.0%) | 1 | 0.999 | ||
| T/T | 208 (90.0%) | 56 (80.0%) | - | ||||
| Log-additive | - | - | - | 2.39 (1.12–5.08) | 0.023* | ||
| Age, > 45 | rs7318578 | Codominant | C/C | 17 (7.4%) | 10 (14.3%) | 1 | 0.042* |
| C/A | 90 (39.0%) | 23 (32.9%) | 0.82 (0.43–1.55) | ||||
| A/A | 121 (52.4%) | 37 (52.9%) | 2.62 (1.04–6.60) | ||||
| Dominant | C/C | 17 (7.4%) | 10 (14.3%) | 1 | 0.826 | ||
| C/A–A/A | 211 (91.3%) | 60 (85.7%) | 1.07 (0.60–1.91) | ||||
| Recessive | C/C–C/A | 107 (46.3%) | 33 (47.1%) | 1 | 0.022* | ||
| A/A | 121 (52.4%) | 37 (52.9%) | 2.83 (1.61–6.92) | ||||
| Log-additive | - | - | - | 1.30 (0.84–2.01) | 0.238 | ||
| Male | rs34904192 | Codominant | A/A | 24 (5.7%) | 5 (4.3%) | 1 | 0.030* |
| A/G | 171 (40.4%) | 34 (29.3%) | 0.61 (0.39–0.95) | ||||
| G/G | 228 (53.9%) | 77 (66.4%) | 0.62 (0.23–1.69) | ||||
| Dominant | A/A | 24 (5.7%) | 5 (4.3%) | 1 | 0.024* | ||
| A/G–G/G | 339 (80.1%) | 111 (95.7%) | 0.61 (0.40–0.94) | ||||
| Recessive | A/A–A/G | 195 (46.1%) | 39 (33.6%) | 1 | 0.557 | ||
| G/G | 228 (53.9%) | 77(66.4%) | 0.74 (0.27–2.00) | ||||
| Log-additive | - | - | - | 0.67 (0.47–0.98) | 0.037* | ||
| Female | rs11911469 | Codominant | C/C | 1 (1.2%) | 0 | 1 | 0.034* |
| C/A | 29 (34.9%) | 17 (20.5%) | 0.47 (0.23–0.94) | ||||
| A/A | 53 (63.9%) | 66 (79.5%) | - | ||||
| Dominant | C/C | 1 (1.2%) | 0 | 1 | 0.026* | ||
| C/A–A/A | 82 (98.8%) | 83 (100.0%) | 0.45 (0.22–0.91) | ||||
| Recessive | C/C–C/A | 30 (36.1%) | 17 (20.5%) | 1 | 0.999 | ||
| A/A | 53 (63.9%) | 66 (79.5%) | - | ||||
| Log-additive | - | - | - | 0.45 (0.23–0.88) | 0.021* | ||
| Course, months | rs77218221 | Codominant | C/C | 0 | 0 | 1 | |
| Case, > 25 | C/T | 10 (7.6%) | 13 (19.4%) | - | |||
| Control, ≤ 25 | T/T | 122 (92.4%) | 54 (80.6%) | - | |||
| Dominant | C/C | 0 | 0 | 1 | 0.037* | ||
| C/T–T/T | 132 (100.0%) | 67 (100.0%) | 2.64 (1.06–6.55) | ||||
| Recessive | C/C–C/T | 10 (7.6%) | 13 (19.4%) | 1 | - | ||
| T/T | 122 (92.4%) | 54 (80.6%) | - | ||||
| Log-additive | - | - | - | 2.64 (1.06–6.55) | 0.038* | ||
| Course, months | rs34904192 | Codominant | A/A | 4 (3.0%) | 7 (10.4%) | 1 | 0.043* |
| Case, > 25 | A/G | 43 (32.6%) | 25 (37.3%) | 1.22 (0.63–2.35) | |||
| Control, ≤ 25 | G/G | 85 (64.4%) | 35 (52.2%) | 3.84 (1.04–14.15) | |||
| Dominant | A/A | 4 (3.0%) | 7 (10.4%) | 1 | 0.240 | ||
| A/G–G/G | 128 (97.0%) | 60 (89.6%) | 1.45 (0.78–2.69) | ||||
| Recessive | A/A–A/G | 47 (35.6%) | 32 (47.8%) | 1 | 0.052 | ||
| G/G | 85 (64.4%) | 35 (52.2%) | 3.56 (0.99–12.82) | ||||
| Log-additive | - | - | - | 1.55 (0.95–2.55) | 0.082 |
* p < 0.05 indicates statistical significance
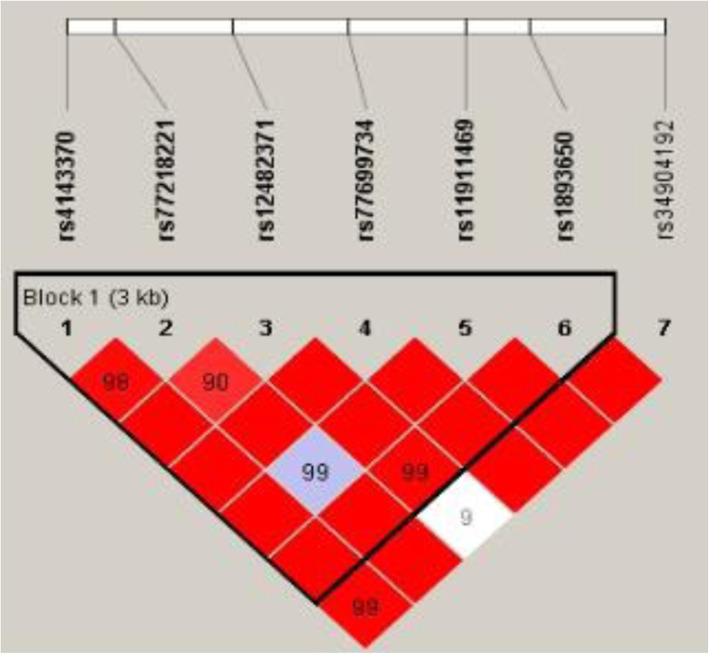
Linkage analysis showed that MIR17HG SNPs (rs75267932, rs7336610, and rs7318578) (Fig. 1) and MIR155HG SNPs (rs4143370, rs77218221, rs12482371, rs77699734, rs11911469, and rs189365) (Fig. 2) exhibited significant linkage disequilibrium.
Fig. 1.
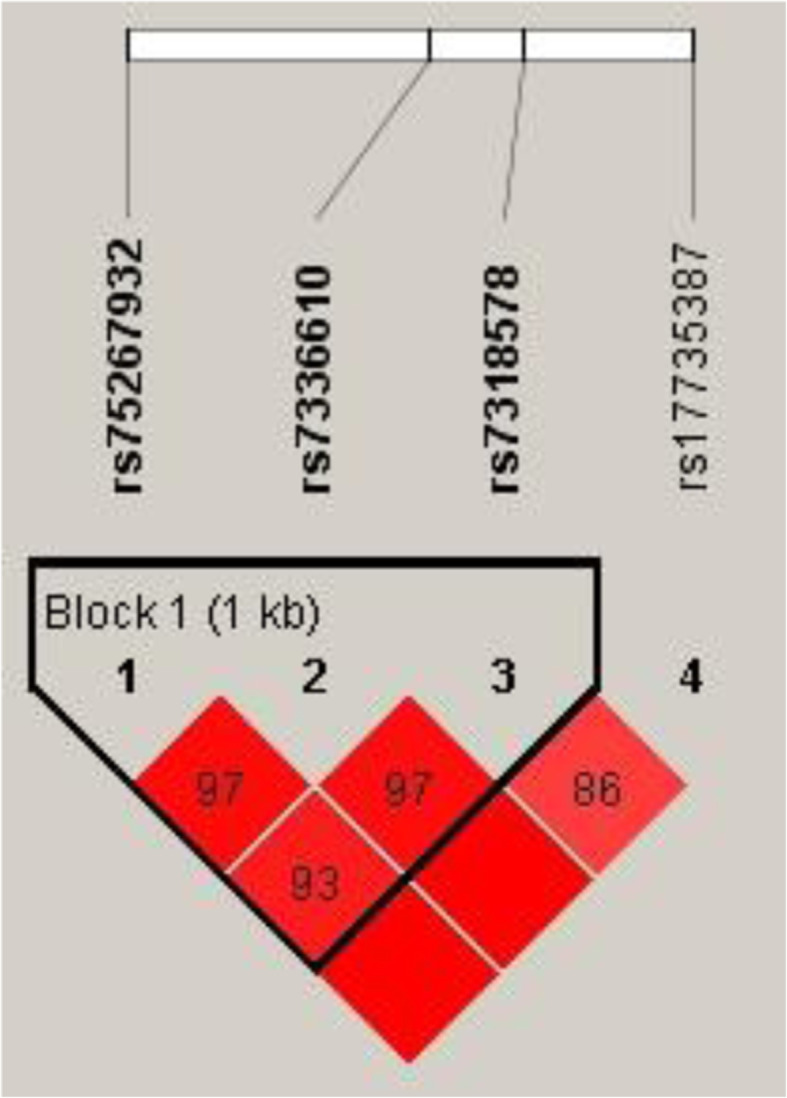
Haplotype block map for the SNPs in MIR17HG gene. The numbers inside the diamonds indicate the D’ for pairwise analyses
Fig. 2.
Haplotype block map for the SNPs in MIR155HG gene. The numbers inside the diamonds indicate the D’ for pairwise analyses
Analysis of clinical indicators revealed significant differences in HDL-C levels between the steroid-induced ONFH group and the control group (p = 0.005), as shown in Fig. 3A. The correlation analysis between rs77699734 and rs34904192 genotypes and serum lipid levels showed that there were differences in serum LDL-C levels among the three genotypes of rs77699734 GG, GC, and CC (p = 0.018), and the serum LDL-C level of ONFH patients with the GG genotype was significantly higher than that of patients with the GC or CC genotype. There were relevant differences in serum HDL-C levels among ONFH patients with rs34904192 AA, AG, and GG genotypes (p = 0.022), as shown in Fig. 3B.
Fig. 3.
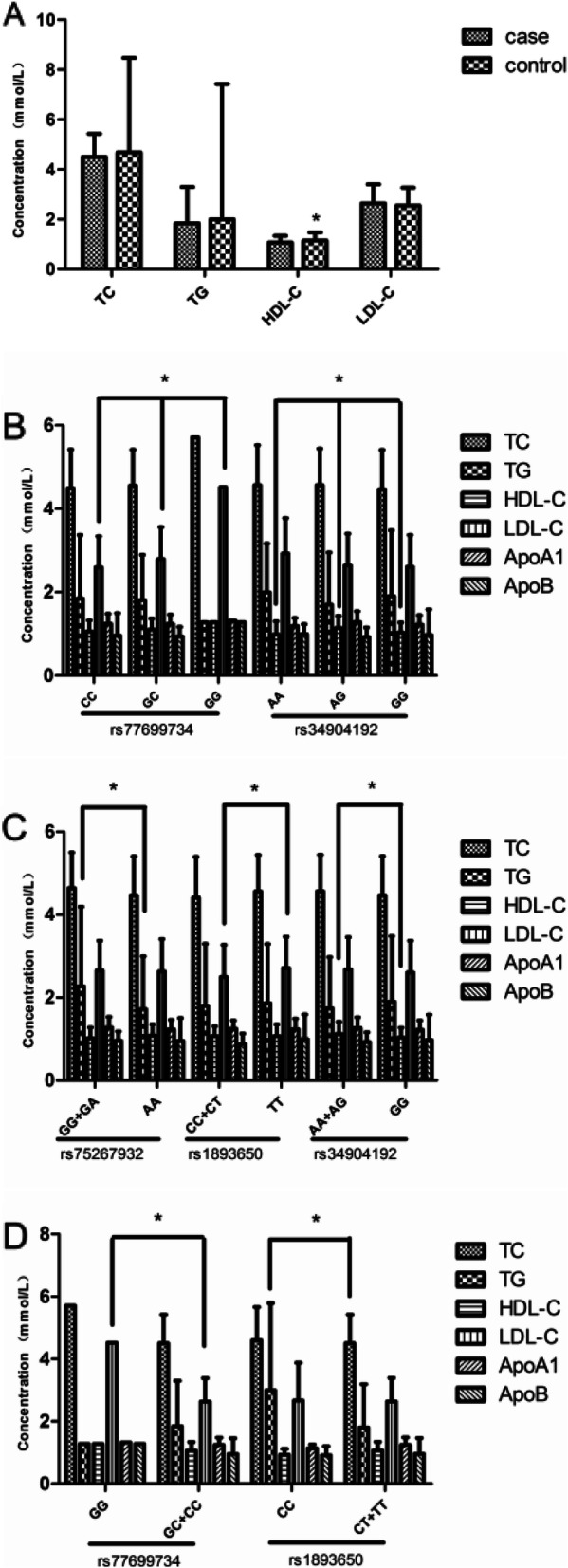
The serum lipid levels between the ONFH and control groups, and the association of MIR17HG and MIR155HG genotypes with the serum lipid levels of ONFH patients. A The serum levels of TC, TG, HDL-c, and LDL-c between the ONFH and control groups. *p < 0.005. B The association of MIR17HG and MIR155HG genotypes with the serum lipid levels of the ONFH group. Rs77699734, *p < 0.018; rs34904192, *p < 0.022 as indicated. Data are presented as mean ± standard deviation. ONFH, osteonecrosis of the femoral head; TC, total cholesterol; TG, triglyceride; HDL, high-density lipoprotein; LDL, low-density lipoprotein; ApoA1, apolipoprotein A1; ApoB, apolipoprotein B. The relationship between MIR17HG and MIR155HG genotypes in the ONFH group and blood lipid levels in ONFH patients. (C) Compare the relationship of blood lipid levels between dominant models of patients' genotypes. TG in Rs75267932, *p<0.027; LDL-C in rs1893650, *p<0.048; HDL-C in rs34904192, *p<0.035. (D) Compare the relationship between blood lipid levels in recessive models of patients' genotypes LDL-C in rs77699734, *p<0.013; TG in rs1893650, *P<0.047; as shown in the figure. Data are expressed as mean ± standard deviation. ONFH, femoral head necrosis; TC, total cholesterol; TG, triglycerides; HDL, high density lipoprotein; LDL, low density lipoprotein; apolipoprotein A1 (ApoA1); apolipoprotein B (apoB)
There were significant differences in serum TG between GG + GA and AA (p = 0.027) in the rs75267932 governing model, in the LDL-C level between CC + CT and TT in the rs1893650 governing model (p = 0.048), and in the HDL-C level in three genotypes of ONFH patients with AA + AG and GG in the rs34904192 dominant model (p = 0.035), as shown in Fig. 3C. In the recessionary model carrying rs77699734, the serum LDL-C levels of patients with ONFH between GG and GC + CC were unusual (p = 0.013), and the serum TG levels of patients with ONFH between CC and CT + TT in the rs1893650 recessive models were distinct (p = 0.047), as shown in Fig. 3D.
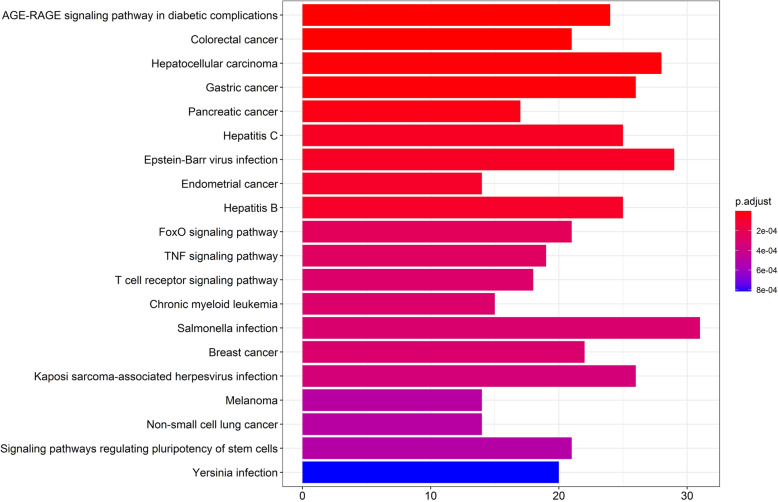
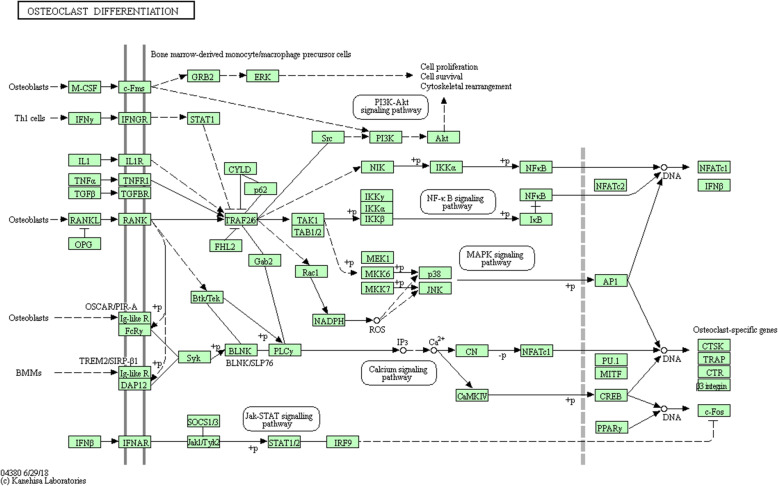
In the database (http://www.bio-bigdata.net/LncACTdb/index.html), we found that MIR155HG has a regulatory effect on miR-155-5p. In the (http://mirtarbase.cuhk.edu.cn/) database, we found 919 target genes regulated by miR-155-5p. Through KEGG analysis, we found that the signal pathway that regulates the pluripotency of stem cells and osteoclast differentiation pathway is involved in the occurrence of ONFH has an important role in it. Among them, six miR-155-5p regulated genes (IFNGR1, AKT1, TAB2, MITF, SOCS1, and SOCS3) were found in the osteoclast differentiation pathway, as shown in Figs. 4 and 5.
Fig. 4.
KEGG analysis of downstream genes regulated by MIR155HG
Fig. 5.
Osteoclast differentiation pathway
Discussion
This study is the first to investigate the selective SNPs in MIR17HG and MIR155HG and their relationship with steroid-induced ONFH. In the population of northern China, we found that MIR17HG SNP (rs7318578) was associated with an increased risk of steroid-induced ONFH. MIR17HG SNP (rs7318578) and MIR155HG SNPs (rs77218221, rs11911469, rs34904192, and rs4143370) were closely related to distinctive clinical phenotypes of steroid-induced ONFH. MIR17HG SNP (rs75267932) and MIR155HG SNPs (rs77699734, rs1893650, and rs34904192) were correlated with different lipid indexes.
MIR17HG is located on chromosome 13 and has been found to play a prominent role in a variety of human diseases. Studies have confirmed that abnormal expression of miR-17 has been detected in the local tissues or serum of patients with ONFH [8, 29]. MiR-17-5p (located in the miR-17-92 cluster) delayed the differentiation and proliferation of OBs by downregulating the partial expression of SMAD7, leading to the occurrence of nontraumatic ONFH [8, 24]. Studies also found that the miR-17-92 cluster was downregulated in the OB differentiation of ES cells and the bone progenitor cell line Mc3t3-e1 [16]. In the analysis of miRNA expression in OBs exposed to apoptotic inducers, it was found that miR-17-92a played a consequential role in protecting estrogen anti-bone loss by regulating Bim expression [6]. These findings suggest that the miR-17-92 cluster plays a key role in the occurrence and development of steroid-induced ONFH. Previous studies have shown an increased risk of multiple myelomas in patients with the rs7336610 allele of MIR17HG, and the three SNPs of MIR17HG (rs7336610, rs7318578, and rs17735387) were associated with a risk of colorectal cancer [2, 28]. In this study, rs7318578 was identified as a genetic susceptibility factor for steroid-induced ONFH. To date, only the SNPs (rs7336610, rs7318578, rs17735387, and rs75267932) of MIR17HG and the risk of steroid-induced ONFH have been investigated. Therefore, more samples are needed for correlation studies to confirm the results.
OCs are one of the most important cells in maintaining homeostasis, and their excessive bone resorption often leads to bone loss. According to the literature, miR-155 is involved in the regulation of OCs. In osteoporotic mice, miR-155 inhibited OC activation by targeting the leptin receptor (LEPR) via the AMPK pathway [12]. MiR-155 can target proto-oncogene SPI1, microsomia-related transcription factor, and cytokine signal transduction inhibitor protein 1 to inhibit macrophage activation and OC differentiation, thus inhibiting the process of bone resorption [16]. As an intron gene with higher transcription levels, miRNA is involved in physiological processes such as development, cell proliferation, differentiation, and metabolism [9]. Research on mesenchymal stem cells of patients with steroid-induced femoral necrosis showed that the overexpression of miR-155-5p in their cells can directly inhibit GSK3B to promote cell proliferation and bone differentiation [25, 26]. MiR-155 is the transcription product of the host gene MIR155HG, and its expression may be affected by genetic variation in the MIR155HG gene. In a study of SNPs in MIR155HG and colorectal cancer, rs12482371, rs1893650, rs92888, rs11911469, and rs34904192 increased the risk of colorectal cancer [27]. In our study, the four SNPs of MIR155HG (rs77218221, RS11911469, rs34904192, and rs4143370) were associated with a risk of steroid-induced ONFH. Our study suggests that MIR155HG may be a new susceptibility gene for steroid-induced ONFH, providing new evidence for the early detection of ONFH.
In a study of the pathogenesis of femoral head necrosis, the lipid metabolism theory is currently the most concerning. Hyperlipidemia can affect the microcirculation of the femoral head from multiple sources (coagulation system, bone fat embolism, and bone microthrombosis), and then lead to femoral head necrosis [6, 14]. With further research, increasing the number of genes have been found to participate in the regulation of lipid metabolism. SREBP2 can activate target gene transcription and gene expression of cholesterol biosynthesis pathways, which play an important role in lipid homeostasis. Through the study of the relationship between SREBP2 gene polymorphism and ONFH, it was concluded that SREBP2 gene polymorphism and function could cause lipid metabolism disorder in ONFH patients [17]. Apolipoprotein is considered to be a sensitive indicator to estimate the disorder of lipid metabolism in the ONFH populatio n[7]. The study found that the -75G/A polymorphism of the ApoAI gene may be associated with susceptibility to osteonecrosis in the Chinese population [31]. With the deepening of research, it has been found that miRNA is also involved in the expression of genes regulating lipid transport and metabolism [19, 22]. MiR-122 is the most expressed miRNA in the liver, which regulates the liver cholesterol network either directly by regulating cholesterol synthesis genes or indirectly by another molecular pathway [4]. Studies in primates have shown that inhibition of miR-33a/b can increase plasma HDL-C and decrease VLDL-C and TG [15]. In coronary artery disease, the higher the expression of miR-17 in patients, the higher the levels of TC, LDL-C, and ApoB in vivo. The lower the expression of miR-92a and miR-106b in patients with coronary artery disease, the lower the levels of HDL-C and ApoA-1 in vivo, and these miRNAs may play an independent role [10, 11]. Wan et al. [20] found that miR-17-92 clusters can play an important role in bone, lipid, and glucose metabolism through a variety of signaling pathways. We studied the genotypes and lipid levels of ONFH patients, and found their relationship with clinical phenotype and the development of ONFH. MIR17HG and MIR155HG may have some effects on lipid metabolism.
Conclusion
In summary, our study demonstrates that MIR17HG SNP (rs7318578) is significantly associated with an increased risk of steroid-induced ONFH in the population of northern China. A MIR17HG SNP (rs75267932) and MIR155HG SNPs (rs77699734, rs1893650, and rs34904192) were associated with abnormal lipid metabolism. These findings provide evidence for the early screening and prevention of femoral head necrosis. However, more samples from different regions are needed to confirm the results.
Acknowledgments
We gratefully acknowledge all of the participants, the clinicians, and all the hospital staff for their contributions to this study. We are also thankful to Zhengzhou TCMT Traumatology Hospital, Zhengzhou, Henan Province, China, for their support.
Authors’ contributions
WJZ and DQM conceived and designed the project. WTT, HCX, and SMH recruited and collected study samples. CYJ and AFM performed the experiments. MC and LTT selected the SNPs and designed primers. WJZ and HCX performed and analyzed the data. LTT and WJZ wrote and revised the manuscript. The authors read and approved the final manuscript.
Funding
This work received the support of the National Natural Science Foundation of China (nos. 81160228, 81260284, 81660378, and 81860401) and Natural Science Foundation of Inner Mongolia Autonomous Region(2017MS08119).
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
Ethics committee approval was received for this study from Zhengzhou Traditional Chinese Medicine Traumatology Hospital. All participants provided written informed consent.
Competing interests
The authors declare there are no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tingting Liu, Yuju Cao and Changxu Han are co-first author.
Contributor Information
Qiumei Dong, Email: dongqiumei18@163.com.
Jianzhong Wang, Email: wanhjianzhongwj@163.com.
References
- 1.Chacon-Cortes D, Smith RA, Lea RA, Youl PH, Griffiths LR. Association of microRNA 17-92 cluster host gene (MIR17HG) polymorphisms with breast cancer. Tumour Biol. 2015;36(7):5369–5376. doi: 10.1007/s13277-015-3200-1. [DOI] [PubMed] [Google Scholar]
- 2.Chen P, Bai Y, Li Y, Yuan Y, Cheng Y, Pang J, Zhu H, Chen C. Association between polymorphisms of MIR17HG and risk of colorectal cancer in the Chinese Han population. Mol Genet Genomic Med. 2019;7:e667. doi: 10.1002/mgg3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui L, Zhuang Q, Lin J, Jin J, Zhang K, Cao L, Lin J, Yan S, Guo W, He W, Pei F, Zhou Y, Weng X. Multicentric epidemiologic study on six thousand three hundred and ninety five cases of femoral head osteonecrosis in China. Int Orthop. 2016;40(2):267–276. doi: 10.1007/s00264-015-3061-7. [DOI] [PubMed] [Google Scholar]
- 4.Esau C, Davis S, Murray S, Yu X, Pandey S, Pear M, Watts L, Booten S, Graham M, McKay R, Subramaniam A, Propp S, Lollo B, Freier S, Bennett C, Bhanot S, Monia B. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Fukushima W, Fujioka M, Kubo T, Tamakoshi A, Nagai M, Hirota Y. Nationwide epidemiologic survey of idiopathic osteonecrosis of the femoral head. Clin Orthop Relat Res. 2010;468(10):2715–2724. doi: 10.1007/s11999-010-1292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo L, Xu J, Qi J, Zhang L, Wang J, Liang J, Qian N, Zhou H, Wei L, Deng L. MicroRNA-17-92a upregulation by estrogen leads to Bim targeting and inhibition of osteoblast apoptosis. J Cell Sci. 2013;126(Pt 4):978–988. doi: 10.1242/jcs.117515. [DOI] [PubMed] [Google Scholar]
- 7.Hao Y, Guo H, Xu Z, Qi H, Wang Y, Lu C, Liu J, Yuan P. The relationship between apolipoprotein genes polymorphisms and susceptibility to osteonecrosis of the femoral head: a meta-analysis. Lipids Health Dis. 2018;17(1):192. doi: 10.1186/s12944-018-0827-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia J, Feng X, Xu W, Yang S, Zhang Q, Liu X, Feng Y, Dai Z. MiR-17-5p modulates osteoblastic differentiation and cell proliferation by targeting SMAD7 in non-traumatic osteonecrosis. Exp Mol Med. 2014;46(7):e107. doi: 10.1038/emm.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Yang B, Weng X, Tse G, Chan MTV, Wu WKK. Emerging roles of MicroRNAs in osteonecrosis of the femoral head. Cell Prolif. 2018;51. [DOI] [PMC free article] [PubMed]
- 10.Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15(1):1–19. doi: 10.1093/bib/bbs075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Li R, Zhang Y, Qiu J, Ling W. Association of plasma MiR-17-92 with dyslipidemia in patients with coronary artery disease. Medicine. 2014;93(23):e98. doi: 10.1097/MD.0000000000000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao Z, Zhu Y, Hao W, Chu C, Su H. MicroRNA-155 inhibition up-regulates LEPR to inhibit osteoclast activation and bone resorption via activation of AMPK in alendronate-treated osteoporotic mice. IUBMB Life. 2019;71(12):1916–1928. doi: 10.1002/iub.2131. [DOI] [PubMed] [Google Scholar]
- 13.Moussa FM, Cook BP, Sondag GR, et al. The role of miR-150 regulates bone cell differentiation and function.[J]. Bone. 2021;145:115470. [DOI] [PubMed]
- 14.Phemister DB. Repair of bone in the presence of aseptic necrosis resulting from fractures, transplantations, and vascular obstruction. 1930. J Bone Joint Surg Am. 2005;87:672. doi: 10.2106/00004623-200503000-00029. [DOI] [PubMed] [Google Scholar]
- 15.Rayner K, Esau C, Hussain F, McDaniel A, Marshall S, van Gils J, Ray T, Sheedy F, Goedeke L, Liu X, Khatsenko O, Kaimal V, Lees C, Fernandez-Hernando C, Fisher E, Temel R, Moore K. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song H, Li X, Zhao Z, Qian J, Wang Y, Cui J, Weng W, Cao L, Chen X, Hu Y, Su J. Reversal of osteoporotic activity by endothelial cell-secreted bone targeting and biocompatible exosomes. Nano Lett. 2019;19(5):3040–3048. doi: 10.1021/acs.nanolett.9b00287. [DOI] [PubMed] [Google Scholar]
- 17.Song Y, Du Z, Chen B, Ren M, Yang Q, Sui Y, Wang Q, Wang A, Zhao H, Qin Y, Zhang G. Association of SREBP2 gene polymorphisms with the risk of osteonecrosis of the femoral head relates to gene expression and lipid metabolism disorders. Mol Med Rep. 2017;16(5):7145–7153. doi: 10.3892/mmr.2017.7473. [DOI] [PubMed] [Google Scholar]
- 18.Tian Y, An F, Wang J, Liu C, Wu H, Cao Y, Wang J. MMP2 and MMP10 Polymorphisms are related to steroid-induced osteonecrosis of the femoral head among Chinese Han population. Biomed Res Int. 2019;2019:8298193. doi: 10.1155/2019/8298193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vickers K, Palmisano B, Shoucri B, Shamburek R, Remaley A. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan S, Chen X, He Y, Yu X. Novel functions of MicroRNA-17-92 cluster in the endocrine system. Curr Drug Targets. 2018;19(2):191–200. doi: 10.2174/1389450118666171117125319. [DOI] [PubMed] [Google Scholar]
- 21.Wang C, Sun W, Ling S, Wang Y, Wang X, Meng H, Li Y, Yuan X, Li J, Liu R, Zhao D, Lu Q, Wang A, Guo Q, Lu S, Tian H, Li Y, Peng J. AAV-Anti-miR-214 prevents collapse of the femoral head in osteonecrosis by regulating osteoblast and osteoclast activities. Mol Ther Nucleic Acids. 2019;18:841–850. doi: 10.1016/j.omtn.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Xia M, Yan X, Li D, Wang L, Xu Y, Jin T, Ling W. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ Res. 2012;111(8):967–981. doi: 10.1161/CIRCRESAHA.112.266502. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Yang X, Shi J, Zhao Y, Pan L, Zhou J, Wang G, Wang J. Combination analysis of NOS3, ABCB1 and IL23R polymorphisms with alcohol-induced osteonecrosis of the femoral head risk in Chinese males. Oncotarget. 2017;8(20):33770–33778. doi: 10.18632/oncotarget.16809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei B, Wei W, Zhao B, Guo X, Liu S. Long non-coding RNA HOTAIR inhibits miR-17-5p to regulate osteogenic differentiation and proliferation in non-traumatic osteonecrosis of femoral head. PLoS One. 2017;12(2):e0169097. doi: 10.1371/journal.pone.0169097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu F, Huang W, Yang Y, Liu F, Chen J, Wang G, et al. miR-155-5p regulates mesenchymal stem cell osteogenesis and proliferation by targeting GSK3B in steroid-associated osteonecrosis. Cell Biol Int. 2020:1065–6995. [DOI] [PubMed]
- 26.Wu F, Huang W, Yang Y, et al. miR-155-5p regulates mesenchymal stem cell osteogenesis and proliferation by targeting GSK3B in steroid-associated osteonecrosis.[J]. Cell Biol Int. 2021;45:83–91. [DOI] [PubMed]
- 27.Wu H, He G, Han H, Xiong W, Song T, Chen H, Chen X, Wu X, Huang G, Zhang Y, Sun C, Zhao C, Chen Y. Analysis of MIR155HG variants and colorectal cancer susceptibility in Han Chinese population. Mol Genet Genomic Med. 2019;7:e778. doi: 10.1002/mgg3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H, Huang T, Ye Z, Fu X, Hu K, Yang X. Correlation of microRNA 17-92 cluster host gene (MIR17HG) polymorphisms with susceptibility and prognosis for multiple myeloma. Clin Lymphoma Myeloma Leuk. 2019;19(7):e359–e366. doi: 10.1016/j.clml.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, Sun W. Noncoding RNAs in steroid-induced osteonecrosis of the femoral head. Biomed Res Int. 2019;2019:8140595. doi: 10.1155/2019/8140595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu B, Hu QH, Zhao B, Zhou ZK. Variation and significance of serum leptin, blood lipid level, adiponectin, NO and TNF-α for patients with non-traumatic ischemic necrosis of the femoral head. Saudi J Biol Sci. 2017;24(8):1763–1766. doi: 10.1016/j.sjbs.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin JM, Liu Z, Zhao SC, Guo YJ, Liu ZT. Relationship between the apolipoprotein AI, B gene polymorphism and the risk of non-traumatic osteonecrosis. Lipids Health Dis. 2014;13(1):149. doi: 10.1186/1476-511X-13-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan L, Li W, Wang X, Yang G, Yu H, Sun S. The relationship between genetic polymorphisms in apolipoprotein E (ApoE) gene and osteonecrosis of the femoral head induced by steroid in Chinese Han population. Genes Genomics. 2018;40(2):225–231. doi: 10.1007/s13258-017-0625-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Zhao H, Chen J, Xia B, Jin Y, Wei W, Shen J, Huang Y. Interferon-β-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett. 2012;586(19):3255–3262. doi: 10.1016/j.febslet.2012.06.047. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, Zheng W, Wang Y, Wang Y, Huang H. Human bone marrow mesenchymal stem cells support the derivation and propagation of human induced pluripotent stem cells in culture. Cell Rep. 2013;15(3):216–223. doi: 10.1089/cell.2012.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Kong X, Wang R, Li S, Niu Y, Zhu L, Chen W, Lin N. Genetic association of the P-glycoprotein gene ABCB1 polymorphisms with the risk for steroid-induced osteonecrosis of the femoral head in Chinese population. Mol Biol Rep. 2014;41(5):3135–3146. doi: 10.1007/s11033-014-3173-y. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Sun R, Zhang L, Feng L, Liu Y. Effect of blood biochemical factors on nontraumatic necrosis of the femoral head: logistic regression analysis. Orthopade. 2017;46(9):737–743. doi: 10.1007/s00132-017-3408-4. [DOI] [PubMed] [Google Scholar]
- 37.Zhou M, Ma J, Chen S, Chen X, Yu X. MicroRNA-17-92 cluster regulates osteoblast proliferation and differentiation. Endocrine. 2014;45(2):302–310. doi: 10.1007/s12020-013-9986-y. [DOI] [PubMed] [Google Scholar]
- 38.Zou W, Li X, Li C, Liu D, Lv Y, Yang Y, Ye N, Guo D, He S. Analysis of the relationship between MIR155HG variants and gastric Cancer susceptibility. BMC Gastroenterol. 2020;20(1):17. doi: 10.1186/s12876-020-1169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.