Abstract
The DAX-1 (NR0B1) gene encodes an unusual member of the nuclear hormone receptor superfamily which acts as a transcriptional repressor. Mutations in the human DAX-1 gene cause X-linked adrenal hypoplasia congenita (AHC) associated with hypogonadotropic hypogonadism (HHG). We have studied the intracellular localization of the DAX-1 protein in human adrenal cortex and mouse Leydig tumor cells and found it to be both nuclear and cytoplasmic. A significant proportion of DAX-1 is associated with polyribosomes and is found complexed with polyadenylated RNA. DAX-1 directly binds to RNA, two domains within the protein being responsible for cooperative binding activity and specificity. Mutations in DAX-1 found in AHC-HHG patients significantly impair RNA binding. These findings reveal that DAX-1 plays multiple regulatory roles at the transcriptional and posttranscriptional levels.
DAX-1 (NR0B1) (32) is an unusual member of the nuclear hormone receptor superfamily whose mutations cause the X-linked form of adrenal hypoplasia congenita (AHC), which is constantly associated with hypogonadotropic hypogonadism (HHG). In addition, the DAX-1 gene locus is mapped inside the minimal region on the X chromosome whose duplication causes male-to-female sex reversal in persons with an intact SRY gene (dosage-sensitive sex reversal) (2, 22, 43, 53).
The human DAX-1 gene encodes a 470-amino-acid (aa) protein whose C terminus is similar to the ligand-binding domain (LBD) of nuclear hormone receptors, while its N terminus is composed of three repeats of 67 to 69 aa with no significant similarity to any other known protein (20, 53). All mutations found in AHC-HHG kindreds have in common the characteristic of altering the structure of the DAX-1 C-terminal domain. We and others have shown that DAX-1 is endowed with transcriptional repressor activity (16, 20, 54). This property is invariably abolished in mutated DAX-1 proteins from AHC-HHG patients (16, 20). This finding suggests that the impairment of the DAX-1 transcriptional activity is directly linked to the pathogenesis of AHC-HHG.
DAX-1 expression is restricted to steroidogenic tissues and to some critical sites in the reproductive axis (15, 45). When introduced in steroidogenic Y-1 cells, DAX-1 blocks steroid biosynthesis by impairing the expression of the steroidogenic acute regulatory protein (StAR) and of the enzymes required to convert cholesterol into pregnenolone and progesterone (21, 54). We have shown that the block of StAR expression is dependent on the binding of DAX-1 to a DNA hairpin site in the StAR promoter, which allows the recruitment to the promoter of the powerful transcriptional repression activity present in the DAX-1 C terminus (54). Using a transgenic mouse model, dax-1 overexpression has also been shown to produce sex reversal in male animals harboring a weak sry allele (42). This phenotype is likely caused by inappropriate repression of testosterone biosynthesis in Leydig cells in the developing male gonad by the overexpressed dax-1 and by a direct repressive effect on the Müllerian inhibiting substance gene promoter (31). Surprisingly, inactivation of the dax-1 gene in the mouse by homologous recombination produces only a very mild adrenal phenotype, while the major consequence is a progressive degeneration of the male germinal epithelium (52).
To get better insight into DAX-1's biological role, we have studied its subcellular distribution and found that the DAX-1 protein is localized in both the nucleus and cytoplasm of human adrenal cortex and mouse Leydig tumor cells. A significant proportion of the DAX-1 protein is associated with polyribosomes and is found complexed to polyadenylated [poly(A)+] RNA in the cell. The three N-terminal repeats appear to act cooperatively in directing DAX-1 binding to RNA. Surprisingly, we found that the DAX-1 C terminus, which consists of the putative LBD, can also function as an autonomous RNA-binding domain reinforcing and modulating the activity of the N-terminal repeats. Importantly, the RNA-binding capacity of DAX-1 mutants found in AHC-HHG kindreds is significantly impaired. These findings show that RNA binding is an essential biological property of DAX-1 and underscore intriguing links between the mechanisms of gene regulation at the transcriptional and posttranscriptional levels.
MATERIALS AND METHODS
Cell lines.
MA-10 mouse Leydig cells were cultured in Waymouth's medium supplemented with 15% horse serum, 20 mM HEPES, and gentamicin. Human adrenocortical H295R cells were cultured in F12-Dulbecco's modified Eagle's medium supplemented with 2% Nuserum (Collaborative Research), 1% ITS Plus (Collaborative Research), and penicillin-streptomycin.
Cell fractionation using sucrose gradient sedimentation.
Cell fractionation was performed at 4°C, as described (10), with slight modifications. A total of 6 × 107 cells were resuspended in a hypotonic (H) buffer (0.25 M sucrose, 50 mM Tris [pH 7.5], 25 mM KCl, 5 mM MgCl2), and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors were added just before fractionation. Cells were disrupted by 30 strokes of a Dounce homogenizer using the A pestle. The lysate was centrifuged at 500 × g for 5 min to pellet the crude nuclear fraction. This pellet was resuspended in 0.8 M sucrose and centrifuged through a 1.6 M sucrose pad at 150,000 × g for 1 h to obtain a cytoplasm-free nuclear fraction (B1). The supernatant obtained after the 500 × g centrifugation was subsequently centrifuged at 10,000 × g for 5 min to yield the heavy membrane pellet (B2) and the postnuclear supernatant. The postnuclear supernatant was centrifuged at 130,000 × g for 1 h to pellet the fraction containing light membranes and polysomes (B3). The supernatant was centrifuged further at 180,000 × g for 3 h to obtain the insoluble cytoplasm (pellet) (B4) and the soluble cytoplasm (supernatant) (B5). Each fraction was resuspended in 1× Laemmli buffer–4 M urea, the protein concentration was measured by the Bradford assay, and an equal amount of protein (15 μg) was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for Western blot analysis, using the anti-DAX-1 2F4 monoclonal antibody (45), anti-L7a ribosomal protein antiserum (8), and the anti-SF-1 antiserum (Upstate Biotechnology). Continuous sucrose gradients (15 to 45%) in C buffer (25 mM Tris [pH 7.5], 100 mM KCl, 5 mM MgCl2) were prepared using a gradient mixer. The following procedures were performed at 4°C unless otherwise mentioned. Cycloheximide (10 μM) was added to the medium for 10 min prior to homogenization in C buffer containing 0.5 mM PMSF and protease inhibitors as described before. The cytoplasmic fraction was obtained as the supernatant after a 500 × g centrifugation of the cell extract for 5 min. This cytoplasmic fraction was carefully placed on top of the sucrose gradient for a 200,000 × g (Beckman SW41 rotor) centrifugation for 90 min. Twenty-four 500-μl fractions were collected from the top, and the absorbance at 260 nm was measured; 60 μl of each fraction was subjected to SDS-PAGE for Western blot analysis. For EDTA treatment, the MgCl2 in the buffer was replaced with 30 mM EDTA and incubated on ice for 15 min before centrifugation in sucrose gradients containing 30 mM EDTA. For RNase treatment, the cytoplasmic fraction was incubated with 1.2 mg of RNase A and 30 U of RNase T1 per ml at 37°C for 15 min. At the end of the incubation, RNasin (10 U/ml) and 0.5% NP-40 were added.
Oligo(dT) chromatography.
H295R cells growing exponentially in 14-cm-diameter culture dishes were washed once with phosphate-buffered saline (PBS) and irradiated with a short-wavelength UV lamp. The UV dose administered was about 104 erg/mm2. Cells were then lysed in 1 ml of buffer containing 20 mM Tris (pH 7.4), 10 mM KCl, 5 mM MgCl2, 0.3% Triton X-100, 100 U of RNasin per ml, 0.5 mM PMSF, and protease inhibitors. After 5 min on ice, cell lysates were centrifuged at 10,000 × g for 10 min at 4°C, to yield the postmitochondrial supernatant. This was then subjected to oligo(dT) spun column chromatography following the manufacturer's (Pharmacia, Uppsala, Sweden) instructions. Flowthrough, high-salt wash, low-salt wash, and eluate fractions were collected from the column. For RNase treatment, cell lysate was incubated with 1.2 mg of RNase A and 30 U of RNase T1 per ml for 15 min at 37°C before centrifugation. Fractions were concentrated to the volume of 100 μl using Centricon 30 concentrators. Then 10 μl of the postmitochondrial supernatant, 12 μl of the flowthrough and high- and low-salt washes, and 60 μl of the eluate were subjected to SDS-PAGE and analyzed by Western blot using the anti-DAX-1 2F4 antibody (45) and the anti-LDH antibody (Sigma). For oligo(dT) chromatography after sedimentation on a continuous sucrose gradient, the gradient was prepared in a low-salt buffer (25 mM Tris [pH 7.5], 10 mM KCl, 5 mM MgCl2) and fractions 1 to 3, 4 to 10, and 11 to 24 from MA-10 cell extracts were pooled. Poly(A)+ RNA was purified by oligo(dT) spun column chromatography in native conditions as described above. The elution fractions were concentrated and analyzed by Western blot.
DNA constructs and RNA homopolymer binding assay.
35S-labeled proteins were produced using the TNT rabbit reticulocyte lysate or wheat germ systems (Promega). DAX-1 was transcribed-translated from pSV.DAX-1, which contains the complete human DAX-1 cDNA inserted in the EcoRI and BamHI sites of pSG5 (14). DAX-1 mutants R267P, ΔV269, N440I, and 1-451 were obtained from pSV.DAX-1 by site-directed mutagenesis, using the QuickChange kit (Stratagene). All mutants were verified by sequencing. pSV.mDAX-1, which contains the sequence corresponding to nucleotides 75 to 1952 of the mouse dax-1 cDNA (41), was used to express the mouse dax-1 protein. Sequences encoding human DAX-1 aa 1 to 69, 1 to 135, and 1 to 204 were PCR-amplified from pSV.DAX-1 and cloned in the NdeI and BamHI sites of pET.15b (Novagen). The sequence encoding DAX-1 aa 205 to 470 was PCR-amplified from pSV.DAX-1 and cloned in the EcoRI and BamHI sites of pSG5. All clones were verified by sequencing. Human retinoic acid receptor gamma (RARγ) LBD was obtained by in vitro transcription-translation of pET/h RARγ D3E (34), encoding human RARγ aa 178 to 423. Bacterially expressed RARα was a gift from M.-P. Gaub (35). The RNA homopolymer binding assay was performed as described (39), except that the buffer used was 10 mM Tris (pH 8). RNA homopolymer-conjugated agarose beads were obtained from Pharmacia [poly(A) and poly(U)] and from Sigma [poly(C) and poly(G)]. 35S-labeled proteins eluted from beads were run on SDS-PAGE gels. The gels were subjected to fluorography, and radioactivity corresponding to the amount of bound proteins was quantified using a BAS2000 (Fuji) phosphoimager. After elution from beads, DAX-1 expressed in a baculovirus system was detected by Western blot using the 2F4 antibody. RARα was detected by Western blot using a specific rabbit polyclonal antibody, as described (12).
DAX-1 expression in Sf9 cells.
DAX-1 cDNA was cloned into the pAcSG His NT-A vector (Pharmingen), recombinant baculovirus was obtained, and the protein was expressed in Sf9 cells, as described (36). Lysates were prepared from 109 infected cells in 20 mM Tris (pH 7.4)–150 mM NaCl–20% glycerol–5 mM β-mercaptoethanol–0.1% NP-40–0.5 mM PMSF–protease inhibitors. The lysate was frozen and thawed three times, NaCl was added to a final concentration of 0.5 M, and the lysate was centrifuged at 16,000 × g for 30 min at 4°C. The pellet was frozen in liquid nitrogen and stored at −80°C. Most of the baculovirus-expressed DAX-1 protein is found associated with this fraction. The supernatant was dialyzed against a buffer containing 50 mM sodium phosphate (pH 8), 500 mM NaCl, 5 mM imidazole, 5 mM β-mercaptoethanol, and 0.5 mM PMSF and then loaded onto a Ni-nitrilotriacetic acid column (Qiagen). The column was washed and eluted following the manufacturer's instructions. The elution fractions were pooled and loaded on a 2F4 antibody-immunoaffinity column. The column was washed with 50 mM sodium phosphate (pH 8)–1 M NaCl–0.5 mM PMSF and eluted with 200 μg of a peptide corresponding to DAX-1 aa 135 to 166, against which the 2F4 antibody was raised, per ml. The eluate was dialyzed against PBS containing 10% glycerol and 0.5 mM PMSF, concentrated using Centricon 30, and stored at −80°C.
Northwestern blot analysis.
Northwestern blotting was performed as described (4), using DAX-1 protein present in the insoluble fraction of recombinant baculovirus-infected Sf9 cells.
Immunofluorescence.
Immunofluorescence assays were performed as described (30), using the anti-DAX-1 2F4 antibody at a 1:500 concentration. Cells were analyzed with a Leica confocal microscope or with a Leica fluorescence microscope.
Heterokaryon assay.
COS cells were transfected with pSV.DAX-1, trypsinized 24 h after transfection, and transferred into chamber slides (Nunc) containing mouse NIH 3T3 cells. COS cells were allowed to attach overnight. On the following day, cells were incubated for 30 min in medium containing cycloheximide (10 μg/ml) and then they were fused by polyethylene glycol 1500 (Boehringer, Mannheim, Germany) treatment for 4 min. Cells were washed three times with PBS, incubated in complete medium containing 10 μg of cycloheximide per ml and 10 μM cytosine arabinoside for 4 h, and then fixed and processed for immunofluorescence.
Immunoelectron microscopy.
Cells were grown on Permanox chamber slides (Nunc), washed with PBS, fixed in 4% paraformaldehyde–0.1% glutaraldehyde–PBS (pH 7.4) for 30 min at 4°C, rinsed in PBS, dehydrated in a graded ethanol series, and embedded in an Araldite-Epon mixture.
For immunogold electron microscopy, ultrathin sections (60 to 70 nm) collected on Maxtaform grids were blocked with 1% normal goat serum diluted in 0.01 M PBS–0.5% Tween 20 (pH 7.4), incubated for 2 h at room temperature with the anti-DAX-1 2F4 antibody diluted 1:250 to 1:500 in PBS-Tween 20, rinsed three times for 10 min in the same buffer, and incubated for 1 h at room temperature with anti-mouse immunoglobulin antibody conjugated to colloidal gold particles (10 to 12 nm; Chemicon GAB264; Jackson Immunoresearch), diluted 1:30 in the same buffer. For some samples, 1-nm gold labeling followed by silver amplification was performed with the R-Gent silver enhancement kit (Aurion) following the manufacturer's protocol. After two washes in PBS, the ultrathin sections were postfixed with 2.5% buffered glutaraldehyde, rinsed in distilled water, contrasted for 15 min with 5% uranyl acetate, washed in distilled water, and examined under an EM 208 Philips electron microscope.
After immunogold labeling, some sections were treated with EDTA, according to the Bernhard method (3), to selectively visualize ribonucleoprotein structures in the cell. To test the antibody specificity, two types of controls were performed: (i) an anti-CD81 mouse monoclonal antibody which recognizes a membrane antigen expressed by lymphocytes and monocytes (Immunotech) was used as the primary antibody; and (ii) the anti-DAX-1 2F4 antibody was preadsorbed with a 1,000-fold peptide excess.
RESULTS
DAX-1 is localized both in the nucleus and in the cytoplasm in steroidogenic cells.
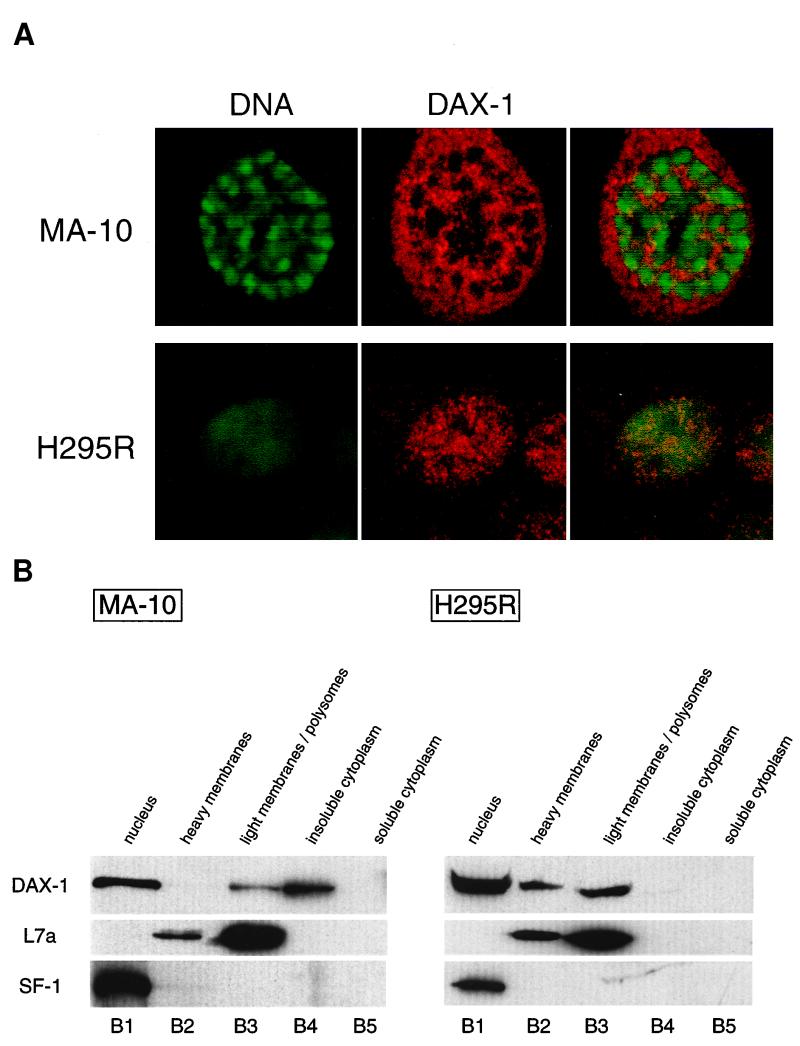
In the H295R human adrenocortical tumor cell line and in the MA-10 mouse Leydig tumor cell line, the endogenous DAX-1 protein is both nuclear and cytoplasmic, as revealed by immunofluorescence (Fig. 1A). The same distribution was found in COS and HeLa cells transfected with a DAX-1 expression vector (not shown). To define the localization of DAX-1 in various subcellular compartments, we fractioned H295R and MA-10 cell lysates on discontinuous and continuous sucrose gradients, and the distribution of the DAX-1 protein was studied by immunoblotting using a specific anti-DAX-1 monoclonal antibody (45). After fractionation of H295R cell extracts on a discontinuous sucrose gradient (10), DAX-1 is mainly found in the nucleus but is also readily detected in fractions containing heavy membranes and polyribosomes (B2 and B3 in Fig. 1B). In MA-10 cells, cytoplasmic DAX-1 is more abundant than in H295R cells and localizes in the fraction containing polyribosomes and in the insoluble cytoplasmic fraction (B3 and B4 in Fig. 1B). To test the quality of the fractionation procedure, nitrocellulose membranes were stripped off and immunoblotting was performed using antibodies directed against specific compartmentalized proteins. SF-1, a member of the nuclear hormone receptor superfamily which colocalizes with DAX-1 in various tissues (15), was found to be exclusively nuclear in both cell types (Fig. 1B). Ribosomal L7a protein (8) was detected in the B2 and B3 fractions (Fig. 1B), and lactate dehydrogenase, a cytosolic protein, was detected only in the B5 fraction (not shown), as expected.
FIG. 1.
Subcellular localization of DAX-1. (A) Confocal microscopy analysis of DAX-1 distribution in steroidogenic mouse MA-10 Leydig cells (top) and human adrenocortical H295R cells (bottom). DNA is shown in green, and DAX-1 is in red. (B) Fractionation of H295R and MA-10 cell extracts on a discontinuous sucrose gradient (10). Proteins from the nuclear (B1), heavy membrane (B2), light membrane-polysome (B3), insoluble cytoplasm (B4), and soluble cytoplasm (B5) fractions were run on an SDS–10% PAGE gel and transferred to nitrocellulose. The membrane was sequentially probed with the anti-DAX-1 (top), anti-L7a ribosomal protein (center), and anti-SF-1 (bottom) antibodies.
Cytoplasmic DAX-1 protein is found associated with actively translating polyribosomes.
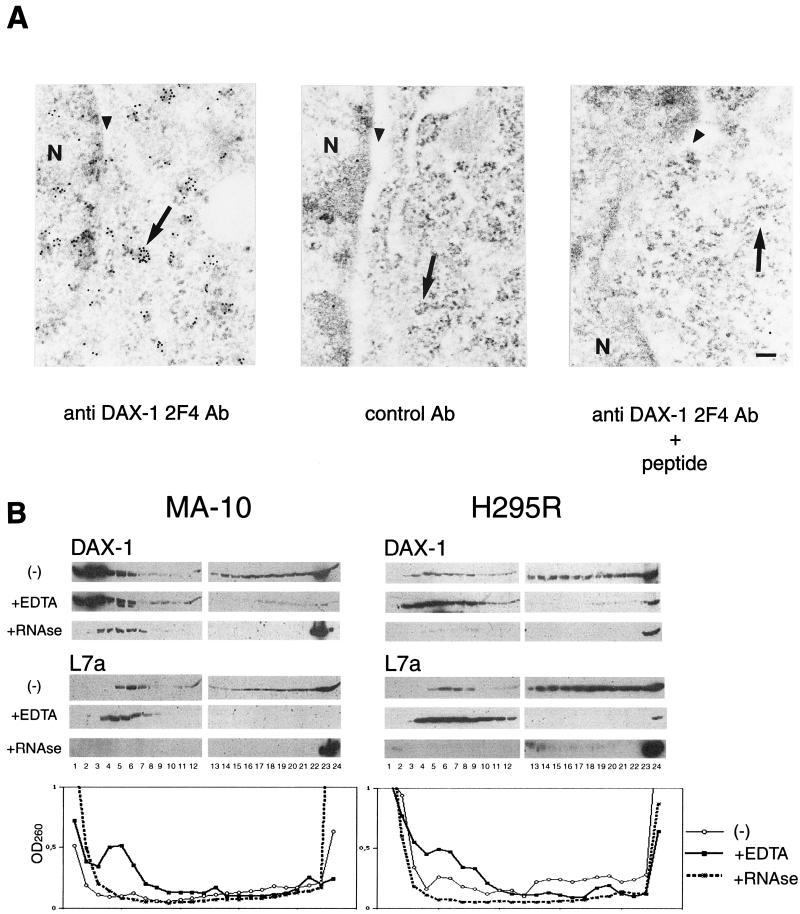
According to our fractionation procedure, a large proportion of the cytoplasmic DAX-1 protein appears localized in a fraction containing polyribosomes in both adrenal cortex and Leydig cells. We performed immunoelectron microscopy to confirm the association of DAX-1 with polyribosomes. In the cytoplasm of MA-10 cells, immunogold particles are mainly localized in small clusters associated with rough endoplasmic reticulum and free ribosomes. No significant staining was obtained when the anti-DAX-1 primary antibody was replaced with a nonrelevant monoclonal antibody or when the anti-DAX-1 antibody was preadsorbed with an excess of the peptide against which the antibody was raised (Fig. 2A). To verify that DAX-1 not only colocalizes but also associates with polyribosomes, we fractioned cell lysates on a 15 to 45% continuous sucrose gradient after low-speed centrifugation to remove nuclei. In MA-10 cells, a large amount of the DAX-1 protein localizes in fractions at the top of the gradient. However, a significant proportion is also found in fractions at the bottom of the gradient, which, as shown by staining with the anti-L7a antibody, contain polyribosomes (Fig. 2B). In H295R cells, DAX-1 is to a large extent distributed in fractions containing polyribosomes (Fig. 2B). EDTA treatment is known to cause the dissociation of translating polyribosomes into subunits and the release of messenger ribonucleoproteins (mRNPs) associated with polyribosomes (11). When added to H295R and MA-10 cytoplasmic lysates, EDTA causes a shift of the DAX-1 protein cosedimenting with polyribosomes towards the upper part of the gradient, while it leaves unaltered the localization of the subset partitioning in the lighter fractions. Staining with the anti-L7a antibody and monitoring the optical density at 260 nm of the gradient fractions (Fig. 2B) show that EDTA treatment was indeed effective in inducing polyribosome dissociation. RNase treatment of cell lysates before loading on the gradients also modified the localization of DAX-1, which was in part shifted to the top fractions in the gradient and in part found associated with the pellet (Fig. 2B). Taken together, these data show that a significant fraction of the cytoplasmic DAX-1 pool is associated with actively translating polyribosomes.
FIG. 2.
DAX-1 is associated with polyribosomes. (A) DAX-1 immunogold labeling in MA-10 cells. Gold particles corresponding to DAX-1 (left) are found in the nucleus localized at the periphery of chromatin, in the interchromatin space, and in the cytoplasm, mostly colocalized with polyribosomes, which appear as electron-dense clusters. No significant staining was observed when the anti-DAX-1 2F4 antibody was replaced with a control antibody (Ab) at the same dilution (center) or when the anti-DAX-1 antibody was preadsorbed with the specific peptide (right). N, nucleus. Perinuclear cisternae are indicated with an arrowhead, and polyribosomes are marked with an arrow. Bar, 0.1 μm. (B) Fractionation of MA-10 (left) and H295R (right) cell extracts on a 15 to 45% continuous sucrose gradient. Twenty-four fractions were collected from the top and analyzed by Western blot using the anti-DAX-1 2F4 antibody and the anti-L7a ribosomal protein antiserum. Distribution of DAX-1 and L7a in the gradients is also shown for extracts treated with EDTA and RNase, as described in Materials and Methods. Optical density profiles of the gradient fractions at 260 nm are shown for each treatment in the graphs at the bottom of the figure. A large proportion of the cytoplasmic DAX-1 protein is found in fractions containing polyribosomes, as shown by staining with the anti-L7a antibody. Polyribosome dissociation by both EDTA and RNase treatment modifies DAX-1 localization in the gradient fractions.
DAX-1 is associated with mRNP particles in the cytoplasm.
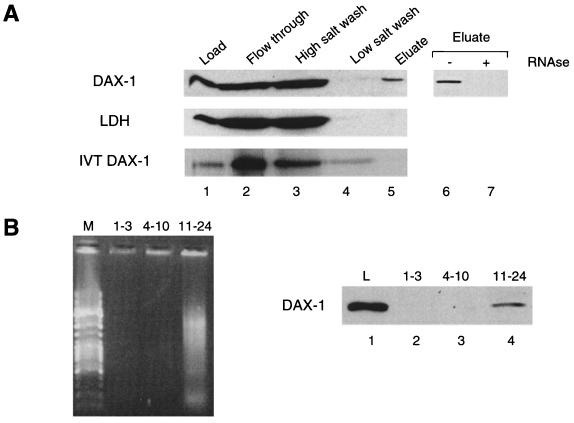
A shift of localization in sucrose gradients caused by EDTA has been reported previously for factors present in polyribosome-associated mRNP particles (11). To test the possibility that DAX-1 may be directly associated with mRNPs, we performed oligo(dT)-cellulose chromatography on postmitochondrial supernatants from H295R cells after in vivo UV cross-linking. While most of the protein is found in the flowthrough and wash fractions, a subset of the DAX-1 protein coelutes with poly(A)+ RNA (Fig. 3). Identical results were obtained with MA-10 lysates (not shown). This situation closely mirrors the behaviour of FMRP, the protein encoded by the FMR1 gene, which is known to directly bind RNA and is found associated with polyribosomes as part of an mRNP complex (8, 11). Conversely, lactate dehydrogenase, which is localized in the soluble fraction of the cytoplasm, is not retained on the oligo(dT)-cellulose column (Fig. 3A). RNase treatment of the extracts abrogates binding of DAX-1 to the oligo(dT) column, showing that the binding is indeed mediated by poly(A)+ RNA (Fig. 3A). In addition, in vitro-translated DAX-1 does not bind to the oligo(dT) column by itself under the conditions used for poly(A)+ RNA purification. These findings show that a subset of the cytoplasmic DAX-1 pool is associated with mRNA in the cell. The possibility that DAX-1 eluting in the flowthrough or the wash fractions may associate with poly(A)+ RNA in the cell cannot be ruled out, since poly(A)-containing mRNA 3′ extremities could be hindered from binding to the column by complexed proteins during purification performed under our experimental conditions. In addition, we performed oligo(dT) chromatography on pooled fractions from the top (fractions 1 to 3), the middle (fractions 4 to 10, corresponding to the isolated ribosomal subunits), and the bottom (fractions 11 to 24, corresponding to polyribosomal particles with a buoyant density higher than the 80S monosomes) (see Fig. 2B) of a continuous sucrose gradient. Selectively, the population of DAX-1 associated with polyribosomes is found to be complexed with poly(A)+ RNA (Fig. 3B).
FIG. 3.
DAX-1 associates with poly(A)+ RNA. (A) Extracts from UV-irradiated H295R cells were subjected to oligo(dT) chromatography under native conditions. Aliquots from total cell extract (lane 1), flowthrough (lane 2), high-salt (lane 3), and low-salt (lane 4) washes and eluate (lane 5) fractions were subjected to SDS-PAGE and analyzed by Western blot using the anti-DAX-1 2F4 antibody (top) and the anti-LDH antibody (center). In vitro-translated DAX-1 from a rabbit reticulocyte lysate was assayed for direct association with the chromatographic column under the same purification conditions (bottom). Previous RNase treatment of cell extracts (1.2 mg of RNase A and 30 U of RNase T1 per ml for 15 min at 37°C) abolishes DAX-1 fractionation in the eluate (lanes 6 and 7). (B) Extracts from MA-10 cells were fractioned on a 15 to 45% continuous sucrose gradient. Fractions 1 to 3, 4 to 10, and 11 to 24, corresponding to the top of the gradient, isolated ribosomal subunits, and polyribosomal particles with a density higher than the 80S monosomes, respectively (see Fig. 2B), were pooled and subjected to oligo(dT) chromatography under native conditions. On the left side is an ethidium bromide-stained agarose gel showing that poly(A)+ RNA is selectively eluted from gradient fractions 11 to 24. Lane M, DNA molecular size markers. On the right side, oligo(dT) column eluates from each pool (lanes 2 to 4) were analyzed for DAX-1 content by Western blot. Lane L, DAX-1 protein present in 5% of the volume of the cell extract loaded on the sucrose gradient.
DAX-1 binds to RNA.
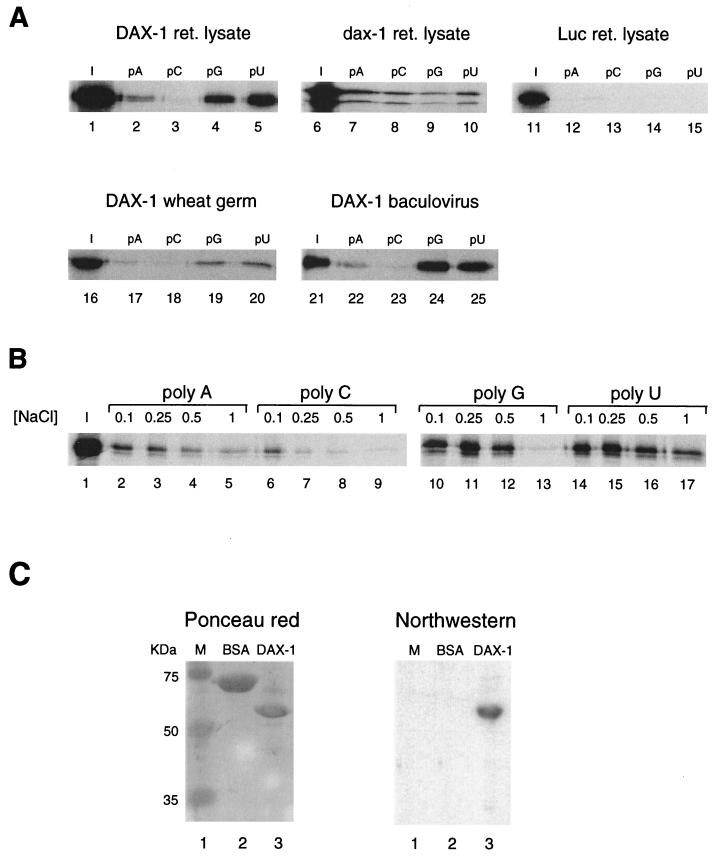
Since DAX-1 is found associated with mRNA within the cell, and since we have shown that DAX-1 can bind to nucleic acid hairpins (54), a structure frequently found in RNA molecules, we questioned whether DAX-1 may directly bind to RNA. To this purpose, we tested the binding of 35S-labeled DAX-1 translated in vitro using rabbit reticulocyte lysate to the four ribonucleotide homopolymers poly(A), poly(C), poly(G), and poly(U) immobilized on agarose beads (44). Differential binding specificity for different homopolymers is a common feature of many RNA-binding proteins (44). DAX-1 binds to RNA homopolymers with different affinities in the order poly(U) > poly(G) >> poly(A) > poly(C) (Fig. 4A). Conversely, luciferase, which was used as a negative control, did not bind to any of the four RNA homopolymers. Also, dax-1, the mouse homologue of human DAX-1, binds to RNA homopolymers (Fig. 4A). Two protein products are generated by in vitro translation of the mouse dax-1 clone, which are most probably derived from translation initiation from two in-frame Kozak ATGs, the most 3′ of which is the one utilized in vivo (1). Both translation products show the same binding specificity, which is different from human DAX-1, as follows: poly(A) ≈ poly(C) ≈ poly(U) > polyG (Fig. 4A). Human DAX-1 shows tenacious binding to poly(U) even in the presence of 1 M NaCl, while binding to poly(G) and to poly(A) is more salt sensitive, with binding to poly(C) being significantly dissociated even at 250 mM NaCl (Fig. 4B).
FIG. 4.
DAX-1 binds to RNA. (A) RNA homopolymer binding assay. Binding to agarose beads coupled to poly(A), poly(C), poly(G), and poly(U) is shown for human (lanes 2 to 5) and mouse (lanes 7 to 10) DAX-1 and for luciferase (Luc) (lanes 12 to 15) translated in a rabbit reticulocyte (ret.) lysate. Binding to RNA homopolymers is also shown for DAX-1 translated in a wheat germ system (lanes 17 to 20) and expressed in insect cells (lanes 22 to 25). A 1:10 ratio of the input protein is shown in each case (lanes 1, 6, 11, 16, and 21). The assay was performed in a buffer containing 0.25 M NaCl. 35S-labeled proteins were detected by fluorography, and DAX-1 expressed in a baculovirus system was detected by Western blot. (B) Salt sensitivity of the binding of DAX-1 to poly(A) (lanes 2 to 5), poly(C) (lanes 6 to 9), poly(G) (lanes 10 to 13), and poly(U) (lanes 14 to 17) was tested in buffers containing NaCl concentrations of 0.1, 0.25, 0.5, and 1 M. Lane I, 1:10 of the input protein (lane 1). (C) Northwestern binding assay using a 32P-labeled 240-nucleotide riboprobe transcribed from PvuII-linearized pBluescript and proteins immobilized on a nitrocellulose membrane. The Ponceau red-stained membrane is also shown on the left. Lanes show molecular size markers (lane 1), BSA (10 μg, lane 2), and DAX-1 (10 μg, lane 3).
To rule out the possibility that RNA binding by DAX-1 may be mediated by association with other proteins present in the reticulocyte lysate, we also expressed DAX-1 in a wheat germ translation system, which should be devoid of factors interacting with mammalian proteins. DAX-1 retains the specificity of binding for poly(U) and poly(G) (Fig. 4A). In addition, we expressed DAX-1 in insect cells, using recombinant baculovirus infection. The soluble DAX-1 protein was purified by affinity chromatography on an anti-DAX-1 antibody column followed by peptide elution. Baculovirus-expressed DAX-1 was used in the RNA-binding assay using homopolymers immobilized on agarose beads, and the protein bound to beads was revealed by Western blot. The baculovirus-expressed DAX-1 shows the same specificity of binding to RNA homopolymers as the protein expressed in in vitro translation systems (Fig. 4A). Finally, we performed Northwestern analysis using a 32P-labeled 240-nucleotide riboprobe transcribed from PvuII-linearized pBluescript using T3 polymerase (4). Binding of DAX-1 to the riboprobe is readily detectable, while bovine serum albumin (BSA) and the proteins used as molecular size markers did not bind to the riboprobe (Fig. 4C). DAX-1 also binds to a variety of other different riboprobes tested in the Northwestern assay (not shown). These results confirm that DAX-1 has intrinsic RNA-binding capacity.
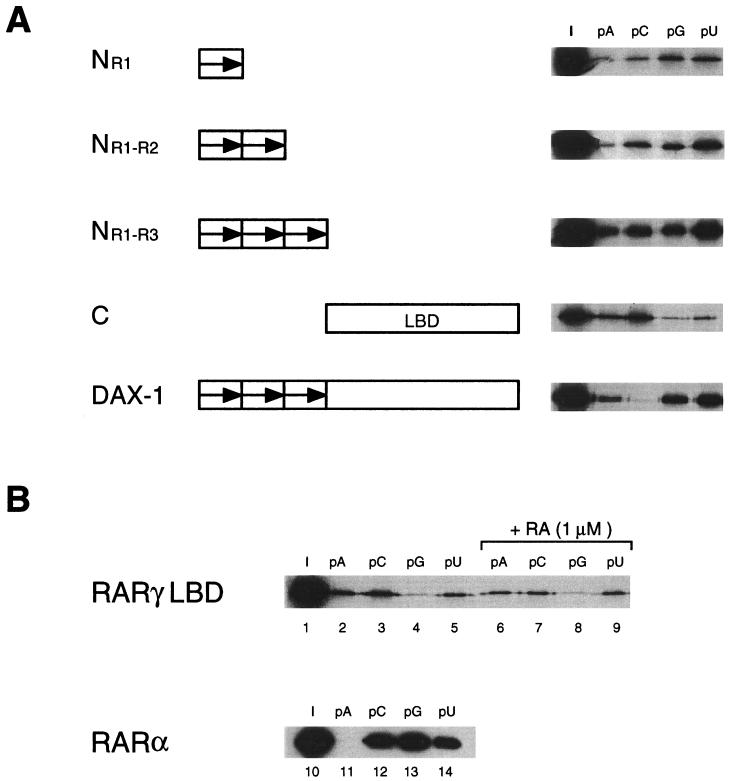
DAX-1 protein domains involved in RNA binding.
To identify the protein domains involved in binding to RNA, we tested N- and C-terminally truncated mutants of the DAX-1 protein in the homopolymer binding assay. A truncated protein encompassing only the sequence of the first N-terminal repeat (aa 1 to 69; NR1 in Fig. 5A) is capable of binding to all four RNA homopolymers, showing a reduced selectivity for poly(G) and poly(U) compared with the full-length protein. A protein containing the sequence corresponding to the first two N-terminal repeats (aa 1 to 135; NR1-R2 in Fig. 5A) shows quantitatively increased binding to RNA homopolymers compared to NR1 but similar reduced selectivity compared to the full-length protein. The NR1-R3 construct (aa 1 to 204) shows strong binding to all four RNA homopolymers, with only a marginal preference for binding to poly(U) (Fig. 5A). Surprisingly, the C construct, encompassing aa 205 to 470 and corresponding to the region in the DAX-1 protein similar to the LBD of nuclear receptors, is also capable of binding to RNA homopolymers, showing relative selectivity for poly(A) and poly(C) (Fig. 5A). As also shown in Fig. 4, the full-length DAX-1 protein binds strongly to poly(U) and poly(G), less efficiently to poly(A), and poorly to poly(C) (Fig. 5A). In order to verify whether binding to RNA homopolymers is a peculiar feature of the DAX-1 putative LBD or is a property which is shared by other nuclear receptors' LBDs, we tested the human RARγ LBD in the RNA homopolymer binding assay in the presence and in the absence of the specific ligand all-trans-retinoic acid (Fig. 5B). RARγ LBD also binds to RNA homopolymers in a ligand-independent fashion, with a specificity which closely matches the one exhibited by the DAX-1 C-terminal domain. Moreover, the closely related RARα binds strongly to poly(C), poly(G), and poly(U) (Fig. 5B).
FIG. 5.
DAX-1 domains involved in RNA binding. (A) Full-length DAX-1 and truncated proteins corresponding to aa 1 to 69 (NR1), 1 to 135 (NR1-R2), 1 to 204 (NR1-R3), and 205 to 470 (C, corresponding to the DAX-1 putative LBD) were translated in the rabbit reticulocyte system and tested in the RNA homopolymer binding assay. Lane I, 1:10 of the input protein. The assay was performed in a buffer containing 0.25 M NaCl. (B) RARγ LBD (aa 178 to 423) was translated in the rabbit reticulocyte system, and binding to RNA homopolymers was tested in the presence (lanes 6 to 9) and in the absence (lanes 2 to 5) of 1 μM all-trans-retinoic acid. Lane I, 1:10 of the input protein (lane 1). Bacterially expressed RARα was used in the RNA homopolymer binding assay and detected by Western blot, as described (12) (lanes 11 to 14). Lane I, 1:10 of the input protein (lane 10). The assay was performed in a buffer containing 0.25 M NaCl.
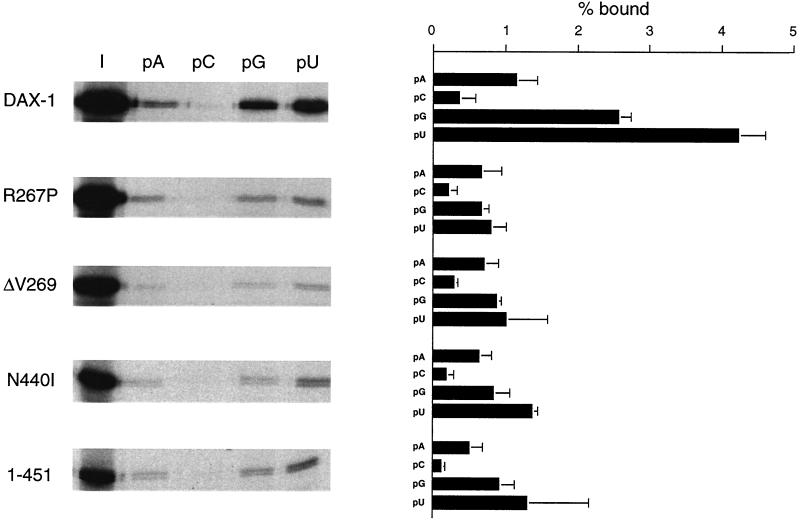
DAX-1 binding to RNA is impaired by mutations found in AHC-HHG patients.
Since we have shown that the DAX-1 C-terminal domain can cooperate with the N-terminal repeats in imparting specificity in RNA binding, we tested the effect of three single-amino-acid mutants found in AHC-HHG patients (R267P, ΔV269, and N440I) and of a C-terminal truncation of the DAX-1 protein (DAX-1 1-451). All DAX-1 mutations found in AHC-HHG have the common feature of altering the protein's C terminus, most frequently by nucleotide insertions or deletions causing a frameshift or introducing a premature stop codon (55). Only a very few cases are known of patients bearing a missense mutation, and only one case is known of a patient with an in-frame deletion (ΔV269) (55). We and others have shown that DAX-1 mutations found in AHC-HHG impair the transcriptional repression activity of the protein (16, 20). Importantly, RNA-binding activity is also impaired in all AHC mutants studied (Fig. 6). While differential specificity of binding to the four RNA homopolymers is not affected in the mutants, the affinity of binding, especially to poly(G) and poly(U), is significantly diminished. Conversely, the R267A mutation has no effect on the RNA-binding properties of DAX-1 (not shown). This mutation also does not impair transcriptional repression (20). These results allow us to establish a close correlation between impairment of transcriptional repression and RNA binding by AHC-causing DAX-1 mutations.
FIG. 6.
DAX-1 binding to RNA is impaired by mutations found in AHC-HHG patients. Wild-type DAX-1 and the R267P, ΔV269, N440I, and 1-451 mutants were expressed in the rabbit reticulocyte lysate and tested in the RNA homopolymer binding assay. Proteins eluted from beads were run on an SDS-PAGE gel and subjected to fluorography. The amount of radioactivity retained on beads was measured by phosphoimaging. Lane I, 1:10 of the input protein. The assay was performed in a buffer containing 0.25 M NaCl. Data are expressed as a percentage of the input protein retained on beads and represent the averages of three independent experiments + standard error of the mean.
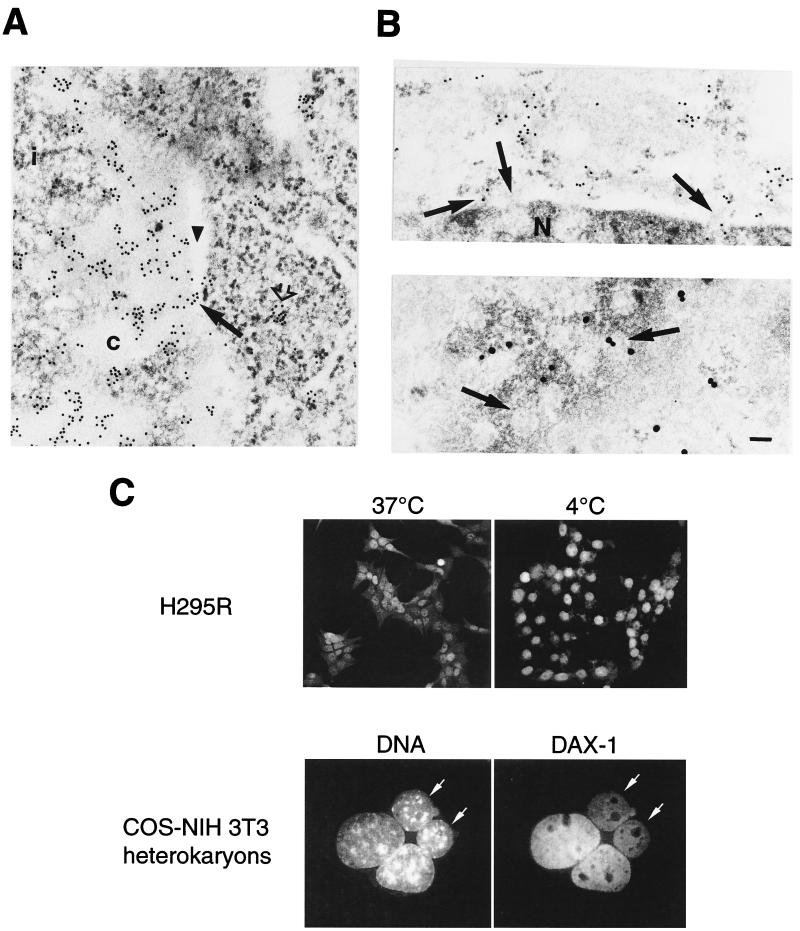
DAX-1 is associated with RNP structures in the nucleus and is actively exported by a temperature-sensitive pathway.
Our results raised the question of a possible role for DAX-1 in the nucleus linked to RNA metabolism. When immunoelectron microscopy was performed on MA-10 cells using the EDTA-regressive staining technique (allowing the visualization of ribonucleoprotein structures [3]), we found nuclear localization of the DAX-1 protein in association with RNP fibrils in the perichromatin region and in the interchromatin space (Fig. 7A). Even inside chromatin-rich regions, which appear less electron dense than RNPs after EDTA treatment, gold particles are constantly found associated with fibrillar constituents, which are readily detectable on the background of the negatively stained chromatin. Cytoplasmic labeling was also confirmed in areas rich in ribosomes by this technique (Fig. 7A). Both in conventionally processed samples and in EDTA-treated samples, we observed gold particles closely associated with nuclear pore structures (Fig. 2 and Fig. 7A and B). In addition, block of the energetic metabolism by incubation of the cells at 4°C in the presence of cycloheximide produces accumulation of DAX-1 in the nucleus (Fig. 7C). Taken together, these findings indicate that DAX-1 is actively being transported out of the nucleus.
FIG. 7.
DAX-1 associates with nuclear RNP and pore structures and is exported from and reimported to the nucleus. (A) DAX-1 immunogold labeling in MA-10 cells processed with the EDTA-regressive staining technique to selectively visualize RNP structures. In the nucleus, gold particles are found in the nucleoplasm associated with perichromatin fibrils and in the interchromatin space. No particles are found in correspondence with the bleached chromatin. Cytoplasmic labeling is found associated with ribosome-rich areas (open arrow; see also Fig. 2A). An image of a cluster of gold particles associated with a nuclear pore structure is indicated with an arrow. c, chromatin; i, interchromatin space. A perinuclear cisterna is indicated with an arrowhead. (B) Gold particles associated with nuclear pore structures. Top: side view. Bottom: tangential view. Nuclear pore structures are indicated with an arrow. Gold particles are larger in the bottom panel because 1-nm immunogold particles were used, followed by silver enhancement. N, nucleus. Bar, 0.1 μm. (C) Top: anti-DAX-1 immunofluorescence in H295R cells cultured at 37°C (left) or kept at 4°C for 4 h (right). Bottom: COS monkey cells transfected with a DAX-1 expression vector were fused to NIH 3T3 mouse cells using the method described in Materials and Methods. Four hours after fusion, cells were fixed with paraformaldehyde, and DAX-1 distribution was detected by immunofluorescence (right). DNA was stained with Hoechst 33342 (left). NIH 3T3 cells (arrows) are easily recognized by their heterochromatin clumps brightly stained by the Hoechst dye. DAX-1 transfer into the NIH 3T3 cell nucleus is readily detectable (arrows). No DAX-1 staining was detected in the nucleus of NIH 3T3 cells mock-fused to COS cells (not shown).
Temperature-dependent export has also been described for human nuclear RNP (hnRNP) A1 (33). In DAX-1, however, no region of similarity to M9 (the motif shown to mediate both nuclear import and export in hnRNP A1 [29]) can be found. In addition, DAX-1 localization was not affected by actinomycin D when it was used at doses that selectively inhibit RNA polymerase I or at higher doses that inhibit polymerases I and II (not shown). Conversely, hnRNP A1 was shown to be redistributed from the nucleus to the cytoplasm after actinomycin D treatment (33). To assess whether DAX-1 can also be newly imported into the cell nucleus, we fused monkey COS cells transfected with a DAX-1 expression vector and mouse NIH 3T3 cells, which do not express endogenous DAX-1. DAX-1 distribution is heterogeneous in transfected COS cells, as the protein may be localized in the nucleus, in the cytoplasm, or in both compartments. Four hours after cell fusion, a large number of heterokaryons were found in which DAX-1 is also localized in the NIH 3T3 cell nucleus, in addition to the COS cell nucleus (Fig. 7C). The DAX-1 protein imported into the NIH 3T3 cell nucleus represents either protein shuttling in and out of the nucleus or protein present in the cytoplasm of COS cells at the time of the fusion that is successively translocated into the NIH 3T3 nucleus. In either case, however, these results show that DAX-1 can be newly imported into the nucleus from a previous nuclear or cytoplasmic localization.
DISCUSSION
Two DAX-1 domains cooperatively specify RNA binding.
Our results show that the motif shared by the three DAX-1 N-terminal repeats constitutes a novel RNA-binding domain, which can bind to RNA homopolymers with little selectivity by itself. Multimerization of this motif in cis cooperatively increases binding affinity but does not impart binding selectivity. Surprisingly, we have found that the DAX-1 and RARγ C-terminal domains constitute autonomous RNA-binding modules. As the LBDs of all nuclear receptors have a common antiparallel α-helical fold (49), it is conceivable that they may share this property. Interestingly, all-helical structures have been found in various other RNA-binding domains (9). Thus, RNA binding is a new function ascribed to the nuclear receptor LBD, in addition to the known ligand binding, dimerization, ligand-dependent transactivation, and, for some members of the family, ligand-independent transcriptional repression (6, 26, 50). In the case of DAX-1, the putative LBD moiety increases the affinity and modulates the selectivity of the N-terminal RNA-binding domain in the context of the full-length protein. This situation is reminiscent of proteins provided with multiple RNP domains, where binding affinity is not simply the sum of the affinities of individual domains and the specificity of the intact protein is generally different from that of the single domains (13, 37, 47). Mouse dax-1 has only 76.3% aa similarity and 66.5% aa identity with its human homologue (41). Nevertheless, as we have shown here, RNA binding is a conserved property of both mouse and human DAX-1.
Since RNA-binding specificity is different for the two homologues, it is possible that they bind to distinct RNA species in the cell and probably do not have completely identical biological functions. This hypothesis is consistent with the remarkably different phenotype observed in dax-1 null mice compared to the human disease caused by mutations in DAX-1 (52). RNA binding is a feature which is most probably shared by other members of the nuclear hormone receptor superfamily, as we have shown in the case of RARα. Interestingly, early work indicated that several steroid-receptor complexes are found associated with RNPs in vivo, and some studies showed that RNA can compete for binding of these complexes to DNA (7, 24, 25). We suggest that also in the case of classical nuclear receptors, provided with two zinc finger modules in their DNA-binding domain, the LBD can synergize and modulate RNA binding by the receptor's N-terminal domain. Zinc finger motifs are known to be able to mediate binding to RNA in addition to DNA, the classical example being TFIIIA (19, 38). Noteworthy are RNA-binding proteins which associate a zinc finger motif with another RNA-binding motif, e.g., WT1 (5, 18) and proteins belonging to the NP220 family (27). Further experiments will determine whether zinc finger motifs found in nuclear receptors have intrinsic RNA-binding properties.
Link between a defect in RNA binding and transcriptional repression in AHC mutants.
The effect of the DAX-1 mutations found in AHC-HHG patients on RNA-binding properties is striking, since they are located in different subdomains in the C terminus and their effect on the fold of the C terminus has been predicted to be different, based on structural modeling (20, 22, 55). Nevertheless, we and others have shown that the same mutations impair the transcriptional repression activity of DAX-1 when they are present both in the context of the full-length protein and in a GAL4–DNA-binding domain fusion of the DAX-1 C terminus (16, 20, 54). Thus, transcriptional repression and RNA binding appear to be two undissociable activities of DAX-1, and this suggests that modulation of gene expression at the transcriptional level possibly requires interaction with RNA cofactors. Recently, an RNA coactivator for steroid receptors (SRA) has been described which functions without being translated into protein (23). The mechanism utilized by SRA to mediate ligand-dependent transcriptional activation by steroid receptors is unknown, but one hypothesis is that it may be involved in the stabilization and scaffolding of a specific coregulator complex containing the coactivator SRC-1 (23). We have shown that DAX-1 requires corepressor factors for its transcriptional silencing activity (20). In the light of the RNA-binding activity of DAX-1, it could be speculated that an RNA species might facilitate DAX-1-mediated transcriptional repression via interaction with corepressors. In keeping with this scenario, mutations found in AHC-HHG patients impairing RNA binding also block repression function. In addition, it has been shown that WT1-KTS isoforms can function as coactivators for SF-1 in activating the Müllerian inhibiting substance gene promoter and that DAX-1 antagonizes WT1 effect (31). Intriguingly, WT1 is also an RNA-binding protein, and the KTS aa sequence lies between the third and fourth zinc fingers, which are present in the domain required for RNA binding (5). The possibility exists then that distinct and antagonistic complexes, one mediating transcriptional activation and the other repression, can be recruited to SF-1 by WT1 and DAX-1, respectively, via binding to specific RNA species.
Role for DAX-1 in RNA metabolism in the nucleus.
The association of DAX-1 with polyribosomes via mRNPs is closely reminiscent of FMRP and related FXR1P and FXR2P proteins (8, 11). These are RNA-binding proteins which shuttle between the nucleus and cytoplasm, and a distinct function has been attributed to each of them in the transport of different RNAs (39, 46). In the nucleus, DAX-1 is found associated with RNP components and nuclear pore structures. Our observations of DAX-1 at the level of nuclear pores are closely reminiscent of Balbiani ring RNP particles being translocated through the nuclear pore (28). In addition, incubation of the cells at 4°C induces nuclear accumulation of DAX-1 in the presence of cycloheximide, indicating that energy is required for export of the presynthesized protein from the nucleus. Conversely, our experiments using monkey-mouse heterokaryons show that DAX-1 is also capable of being newly imported into the nucleus within a short time scale, since it can be detected inside the nucleus of the mouse cells only 4 h after cell fusion.
All our findings are strongly suggestive that DAX-1 is implicated in the process of RNA export from the nucleus to the cytoplasm and that the protein rapidly shuttles between the two compartments. RNA export from the nucleus is a process whose mechanisms remain largely unknown, even if considerable progress has been made recently in elucidating the components of several export pathways (for reviews, see references 17 and 40). At this stage, it is unknown whether DAX-1 associates with specific RNA species during the process of export from the nucleus. However, mRNA, to which DAX-1 is found complexed in the cytoplasm, is a likely candidate. The export of different mRNA species is known to be dependent on different pathways, since distinct hnRNP proteins associate with different transcripts and the export of different mRNAs has distinct requirements for GTP hydrolysis (40). Various RNA-binding proteins have been shown to accompany transcripts from the gene into polyribosomes (48), where they are probably involved in translational control and in the delivery of mRNPs to the translational machinery. DAX-1 may function in a similar fashion. As many steroid receptors undergo ligand-dependent nucleocytoplasmic shuttling (for a review, see reference 51) and since we have shown here that at least one other member of the family in addition to DAX-1 can bind to RNA, it is tempting to speculate on the role played by these proteins in RNA export from the nucleus.
ACKNOWLEDGMENTS
We thank J.-P. Renaud for critical reading of the manuscript; B. Bardoni, J.-P. Renaud, E. Puvion, F. Puvion, J.-L. Vonesch, N. Messaddeq, M.-P. Gaub, G. Lesa, and A. Ziemiecki for discussions, help, and gifts of reagents; and E. Heitz, M. Rastegar, J.-L. Weickert, I. Kolb-Cheynel, P. Eberling, and the IGBMC oligonucleotide synthesis, sequencing, and cell culture services for technical help.
K. Ohe is supported by a postdoctoral fellowship from the Fondation de la Recherche Médicale. This study was funded by grants from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Universitaire Régional, Fondation de la Recherche Médicale, and Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Bae D S, Schaefer M L, Partan B W, Muglia L. Characterization of the mouse DAX-1 gene reveals evolutionary conservation of a unique amino-terminal motif and widespread expression in mouse tissue. Endocrinology. 1996;137:3921–3927. doi: 10.1210/endo.137.9.8756567. [DOI] [PubMed] [Google Scholar]
- 2.Bardoni B, Zanaria E, Guioli S, Floridia G, Worley K C, Tonini G, Ferrante E, Chiumello G, McCabe E R B, Fraccaro M, Zuffardi O, Camerino G. A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat Genet. 1994;7:497–501. doi: 10.1038/ng0894-497. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard W. A new staining procedure for electron microscopical cytology. J Ultrastruct Res. 1969;27:250–265. doi: 10.1016/s0022-5320(69)80016-x. [DOI] [PubMed] [Google Scholar]
- 4.Bertrandy S, Burlet P, Clermont O, Huber C, Fondrat C, Thierry-Mieg D, Munnich A, Lefebvre S. The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution. Hum Mol Genet. 1999;8:775–782. doi: 10.1093/hmg/8.5.775. [DOI] [PubMed] [Google Scholar]
- 5.Caricasole A, Duarte A, Larsson S H, Hastie N D, Little M, Holmes G, Todorov I, Ward A. RNA binding by the Wilms tumor suppressor zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:7562–7566. doi: 10.1073/pnas.93.15.7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 7.Chong M T, Lippman M E. Effects of RNA and ribonuclease on the binding of estrogen and glucocorticoid receptors from MCF-7 cells to DNA-cellulose. J Biol Chem. 1982;257:2996–3002. [PubMed] [Google Scholar]
- 8.Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F, Khandjian E W. The fragile X mental retardation protein is associated with poly(A)+ RNA in actively translating polyribosomes. Hum Mol Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- 9.Cusack S. RNA-protein complexes. Curr Opin Struct Biol. 1999;9:66–73. doi: 10.1016/s0959-440x(99)80009-8. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Gutekunst C-A, Eberhart D E, Yi H, Warren S T, Hersch S M. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Absher D, Eberhart D E, Brown V, Malter H E, Warren S T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 12.Gaub M-P, Rochette-Egly C, Lutz Y, Ali S, Matthes H, Scheuer I, Chambon P. Immunodetection of multiple species of retinoic acid receptor α: evidence for phosphorylation. Exp Cell Res. 1992;201:335–346. doi: 10.1016/0014-4827(92)90282-d. [DOI] [PubMed] [Google Scholar]
- 13.Görlach M, Burd C G, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 14.Green S, Issemann I, Scheer E. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda Y, Swain A, Weber T J, Hentges K E, Zanaria E, Lalli E, Tamai K, Sassone-Corsi P, Lovell-Badge R, Camerino G, Parker K L. Steroidogenic factor 1 and DAX-1 colocalize in multiple cell lineages: potential links in endocrine development. Mol Endocrinol. 1996;10:1261–1272. doi: 10.1210/mend.10.10.9121493. [DOI] [PubMed] [Google Scholar]
- 16.Ito M, Yu R, Jameson J L. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol. 1997;17:1476–1483. doi: 10.1128/mcb.17.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izaurralde E, Mattaj I M. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy D, Ramsdale T, Mattick J, Little M. An RNA recognition motif in WT1 revealed by structural modelling. Nat Genet. 1996;12:329–332. doi: 10.1038/ng0396-329. [DOI] [PubMed] [Google Scholar]
- 19.Ladomery M. Multifunctional proteins suggest connections between transcriptional and post-transcriptional processes. BioEssays. 1997;19:903–909. doi: 10.1002/bies.950191010. [DOI] [PubMed] [Google Scholar]
- 20.Lalli E, Bardoni B, Zazopoulos E, Wurtz J-M, Strom T M, Moras D, Sassone-Corsi P. A transcriptional silencing domain in DAX-1 whose mutation causes adrenal hypoplasia congenita. Mol Endocrinol. 1997;11:1950–1960. doi: 10.1210/mend.11.13.0038. [DOI] [PubMed] [Google Scholar]
- 21.Lalli E, Melner M H, Stocco D M, Sassone-Corsi P. DAX-1 blocks steroid production at multiple levels. Endocrinology. 1998;139:4237–4243. doi: 10.1210/endo.139.10.6217. [DOI] [PubMed] [Google Scholar]
- 22.Lalli E, Sassone-Corsi P. DAX-1 and the adrenal cortex. Curr Opin Endocrinol Diabetes. 1999;6:185–190. [Google Scholar]
- 23.Lanz R B, McKenna N J, Onate S A, Albrecht U, Wong J, Tsai S Y, Tsai M-J, O'Malley B W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 24.Liang T, Liao S. Association of the uterine 17β-estradiol-receptor complex with ribonucleoprotein in vitro and in vivo. J Biol Chem. 1974;249:4671–4678. [PubMed] [Google Scholar]
- 25.Liao S, Smythe S, Tymoczko J L, Rossini G P, Chen C, Hiipakka R A. RNA-dependent release of androgen- and other steroid-receptor complexes from DNA. J Biol Chem. 1980;255:5545–5551. [PubMed] [Google Scholar]
- 26.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsushima Y, Matsumura K, Kitagawa Y. Zinc finger-like motif conserved in a family of RNA binding proteins. Biosci Biotechnol Biochem. 1997;61:905–906. doi: 10.1271/bbb.61.905. [DOI] [PubMed] [Google Scholar]
- 28.Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 29.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 30.Moog-Lutz C, Tomasetto C, Régnier C H, Wendling C, Lutz Y, Muller D, Chenard M-P, Basset P, Rio M-C. MLN64 exhibits homology with the steroidogenic acute regulatory protein (StAR) and is over-expressed in human breast carcinomas. Int J Cancer. 1997;71:183–191. doi: 10.1002/(sici)1097-0215(19970410)71:2<183::aid-ijc10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 31.Nachtigal M W, Hirokawa Y, Enyeart-VanHouten D L, Flanagan J N, Hammer G D, Ingraham H A. Wilms' tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell. 1998;93:445–454. doi: 10.1016/s0092-8674(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 32.Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 33.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 34.Rochel N, Renaud J-P, Ruff M, Vivat V, Granger F, Bonnier D, Lerouge T, Chambon P, Gronemeyer H, Moras D. Purification of the human RARγ ligand-binding domain and crystallization of its complex with all-trans retinoic acid. Biochem Biophys Res Commun. 1997;230:293–296. doi: 10.1006/bbrc.1996.5787. [DOI] [PubMed] [Google Scholar]
- 35.Rochette-Egly C, Oulad-Abdelghani M, Staub A, Pfister V, Scheuer I, Chambon P, Gaub M-P. Phosphorylation of the retinoic acid receptor-alpha by protein kinase A. Mol Endocrinol. 1995;9:860–871. doi: 10.1210/mend.9.7.7476969. [DOI] [PubMed] [Google Scholar]
- 36.Rossignol M, Kolb-Cheynel I, Egly J-M. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J. 1997;16:1628–1637. doi: 10.1093/emboj/16.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shamoo Y, Abdul-Manam N, Williams K R. Multiple RNA binding domains (RBDs) just don't add up. Nucleic Acids Res. 1995;23:725–728. doi: 10.1093/nar/23.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shastry B S. Transcription factor IIIA (TFIIIA) in the second decade. J Cell Sci. 1996;109:535–539. doi: 10.1242/jcs.109.3.535. [DOI] [PubMed] [Google Scholar]
- 39.Siomi H, Siomi M C, Nussbaum R L, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 40.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 41.Swain A, Zanaria E, Hacker A, Lovell-Badge R, Camerino G. Mouse Dax1 expression is consistent with a role in sex determination as well as in adrenal and hypothalamus function. Nat Genet. 1996;12:404–409. doi: 10.1038/ng0496-404. [DOI] [PubMed] [Google Scholar]
- 42.Swain A, Narvaez V, Burgoyne P, Camerino G, Lovell-Badge R. Dax1 antagonizes Sry action in mammalian sex determination. Nature. 1998;391:761–767. doi: 10.1038/35799. [DOI] [PubMed] [Google Scholar]
- 43.Swain A, Lovell-Badge R. Mammalian sex determination: a molecular drama. Genes Dev. 1999;13:755–767. doi: 10.1101/gad.13.7.755. [DOI] [PubMed] [Google Scholar]
- 44.Swanson S M, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamai K T, Monaco L, Alastalo T-P, Lalli E, Parvinen M, Sassone-Corsi P. Hormonal and developmental regulation of DAX-1 expression in Sertoli cells. Mol Endocrinol. 1996;10:1561–1569. doi: 10.1210/mend.10.12.8961266. [DOI] [PubMed] [Google Scholar]
- 46.Tamanini F, Bontekoe C, Bakker C E, van Unen L, Anar B, Willemsen R, Yoshida M, Galjaard H, Oostra B A, Hoogeveen A T. Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Hum Mol Genet. 1999;8:863–869. doi: 10.1093/hmg/8.5.863. [DOI] [PubMed] [Google Scholar]
- 47.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 48.Visa N, Alzhanova-Ericsson A T, Sun X, Kiseleva E, Björkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- 49.Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. . (Erratum, 3:206.) [DOI] [PubMed] [Google Scholar]
- 50.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 51.Yamashita S. Localization and functions of steroid hormone receptors. Histol Histopathol. 1998;13:255–270. doi: 10.14670/HH-13.255. [DOI] [PubMed] [Google Scholar]
- 52.Yu R N, Ito M, Saunders T L, Camper S A, Jameson J L. Role of Ahch in gonadal development and gametogenesis. Nat Genet. 1998;20:353–357. doi: 10.1038/3822. [DOI] [PubMed] [Google Scholar]
- 53.Zanaria E, Muscatelli F, Bardoni B, Strom T M, Guioli S, Guo W, Lalli E, Moser C, Walker A P, McCabe E R B, Meitinger T, Monaco A P, Sassone-Corsi P, Camerino G. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature. 1994;372:635–641. doi: 10.1038/372635a0. [DOI] [PubMed] [Google Scholar]
- 54.Zazopoulos E, Lalli E, Stocco D M, Sassone-Corsi P. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature. 1997;390:311–315. doi: 10.1038/36899. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y-H, Guo W, Wagner R L, Huang B-L, McCabe L, Vilain E, Burris T P, Anyane-Yeboa K, Burghes A H M, Chitayat D, Chudley A E, Genel M, Gertner J M, Klingensmith G J, Levine S N, Nakamoto J, New M I, Pagon R A, Pappas J G, Quigley C A, Rosenthal I M, Baxter J D, Fletterick R J, McCabe E R B. DAX1 mutations map to putative structural domains in a deduced three-dimensional model. Am J Hum Genet. 1998;62:855–864. doi: 10.1086/301782. [DOI] [PMC free article] [PubMed] [Google Scholar]