Abstract
The significant advances made by the global scientific community during the COVID-19 pandemic, exemplified by the development of multiple SARVS-CoV-2 vaccines in less than 1 y, were made possible in part because of animal research. Historically, animals have been used to study the characterization, treatment, and prevention of most of the major infectious disease outbreaks that humans have faced. From the advent of modern ‘germ theory’ prior to the 1918 Spanish Flu pandemic through the more recent Ebola and Zika virus outbreaks, research that uses animals has revealed or supported key discoveries in disease pathogenesis and therapy development, helping to save lives during crises. Here we summarize the role of animal research in past pandemic and epidemic response efforts, as well as current and future considerations for animal research in the context of infectious disease research.
Abbreviations: ACE2, angiotensin-converting enzyme 2; CEPI, Coalition for Epidemic Preparedness Innovations; RSV, respiratory syncytial virus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane protease serine 2
From the moment it began in late 2019, the COVID-19 pandemic has been met with remarkable scientific effort. In less than 1 y, substantial progress has been made in understanding the behavior of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, characterizing the damage it inflicts on the body, and developing safe and effective vaccines. Research in animals has provided many breakthroughs, as it has for most significant outbreaks in the past. Animals have been used to study infectious diseases long before disease-causing microorganisms were known to exist. Animal research in response to pandemics, past and present, provides a clear example of how such research can best serve the scientific community in the event of future outbreaks and other disease conditions. Response to a pandemic requires quick action to identify the emerging diseases, characterize transmission and pathogenesis, and develop preventative measures and therapies. Ideally, through the surveillance of environments and animal populations that may harbor pathogens with pandemic potential and through preclinical and basic science research in virology and vaccinology for diseases that are suspected to be potential threats, the pandemic response should begin before a disease gains the ability to spread easily through a population. In this article, we discuss the importance of animal research in all aspects of pandemic research response and the vital role it continues to play today.
Early Pandemic Research
The 1918 ‘Spanish flu’ influenza outbreak is one of the deadliest infectious disease events in modern history. Roughly a third of the world’s population was infected with the 1918 H1N1 influenza A virus, and an estimated 50 million people died from the infection.4,11 Although Spanish flu was not the first pandemic that humans faced, its outbreak occurred when science and medicine had been revolutionized by new knowledge about the cause and treatment of disease. Public health officials at the time recommended wearing masks and practicing good hygiene to prevent the spread of illness.88 These measures were instituted even though the etiologic agent of influenza had not been identified.4,88
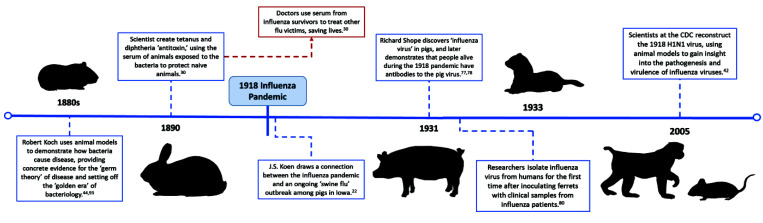
The prevailing theory of previous centuries was that ‘bad air’ from rotting food, waste, or other organic materials created a noxious ‘miasma’ that led to epidemics.32 The ‘germ theory of disease’—the now-accepted idea that microorganisms such as bacteria and viruses cause illness—superseded the ‘miasma theory’ in the late 19th century, precipitating a great search for disease-causing organisms.4,44 Leading this hunt was Robert Koch, a physician–scientist who developed a set of postulates that are still used to demonstrate the cause-and-effect relationship between infection with a specific agent and clinical disease. In the 1880s, Koch and fellow researcher Friedrich Loeffler used animal models to prove that bacteria caused tuberculosis, cholera, anthrax, and more.44,93 These models were fundamental to Koch’s postulates, allowing scientists to systematically link infection with a microbe to subsequent disease in the model organism (Figure 1).
Figure 1.
1918 influenza pandemic: timeline of significant contributions from animal research.
The groundbreaking work done by Koch and colleagues led to a golden age of bacteriology.5 When the 1918 H1N1 outbreak emerged, scientists immediately began searching for the causative agent. Viruses were challenging to isolate, and little was known about them; therefore, bacteria were theorized to be the cause of influenza. At the time, many scientists mistakenly believed the bacteria Haemophilus influenzae—an opportunistic pathogen often found in flu-ravaged lungs—lay behind the disease, but they had trouble consistently isolating it from sick patients.4,26,84 The actual etiologic agent of influenza remained a mystery throughout the pandemic.
In 1918, the veterinarian JS Koen, who worked for the US Bureau of Animal Industry in Iowa, noted that pigs at the National Swine Show and Exposition in Cedar Rapids were experiencing an influenza-like illness similar to the one concurrently affecting humans (Figure 1). He called this new illness ‘swine influenza’ and, a year later, wrote:
“The similarity of the epidemic among people and the epizootic among pigs was so close…as to present a most striking coincidence, if not suggesting a close relation between the two conditions. It looked like ‘flu,’ it presented the identical symptoms of ‘flu,’ and until proved it was not ‘flu,’ I shall stand by that diagnosis.”22
This diagnosis was not popular among swine farmers, who feared that the negative connotation would make their product seem unhealthy, but years later Koen’s observation garnered the attention of another scientist. In 1931, Richard Shope, a virologist working with the Rockefeller Institute, investigated Koen’s ‘swine flu’ on farms in Iowa.22 Previous investigators had failed to isolate the specific agent behind the swine flu, as they had often inadvertently filtered out the small viral particles in their attempt to isolate bacterium. Shope repeated these experiments in pigs, taking filtrate from mucus collected from sick pigs and inoculating healthy swine. When the pigs became sick, Shope concluded that a ‘filterable agent,’ such as a virus, was the cause of swine influenza (Figure 1).77 He also concluded that H. influenzae, which also had been isolated from pigs, worked with the virus to cause more severe disease—exactly what had been seen in humans during the 1918 pandemic. Shope went on to show that people old enough to have experienced the 1918 pandemic carried antibodies against the swine virus, drawing a concrete connection between the swine flu and the Spanish Influenza.78
Inspired by Shope’s work, in 1933, British physician and scientist Dr. Wilson Smith and colleagues obtained filtered throat washings from human influenza patients and exposed a variety of animals to the material.80,84 The investigators had little success infecting animals until they turned to ferrets, which were available at their institution because of ongoing canine distemper virus research.25 Ferrets were the only animals that became sick and consequently they proved to be an excellent model for human disease in regard to both the clinical progression and transmission of influenza (Figure 1). In addition, the researchers used convalescent human sera protect ferrets against subsequent passage of the virus and showed that ferrets inoculated with Shope’s swine influenza virus were immune to the human strain of the virus. When accidental ferret-to-human transmission of influenza occurred after a sick animal sneezed near an investigator’s face, Smith and colleagues further demonstrated that the virus grown in ferrets caused disease in humans and could subsequently be isolated again to infect other ferrets.81
With these early studies, the scientists isolated the influenza A virus from humans and developed an animal model—still critical to modern influenza research—with which laboratory research could be performed. Smith and colleagues noted that “direct experiments on man are fraught with difficulties” and discussed the potential value of parallel clinical and laboratory research during an ongoing epidemic.25,82 Dr Smith, along with scientists CH Andrewes and CH Stuart-Harris, went on to study influenza immunity and develop vaccines in ferrets and mice, followed by human volunteers, with the intention to deploy larger-scale clinical trials during the inevitable next epidemic.81,82 This foresight to initiate preclinical trials in advance of anticipated disease outbreaks is notable and reflects modern goals for animal research on pandemic diseases.
The breakthroughs achieved with animals before, during, and after the 1918 influenza pandemic exemplify the fundamental role animal research has played in humankind’s fight against emerging infectious diseases. Scientists have expanded on this research throughout subsequent decades of epidemic response.
Identifying Etiology
Despite having modern techniques to help identify disease-causing organisms today, animals remain instrumental in understanding how pathogens infect, replicate, spread, and cause disease in living organisms. Categorizing the type of agent causing disease is an obvious critical first step in an outbreak response. As evidenced by the work of researchers on the bacteria H. influenzae during and after 1918 influenza pandemic, identifying the primary etiologic agent of a disease can be difficult when other factors such as secondary infections, coinfections, host immune response, and comorbidities complicate data interpretation.4,26,84 Animal model systems allow researchers to conduct highly controlled experiments with well-defined subjects in order to isolate and identify novel infectious pathogens in the face of an outbreak.
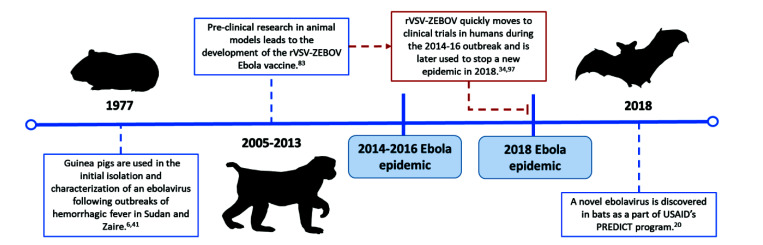
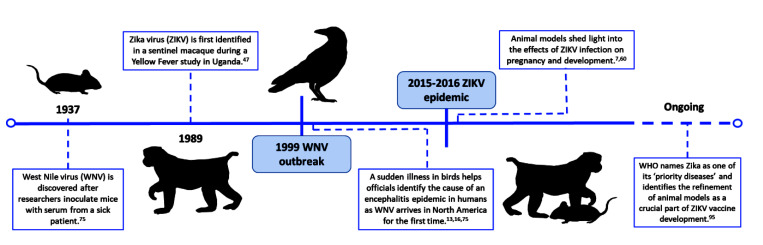
Koch’s postulates served as guidelines for the identification of disease-causing agents for years after his original research was published.44,93 Shope found that although H. influenzae could be consistently isolated from pigs sick with swine influenza, the bacteria did not induce disease unless the animals were also inoculated with the influenza virus.77 During an epidemic in Hong Kong, Alexandre Yersin, a Swiss–French bacteriologist and physician, discovered the etiologic agent of the bubonic plague after isolating bacteria from the swollen lymph node of a patient, injecting it into mice and guinea pigs, and observing disease. His careful study of the bacillus found in the buboes of deceased plague patients and infected rodents, which developed similar clinical signs and dissemination of the bacteria, led to advances relevant to both transmission and treatment.8 Guinea pigs were also used to identify and characterize Ebola viruses after an outbreak of hemorrhagic fever among people in Sudan and Zaire (now the Democratic Republic of the Congo) in 1976 (Figure 2).6,41 Guided by epidemiologic and clinical data, the study teams working on the hemorrhagic fever outbreaks collected various pre- and postmortem samples and inoculated guinea pigs intraperitoneally with sample suspensions. The animals developed a 5-d course of febrile illness, and successive passage of infected guinea pig blood into additional guinea pigs produced a fatal febrile disease. The Ebola virus from these animals was isolated and cultured in Vero cells and was further characterized using immunofluorescent and electron microscopy.6,72 Similarly, West Nile virus was discovered in 1937 after mice were inoculated with serum isolated from a febrile patient in Uganda (Figure 3). Although the patient initially was assumed to have Yellow Fever, the mouse model and subsequent studies in NHP identified the etiologic agent as a novel virus with distinct pathogenicity and immunogenicity.75
Figure 2.
Ebola virus epidemics: timeline of significant contributions from animal research.
Figure 3.
West Nile virus and Zika virus outbreaks: timeline of significant contributions from animal research.
The discovery of HIV, the virus which causes AIDS, is a more recent example of how animals have been used to study connections between pathogens and human disease. HIV remains a global health threat: approximately 38 million people worldwide were reported to be infected in 2019.35 AIDS was first observed in the United States in 1981, after outbreaks of typically rare opportunistic infections among otherwise healthy men. In 1983, French and American research groups independently uncovered a novel retrovirus in patients with AIDS.76 Retroviruses had first been described by the physician–veterinarian team of Vilhelm Ellermann and Oluf Bang in 1908. They found that chickens developed a form of leukemia caused by viral infection.18 Many other animal viruses were subsequently discovered to be retroviruses, including the Visna–Maedi virus, which causes a progressive immunosuppressive disease in sheep.87 In 1985, researchers Janice Clements and HIV codiscoverer Robert Gallo published research that compared HIV and Visna–Maedi virus.29 This research helped to establish the identity of HIV as a lentivirus and provided initial clues regarding the mechanism of HIV infection. In addition, the sheep Visna–Maedi infection model was recognized as a model system for in vivo testing of antiHIV drugs.87
Animal research provides a foundation for current science on pathogenicity. In 2002, an outbreak of unidentified severe respiratory disease began in the Guangdong province of China. The illness was initially termed Severe Acute Respiratory Syndrome (SARS) because the microorganism causing the clinical signs was unknown.68 After a promising viral candidate was identified by microbiologic studies,50,68,69 scientists at Erasmus University fulfilled Koch’s postulates by verifying that the newly isolated coronavirus (SARS-CoV-1) caused disease in macaques and was likely the primary etiologic agent of SARS.27 When the COVID-19 pandemic began in December 2019, previous research with animals and animal viruses permitted scientists to rapidly place the novel coronavirus within the Coronaviridae family tree, suggesting a probable zoonotic origin for SARS-CoV-2 and a likely mechanism of host-cell entry by the virus.101 Research with sophisticated animal models and modern genetic and molecular techniques will continue to provide key information about the relationship between pathogens and disease.
Describing Pathogenesis
Close scrutiny of COVID-19 over the past year has revealed a complex constellation of symptoms and conditions beyond what is typically associated with seasonal coronaviruses. SARS-CoV-2 has been implicated in neurologic, cardiovascular, and autoimmune pathology. The complex interplay between the novel coronavirus and various systems of the body is not yet well understood and raises many questions about disease pathogenesis. The advent of organoid and other complex tissue model systems has allowed the study of organ-specific problems,45 but these systems do not model the entire body. Animal models of SARS-CoV-2 are important in answering these questions.
Animal research has contributed to our knowledge of disease pathogenesis, transmission, virulence factors, and immune response for most major infectious disease outbreaks. Research with rats conducted by Paul-Louis Simond in 1898 illustrated for the first time that flea-borne transmission was one of the main mechanisms behind the spread of bubonic plague.33,71 Although not well accepted at the time, Simond’s experimental work with animal models of disease transmission eventually changed the way plague prevention and control was approached.21 Studies of influenza in ferrets, birds, and pigs have clarified viral tropism in the respiratory or gastrointestinal tracts and improved our understanding of the pathogenesis, transmission, and the epidemic potential of various flu strains.66 Animal models have been essential for investigating deadly human diseases such as Ebola and Nipah, which occur only sporadically and with high mortality in people and are therefore challenging to study.
The 1918 H1N1 influenza pandemic was known from historical records to have an unusual characteristic: the virus was especially deadly among healthy adults from 15 to 34 y of age. This feature is in contrast to other seasonal or pandemic flu strains, which typically kill the very young or old.42 To understand the behavior of this virus, in 1997, a research team excavated a mass grave in Brevig Mission, Alaska, where nearly 80 of the village’s inhabitants had been buried after dying from the 1918 influenza virus. Scientists were able to extract 1918 virus genetic material from the lungs of a deceased woman preserved in the permafrost and subsequently sequenced the hemagglutinin (HA) gene. This innovative work ultimately allowed a team at the Centers for Disease Control and Prevention to reconstruct a recombinant pandemic flu.42 A mouse model was used to characterize infection with 1918 H1N1, which confirmed that the influenza strain was incredibly adept at replicating in the lungs and over-activating the host immune response. The virus was highly lethal to the mice and potentially explained why healthy adults with robust immune systems fared worse during the pandemic.43 Because the original 1918 strain no longer circulates in humans, much of what we know about its virulence is based on animal research. This research gave us insight into the complicated interplay between virus and immune system, including the role of specific genes and cytokines in triggering severe disease (Figure 1).17,43 Such knowledge, in turn, can help us identify danger signals in future flu strains.
Basic virology research in animals is also important during an outbreak. During the 2015–2016 Zika virus epidemic, animal models facilitated the progression from epidemiologic observations into viral pathogenesis research (Figure 3). Mouse and NHP models were used to correlate Zika-associated neurologic disease in humans with the neurotropic behavior of the virus in vivo.7,58,60 Both mice and NHP have similar placentation to humans, thus serving as useful models for intrauterine Zika virus infection and developmental studies.58 These models offered crucial insight into infection during pregnancy and subsequent fetal CNS lesions.7,58,60,74 Mouse models of infection also established that the virus replicated in the testicles and female reproductive tract and therefore helped to explain reports of sexual transmission in humans.58,60,73 Ongoing Zika vaccine research uses animals to study acquired immunity and cross-protection in the context of distinct Zika virus lineages and other arboviruses, such as Dengue and Chikungunya (Figure 3).23 Given that overlap between these insect-borne diseases is common in the endemic range of Zika, animal studies are necessary to unravel the complexity of the immune response to Zika in association with prior or concurrent infections with other viruses. For example, a mouse model has been used to generate Zika vaccine candidates that protect against both antibody-dependent disease enhancement and disease exacerbation in vaccinated persons who produce deleterious antibodies that enhance subsequent natural infection.23 Investigation of these problems in preclinical models is imperative before vaccine candidates are tested in virus-endemic regions.
COVID-19 pathobiology research has capitalized on animal studies performed with regard to the 2002 SARS outbreak. Like SARS-CoV-2, SARS-CoV-1 uses angiotensin-converting enzyme 2 (ACE2) as a receptor and means of entry into host cells.68 Studies in rodents, combined with sequencing data from human tissues, elucidated the expression pattern of ACE2 through the body, explaining viral tropism and subsequent clinical disease.55,68 ACE2 knockout studies in mice proved that the protein was necessary for SARS-CoV-1 infection.51 This earlier work was important when SARS-CoV-2 was first described; researchers were quickly able to culture, isolate, and characterize the virus by using ACE2-expressing cells.101 The cell-surface transmembrane protease serine 2 (TMPRSS2) is another protein that facilitates the entry of SARS-CoV-1 and -2 into host cells.36,37 TMRPSS2-knockout mice infected with SARS-CoV-1 fail to develop pneumonia or weight loss, whereas wild-type mice develop significant clinical disease.40 A TMPRSS2 inhibitor, camostat mesylate, has been shown to block SARS-CoV-1 infection in mice and is being investigated as a potential therapeutic agent for COVID-19.36,102
Animal studies have also been used to investigate COVID-19 transmission and the behavior of viral variants. The scientific community has debated the contribution of airborne transmission (i.e., the dissemination of virus through small-particle aerosols suspended in the air) in COVID-19 spread.3,31 Studies in ferrets demonstrated that airborne exposure alone could result in transmission of SARS-CoV-2.46,52 Research such as that supports epidemiologic observations and facilitates controlled investigation into the mechanism of COVID-19 spread. Although next-generation genetic sequencing has given researchers the ability to closely monitor changes in circulating SARS-CoV-2 variants, the significance of a new mutation in the virus cannot necessarily be determined based on sequence alone. However, validated animal models can be used to test the effects of novel mutations on virulence or transmissibility, thus providing data to inform public health actions. Animal models of SARS-CoV-2 infection were used in concert with population genetic modeling to study the D614G mutation of the SARS-CoV-2 spike protein, thereby offering initial evidence that the variant had greater transmissibility.38,54 Animal models are already being used to test mutations found in the novel, fast-spreading ⟨ variant and other variants.9 Similar studies will likely remain useful as the virus continues to circulate and mutate.
Developing Vaccines and Therapeutics
In addition to revealing the nature of the agent causing an outbreak, animal models have long been important in the development of treatments and vaccines. Antibody therapy—whether convalescent serum therapy or monoclonal antibody therapy—has been a mainstay of infectious disease treatment since the late 1800s.30 The idea of ‘passive immunization’ began with the work of scientists Emil von Behring and Shibasaburo Kitasato, who showed that serum collected from rabbits that had been inoculated with tetanus or diphtheria toxin (i.e., convalescent serum) could prevent illness when administered to naive animals (Figure 1). After this breakthrough, standardized diphtheria and tetanus antitoxin for human use were produced in horses and cattle. Passive immunization via the administration of convalescent sera was also used during the 1918 influenza pandemic, thus reducing fatality rates in newly infected persons by nearly 50% (Figure 1).7,30,100 Convalescent serum and monoclonal antibody products have been used therapeutically in the treatment of several recently emergent infectious diseases, including SARS, Ebola, and COVID-19, and through genetic engineering, animals can be used to produce highly specific human monoclonal antibodies.24,61,63,85
For cases in which human challenge studies would not be ethical or feasible, the FDA Animal Rule may be invoked to facilitate the approval of a medical product through the use of animal studies alone.89 Such measures are important in preparing for events such as biologic terror attacks in which potential biologic weapons such as anthrax or smallpox cannot safely be given to volunteers and when natural disease occurs too sporadically to assess vaccines or therapeutics in the human population. To achieve approval through the Animal Rule, the effect of the product must be established in either multiple animal species or a single well-characterized animal model, and additional requirements surrounding pharmacokinetic and pharmacodynamic data must be met.89 The FDA Animal Rule has been used to approve treatments for smallpox, inhaled anthrax, and others.90,91
Animal use is also instrumental in the development of vaccines. The recipe for an effective, safe vaccine is not straightforward, given that pathogen biology, the innate and adaptive immune responses, and the recipient’s health and socioeconomic background can all affect vaccine performance. Although strategies surrounding vaccine design have changed over the years, animal models remain the optimal biologic systems for the investigation of complicated immunologic questions and therefore are a critical part of assessing efficacy and safety of vaccines before they enter human trials. For example, animal models of respiratory syncytial virus (RSV) infection have allowed researchers to determine the mechanisms behind the phenomenon of ‘disease enhancement’ in children vaccinated against RSV and subsequently exposed to the virus. These models are now used to predict the safety of new RSV vaccines under development.53 Animal research has had an important role in the development of vaccines against many diseases, including measles, tuberculosis, polio, and influenza.28
For many emerging and reemerging infectious diseases without known therapeutics, the use of vaccines is essential to interrupt disease transmission and prevent further morbidity and mortality during an epidemic. The response to the 2014–2016 Ebola outbreak in West Africa is an example of how prior animal research facilitated the rapid initiation of a lifesaving vaccine trial.83 At the outset of the public health emergency, WHO experts—in collaboration with public health organizations, pharmaceutical companies, and regulatory agencies—rapidly developed a plan to review and accelerate the development of Ebola vaccine candidates based on efficacy and safety data from NHP research (Figure 2).34 The rVSV-ZEBOV recombinant Ebola virus vaccine was thus able to move rapidly from preclinical research to clinical trials in humans. In 2018, this vaccine was able to effectively end the Ebola outbreak in the Équateur province of the Democratic Republic of the Congo while the outbreak was still in its early stages, and the rVSV-ZEBOV recombinant vaccine remains a vital tool in response to Ebola (Figure 2).94,97
Vaccine technology using mRNA saw its clinical debut during the COVID-19 pandemic. From the publishing of the SARS-CoV-2 genome sequence in January 2020 to the first therapeutic doses given in December 2020, the Pfizer-BioNTech (BNT162b2) and Moderna (mRNA-1273) COVID-19 vaccines were developed in only 11 mo.2,39 Vaccine development timelines typically stretch into several years—on average 10 y—so the speed with which these mRNA vaccines were produced was unprecedented.86 Numerous factors led to the rapid progress in vaccine creation,86 but although the technology had not yet been used widely in a clinical setting, a long history of mRNA vaccine research in animals meant that a robust body of preclinical data was already available before the COVID-19 pandemic began.2,65
Proof-of-concept research for mRNA therapeutics began in the early 1990s with studies in mice and rats. Animals given intramuscular injections of RNA were monitored for production of the associated protein. In vivo studies continued to be essential, because mRNA was quickly found to be challenging to work with: it is easily degraded by extracellular host enzymes, has high innate immunogenicity, and can be difficult to deliver to target cells.65 Over the subsequent decades, studies in both large and small animals supported significant advances in modifying mRNA for stability and immunogenicity. Studies to develop novel vaccine delivery vehicles, such as lipid nanoparticles, were performed in animals to assess safety and efficacy.39,65 Building on this thorough preclinical evidence, vaccine candidate BNT162b moved into rodent and NHP studies just months after the initial publication of the SARS-CoV-2 genome sequence.92 Vaccine candidate mRNA-1273 moved into phase I clinical trials about 2 mo after sequence availability and into phase II and III trials 5 and 6 mo later, respectively.39 Further research into mRNA vaccine technology—and other vaccination platforms—ideally should be performed for emerging disease candidates in advance of an outbreak.
In this vein, the success of the Ebola vaccine development program led WHO to create an R&D Blueprint for Action to Prevent Epidemics,95,96 described as “a global strategy and preparedness plan that allows the rapid activation of research and development activities during epidemics.” The R and D Blueprint outlines a list of priority pathogens (such as novel coronaviruses, Zika virus, and Nipah virus) that present global disease threats.95,96 The Oslo-based Coalition for Epidemic Preparedness Innovations (CEPI), a nonprofit organization that finances research projects, is working from this blueprint to develop vaccines against these priority pathogens before an outbreak or in rapid response to one.15 WHO and CEPI represent major players in the international effort to reduce the time between the identification of an outbreak and approval of diagnostics, therapeutics, and vaccines. Both have emphasized the importance of preclinical studies in animals in laying the groundwork for clinical trials, as was done during the 2014–2016 Ebola epidemic and COVID-19 pandemic. In response to COVID-19, CEPI, the Geneva-based Gavi (the Vaccine Alliance), and WHO are coleading COVAX, with key delivery partner UNIFCEF, to ensure equitable access to COVID-19 vaccines and to end the acute phase of the pandemic by the close of 2021.15
Pandemic Preparedness
Scientists have taken a ‘One Health’ approach to pandemic research throughout history, even before the concept was given a name. When West Nile virus first arrived in the United States in 1999, doctors and scientists initially struggled to identify the agent at the root of the outbreak of encephalitis that erupted in New York City. Diligent investigation of a concurrent avian mortality event by veterinary pathologist Tracy McNamara led to the CDC’s isolation of West Nile virus (Figure 3).13,16,75 Reston ebolavirus was discovered after an outbreak of hemorrhagic fever in a colony of research macaques; blood samples from their human caretakers showed that they had been exposed to the virus, although none became ill.14,59 Alexandre Yersin demonstrated the origin of human plague epidemics for the first time when he noticed dead rats littering the streets of an affected city and subsequently isolated the same bacillus—now eponymously called Yersinia pestis—from both rats and humans.8,33 These examples underscore the importance of considering animal health in the context of emerging infectious diseases. Indeed, the spillover of SARVS-CoV-2 into pets, zoo animals, and farmed mink highlights the pervasiveness of the human–animal interface.49,56,57,67
An estimated 75% of emerging and reemerging infectious diseases are zoonotic.12 Wild animals, livestock, and even pets can act as sentinels, alerting us to impending outbreaks or emerging pathogens. Zika virus, which rapidly spread to the Americas during the 2015–2016 epidemic, was first described in 1947 in a rhesus macaque in a forest of Uganda (Figure 3). Macaques were used as sentinels for the yellow fever virus by being placed in cages built into the tree canopy as part of a study to detect the virus circulating in the forest. When a macaque developed a fever, researchers took a sample of its blood and injected it into the brains of mice, which also fell ill. This newly identified virus was later named ‘Zika’ after the forest in which it was discovered.47
Using animals as sentinels for potentially dangerous novel viruses is not a new idea, but is being increasingly recognized as important. In 2009, the United States Agency for International Development initiated the PREDICT program, a surveillance system designed to identify pathogens with zoonotic or pandemic potential before they caused outbreaks. The program, which ran through 2019, sent researchers to look for animal viruses in ‘hot spots’ of animal–human interaction; the program also provided disease outbreak prevention training and strengthened medical laboratories in developing countries.10 Through the investigation of areas with high biodiversity, dense human population, and environments where animals and humans have close contact, PREDICT identified nearly 1000 new viruses—including a new species of Ebola virus— from animal samples (Figure 2).10,20 Bat coronavirus RaTG13, the closest known relative to SARS-CoV-2, was identified through surveillance of wild bat populations near a mining town in Yunnan province, China. Researchers have discovered that wild horseshoe bats have other circulating coronaviruses that are intrinsically capable of using the ACE2 receptor.48,101
‘Virus hunting’ programs such as PREDICT are valuable for understanding what pathogens are circulating in potential animal or environmental reservoirs, but determining which of these pathogens are an actual threat is more challenging. The emergence of ‘ have identified crucial areas for acceleration in biomedical research and product development to prepare for Disease X, including animal model development, validation, and standardization. One group of authors recognized “insufficient available and standardized animal models” as a challenge facing the development of diagnostics, vaccines, and therapeutics during a Disease X epidemic and recommended expanding and curating databases of existing animal models to help researchers and scientific advisory boards to generate rigorous, reproducible research.79 These principles were implemented during the current pandemic: the WHO R and D Blueprint team established an ad hoc COVID-19 modeling expert working group (WHO-COM), composed of more than 150 scientists from around the world, with the goal of advancing the development of COVID-19 medical countermeasures. WHO-COM’s aim is to provide a platform to share data, coordinate preclinical studies, foster collaboration, and promote the 3Rs principles to guide international efforts in modeling human disease.62,98 The group met in December 2020 to address the emerging SARS-CoV-2 variants, discussing plans for pathogenicity and transmission studies using COVID-19 animal models and the distribution of reagents and sera to group members to facilitate research on virus immune evasion.99 Coordinated action such as this could be leveraged to standardize research on other infectious diseases in preparation for future epidemics and to support rigorous and ethical research during a public health crisis.
Animal research is necessary to elucidate the risks inherent at the human-animal interface and to predict and prevent future pandemics. For example, experimental SARS-CoV-2 infection studies not only helped identify animal models of disease but also revealed potential viral reservoir species. White-tailed deer (Odocoileus virginianus) were permissive to SARS-CoV-2 infection in the lab, shedding virus from their respiratory tissues and transmitting the virus to other deer.64 In light of these data, researchers examined the blood of wild white-tail deer in the northeast United States and found that 40% of the animals had antibodies to SARS-CoV-2.56 These findings are potentially troubling, given that wildlife reservoirs could promote the mutation and adaption of viral strains and even reemergence of disease into new populations. Experimental transmission studies could also aid in the identification of species that may provide sources of future outbreaks, such as livestock or common commensal species that have frequent overlap with humans and human environments. Similar studies will be key to determining characteristics of pathogen or host species that permit movement of disease between animal and human populations, potentially offering critical information about pending outbreaks. These efforts, in concert with improvement and standardization of animal model design, can be a valuable component of pandemic countermeasures.
Conclusion
Comparative medicine has been an integral part of outbreak response research even before the first disease-causing organisms were identified. Over the course of history, people have worked with animals to increase our understanding of emerging infectious diseases, where they come from, and how to prevent them. Although scientific advances over the past century have changed the way that we explore and combat disease, the field of infectious disease research has continuously highlighted the value of animal research inside the lab and out. Animal models allow for rapid, controlled investigation into disease processes, pathogen biology, and the development of therapeutics and vaccines, but these model systems must be used in ways that promote ethical, rigorous, and reproducible science. Rarely will one model perfectly capture all aspects of human disease, so the strengths and limitations for each model must be well defined. International organizations such as WHO and CEPI have led attempts to foster communication and collaboration in validating animal models during the COVID-19 pandemic. Researchers should strive to be thorough and detailed when reporting work with animals. The ARRIVE guidelines have been developed for the purpose of clarifying the reporting of animal studies.70 Such guidelines can help to assure comprehensive descriptions of work with animals even in the fast-paced, global surge of published research that has become the norm during the current pandemic. As always, studies conducted in animals should follow the principles of the 3Rs—replacement, reduction, and refinement. In some circumstances, molecular diagnostics and genetic techniques may reduce or replace the need for in vivo animal work. Additional considerations regarding ethics and the design of animal studies of infectious disease have been covered elsewhere.1,19 When conducted appropriately, animal research can facilitate lifesaving science in the face of a global health crisis. Animal research will no doubt continue to inform our response to future significant infectious disease events, just as it has throughout history.
References
- 1. Acosta A, Norazmi MN, Hernandez-Pando R, Alvarez N, Borrero R, Infante JF, Sarmiento ME. 2011. The importance of animal models in tuberculosis vaccine development. Malays J Med Sci 18: 5– 12. [PMC free article] [PubMed] [Google Scholar]
- 2. Ball P. 2021. The lightning-fast quest for COVID vaccines - and what it means for other diseases. Nature 589: 16– 18. 10.1038/d41586-020-03626-1. [DOI] [PubMed] [Google Scholar]
- 3. Baraniuk C. 2021. Covid-19: What do we know about airborne transmission of SARS-CoV-2? BMJ 373: 1– 3. 10.1136/bmj.n1030. [DOI] [PubMed] [Google Scholar]
- 4. Barry J. 2004. The great influenza: The epic story of the greatest plague in history. New York ( NY): Viking Penguin. [Google Scholar]
- 5. Blevins SM, Bronze MS. 2010. Robert Koch and the ‘golden age’ of bacteriology. Int J Infect Dis 14: e744– e751. 10.1016/j.ijid.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 6. Bowen ET, Lloyd G, Harris WJ, Platt GS, Baskerville A, Vella EE. 1977. Viral haemorrhagic fever in southern Sudan and northern Zaire. Preliminary studies on the aetiological agent. Lancet 309: 571– 573. 10.1016/S0140-6736(77)92001-3. [DOI] [PubMed] [Google Scholar]
- 7. Branswell H. [Internet]. 2016. “ Zika infection causes brain stunting in monkey fetus” STAT. [Cited 20 January 2021]. Available at: https://www.statnews.com/2016/09/12/zika-fetus-brain-monkey/.
- 8. Butler T. 2014. Plague history: Yersin’s discovery of the causative bacterium in 1894 enabled, in the subsequent century, scientific progress in understanding the disease and the development of treatments and vaccines. Clin Microbiol Infect 20: 202– 209. 10.1111/1469-0691.12540. [DOI] [PubMed] [Google Scholar]
- 9. Callaway E. 2021. Could new COVID variants undermine vaccines? Labs scramble to find out. Nature 589: 177– 178. 10.1038/d41586-021-00031-0. [DOI] [PubMed] [Google Scholar]
- 10. Carlson CJ. 2020. From PREDICT to prevention, one pandemic later. Lancet Microbe 1: e6– e7. 10.1016/S2666-5247(20)30002-30011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention. [Internet]. 2019. “1918 Pandemic (H1N1 Virus).” Centers for Disease Control and Prevention. [Cited 28 December 2020]. Available at: www.cdc.gov/flu/pandemic-resources/1918-pandemic-h1n1.html.
- 12. Centers for Disease Control and Prevention. [Internet]. 2017. Zoonotic Diseases. [Cited 19 December 2020]. Available at: www.cdc.gov/onehealth/basics/zoonotic-diseases.html.
- 13. Centers for Disease Control and Prevention. 1999. Outbreak of West Nile-like viral encephalitis–New York, 1999. MMWR Morb Mortal Wkly Rep 48: 845– 849. [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention. 1990. Update: filovirus infection in animal handlers. MMWR Morb Mortal Wkly Rep 39: 221. [PubMed] [Google Scholar]
- 15. CEPI. [Internet]. 2021. COVAX: CEPI’s Response to COVID-19. CEPI. [Cited 20 January 2021]. Available at: https://cepi.net/COVAX/.
- 16. Charatan F. 1999. Eight die in outbreak of virus spread from birds. BMJ 319: 941. 10.1136/bmj.319.7215.941a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cillóniz C, Shinya K, Peng X, Korth MJ, Proll SC, Aicher LD, Carter VS, Chang JH, Kobasa D, Feldmann F, Strong JE, Feldmann H, Kawaoka Y, Katze MG. 2009. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog 5: 1– 12. 10.1371/journal.ppat.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coffin JM, Hughes SH, Varmus HE, editors. 1997. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- 19. Colby LA, Quenee LE, Zitzow LA. 2017. Considerations for infectious disease research studies using animals. Comp Med 67: 222– 231. [PMC free article] [PubMed] [Google Scholar]
- 20. Columbia University Mailman School of Public Health. [Internet]. 2019. “Scientists Discover Ebola Virus in West African Bat, Columbia University. [Cited 19 December 2020]. Available at: www.publichealth.columbia.edu/public-health-now/news/scientists-discover-ebola-virus-west-african-bat.
- 21. Crawford EA, Jr. 1996. Paul-Louis Simond and his Work on Plague. Perspect Biol Med 39: 446– 458. 10.1353/pbm.1996.0031. [DOI] [Google Scholar]
- 22. Currier RW. [Internet]. 2005. People and Pigs: Iowa’s Role in 20th-Century Influenza History. Iowa Heritage Illustrated 86, 80– 85. Available at: https://ir.uiowa.edu/ihi/vol86/iss2/13 [Google Scholar]
- 23. Dai L, Xu K, Li J, Huang Q, Song J, Han Y, Zheng T, Gao P, Lu X, Yang H, Liu K, Xia Q, Wang Q, Chai Y, Qi J, Yan J, Gao GF. 2021. Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch. Nat Immunol 22: 958– 968. 10.1038/s41590-021-00966-6. [DOI] [PubMed] [Google Scholar]
- 24. Duan K, Liu B, Li C, Meng S, Hu Y, Peng C, Yuan M, Huang J, Wang Z, Yu J, Gao X, Wang D, Yu X, Li L, Zhang J, Wu X, Li B, Xu Y, Chen W, Peng Y, Hu Y, Lin L, Liu X, Huang S, Zhou Z, Zhang L, Wang Y, Zhang Z, Deng K, Xia Z, Gong Q, Zhang W, Zheng X, Liu Y, Yang H, Zhou D, Yu D, Hou J, Shi Z, Chen S, Chen Z, Zhang X, Yang X. 2020. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci USA 117: 9490– 9496. 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Evans DG. 1966. Wilson Smith, 1897–1965. Biogr Mem Fellows R Soc 12: 478– 487. 10.1098/rsbm.1966.0023 [DOI] [Google Scholar]
- 26. Eyler JM. 2010. The state of science, microbiology, and vaccines circa 1918. Public Health Rep 125 3 Suppl: 27– 36. 10.1177/00333549101250S306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fouchier RAM, Kuiken T, Schutten M, van Amerongen G, van Doornum GJJ, van den Hoogen B, Peiris M, Lim W, Stöhr K, Osterhaus ADM. 2003. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature 423: 240. 10.1038/4240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Genzel L, Adan R, Berns A, van den Beucken JJJP, Blokland A, Boddeke EHWGM, Bogers WM, Bontrop R, Bulthuis R, Bousema T, Clevers H, Coenen TCJJ, van Dam A-M, Deen PMT, van Dijk KW, Eggen BJL, Elgersma Y, Erdogan I, Englitz B, van Vlissingen JMF, la Fleur S, Fouchier R, Fitzsimons CP, Frieling W, Haagmans B, Heesters BA, Henckens MJAG, Herfst S, Hol E, van den Hove D, de Jonge MI, Jonkers J, Joosten LAB, Kalsbeek A, Kamermans M, Kampinga HH, Kas MJ, Keijer JA, Kersten S, Kiliaan AJ, Kooij TWA, Kooijman S, Koopman WJH, Korosi A, Krugers HJ, Kuiken T, Kushner SA, Langermans JAM, Lesscher HMB, Lucassen PJ, Lutgens E, Netea MG, Noldus LPJJ, van der Meer JWM, Meye FJ, Mul JD, van Oers K, Olivier JDA, Pasterkamp RJ, Philippens IHCHM, Prickaerts J, Pollux BJA, Rensen PCN, van Rheenen J, van Rij RP, Ritsma L, Rockx BHG, Roozendaal B, van Schothorst EM, Stittelaar K, Stockhofe N, Swaab DF, de Swart RL, Vanderschuren LJMJ, de Vries TJ, de Vrij F, van Wezel R, Wierenga CJ, Wiesmann M, Willuhn I, de Zeeuw CI, Homberg JR. 2020. How the COVID-19 pandemic highlights the necessity of animal research. Curr Biol 30: R1014– R1018. 10.1016/j.cub.2020.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gonda MA, Wong-Staal F, Gallo RC, Clements JE, Narayan O, Gilden RV. 1985. Sequence homology and morphologic similarity of HTLV-III and visna virus, a pathogenic lentivirus. Science 227: 173– 177. 10.1126/science.2981428. [DOI] [PubMed] [Google Scholar]
- 30. Graham BS, Ambrosino DM. 2015. History of passive antibody administration for prevention and treatment of infectious diseases. Curr Opin HIV AIDS 10: 129– 134. 10.1097/COH.0000000000000154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Greenhalgh T, Jimenez JL, Prather KA, Tufekci Z, Fisman D, Schooley R. 2021. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet 397: 1603– 1605 10.1016/S0140-6736(21)00869-00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Halliday S. 2001. Death and miasma in Victorian London: an obstinate belief. BMJ 323: 1469– 1471. 10.1136/bmj.323.7327.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hawgood BJ. 2008. Alexandre Yersin (1863-1943): discoverer of the plague bacillus, explorer and agronomist. J Med Biogr 16: 167– 172. 10.1258/jmb.2007.007017. [DOI] [PubMed] [Google Scholar]
- 34. Henao-Restrepo AM, Preziosi MP, Wood D, Moorthy V, Kieny MP, Ebola Research WHO. Development Team. 2016. On a path to accelerate access to Ebola vaccines: The WHO’s research and development efforts during the 2014-2016 Ebola epidemic in West Africa. Curr Opin Virol 17: 138– 144. 10.1016/j.coviro.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. HIV.gov. [Internet]. 2020. Global HIV/AIDS Overview. [Cited 20 January 2021]. Available at: www.hiv.gov/federal-response/pepfar-global-aids/global-hiv-aids-overview.
- 36. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271– 280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoffmann M, Hofmann-Winkler H, Smith JC, Krüger N, Arora P, Sørensen LK, Søgaard OS, Hasselstrøm JB, Winkler M, Hempel T, Raich L, Olsson S, Danov O, Jonigk D, Yamazoe T, Yamatsuta K, Mizuno H, Ludwig S, Noé F, Kjolby M, Braun A, Sheltzer JM, Pöhlmann S. 2021. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine 65: 1– 12. 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH, Leist SR, Schäfer A, Nakajima N, Takahashi K, Lee RE, Mascenik TM, Graham R, Edwards CE, Tse LV, Okuda K, Markmann AJ, Bartelt L, de Silva A, Margolis DM, Boucher RC, Randell SH, Suzuki T, Gralinski LE, Kawaoka Y, Baric RS. 2020. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370: 1464– 1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hou X, Zaks T, Langer R, Dong Y. 2021. Lipid nanoparticles for mRNA delivery. Nat Rev Mater 1–17. 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. 2019. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol 93: e01815– e01818. 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson KM, Lange JV, Webb PA, Murphy FA. 1977. Isolation and partial characterisation of a new virus causing acute haemorrhagic fever in Zaire. Lancet 310: P569– 571. 10.1016/S0140-6736(77)92000-1. [DOI] [PubMed] [Google Scholar]
- 42. Jordan D. [Internet]. 2019. The deadliest flu: The complete story of the discovery and reconstruction of the 1918 pandemic virus. [Cited 28 December 2020]. Available at: www.cdc.gov/flu/pandemic-resources/reconstruction-1918-virus.html
- 43. Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, Basler CF, Palese P, Taubenberger JK, García-Sastre A, Swayne DE, Katze MG. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443: 578– 581. 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaufmann SH, Schaible UE. 2005. 100th anniversary of Robert Koch’s Nobel Prize for the discovery of the tubercle bacillus. Trends Microbiol 13: 469– 475. 10.1016/j.tim.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 45. Kim J, Koo BK, Knoblich JA. 2020. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol 21: 571– 584. 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim YI, Kim SG, Kim SM, Kim E-H, Park S-J, Yu K-M, Chang J-H, Kim EJ, Lee S, Casel MAB, Um J, Song M-S, Jeong HW, Lai VD, Kim Y, Chin BS, Park J-S, Chung K-H, Foo S-S, Poo H, Mo I-P, Lee O-J, Webby RJ, Jung JU, Choi YK. 2020. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 27: 704– 709.e2. 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kindhauser MK, Allen T, Frank V, Santhana RS, Dye C. 2016. Zika: the origin and spread of a mosquito-borne virus. Bull World Health Organ 94: 675– 686C. 10.2471/BLT.16.171082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kohl C, Nitsche A, Kurth A. 2021. Update on potentially zoonotic viruses of European bats. Vaccines (Basel) 9: 690. 10.3390/vaccines9070690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koopmans M. 2021. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect Dis 21: 18– 19. 10.1016/S1473-3099(20)30912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling A-E, Humphrey CD, Shieh W-J, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang J-Y, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348: 1953– 1966. 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 51. Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 11: 875– 879. 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kutter JS, de Meulder D, Bestebroer TM, Lexmond P, Mulders A, Richard M, Fouchier RAM, Herfst S. 2021. SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. Nat Commun 12: 1– 8. 10.1038/s41467-021-21918-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lambert PH, Ambrosino DM, Andersen SR, Baric RS, Black SB, Chen RT, Dekker CL, Didierlaurent AM, Graham BS, Martin SD, Molrine DC, Perlman S, Picard-Fraser PA, Pollard AJ, Qin C, Subbarao K, Cramer JP. 2020. Consensus summary report for CEPI/BC March 12-13, 2020 meeting: Assessment of risk of disease enhancement with COVID-19 vaccines. Vaccine 38: 4783– 4791. 10.1016/j.vaccine.2020.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lauring AS, Hodcroft EB. 2021. Genetic variants of SARS-CoV-2—What do they mean? JAMA 325: 529– 531. [DOI] [PubMed] [Google Scholar]
- 55. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426: 450– 454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mallapaty S. [Internet]. 2021. The coronavirus is rife in common US deer. Nature News, Nature Publishing Group. [Cited 25 September 2021]. Available at: https://www.nature.com/articles/d41586-021-02110-8. 10.1038/d41586-021-02110-8 [Mismatch] [DOI] [PubMed]
- 57. McAloose D, Laverack M, Wang L, Killian ML, Caserta LC, Yuan F, Mitchell PK, Queen K, Mauldin MR, Cronk BD, Bartlett SL, Sykes JM, Zec S, Stokol T, Ingerman K, Delaney MA, Fredrickson R, Ivančić M, Jenkins-Moore M, Mozingo K, Franzen K, Bergeson NH, Goodman L, Wang H, Fang Y, Olmstead C, McCann C, Thomas P, Goodrich E, Elvinger F, Smith DC, Tong S, Slavinski S, Calle PP, Terio K, Torchetti MK, Diel DG. 2020. From people to Panthera: natural SARS-CoV-2 infection in tigers and lions at the Bronx zoo. MBio 11: 1– 13. 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miner JJ, Diamond MS. 2017. Zika virus pathogenesis and tissue tropism. Cell Host Microbe 21: 134– 142. 10.1016/j.chom.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miranda MEG, Miranda NLJ. 2011. Reston ebolavirus in humans and animals in the Philippines: a review. J Infect Dis 204 Suppl 3: S757– S760. 10.1093/infdis/jir296. [DOI] [PubMed] [Google Scholar]
- 60. Morrison TE, Diamond MS. 2017. Animal models of Zika virus infection, pathogenesis, and immunity. J Virol 91:e00009–e00017. 10.1128/JVI.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mulangu S, Dodd LE, Davey RT, Tshiani Mbaya O, Proschan M, Mukadi D, Lusakibanza Manzo M, Nzolo D, Tshomba Oloma A, Ibanda A, Ali R, Coulibaly S, Levine AC, Grais R, Diaz J, Lane HC, Muyembe-Tamfum J-J, PALM Writing Group, Sivahera B, Camara M, Kojan R, Walker R, Dighero- Kemp B, Cao H, Mukumbayi P, Mbala- Kingebeni P, Ahuka S, Albert S, Bonnett T, Crozier I, Duvenhage M, Proffitt C, Teitelbaum M, Moench T, Aboulhab J, Barrett K, Cahill K, Cone K, Eckes R, Hensley L, Herpin B, Higgs E, Ledgerwood J, Pierson J, Smolskis M, Sow Y, Tierney J, Sivapalasingam S, Holman W, Gettinger N, Vallée D, Nordwall J, PALM Consortium Study Team. 2019. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med 381: 2293– 2303. 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Muñoz-Fontela C, Dowling WE, Funnell SGP, Gsell P-S, Riveros-Balta AX, Albrecht RA, Andersen H, Baric RS, Carroll MW, Cavaleri M, Qin C, Crozier I, Dallmeier K, deWaal L, de Wit E, Delang L, Dohm E, Duprex WP, Falzarano D, Finch CL, Frieman MB, Graham BS, Gralinski LE, Guilfoyle K, Haagmans BL, Hamilton GA, Hartman AL, Herfst S, Kaptein SJF, Klimstra WB, Knezevic I, Krause PR, Kuhn JH, Le Grand R, Lewis MG, Liu W-C, Maisonnasse P, McElroy AK, Munster V, Oreshkova N, Rasmussen AL, Rocha-Pereira J, Rockx B, Rodríguez E, Rogers TF, Salguero FJ, Schotsaert M, Stittelaar KJ, Thibaut HJ, Tseng C-J, Vergara-Alert J, Beer M, Brasel T, Chan JFW, García-Sastre A, Neyts J, Perlman S, Reed DS, Richt JA, Roy CJ, Segalés J, Vasan SS, Henao-Restrepo AM, Barouch DH. 2020. Animal models for COVID-19. Nature 586: 509– 515. 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Murphy AJ, Macdonald LE, Stevens S, Karow M, Dore AT, Pobursky K, Huang TT, Poueymirou WT, Esau L, Meola M, Mikulka W, Krueger P, Fairhurst J, Valenzuela DM, Papadopoulos N, Yancopoulos GD. 2014. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc Natl Acad Sci USA 111: 5153– 5158. 10.1073/pnas.1324022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Palmer MV, Martins M, Falkenberg S, Buckley A, Caserta LC, Mitchell PK, Cassmann ED, Rollins A, Zylich NC, Renshaw RW, Guarino C, Wagner B, Lager K, Diel DG. 2021. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. J Virol 95:e00083–e21.[Online ahead of print]. 10.1128/JVI.00083-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pardi N, Hogan M, Porter F, Weissman D. 2018. mRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov 17: 261– 279. 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Park MS, Kim JI, Bae J-Y, Park M-S. 2020. Animal models for the risk assessment of viral pandemic potential. Lab Anim Res 36: 1– 15. 10.1186/s42826-020-00040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Patterson EI, Elia G, Grassi A, Giordano A, Desario C, Medardo M, Smith SL, Anderson ER, Prince T, Patterson GT, Lorusso E, Lucente MS, Lanave G, Lauzi S, Bonfanti U, Stranieri A, Martella V, Solari Basano F, Barrs VR, Radford AD, Agrimi U, Hughes GL, Paltrinieri S, Decaro N. 2020. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat Commun 11: 1– 5. 10.1038/s41467-020-20097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Peiris JS, Guan Y, Yuen KY. 2004. Severe acute respiratory syndrome. Nat Med 10 12 Suppl: S88– S97. 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, Nicholls J, Yee WKS, Yan WW, Cheung MT, Cheng VCC, Chan KH, Tsang DNC, Yung RWH, Ng TK, Yuen KY. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361: 1319– 1325. 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, Emerson M, Garner P, Holgate ST, Howells DW, Karp NA, Lazic SE, Lidster K, MacCallum CJ, Macleod M, Pearl EJ, Petersen O, Rawle F, Peynolds P, Rooney K, Sena ES, Silberberg SD, Steckler T, Wurbel H. 2020. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol 18: 1– 12. 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Perry RD, Fetherston JD. 1997. Yersinia pestis–etiologic agent of plague. Clin Microbiol Rev 10: 35– 66. 10.1128/CMR.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Report of a WHO/International Study Team. 1978. Ebola haemorrhagic fever in Sudan, 1976. Report of a WHO/International Study Team. Bull World Health Organ 56: 247– 270. [PMC free article] [PubMed] [Google Scholar]
- 73. Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC. 2016. Characterization of a novel murine model to study zika virus. Am J Trop Med Hyg 94: 1362– 1369. 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Seferovic M, Sánchez-San Martín C, Tardif SD, Rutherford J, Castro ECC, Li T, Hodara VL, Parodi LM, Giavedoni L, Layne-Colon D, Tamhankar M, Yagi S, Martyn C, Reyes K, Suter MA, Aagaard KM, Chiu CY, Patterson JL. 2018. Experimental Zika virus infection in the pregnant common marmoset induces spontaneous fetal loss and neurodevelopmental abnormalities. Sci Rep 8:6851. 10.1038/s41598-018-25205-1. Correction. 2018. Sci Rep 8: 16131. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sejvar JJ. 2003. West nile virus: an historical overview. Ochsner J 5: 6– 10. [PMC free article] [PubMed] [Google Scholar]
- 76. Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1: 1– 22. 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shope RE. 1931. Swine influenza: III. Filtration experiments and etiology. J Exp Med 54: 373– 385. 10.1084/jem.54.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shope RE. 1936. The incidence of neutralizing antibodies for swine influenza virus in the sera of human beings of different ages. J Exp Med 63: 669– 684. 10.1084/jem.63.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Simpson S, Kaufmann MC, Glozman V, Chakrabarti A. 2020. Disease X: accelerating the development of medical countermeasures for the next pandemic. Lancet Infect Dis 20: e108– e115. 10.1016/S1473-3099(20)30123-30127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Smith W, Andrewes CH, Laidlaw PP, Timbury MC. 1995. A virus obtained from influenza patients. Rev Med Virol 5: 187– 191. 10.1002/rmv.1980050402. [DOI] [Google Scholar]
- 81. Smith W, Stuart-Harris CH. 1936. Influenza infection of man from the ferret. Lancet 228: 121– 123. 10.1016/S0140-6736(00)81750-X. [DOI] [Google Scholar]
- 82. Stuart-Harris CH, Andrewes CH, Smith W. [Internet]. 1938. Study of epidemic influenza with special reference to the 1936–37 epidemic. Medical Research Council. Eep. No. 228. [Cited 28 December 2020]. Available at: https://babel.hathitrust.org/cgi/pt?id=ien.35558005375122&view=1up&seq=3 [Google Scholar]
- 83. Suder E, Furuyama W, Feldmann H, Marzi A, de Wit E. 2018. The vesicular stomatitis virus-based Ebola virus vaccine: From concept to clinical trials. Hum Vaccin Immunother 14: 2107– 2113. 10.1080/21645515.2018.1473698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Taubenberger JK, Hultin JV, Morens DM. 2007. Discovery and characterization of the 1918 pandemic influenza virus in historical context. Antivir Ther 12: 581– 591. [PMC free article] [PubMed] [Google Scholar]
- 85. ter Meulen J, van den Brink EN, Poon LL, Marissen WE, Leung CS, Cox F, Cheung CY, Bakker AQ, Bogaards JA, van Deventer E, Preiser W, Doerr HW, Chow VT, de Kruif J, Peiris JS, Goudsmit J. 2006. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med 3: 1071– 1079. 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thanh Le T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, Saville M, Mayhew S. 2020. The COVID-19 vaccine development landscape. Nat Rev Drug Discov 19: 305– 306. 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 87. Thormar H. 2005. Maedi-visna virus and its relationship to human immunodeficiency virus. AIDS Rev 7: 233– 245. [PubMed] [Google Scholar]
- 88. Tomes N. 2010. “Destroyer and teacher”: Managing the masses during the 1918-1919 influenza pandemic. Public Health Rep 125 Suppl 3: 48– 62. 10.1177/00333549101250S308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. U.S. Food and Drug Administration. [Internet]. 2021. Animal Rule Information. 4 June 2021. FDA.[Cited 21 August 2021] Available at: www.fda.gov/emergency-preparedness-and-response/mcm-regulatory-science/animal-rule-information.
- 90. U.S. Food and Drug Administration. [Internet]. 2018. FDA Approves the First Drug with an Indication for Treatment of Smallpox. 13 July 2018. FDA. [Cited 21 August 2021]. Available at: www.fda.gov/news-events/press-announcements/fda-approves-first-drug-indication-treatment-smallpox.
- 91. U.S. Food and Drug Administration. [Internet]. 2016. FDA Approves New Treatment for Inhalation Anthrax. 21 March 2016. FDA. [Cited 21 August 2021]. Available at: https://www.fda.gov/news-events/press-announcements/fda-approves-new-treatment-inhalation-anthrax
- 92. Vogel AB, Kanevsky I, Che Y, Swanson KA, Muik A, Vormehr M, Kranz LM, Walzer KC, Hein S, Güler A, Loschko J, Maddur MS, Ota-Setlik A, Tompkins K, Cole J, Lui BG, Ziegenhals T, Plaschke A, Eisel D, Dany SC, Fesser S, Erbar S, Bates F, Schneider D, Jesionek B, Sänger B, Wallisch A-K, Feuchter Y, Junginger H, Krumm SA, Heinen AP, Adams-Quack P, Schlereth J, Schille S, Kröner C, de la Caridad Güimil Garcia R, Hiller T, Fischer L, Sellers RS, Choudhary S, Gonzalez O, Vascotto F, Gutman MR, Fontenot JA, Hall-Ursone S, Brasky K, Griffor MC, Han S, Su AAH, Lees JA, Nedoma NL, Mashalidis EH, Sahasrabudhe PV, Tan CY, Pavliakova D, Singh G, Fontes-Garfias C, Pride M, Scully IL, Ciolino T, Obregon J, Gazi M, Carrion R, Jr., Alfson KJ, Kalina WV, Kaushal D, Shi P-Y, Klamp T, Rosenbaum C, Kuhn AN, Türeci Ö, Dormitzer PR, Jansen KU, Sahin U. 2021. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592: 283– 289. 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 93. Walker L, Levine H, Jucker M. 2006. Koch’s postulates and infectious proteins. Acta Neuropathol 112: 1– 4. 10.1007/s00401-006-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wells CR, Pandey A, Parpia AS, Fitzpatrick MC, Meyers LA, Singer BH, Galvani AP. 2019. Ebola vaccination in the Democratic Republic of the Congo. Proc Natl Acad Sci USA 116: 10178– 10183. 10.1073/pnas.1817329116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. World Health Organization. [Internet]. Prioritizing diseases for research and development in emergency contexts. [Cited 15 January 2021]. Available at: https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts
- 96. World Health Organization. [Internet]. 2016. An R&D Blueprint for Action to Prevent Epidemics. World Health Organization. [Cited 15 January 2021]. Available at: https://www.who.int/blueprint/about/r_d_blueprint_plan_of_action.pdf
- 97. World Health Organization. [Cited 26 December 2020]. 2018. “Democratic Republic of Congo: Ebola Virus Disease - External Situation Report 17: Declaration of End of Ebola Virus Disease Outbreak.” ReliefWeb. Retrieved 26 July 2018. [Cited 16 January 2021]. Available at: https://reliefweb.int/report/democratic-republic-congo/democratic-republic-congo-ebola-virus-disease-external-situation-14
- 98. World Health Organization. [Internet]. 2020. WHO R&D Blueprint COVID-19 Animal Models WHO Working Group. World Health Organization. [Cited 16 January 2021]. Available at: https://www.who.int/publications/m/item/who-working-group-animal-models
- 99. World Health Organization. [Internet]. 2020. Statement of the WHO Working Group on COVID-19 Animal Models (WHO-COM) about the UK and South African SARS-CoV-2 new variants. 23 December 2020. World Health Organization. [Cited 16 January 2021]. Available at: https://www.who.int/publications/m/item/statement-of-the-who-working-group-on-covid-19-animal-models-(who-com)-about-the-uk-and-south-african-sars-cov-2-new-variants
- 100. Winau F, Winau R. 2002. Emil von Behring and serum therapy. Microbes Infect 4: 185– 188. 10.1016/S1286-4579(01)01526-X. [DOI] [PubMed] [Google Scholar]
- 101. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270– 273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion R, Jr, Nunneley JW, Barnard D, Pöhlmann S, McKerrow JH, Renslo AR, Simmons G. 2015. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res 116: 76– 84. 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]