Abstract
Fungal pathogens, from phytopathogenic fungus to human pathogens, are able to alternate between the yeast-like form and filamentous forms. This morphological transition (dimorphism) is in close connection with their pathogenic lifestyles and with their responses to changing environmental conditions. The mechanisms governing these morphogenetic conversions are still not fully understood. Therefore, we studied the filamentous growth of the less-known, non-pathogenic dimorphic fission yeast, S. japonicus, which belongs to an ancient and early evolved branch of the Ascomycota. Its RNA sequencing revealed that several hundred genes were up- or down-regulated in the hyphae compared to the yeast-phase cells. These genes belonged to different GO categories, confirming that mycelial growth is a rather complex process. The genes of transport- and metabolic processes appeared especially in high numbers among them. High expression of genes involved in glycolysis and ethanol production was found in the hyphae, while other results pointed to the regulatory role of the protein kinase A (PKA) pathway. The homologues of 49 S. japonicus filament-associated genes were found by sequence alignments also in seven distantly related dimorphic and filamentous species. The comparative genomic analyses between S. japonicus and the closely related but non-dimorphic S. pombe shed some light on the differences in their genomes. All these data can contribute to a better understanding of hyphal growth and those genomic rearrangements that underlie it.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00294-021-01206-y.
Keywords: Schizosaccharomyces japonicus, Dimorphism, Hyphae, Mycelial growth, Transcriptional profiling, Bioinformatic analysis
Introduction
Many microorganisms are able to grow over a wide range of environmental factors because they have sophisticated cell processes, which ensure their cell proliferation and survival. One of these cell processes is dimorphism, which means that cells can form unicellular yeast cells or mycelia depending on nutrient supply or other environmental factors (reviewed in Biswas et al. 2007 and Whiteway and Bachewich 2007). Switching from yeast to hypha-phase means that cells become elongated, grow in a unipolar fashion and produce chains of cells (pseudohyphae) or long true invasive hyphae (Sipiczki et al. 1998a, b; reviewed in Ernst 2000 and Sudbery et al. 2004). Several external factors, such as low glucose concentration, changes in pH or temperature, presence of serum, peptone or certain stress factors can stimulate mycelial growth, which can contribute also to the virulence of these microorganisms (Buffo et al. 1984; Dede and Okungbowa 2009, reviewed in Biswas et al. 2007 or Ernst 2000; Szabo 1999, or Ceccato-Antonini and Sudbery 2004; Alby and Bennet 2009; Lo et al. 1997; Kumamoto and Vinces 2005; Brand 2012).
Previous studies have revealed that one of the fission yeasts, the Schizosaccharomyces japonicus belongs to dimorphic microorganisms (Sipiczki et al. 1998a, b). Its mycelial growth was regulated by the nutritional gradient and depended on alteration of the cAMP level, pH and temperature (Sipiczki et al.1998a, b; Sipiczki et al. 1998a; Papp et al. 2014). The presence of certain amino acids, light or DNA damage could also influence the length of its hyphae (Papp et al. 2014; Okamoto et al. 2013; Furuya and Niki 2010). Although this species is distantly related to budding yeasts, its hyphal growth could be induced by FBS (Fetal Bovin Serum), similarly to Candida albicans or Yarrowia lipolytica (Papp et al. 2014; Mackenzie 1962; Joshi et al. 1973; Perez-Campo and Dominguez 2001; Kim et al. 2000). At the same time, remarkable differences can also be found between the mycelia of fission and budding yeasts. Candida albicans hyphae have Spitzenkörper, while the S. japonicus does not (Crampin et al. 2005; Kinnaer et al. 2019). S. japonicus mycelial growth increased at pH 4–7, while pH 4 inhibited the hyphae formation of Candida (Papp et al. 2014; Konno et al. 2006). Hyphae of the most filamentous fungi are multinuclear, while S. japonicus hyphae remain mononuclear (Sipiczki et al. 1998a; Kinnaer et al. 2019).
Because of the similarities and differences mentioned above and because the fission yeasts have several ancient features (they share important cell processes with metazoans and have remarkably conserved gene content) (Sipiczki 2000; Rhind et al. 2011; Kuramae et al. 2006), the aim of this study was to further investigate S. japonicus’s mycelial growth, to reveal its filament-associated genes, as well as its evolutionarily conserved and species-specific features.
Our study revealed the S. japonicus’s genes involved in hyphal growth. We determined the gene functions and GO categories, which showed the complexity of this process. Our data suggested strong glycolysis and ethanol production in the hyphae and revealed the regulatory role of the PKA pathway. Comparison of the S. japonicus transcriptional data with other dimorphic and filamentous species revealed the conserved genes of hyphae formation, while the comparative genomic analysis of S. japonicus and the closely related but non-dimorphic S. pombe shed some light on those genetic changes which could have contributed to the different dimorphic capacities of the two species.
Materials and methods
Strains
The S. japonicus var. japonicus wild-type yeast strain (7–1) (CCY-44-5-1, CBS354, ATCC 10660) and S. pombe (2-1210) ura4-D18 h90 were used in this study.
Media
The YEA (2% D-glucose, 1% yeast extract (Scharlau), 2% agar) and YPA (2% glucose, 1% yeast extract (Scharlau), 1% (w/v) casein tryptone (Scharlau), 2% agar) were used as a standard culture media. The Petri dishes were incubated at 30 °C.
YPG (YEA solidified with 10% gelatin instead of agar) was used for the culturing of the yeast-phase and hyphal-phase cells necessary for RNA isolation (Papp et al. 2021).
The transformant S. pombe cells were spread onto EMMA + 15 µM thiamin, while the morphology of the transformant cells was investigated on EMMA (Mitchison 1970).
Effect of isoamyl-alcohol, higher glucose concentration or presence of iron on the mycelial growth
YEA medium and YEA supplemented with 5% glucose, or 200 µM FeCl3, or 0.25% or 0.5% isoamyl-alcohol were prepared. The S. japonicus cells were streaked onto the surface of these agar plates and incubated at 30 °C. Length of the hyphae was photographed and measured after 5 days. The results were also checked after 12 days.
Total RNA isolation
RNA was extracted from yeast-phase cells and hyphae. The culture conditions and modifications of the RNA isolation protocol (Lyne et al. 2003) can be found in the previous article (Papp et al. 2021). The most important modifications were the application of glass beads to achieve stronger break of the mycelial wall and RNA was extracted from cytoplasm-filled tips of the true invasive hyphae (instead of whole mycelia) which were grown on gelatin solidified culture medium (step-by-step protocol is in Papp et al. 2021).
RNA sequencing strategy
To obtain global transcriptome data, the high throughput mRNA sequencing analysis was performed on Illumina sequencing platform. Total RNA sample quality was checked on Agilent BioAnalyzer using Eukaryotic Total RNA Nano Kit according to the manufacturer’s protocol. Samples with RNA integrity number (RIN) value 7 were accepted for the library preparation process.
Raw reads were aligned to the reference genome (Schizosaccharomyces japonicus yFS275). Tophat and Cufflinks bioinformatics tools were used for mapping and generating expression values.
The library preparations and the sequencing run were performed by UD-GenoMed Kft. and the Genomic Medicine and Bioinformatics Core Facility of Department of Biochemistry and Molecular Biology, Faculty of Medicine, University of Debrecen, Hungary. The data came from three separate experiments.
RT-PCR analysis
To validate the RNA sequencing data, RT-PCR was performed with a few intron-containing and randomly selected genes. Two genes were up-regulated (SJAG_03283, crp79 polyA binding protein and SJAG_04575, meu6 meiotic chromosome segregation protein) and one gene was down-regulated (SJAG_05398, COX assembly mitochondrial protein) (Table S1).
Total RNA was extracted from yeast-phase cells and hyphae as described previously (Papp et al. 2021). cDNA synthesis was performed with the Thermo Scientific Revert Aid First Strand cDNA Synthesis Kit (K1621).
For the RT-PCR (Bio-Rad IQ5 real-time PCR system) reaction, SsoAdvanced Universal SYBR® Green Supermix (Bio-Rad, 1725272) reagent was used with final primer concentration of 0.2 mM. The primers are listed in Table 1. Serial dilutions of cDNA (1/5, 1/25, 1/125, 1/525) were prepared to generate standard curves for each reaction. All PCR reactions were performed in triplicates. PCR conditions were as follows: 98 °C for 2 min, 40–50 cycles: 98 °C 5 s, 57 °C 20 s. Melt curve was also generated according to the company instructions. The experiments were repeated at least twice using cDNA from different biological repeats. Data were analyzed with the software (Bio-Rad iQ5 2.0) supplied with the qPCR instrument, expression levels were normalized to sce3+ transcription data and outlying data were removed during analysis.
Table 1.
Primers used in this study
| Collection number and name of the primer | Sequence (5′–3′) | Position of the primers |
|---|---|---|
| 1291 Crp79F | TTCTCTCAATACGGAAACG | exon1–exon2 border |
| 1292 Crp79R | GGCTTTGACTGTAATTTTGC | exon3–exon2 border |
| 1293 MEU6F | CGGCATTCTTCCTCATTC | exon1–exon2 border |
| 1294 MEU6R | GACTCGGTCGCTGTTTTATC | exon2 |
| 1299 SJAG_05398R | TCGAGAATCACGTAACGATAC | exon2 |
| 1300 SJAG_05398F | CTTGATGGAAGATCGGAAAG | exon1–exon2 border |
| 197 sce3F | GTCCGAGGGTGAGATTACCA | exon4 |
| 198 sce3R | GAACTCAACGTAGGCGAAGC | exon4 |
| 1310 Nrg1 Rev | GCTCGGATCCTTAGGAGGACAATAGGGATG | nrg1 gene |
| 1311 Nrg1 Forw | TGGTGGTGGTGGTTCTGGTGGTGGTGGTTCTGGTGGTGGTGGTTCTATGAGCGCCTCATTGTGTGT | nrg1 gene |
| 588 pREP F | GTCATTCGGCAATGTGCAGC | nmt1 promoter of the pREP42 vector |
Bioinformatics
Source of the protein sequences
Protein sequences were gained from the Uniprot and the Pombase databases (https://www.uniprot.org/uniprot, http://www.pombase.org).
Identification of orthologous proteins
Orthologous proteins were identified by reciprocal BLASTp analysis performed on the website of NCBI (http://blast.ncbi.nlm.nih.gov). When the reciprocal BLASTp analyses did not give a clear result, the homologous S. pombe genes were also searched using phmmer program (https://www.ebi.ac.uk/Tools/hmmer/search/phmmer) and synteny analyses (Ács-Szabó et al. 2018).
Identification of GO terms
GO numbers and categories were obtained from the S. pombe database (http://www.pombase.org) (Lock et al. 2018) using S. pombe homologous genes.
Identification of evolutionarily conserved genes of hyphal growth
Reciprocal BLASTp analyses were carried out with the S. japonicus protein sequences in https://fungi.ensembl.org. To find the Taphrina deformans homologous sequences we used the https://mycocosm.jgi.doe.gov database. RNA sequencing data of C. albicans and H. capsulatum were obtained from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?-acc=GSE19583 based on Epp et al. (2010) and Gilmore et al. (2015).
Source of the evolutionary rates
Data set of evolutionary rates of the fission yeasts protein sequences was obtained from Rhind et al. (2011).
Investigation of chromosomal localization
Chromosomal localization coordinates of the fission yeast genes were obtained from Pombase and from the fungal ftp server of the Broad Institute (ftp://ftp.broadinstitute.org/pub/annotation/fungi/schizosaccharomyces/). Synteny analyses of certain genes between S. japonicus and S. pombe were performed manually using orthology inference and the genomic coordinates of the concerning genes. Synteny information on S. octosporus and S. cryophilus was obtained from Ács-Szabó et al. (2018).
Small-scale collinearity and gene losses were depicted with the online tool SimpleSynteny (https://www.dveltri.com/simplesynteny/) (Veltri et al. 2016). Localization of the S. japonicus genes with altered expression levels was displayed by using the OrthoClusterDB online platform (http://genome.sfu.ca/cgi-bin/orthoclusterdb/runortho.cgi) (Ng et al. 2009).
Phylogenetic analysis
Phylogenetic tree was created at the website of Phylogeny.fr (http://www.phylogeny.fr/) (Dereeper et al. 2008) using certain protein sequences of high copy number genes of S. japonicus. The sequences were submitted to a manually adjusted workflow consisting of MUSCLE for alignment, GBLOCKS for the curation of the alignment and PhyML with WAG substitution model for phylogeny. The number of substitution rate category was adjusted to 4, gamma distribution parameter and proportions of invariable sites were both estimated. Branch support was estimated with an approximate likelihood-ratio test (aLRT) (Anisimova and Gascuel 2006). The tree was displayed with FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). The protein sequence of SJAG_04799 was used as outgroup.
Statistical analyses
Normal distributions of the data were tested by Shapiro–Wilk and Anderson–Darling tests. Since most of our datasets proved not to be normally distributed, Mann–Whitney U test was performed in the case of pairwise scenarios. Kruskal–Wallis test was used for multiple comparison, followed by pairwise Dunn test as post hoc test with Bonferroni corrections. P values were considered significant below the alpha level 0.05. All statistical analyses were performed in PAST v.3.20 software (https://folk.uio.no/ohammer/past/) (Hammer et al. 2001) and in Microsoft Office Excel 2016.
Cloning of the S. japonicus nrg1 gene
The nrg1 gene was amplified from the S. japonicus genomic DNA with the 1310–1311 primers (Table 1) and the following parameters: 98 °C 1 min, 98 °C 30 s, 58 °C 30 s, 72 °C 30 s (30X), 72 °C 10 min. The PCR fragment (amplified with the NEB Phusion high-fidelity DNA polymerase) was inserted into the NdeI site of the pREP42 (Maundrell 1993). This vector has an inducible nmt1+ promoter which is regulated by thiamine. Orientation of the gene was checked by PCR (primers 588–1310). Morphology and size of the transformant cells cultured on minimal medium EMMA (nmt1+ promoter induced) (30 °C after 1 days) were investigated under an Olympus BX40 microscope. To calculate the ratio of the longer cells, 300 cells were investigated in both transformant cell populations. Localisation of the Nrg1 protein was checked by Olympus BX40 fluorescent microscopy.
Transformation of the S. pombe cells
The uracil auxotrophic cells (2-1210) were transformed by the electroporator method (Gene Pulser Xcell-BioRad), according to the manufacturer’s protocol.
Results
Identification of the genes involved in S. japonicus mycelial growth
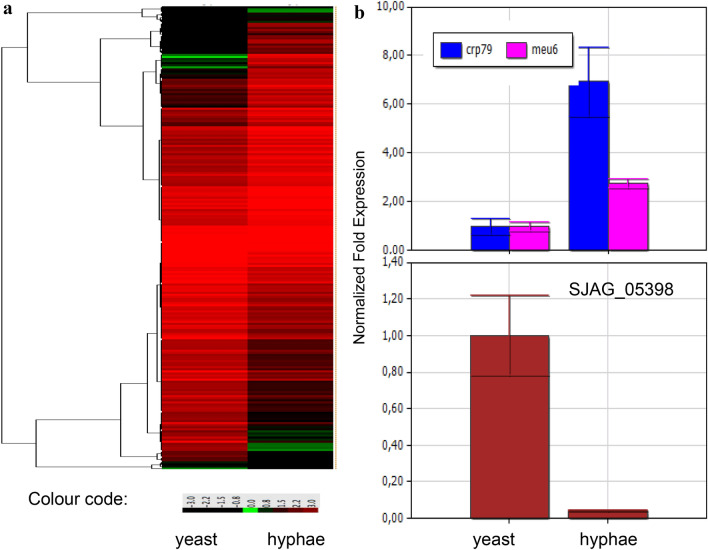
To identify the genes involved in mycelial growth of the fission yeast S. japonicus, we performed RNA sequencing analysis. It revealed that the expression of more than 2000 genes changed in a statistically significant manner. Later we selected and investigated those genes whose log2 fold changes were at least ± 1.5 or higher (Fig. 1a). 191 genes were up-regulated, while 212 genes were down-regulated, compared to the yeast-phase cells. Their functions and GO categories were determined based on the homologous proteins of the closely related S. pombe (Table S1) (Lock et al. 2018). Interestingly, the filament-associated genes belonged to very different GO categories (Table S1, Table 2). Genes of transport and metabolic processes appeared especially in high numbers among them. The genes of vesicle-mediated transport or the catabolic processes were mostly up-regulated, while the genes of mRNA-, tRNA metabolic processes or DNA replication were mostly down-regulated (Table 2). Besides, dozens of hypothetical genes (57 up-regulated, 38 down-regulated) whose function and GO category could not be determined based on their sequence homology were also found among the filament-associated genes (Table S1).
Fig. 1.
Heat map of the gene expression data and RT-PCR validation. Heat map was created from RNA sequencing data and showed differentially expressed genes in the hyphae, compared to the yeast-phase cells (a). The data were obtained from three separate experiments. Normalised expression of crp79, meu6 and SJAG_05398 genes obtained with RT-PCR analyses (b)
Table 2.
Number of the filament-associated genes in the different GO categories
| GO categories | Number of genes | ||
|---|---|---|---|
| Up-regulated | Down-regulated | Total number | |
| GO:0055085 transmembrane transport | 18 | 19 | 37 |
| GO:0016192 vesicle-mediated transport | 7 | 2 | 9 |
| GO:0006913 nucleocytoplasmic transport | 1 | 1 | 2 |
| GO:0006091 generation of precursor metabolites and energy | 6 | 7 | 13 |
| GO:0006629 lipid metabolic process | 8 | 7 | 15 |
| GO:0006520 cellular amino acid metabolic process | 5 | 9 | 14 |
| GO:0016071 mRNA metabolic process | 2 | 12 | 14 |
| GO:0006399 tRNA metabolic process | 1 | 9 | 10 |
| GO:0055086 nucleobase-containing small molecule metabolic process, cofactor | 6 | 4 | 10 |
| GO:0005975 carbohydrate metabolic process | 2 | 2 | 4 |
| GO:0006766 vitamin metabolic process | 1 | 1 | 2 |
| GO:0051186 cofactor metabolic process | 4 | 1 | 5 |
| GO:0019249 lactate biosynthetic process | 0 | 1 | 1 |
| GO:0023052 signalling | 6 | 8 | 14 |
| GO:0006355 regulation of transcription, DNA-templated | 11 | 10 | 21 |
| GO:0006351 transcription, DNA-templated | 1 | 0 | 1 |
| GO:1901990 regulation of mitotic cell cycle phase transition | 0 | 3 | 3 |
| GO:0000070 mitotic sister chromatid segregation | 1 | 1 | 2 |
| GO:0140013 meiotic nuclear division | 1 | 4 | 5 |
| GO:0007163 establishment or maintenance of cell polarity | 2 | 0 | 2 |
| GO:0030036 actin cytoskeleton organization | 3 | 1 | 4 |
| GO:0000226 microtubule cytoskeleton organization | 0 | 2 | 2 |
| GO:0071554 cell wall organization or biogenesis | 4 | 3 | 7 |
| GO:0061024 membrane organization | 5 | 3 | 8 |
| GO:0042254 ribosome biogenesis | 1 | 3 | 4 |
| GO:0032200 telomere organization | 2 | 1 | 3 |
| GO:0006325 chromatin organization | 4 | 5 | 9 |
| GO:0007005 mitochondrion organization | 2 | 5 | 7 |
| GO:0140053 mitochondrial gene expression | 3 | 6 | 9 |
| GO:0005739 mitochondrion | 1 | 2 | 3 |
| GO:0005783 endoplasmic reticulum | 0 | 1 | 1 |
| GO:0005794 Golgi apparatus | 0 | 1 | 1 |
| GO:0140056 organelle localization by membrane tethering | 0 | 1 | 1 |
| GO:0098754 detoxification | 2 | 4 | 6 |
| GO:0007155 cell adhesion | 1 | 0 | 1 |
| GO:0006914 autophagy | 1 | 1 | 2 |
| GO:0006260 DNA replication | 0 | 7 | 7 |
| GO:0006281 DNA repair | 0 | 3 | 1 |
| GO:0006310 DNA recombination | 1 | 2 | 3 |
| GO:0003677 DNA binding | 1 | 0 | 1 |
| GO:0003723 RNA binding | 0 | 1 | 1 |
| GO:0002181 cytoplasmic translation | 6 | 1 | 7 |
| GO:0006457 protein folding | 8 | 0 | 8 |
| GO:0051604 protein maturation | 0 | 4 | 4 |
| GO:0030163 protein catabolic process | 5 | 0 | 5 |
| GO:0065003 protein-containing complex assembly | 0 | 1 | 1 |
| GO:0006486 protein glycosylation | 0 | 2 | 2 |
| GO:0070647 protein modification by small protein conjugation or removal | 0 | 5 | 5 |
| GO:0055065 metal ion homeostasis | 1 | 0 | 1 |
| GO:0016491 oxidoreductase activity | 2 | 3 | 5 |
| GO:0016616 oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 0 | 1 | 1 |
| GO:0004145 diamine N-acetyltransferase activity | 0 | 1 | 1 |
| GO:0008168 methyltransferase activity | 0 | 2 | 2 |
| GO:0003959 NADPH dehydrogenase activity | 1 | 0 | 0 |
| GO:0071164 RNA trimethylguanosine synthase activity | 0 | 1 | 1 |
| GO:0036361 racemase activity, acting on amino acids and derivatives | 0 | 1 | 1 |
| GO:0008080 N-acetyltransferase activity | 0 | 2 | 2 |
To validate our RNA sequencing results, RT-PCR analyses of intron-containing and randomly selected genes were performed (crp79 and meu6 were up-regulated, while the SJAG_05398 was down-regulated) (Table S1). Their RT-PCR data (Fig. 1b) were in good agreement with the RNA sequencing results. Besides, the appearance of the famous mycelial regulators nrg1 and fkh2 (SJAG_00124, SJAG_04813) among the up-regulated genes can also indicate reliability of the RNA sequencing data (Murad et al. 2001; Bensen et al. 2002).
High expression of ethanol production-related genes in hyphae
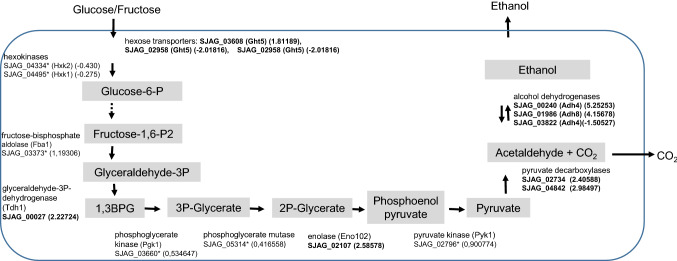
Investigating the genes and their functions, we noticed that several glycolytic genes had altered mRNA levels in the S. japonicus hyphae. One hexose transporter (SJAG_03608), the glyceraldehyde-3P-dehydrogenase (SJAG_00027) and enolase (SJAG_02107) were strongly up-regulated, while further glycolytic genes also showed significantly elevated mRNA levels compared to yeast cells. However, their RNA values were lower than log2 1.5 (indicated with * in the Fig. 2). Interestingly, pyruvate decarboxylases (SJAG_02734, SJAG_04842) and the alcohol dehydrogenases (SJAG_00240, Adh4), (SJAG_01986, Adh8) also had increased mRNA values (Table S1, Fig. 2).
Fig. 2.
Gene expression of glycolytic and ethanol production genes in S. japonicus hyphae. The log2 fold changes are indicated in brackets. The genes whose mRNA level changed significantly in the hyphae in contrast to the yeast-phase cells, but their log2 value was lower than 1.5 are indicated with *
The PKA pathway can regulate a set of the filament-associated genes
Our earlier data suggested that high cAMP level inhibited yeast-to-mycelium transition, while the pka1 deleted cells produced shorter hyphae compared to the wild-type cells (Sipiczki et al. 1998a,b; Papp et al. 2017). Thus, we assumed that at least a portion of the mycelial genes might be regulated by the Pka1 protein. Comparison of the RNA sequencing data obtained from hypha and the pka1 deleted strain revealed that there is an overlap between them. 75 filament-associated genes were found in the transcriptional profile of the pka1 mutant strain (Papp et al. 2017) (Table S2). These genes could be directly or indirectly under the regulation of the PKA pathway.
Different dimorphic yeast species share several filament-associated genes
Since mycelial growth is characteristic of different species, we wanted to learn whether there are common evolutionarily conserved filament-associated genes in these species or not. Thus, orthologous sequences of S. japonicus genes, whose mRNA level altered in the hyphae, were identified by reciprocal BLASTp analyses. As a result, orthologues of 49 filament-associated genes could be identified in seven dimorphic and filamentous species (S. cerevisiae, C. albicans, Y. lipolytica, H. capsulatum, C. neoformans, U. maydis T. deformans) (Table S3). These common genes belonged to different GO categories and were often differentially regulated in the hyphae of S. japonicus, C. albicans and H. capsulatum (Table S3, Table 3) (Epp et al. 2010; Gilmore et al. 2015).
Table 3.
mRNA levels of orthologous mycelial genes in S. japonicus, C. albicans and H. capsulatum hyphae compared to the yeast-phase cells
| Gene identifier in S. japonicus | mRNA level in hyphae | Description | Gene identifier in C. albicansa | mRNA level in hyphaea | Gene identifier in H. capsulatumb | mRNA level in hyphaeb |
|---|---|---|---|---|---|---|
| SJAG_04352 | + | Cyclophilin family peptidyl-prolyl cis–trans isomerase Wis2 | AOW31715 (orf19.7654) | + | HCBG_08524 | + |
| SJAG_05015 | + | NADPH dehydrogenase | orf19.3131 | + | HCBG_03022 | + |
| SJAG_04842 | + | Pyruvate decarboxylase | AOW29380 (orf19.2877) | + | HCBG_06694 | + |
| SJAG_00413 | + | Acetyl-CoA C-acetyltransferase Erg10 | orf19.1591 | + | HCBG_07982 | + |
| SJAG_02107 | + | Enolase | AOW26488 | + | HCBG_00056 | + |
| SJAG_02734 | + | Pyruvate decarboxylase | AOW29380 (orf19.2877) | + | HCBG_06694 | + |
| SJAG_04715 | – | Membrane transporter | orf19.1308 | – | HCBG_06390 | – |
| SJAG_03432 | – | WDR44 family WD repeat protein | orf19.7235 | – | HCBG_01676 | – |
| SJAG_01147 | – | Eukaryotic protein | orf19.6585 | – | HCBG_02667 | – |
| SJAG_00308 | – | Peptide release factor | orf19.5488 | – | HCBG_02802 | – |
| SJAG_05182 | – | Allantoate permease | orf19.5023 | – | HCBG_02854 | – |
| SJAG_00179 | + | Glutathione S-transferase Gst2 | AOW27495 (orf19.155) | + | HCBG_01438 | – |
| SJAG_02192 | + | Glucan 1,3-beta-glucosidase Bgl2 | AOW28996 (orf19.4565) | + | HCBG_03629 | – |
| SJAG_02233 | + | G-protein alpha subunit | orf19.4015 | + | HCBG_02983 | – |
| SJAG_02313 | + | D-3 phosphoglycerate dehydrogenase | orf19.5263 | + | HCBG_02183 | – |
| SJAG_00027 | + | Glyceraldehyde-3-phosphate dehydrogenase Tdh1 | orf19.6814 | + | HCBG_05811 | – |
| SJAG_03204 | + | Phospholipase | orf19.6594 | + | HCBG_09211 | – |
| SJAG_02017 | + | Translation elongation factor EF-1 gamma subunit | orf19.7382 | + | HCBG_08684 | – |
| SJAG_02896 | + | Hydroxy-methylbilane synthase | AOW28013 (orf19.1742) | – | HCBG_01754 | – |
| SJAG_02827 | + | ER oxidoreductin Ero1a | orf19.4871 | – | HCBG_01882 | – |
| SJAG_00259 | + | gar2 hipothetical protein | orf19.6090 | – | HCBG_03744 | – |
| SJAG_01768 | + | Ubiquitin-specific protease | orf19.2933 | – | HCBG_03115 | – |
| SJAG_02581 | + | Parasitic phase-specific protein PSP-1 | orf19.24 | – | HCBG_01945 | – |
| SJAG_04268 | + | HAL protein kinase Oca2 | orf19.6232 | – | HCBG_01745 | – |
| SJAG_03809 | + | Cdc20/Fizzy family WD repeat protein | orf19.2084 | – | HCBG_03481 | – |
| SJAG_01690 | + | NADP-dependent L-serine/L-allo-threonine dehydrogenase ydfG | orf19.4633 | – | HCBG_04866 | + |
| SJAG_03794 | + | DNAJ domain-containing protein Psi1 | orf19.3861 | – | HCBG_05481 | + |
| SJAG_04185 | + | ZIP zinc transporter Zrt1 | orf19.1585 | – | HCBG_07321 | + |
| SJAG_01475 | + | SAGA complex/transcription initiation factor Taf9 | orf19.1111 | – | HCBG_01443 | + |
| SJAG_01725 | + | Transcription factor Atf21 | AOW26054 | – | HCBG_06790 | + |
| SJAG_04008 | + | Cytochrome c heme lyase | orf19.4578 | – | HCBG_08300 | + |
| SJAG_03671 | + | Cystathionine beta-lyase | orf19.2092 | – | HCBG_07173 | + |
| SJAG_02199 | – | DNA replication ATPase | orf19.3019 | – | HCBG_03457 | + |
| SJAG_02615 | – | Phenylalanyl-tRNA synthetase | orf19.2039 | – | HCBG_00956 | ? |
| SJAG_01492 | – | NADP-dependent L-serine/L-allo-threonine dehydrogenase yd | orf19.4633 | – | HCBG_04866 | + |
| SJAG_00307 | – | Centromere-specific histone H3 CENP-A | orf19.6163 | – | HCBG_02786 | + |
| SJAG_00799 | – | Ribosomal protein subunit L23 | orf19.3350 | – | HCBG_01756 | + |
| SJAG_00238 | – | Glutathione S-transferase Gst1 | AOW27495 (orf19.155) | + | HCBG_01438 | – |
| SJAG_04183 | – | DNA replication endonuclease-helicase Dna2 | orf19.1192 | + | HCBG_03329 | – |
| SJAG_02139 | – | Allantoate permease//membrane transporter | orf19.5535 | + | HCBG_02854 | – |
| SJAG_02091 | – | Phospholipase B Plb1 | orf19.6594 | + | HCBG_09211 | – |
| SJAG_00645 | – | Ubiquitin-like conjugating enzyme Atg7 | orf19.707 | + | HCBG_05286 | – |
| SJAG_00674 | – | Succinate-semialdehyde dehydrogenase | orf19.4543 | + | HCBG_07054 | – |
| SJAG_04085 | – | Diphthamide biosynthesis protein | orf19.4173 | + | HCBG_08193 | + |
| SJAG_02958 | – | Hexose transporter Ght5 | orf19.2023 | + | HCBG_04231 | + |
| SJAG_03606 | – | Hexose transporter Ght6 | orf19.2023 | + | HCBG_04231 | + |
| SJAG_02883 | – | Potassium ion transporter Trk2 | orf19.600 | + | HCBG_06373 | + |
| SJAG_05329 | – | Anaphase-promoting complex subunit Apc1 | orf19.6046 | + | HCBG_07750 | + |
| SJAG_00493 | – | Fumarylacetoacetate hydrolase | orf19.2184 | + | HCBG_00180 | ? |
Effect of environmental factors on S. japonicus mycelial growth
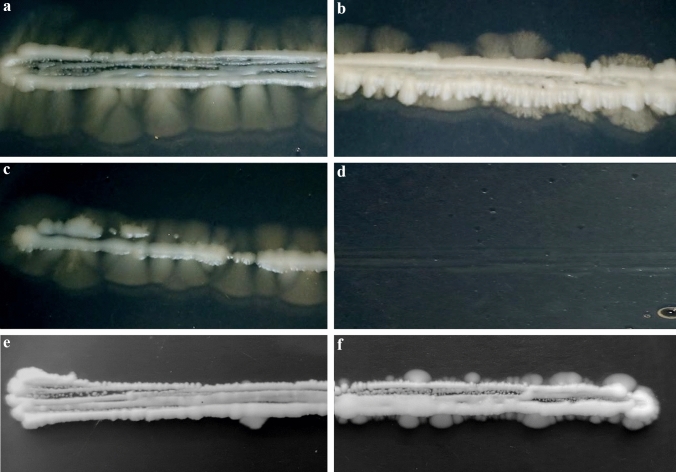
Since mRNA levels of several genes were different in the different dimorphic species (Table 3) (Epp et al. 2010; Gilmore et al. 2015), the question arose whether these differences could originate from the different responses to the environmental factors or not. Thus, mycelial growth and length of the S. japonicus hyphae were investigated on YEA culture medium (control) and YEA supplemented with isoamyl-alcohol or FeCl3 or higher concentration of glucose. Our result showed that higher glucose concentration (Fig. 3b) and presence of 0.25% isoamyl-alcohol (Fig. 3c) decreased length of the hyphae, compared to control plates (Fig. 3a) (average length of hyphae were 9 mm on YEA, 4 and 6 mm were on 5% glucose and 0.25% alcohol containing media). 0.5% alcohol inhibited even the cell division of the yeast-phase cells (Fig. 3d). Interestingly, the presence of 200 µM FeCl3 accelerated the yeast-to-hyphae transition and little hyphae appeared already after 5 days of incubation (Fig. 3f), in contrast to the YEA control plates (Fig. 3e). However, later (after 12 days) length of the hyphae was almost similar on the iron-containing and control media.
Fig. 3.
Mycelial growth of S. japonicus cells on different agar plates. a YEA (control), b YEA + 5% glucose, c YEA + 0.25% isoamyl-alcohol, (d) YEA + 0.5% isoamyl-alcohol. The agar plates were incubated at 30 °C for 12 days. e YEA and f YEA + 200 µM FeCl3 were photographed after 5 days
Genome comparison between dimorphic S. japonicus and non-dimorphic S. pombe revealed some differences
Since we failed to identify the homologues of several S. japonicus filament-associated genes in the non-dimorphic but closely related S. pombe sequence by reciprocal BLASTp analyses, we wanted to know whether S. pombe homologous genes were missing or not. Since gene content and -structure seem to be remarkably conserved in the fission yeast clade, a synteny analysis was performed between the S. pombe and S. japonicus genomes (Rhind et al. 2011; Ács-Szabó et al. 2018). This analysis confirmed the lack of several genes in the non-dimorphic S. pombe genome (Table S4), including the known regulator of hyphae production, the nrg1 (Gómez-Gil et al. 2019; Braun et al. 2001; Murad et al. 2001; Kuchin et al. 2002) (Table S4). The synteny analysis clearly showed that the chromosome fragment, which contains the nrg1 (SJAG_00124) gene and the neighbour SJAG_00121.5 ORF, are missing from the non-dimorphic S. pombe chromosome, while their 5′ and 3′ adjacent genes are present in it (Fig. 4a) (Table S5). Further analyses showed that this chromosome fragment is also missing from the other related and non-dimorphic fission yeast species, S. octosporus and S. cryophilus (Fig. 4a) (Table S5).
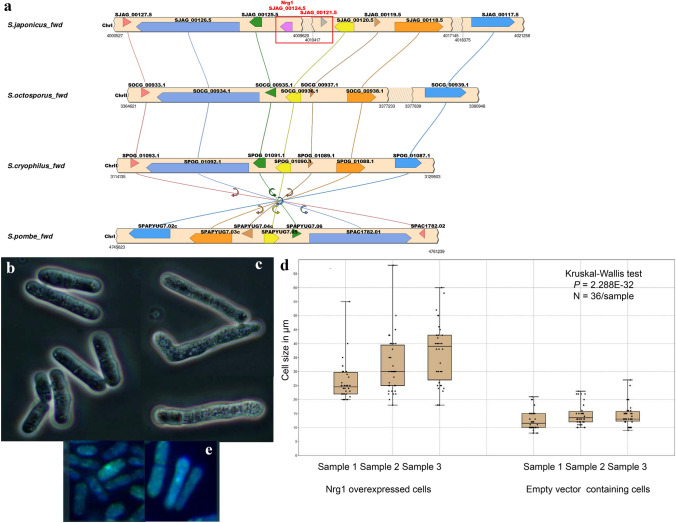
Fig. 4.
S. japonicus nrg1 gene. S. japonicus nrg1 gene and its neighbour SJAG_00121.5 ORF are missing from the chromosomes of the non-dimorphic Schizosaccharomyces species (a). Cell morphology of the S. pombe cells transformed with pREP42 empty vector (b) and pREP42 + S. japonicus nrg1 gene (c) (EMMA, at 30 °C, after 1 day). Cell size of the transformant cells (d). Localisation of the Nrg1-GFP protein (e)
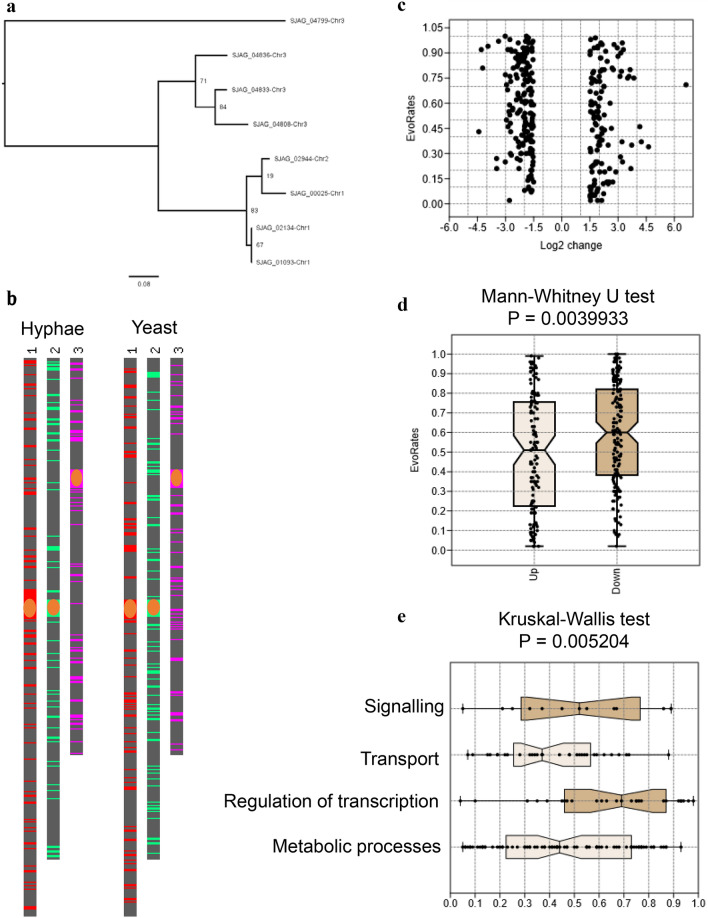
Besides the missing genes, we found also a multicopy gene (SJAG_04836) among the filament-associated genes, too, which had several paralogs in the S. japonicus genome. Interestingly, all paralogs showed elevated mRNA levels in the hyphae, compared to the yeast cells (Table 4). These paralogous genes were located on all three chromosomes (Table 4) (Fig. 5a), similarly to other mycelial genes (Fig. 5b). However, their filogenetic analysis showed that genes localised on the same chromosome had higher sequence similarity (Fig. 5a).
Table 4.
Paralogous mycelial genes in the S. japonicus genome
| Paralogs in the S. japonicus genome | Log2 (fold_change) in hyphae | Significant | S. pombe homologues | Localisation on the S. japonicus chromosomes |
|---|---|---|---|---|
| SJAG_04836 hypothetical protein | 5.92286 | Yes | No | 3 |
| SJAG_01093 hypothetical protein | 4.59153 | Yes | No | 1 |
| SJAG_04808 hypothetical protein | 3.76076 | Yes | No | 3 |
| SJAG_00025 hypothetical protein | 3.23770 | Yes | No | 1 |
| SJAG_02944 hypothetical protein | 2.17468 | Yes | No | 2 |
| SJAG_04833 hypothetical protein | 1.72023 | Yes | No | 3 |
| SJAG_02134 hypothetical protein | 1.37 | Yes | No | 1 |
| SJAG_04799 hypothetical protein | 0.66 | Yes | No | 3a |
aReverse orientation https://fungi.ensembl.org/
Fig. 5.
Bioinformatic analyses of filament-associated genes. Phylogenetic analysis of paralogous genes showed that the genes localised on the same chromosome had higher sequence similarity (a). Filament-associated genes localised equally to all three S. japonicus chromosomes (b). (Orange ellipses represent centromers) Evolutionary rates of differentially expressed genes were varied (c). Higher evolutionary rates were found in the case of the down-regulated genes compared to up-regulated mycelial genes (d). Transcriptional regulators had significantly higher evolutionary rates than those genes which were involved in transport and metabolic processes (e). (Dunn’s post hoc test with Bonferroni correction, P = 0.0051 and P = 0.01203, respectively)
Transformation of the S. japonicus nrg1 gene into the S. pombe cells
Since mRNA level of the nrg1 was high in the S. japonicus hyphae (Table S4) and at the same time the nrg1 homologous gene was missing from the S. pombe genome (Tables S4, S5), we assumed that its absence could have greatly contributed to the fact that S. pombe cells are not able to produce true hyphae. To test this assumption, the S. japonicus nrg1 gene was cloned into an S. pombe-specific vector (pREP42-GFP) and this DNA construction was transformed into a uracil auxotrophic S. pombe strain (2-1210). Morphology of the transformant cells was investigated on EMMA minimal medium, where the nmt promoter was induced (30 °C). However, the transformed cells were not able to form true hyphae, the S. pombe cells having the nrg1-containing vector showed frequently longer cell shape (Fig. 4c) compared to the control cells (Fig. 4b). Ratio of the longer cells was 47% in the cell population having the pREP42 + nrg1 vector, and 8% in the empty vector containing cells. The average size was 12–13 µm in the control cells and more than 20 µm in the nrg1 over-expressed cells (Fig. 4d). The Nrg1 protein was localised to the nucleus (Fig. 4e).
Evolutionary rate of mycelial genes
We assumed that not only the presence of certain hyphae-specific genes, but their greater flexibility could also have contributed to the greater morphological complexity of S. japonicus. To prove this assumption, evolutionary rate of the filament-associated genes was collected and analysed (Rhind et al. 2011). The analyses suggested that although a wide range of evolutionary rates is typical of mycelial genes (Fig. 5c), a little bit higher evolutionary rate was found in the case of down-regulated genes (Fig. 5d). Interestingly, further analyses also revealed that regulators, especially transcriptional regulators could have significantly higher evolutionary rate than those genes which are involved in the transport and metabolic processes (Fig. 5e).
Discussion
Dimorphic S. japonicus belongs to a divergent and early separated branch of the Ascomycete fungi, which is only distantly related to the budding yeasts (Sipiczki 2000; Kuramae et al. 2006). In addition, this clade contains three additional non-dimorphic species (S. pombe, S. octosporus, S. cryophylus), which have remarkably conserved gene- content and structure (Rhind et al. 2011; Ács-Szabó et al. 2018). Taking advantage of these opportunities, our aim was to reveal similarities and differences of mycelial growth of the different yeast species and find deviations, if there are, in the genomes of the dimorphic and non-dimorphic fission yeasts.
To obtain insight into the mycelial genes of S. japonicus, RNA sequencing was performed in wild-type hyphae and yeast-phase cells. 403 genes (which had at least ± 1.5 or higher average log2 value) were differentially expressed in hyphae compared to yeast-phase cells. These genes had varied functions and belonged to very different GO categories, similar to the Candida mycelial genes (Nantel et al. 2002; Kadosh and Johnson 2005; Carlisle and Kadosh 2013; Wu et al. 2016), suggesting that production of hyphae can be a quite complex process. Although Candida and S. japonicus are distantly related species, comparison of their transcriptional profiling data showed that the transport- and metabolic genes were involved in high number in their hyphal growth (Kadosh and Johnson 2005; Wu et al. 2016). A further similarity is that the nrg1 and fkh2 genes can be important mycelial regulators in both species (Murad et al. 2001; Bensen et al. 2002).
We also noticed that the pyruvate decarboxylases (SJAG_02734, SJAG_04842) or alcohol dehydrogenases (SJAG_00240, Adh4), (SJAG_01986, Adh8) had elevated mRNA levels in the S. japonicus hyphae. These data suggest that strong glycolysis, ethanol and CO2 production may characterise mycelial growth. This result can be in good agreement with the phenotype of the C. albicans TYE7∆ strain, which suggests a relationship between expression of glycolytic genes and biofilm formation (Bonhomme et al. 2011). The strong fermentative processes of the hyphae can be in connection with the fact that S. japonicus frequently produce true invasive hyphae, which penetrate the medium, where there are oxygen-poor conditions (Sipiczki et al. 1998a).
Comparison of our data with further distantly related dimorphic species (S. cerevisiae, C. albicans, Y. lipolytica, H. capsulatum, C. neoformans, U. maydis T. deformans) revealed that these species shared 49 common filament-associated genes. Regulation of these evolutionarily conserved genes can partly be different in S. japonicus, C. albicans and H. capsulatum mycelia (Epp et al. 2010 and Gilmore et al. 2015), because their mRNA levels ran sometimes in opposite directions. However, these differences can also originate from the different responses to environmental factors. Since the environmental and stress factors have a strong impact on hyphae production (reviewed in Biswas et al. 2007), we tested the effect of some culture factors on the S. japonicus mycelial growth. The presence of isoamyl-alcohol did not induce hyphal development in S. japonicus, unlike budding yeasts, instead it slightly decreased length of the hyphae (Ceccato-Antonini and da Silva 2002; Dickinson 1996). We also obtained different results from budding yeasts when we applied FeCl3 supplementation in the medium. Iron supplementation prevented the mycelial growth of Candida (Hameed et al. 2008), but increased the yeast-to-hyphae transition in the S. japonicus cells, which, however, produced a similar length of hyphae after longer incubation time as control cells. In contrast, the lower glucose concentration favoured mycelial growth, similarly to the budding yeasts (Cullen and Sprague 2000; Buu and Chen 2014). These findings were consistent with the previous results, which showed that S. japonicus cells responded to environmental changes partly similarly (FBS induction), partly differently (pH), compared to the Candida species (Mackenzie 1962; Joshi et al. 1973; Perez-Campo and Dominguez 2001; Kim et al. 2000; Papp et al. 2014; Konno et al. 2006).
Further comparisons confirmed the role of the PKA pathway in the morphological transition. Comparison of the transcriptional profiling data obtained from S. japonicus hyphae and pka1 deleted cells showed that there was an overlap between them (Papp et al. 2017). This is in good agreement with our previous observations that cAMP level and the pka1 mutation can influence S. japonicus hyphae production (Sipiczki et al. 1998a,b; Papp et al. 2017), and that PKA pathway is involved in the filamentous growth of several yeast species (reviewed in Biswas et al. 2007; Giacometti et al. 2011; Pan and Heitman 1999).
Our further studies focused on the differences between the genomes of S. japonicus and its non-dimorphic cousin, S. pombe. Sequence alignments and synteny analyses were carried out and they showed that one group of filament–associated genes (almost one hundred genes) was missing from the S. pombe genome. Unexpectedly, the known regulator of mycelial growth, the nrg1 (Murad et al. 2001; Braun et al. 2001) and its adjacent ORF (SJAG_00121.5) were also among the missing genes, while their 5’ and 3’ neighbour genes could be found in the S. pombe chromosome. The synteny analyses also pointed to the fact that these two genes have been eliminated from the chromosome early, because they were also missing from the genomes of their other non-dimorphic cousins, S. cryophylus, S. octosporus.
As for the role of nrg1 gene, our RNA sequencing data seems to confirm that it can have a strong role as an activator in the S. japonicus filamentous growth (its log2 value was 4.05163 in the hyphae), as it was suggested by the previous results of Gomez-Gil et al. (2019), in contrast to Candida NRG1, which has a repressor effect (Braun et al. 2001; Murad et al. 2001). However, this gene alone was not able to induce filamentous growth in non-dimorphic S. pombe cells when we cloned and transformed it into this closely related species. We assume that other missing mycelial genes could also be necessary for dimorphic capacity. This assumption is supported by the fact that we found, for example, a japonicus-specific gene (SJAG_04836) which had seven paralogs in the S. japonicus genome and all of them showed elevated mRNA levels in the hyphae. Expansion of certain genes can be in good agreement with the findings that genomes of filamentous fungi can contain more genes than non-dimorphic species (Soanes et al. 2008). However, fission yeast species have quite similar gene content (Rhind et al. 2011). Besides the chromosomal mutations, the higher evolutionary rate of the down-regulated mycelial genes and the regulator genes could also contribute to the higher morphological capacity of S. japonicus.
Taken together, this study revealed the filament-associated genes of the wild-type S. japonicus strain. We identified the common mycelial genes of different dimorphic yeast species and shed light on some similar features of the hyphae production of budding and fission yeasts. We confirmed the regulatory role of the PKA pathway in the hyphal growth and pointed to the fact that chromosomal changes could have contributed to loss of filamentous growth in non-dimorphic species and preservation of S. japonicus dimorphic capacity. We believe that all these results can provide valuable information about the dimorphic capacity of yeasts and the genomic background behind it.
Supplementary Information
Below is the link to the electronic supplementary material.
Table S1 The S. japonicus mycelial genes, their functions and GO categories (XLSX 59 KB)
Table S2 The PKA pathway regulated mycelial genes (DOCX 21 KB)
Table S3 The conserved filament-associated genes and their orthologues in the different dimorphic yeast species (XLSX 21 KB)
Table S4 List of the filament-associated genes which were missing in S. pombe genome based on Blastp and synteny analyses (XLSX 1073 KB)
Table S5 The nrg1 gene and its adjacent ORF are missing from genomes of the non-dimorphic fission yeasts (S. pombe, S. octosporus, S. cryophylus) based on Blastp and synteny analyses (XLSX 11 KB)
Acknowledgements
We thank Ilona Lakatos for technical assistance.
Author contributions
LAP: performed the molecular experiments and participated in study design. LÁS: carried out the bioinformatic analyses and participated in study design. GB: performed the RT-PCR analysis. IM: wrote the manuscript and participated in study design. All authors read and approved the manuscript.
Funding
Open access funding provided by University of Debrecen. This work was supported by the Hungarian National Research Fund (OTKA K10172), the Thematic Excellence Programme projects of the Ministry for Innovation and Technology in Hungary (FIK 20428-3/2018, TKP2020-IKA-04) and EFOP-3.6.1-16-2016-00022 project.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
University of Debrecen.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ács-Szabó L, Papp LA, Antunovics Z, Sipiczki M, Miklós I. Assembly of Schizosaccharomyces cryophilus chromosomes and their comparative genomic analyses revealed principles of genome evolution of the haploid fission yeasts. Sci Rep. 2018;8(1):14629. doi: 10.1038/s41598-018-32525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K, Bennett RJ. Stress-induced phenotypic switching in Candida albicans. Mol Biol Cell. 2009;20:3178–3191. doi: 10.1091/mbc.E09-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood ratio test for branchs: a fast, accurate and powerful alternative. Syst Biol. 2006;55(4):539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Bensen ES, Filler SG, Berman J. A Forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Am Soc Microbiol. 2002;1(5):787–798. doi: 10.1128/EC.1.5.787-798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme J, Chauvel M, Goyard S, Roux P, Rossignol T, d'Enfert C. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol Microbiol. 2011;80(4):995–1013. doi: 10.1111/j.1365-2958.2011.07626.x. [DOI] [PubMed] [Google Scholar]
- Brand A. Hyphal growth in human fungal pathogens and its role in virulence. Int J Microbiol. 2012;2012:517–529. doi: 10.1155/2012/517529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20(17):4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffo J, Herman MA, Soll DR. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia. 1984;85:21–30. doi: 10.1007/BF00436698. [DOI] [PubMed] [Google Scholar]
- Buu LM, Chen YC (2014) Impact of glucose levels on expression of hypha-associated secreted aspartyl proteinases in Candida albicans. J Biomed Sci 21:22–31 [DOI] [PMC free article] [PubMed]
- Carlisle PL, Kadosh D. A genome-wide transcriptional analysis of morphology determination in Candida albicans. Mol Biol Cell. 2013;24(3):246–260. doi: 10.1091/mbc.E12-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccato-Antonini SR, da Silva PC. Hyphal-like extension and pseudohyphal formation in industrial strains of yeasts induced by isoamyl alcohol. Braz J Microbiol. 2002;33:209–212. [Google Scholar]
- Ceccato-Antonini SR, Sudbery PE. Filamentous growth in Saccharomyces cerevisiae. Braz J Microbiol. 2004;35:173–181. [Google Scholar]
- Crampin H, Finley K, Gerami-Nejad M, Court H, Gale C, Berman J, Sudbery P. Candida albicans hyphae have a Spitzenkörper that is distinct from the polarisome found in yeast and pseudohyphae. J Cell Sci. 2005;118(13):2935–2947. doi: 10.1242/jcs.02414. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF., Jr Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci USA. 2000;97(25):13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dede A, Okungbowa FT. Effect of pH on in vitro yeast-mycelial dimorphism in genitourinary Candida spp. Biosci Res Comms. 2009;21:177–181. [Google Scholar]
- Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson JR. 'Fusel' alcohols induce hyphal-like extensions and pseudohyphal formation in yeasts. Microbiology. 1996;142(6):1391–1397. doi: 10.1099/13500872-142-6-1391. [DOI] [PubMed] [Google Scholar]
- Epp E, Walther A, Lépine G, Leon Z, Mullick A, Raymond M, Wendland J, Whiteway M. Forward genetics in Candida albicans reveals the Arp2/3 complex is required for hyphal formation, but not endocytosis. Mol Microbiol. 2010;75(5):1182–1198. doi: 10.1111/j.1365-2958.2009.07038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JF. Transcription factors in Candida albicans-environmental control of morphogenesis. Microbiology. 2000;146(8):1763–1774. doi: 10.1099/00221287-146-8-1763. [DOI] [PubMed] [Google Scholar]
- Furuya K, Niki H. The DNA damage checkpoint regulates a transition between yeast and hyphal growth in Schizosaccharomyces japonicus. Mol Cell Biol. 2010;30(12):2909–2917. doi: 10.1128/MCB.00049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti R, Kronberg F, Biondi RM, Passeron S. Candida albicans Tpk1p and Tpk2p isoforms differentially regulate pseudohyphal development, biofilm structure, cell aggregation and adhesins expression. Yeast. 2011;28:293–308. doi: 10.1002/yea.1839. [DOI] [PubMed] [Google Scholar]
- Gilmore SA, Voorhies M, Gebhart D, Sil A. Genome-wide reprogramming of transcript architecture by temperature specifies the developmental states of the human pathogen Histoplasma. PLoS Genet. 2015;11(7):e1005395. doi: 10.1371/journal.pgen.1005395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gil E, Franco A, Madrid M, Vázquez-Marín B, Gacto M, Fernández-Breis J, Vicente-Soler J, Soto T, Cansado J. Quorum sensing and stress-activated MAPK signaling repress yeast to hypha transition in the fission yeast Schizosaccharomyces japonicus. PLoS Genet. 2019;15(5):e1008192. doi: 10.1371/journal.pgen.1008192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed S, Prasad T, Banerjee D, Chandra A, Mukhopadhyay CK, Goswami SK, Lattif AA, Chandra J, Mukherjee PK, Ghannoum MA, Prasad R. Iron deprivation induces EFG1-mediated hyphal development in Candida albicans without affecting biofilm formation. FEMS Yeast Res. 2008;5:744–755. doi: 10.1111/j.1567-1364.2008.00394.x. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PDPAST. Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):9–18. [Google Scholar]
- Joshi KR, Bremner DA, Gavin JB, Herdson PB, Parr DN. The formation of germ tubes by Candida albicans in sheep serum and trypticase soya broth. Am J Clin Pathol. 1973;60:839–842. doi: 10.1093/ajcp/60.6.839. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cheon SA, Park S, Song Y, Kim JY. Serum-induced hypha formation in the dimorphic yeast Yarrowia lipolytica. FEMS Microbiol Lett. 2000;190:9–12. doi: 10.1111/j.1574-6968.2000.tb09254.x. [DOI] [PubMed] [Google Scholar]
- Kinnaer C, Dudin O, Martin SG. Yeast-to-hypha transition of Schizosaccharomyces japonicus in response to environmental stimuli. Mol Biol Cell. 2019;30:975–991. doi: 10.1091/mbc.E18-12-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno N, Ishii M, Nagai A, Watanabe T, Ogasawara A, Mikami T, Matsumoto T. Mechanism of Candida albicans transformation in response to changes of pH. Biol Pharm Bull. 2006;29(5):923–926. doi: 10.1248/bpb.29.923. [DOI] [PubMed] [Google Scholar]
- Kuchin S, Vyas VK, Marian C. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth and diploid pseudohyphal differentiation. Mol Cell Biol. 2002;22(12):3994–4000. doi: 10.1128/MCB.22.12.3994-4000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7(11):1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Kuramae EE, Robert V, Snel B, Boekhout T. Conflicting phylogenetic position of Schizosaccharomyces pombe. Genomics. 2006;88(4):387–393. doi: 10.1016/j.ygeno.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous Candida albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lock A, Rutherford K, Harris MA, Hayles J, Oliver SG, Bähler J, Wood V. PomBase 2018: user-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bähler J. Whole-genome microarrays of Fission yeast: characteristics, accuracy, reproducibility and processing of array data. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie DW. Serum tube identification of Candida albicans. J Clin Pathol. 1962;15:563–565. doi: 10.1136/jcp.15.6.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Mitchison JM. Physiological and cytological methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970;4:131–165. [Google Scholar]
- Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, d'Enfert C, Gaillardin C, Odds FC, Brown AJ. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20(17):4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, Sensen CW, Hogues H, van het Hoog M, Gordon P, Rigby T, Benoit F, Tessier DC, Thomas DY, Whiteway M. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MP, Vergara IA, Frech C, et al. OrthoClusterDB: an online platform for synteny blocks. BMC Bioinform. 2009;10:192–200. doi: 10.1186/1471-2105-10-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Furuya K, Nozaki S, Aoki K, Nikia H. Synchronous activation of cell division by light or temperature stimuli in the dimorphic yeast Schizosaccharomyces japonicus. Eukaryot Cell. 2013;12(9):1235–1243. doi: 10.1128/EC.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol and Cell Biol. 1999;19(7):4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp L, Sipiczki M, Holb IJ, Miklós I. Optimal conditions for mycelial growth of Schizosaccharomyces japonicus cells in liquid medium: it enables the molecular investigation of dimorphism. Yeast. 2014;31(12):475–482. doi: 10.1002/yea.3048. [DOI] [PubMed] [Google Scholar]
- Papp L, Sipiczki M, Miklós I. Expression pattern and phenotypic characterization of the mutant strain reveals target genes and processes regulated by pka1 in the dimorphic fission yeast Schizosaccharomyces japonicus. Curr Genet. 2017;63(3):487–497. doi: 10.1007/s00294-016-0651-x. [DOI] [PubMed] [Google Scholar]
- Papp LA, Ács-Szabó L, Sz P, Miklós I. A modified culture medium and hyphae isolation method can increase quality of the RNA extracted from mycelia of a dimorphic fungal species. Curr Genet. 2021 doi: 10.1007/s00294-021-01181-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Campo FM, Dominguez A. Factors affecting the morphogenetic switch in Yarrowia lipolytica. Curr Microbiol. 2001;43:429–433. doi: 10.1007/s002840010333. [DOI] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332(6032):930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. Where does fission yeast sit on the tree of life? Genome Biol. 2000 doi: 10.1186/gb-2000-1-2-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M, Takeo K, Grallert A. Growth polarity transitions in a dimorphic fission yeast. Microbiology. 1998;144(12):3475–3485. doi: 10.1099/00221287-144-12-3475. [DOI] [PubMed] [Google Scholar]
- Sipiczki M, Takeo K, Yamaguchi M, Yoshida S, Miklos I. Environmentally controlled dimorphic cycle in a fission yeast. Microbiology. 1998;144(5):1319–1330. doi: 10.1099/00221287-144-5-1319. [DOI] [PubMed] [Google Scholar]
- Soanes DM, Alam I, Cornell M, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Oliver SG, Talbot NJ. Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS ONE. 2008;3(6):e2300. doi: 10.1371/journal.pone.0002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Szabo R. Dimorphism in Yarrowia lipolytica: filament formation is suppressed by nitrogen starvation and inhibition of respiration. Folia Microbiol. 1999;44:19–24. doi: 10.1007/BF02816215. [DOI] [PubMed] [Google Scholar]
- Veltri D, Malapi-Wight M, Crouch JA. Simple Synteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res. 2016;44(W1):W41–W45. doi: 10.1093/nar/gkw330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Bachewich M. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li YH, Yu SB, Li WG, Liu XS, Zhao L, Lu JX. A genome-wide transcriptional analysis of yeast-hyphal transition in Candida tropicalis by RNA-Seq. PLoS ONE. 2016;11(11):e0166645. doi: 10.1371/journal.pone.0166645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 The S. japonicus mycelial genes, their functions and GO categories (XLSX 59 KB)
Table S2 The PKA pathway regulated mycelial genes (DOCX 21 KB)
Table S3 The conserved filament-associated genes and their orthologues in the different dimorphic yeast species (XLSX 21 KB)
Table S4 List of the filament-associated genes which were missing in S. pombe genome based on Blastp and synteny analyses (XLSX 1073 KB)
Table S5 The nrg1 gene and its adjacent ORF are missing from genomes of the non-dimorphic fission yeasts (S. pombe, S. octosporus, S. cryophylus) based on Blastp and synteny analyses (XLSX 11 KB)
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.
Not applicable.