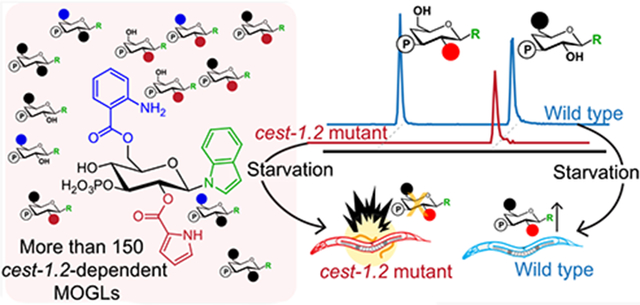
Abstract
The recently discovered modular glucosides (MOGLs) form a large metabolite library derived from combinatorial assembly of moieties from amino acid, neurotransmitter, and lipid metabolism in the model organism C. elegans. Combining CRISPR-Cas9 genome editing, comparative metabolomics, and synthesis, we show that the carboxylesterase homologue Cel-CEST-1.2 is responsible for specific 2-O-acylation of diverse glucose scaffolds with a wide variety of building blocks, resulting in more than 150 different MOGLs. We further show that this biosynthetic role is conserved for the closest homologue of Cel-CEST-1.2 in the related nematode species C. briggsae, Cbr-CEST-2. Expression of Cel-cest-1.2 and MOGL biosynthesis are strongly induced by starvation conditions in C. elegans, one of the premier model systems for mechanisms connecting nutrition and physiology. Cel-cest-1.2-deletion results in early death of adult animals under starvation conditions, providing first insights into the biological functions of MOGLs.
Graphical Abstract
INTRODUCTION
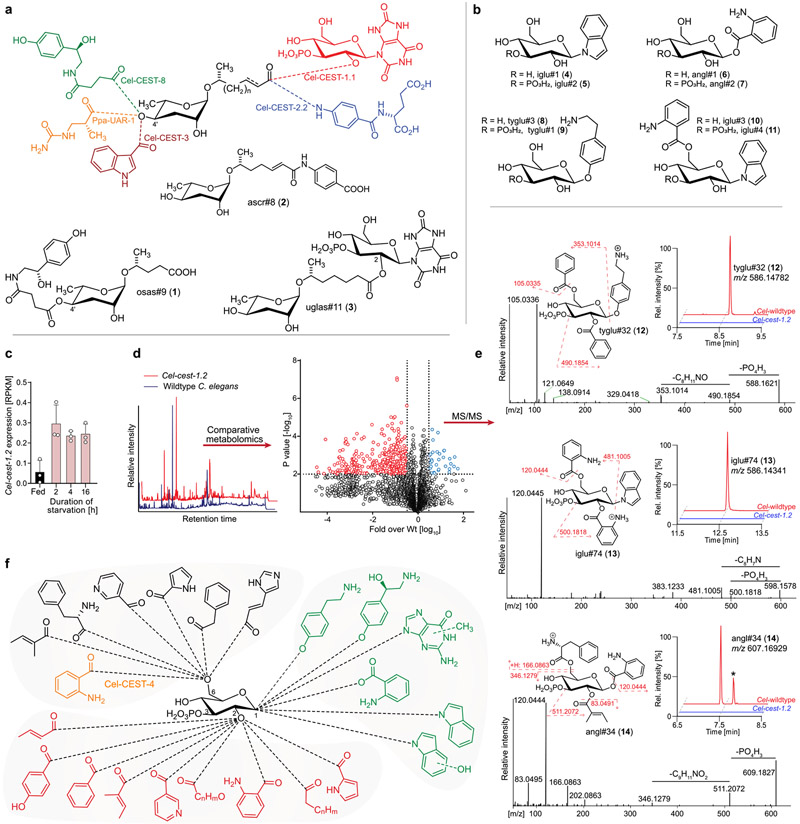
The nematode C. elegans has become an important model system for metabolomics and small molecule signaling in animals. These efforts have led to the identification of a large, structurally diverse library of signaling molecules derived from glycosides of the dideoxysugar ascarylose (Figure 1a).1-4 Ascarosides play a central role in the regulation of development and behavior in C. elegans and other nematodes and mediate interactions of nematodes with animals, plants, and microbiota.5,6 Examples include the dispersal signal osas#9 (1), in which N-succinylated octopamine is attached to the 4′-position of the ascarylose, the dauer pheromone component ascr#8 (2), incorporating a folate-derived p-aminobenzoic acid moiety, and uglas#11 (3), featuring an N3-glucosylated uric acid moiety (Figure 1a). Several recent studies demonstrated that carboxylesterase (cest) homologues are responsible for the ester and amide bonds connecting other building blocks to the ascaroside scaffold (Figure 1a).7-9 CEST enzymes belong to the α/β-hydrolase superfamily of serine hydrolases, which includes more than 200 other members in C. elegans and a similar number in mouse and humans, many of which have no characterized function.10,11
Figure 1.
Modularity of C. elegans biosynthesis pathways and comparative metabolomics of Cel-cest-1.2 mutants. (a) Assembly of modular ascarosides via CEST enzymes attaching, e.g., glucosyl uric acid (Cel-CEST-1.1), p-aminobenzoic acid (Cel-CEST-2.2), indole-3-carboxylic acid (Cel-CEST-3), succinylated octopamine (Cel-CEST-8), and ureidoisobutyric acid (Ppa-UAR-1). (b) Structures of MOGL scaffolds and example MOGLs iglu#3 (10) and iglu#4 (11). (c) Expression levels for Cel-cest-1.2 under fed and starvation conditions.15 (d) Representative ESI− total ion chromatograms (left) and volcano plot (right) of comparative analysis of the endo-metabolomes of wildtype and Cel-cest-1.2 mutants. (e) Example ESI+ MS/MS spectra, ESI− ion chromatograms, and putative structures of MOGLs from three main scaffold families, tyglu#32 (12), iglu#74 (13), and angl#34 (14). *Cel-cest-1.2-dependent isomer of angl#34 (14). (f) Schematic overview of Cel-cest-1.2 dependent metabolites. Points of attachment of the octopamine, methylguanine or hydroxyindole moieties are not known. New metabolites were named using SMIDs (see Methods and Table S4).
The biosynthesis of most cest-dependent ascarosides further depends on the activity of Cel-GLO-1, a Rab GTPase that is required for the formation of lysosome-related organelles (LROs), cellular compartments similar to mammalian melanosomes.7,12 Recent comparative metabolomic studies of Cel-glo-1 mutants and wildtype C. elegans led to the discovery of a previously undescribed class of metabolites, a large library of over one hundred modular glucosides (MOGLs).7 The MOGLs are derived from combinatorial attachment of a wide range of metabolic building blocks to several different core scaffolds, e.g., indole glucoside (iglu#1 (4), iglu#2 (5)), anthranilic acid glucoside (angl#1 (6), angl#2(7)), or tyramine glucoside (tyglu#3 (8), tyglu#1 (9), Figure 1b).7,13,14 These scaffolds are decorated with one or two additional building blocks and usually bear a phosphate at the 3 position of the glucose, although smaller amounts of nonphosphorylated derivatives are also found.7,14 The biosynthesis of most MOGLs is abolished in Cel-glo-1 mutants, indicating that, like modular ascarosides, their biosynthesis requires the LROs. In contrast to ascarosides, which are excreted into the growth media, MOGLs are primarily retained in the worm body, suggesting that they serve intraorganismal functions.7
In the MOGLs, other building blocks are linked to the core scaffolds via ester bonds, suggesting that MOGL biosynthesis may also be mediated by cest homologues. Comparative metabolomic analysis of Cel-cest-4 mutants recently showed that Cel-CEST-4 is required for 6-O-attachment of anthranilic acid in two MOGLs, iglu#3 (10) and iglu#4 (11) (Figure 1b); however, it remained unclear how enzymatic pathways could furnish a library of over a hundred MOGLs.7
RESULTS
Cel-CEST-1.2 Contributes to Biosynthesis of >150 MOGLs.
Following the initial discovery of the MOGLs,7,14,16 we noted that their production is greatly increased under starvation conditions. Surveying published transcriptomic data sets for starvation-induced cest-homologues, we noted that Cel-cest-1.2 expression is rapidly induced 4–5-fold by starvation (Figure 1c).15 Cel-cest-1.2 is a close paralog of Cel-cest-1.1, which we had recently shown to be required for attachment of the ascaroside side chain to the 2-position of the gluconucleoside moiety in uglas#11 (3) (Figure 1a). Therefore, we hypothesized that Cel-cest-1.2, may be required for the production of 2-O-acylated MOGLs. Like Cel-CEST-1.1, Cel-CEST-1.2 features a conserved C-terminal transmembrane domain and is predicted to be expressed primarily in the intestine (Figure S1 of the Supporting Information, SI).17
To investigate the biosynthetic role of Cel-cest-1.2, we obtained a mutant lacking the first 1500 bp of the coding sequence, including the serine at the putative active site (Figure S2). Using HPLC-HRMS followed by comparative analysis utilizing the Metaboseek platform, we analyzed the endo-metabolome (compounds extractable from the worm bodies) and exo-metabolome (compounds secreted into the media) of Cel-cest-1.2 mutants for compounds whose production was more than 50-fold reduced compared to C. elegans wildtype (Figure 1d, Table S4).18 These analyses revealed that Cel-cest-1.2 deletion has a dramatic impact on the C. elegans metabolome, as we detected >150 distinct metabolites whose production were strongly reduced or abolished in Cel-cest-1.2 mutants (Figure 1d, Table S4). Most of the Cel-cest-1.2-dependent compounds were detected in the endo-metabolome, whereas comparatively few differences were observed in the exo-metabolomes. MS/MS fragmentation indicated that most of the detected Cel-cest-1.2-dependent metabolites are based on the recently described MOGL scaffolds and are further modified with a wide variety of acyl moieties, primarily derived from amino acid and fatty acid metabolism (Figures 1e,f and S3, Table S4). In contrast, production of the metabolites previously shown to be Cel-cest-1.1-dependent (e.g., uglas#11, 3) or Cel-cest-4 dependent (e.g., iglu#4, 11) was not affected in the Cel-cest-1.2 mutant (Figure S4). Similarly, abundances of ascarosides were largely unchanged in Cel-cest-1.2 mutants (Figure S5). Conversely, none of the Cel-cest-1.2-dependent compounds were abolished in mutants of Cel-daf-22, which codes for a peroxisomal 3-ketoacylthiolase required for ascaroside biosynthesis (Figure S6a).19,20 Consistent with previous results for the role of LROs in MOGL biosynthesis, production of Cel-cest-1.2-dependent compounds was also strongly reduced or abolished in LRO-defective Cel-glo-1 mutants (Figure S6b).
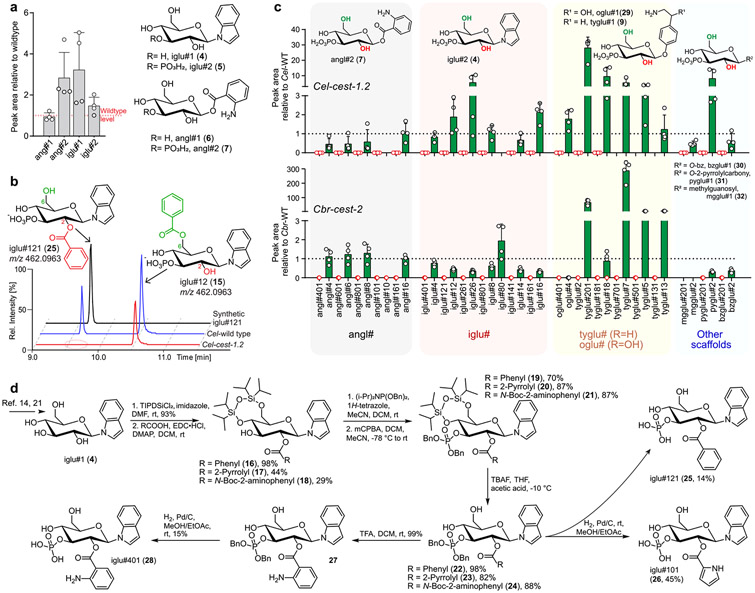
Next, we categorized the large number of Cel-cest-1.2-dependent metabolites based on their MS/MS fragmentation patterns, which enabled putative assignment to families of MOGLs based on several different scaffolds, e.g., N- or O-glucosylated indole and anthranilic acid (Figures 1e and S3, Table S4). Importantly, biosynthesis of the unmodified parent scaffolds, e.g., iglu#1 (4) or angl#2 (7), is not abolished in Cel-cest-1.2 mutants (Figure 2a). Instead, abundances of these parent scaffolds are slightly increased relative to wildtype C. elegans, suggesting that they may accumulate as shunt metabolites. Detailed analysis of the MS/MS fragmentation patterns further suggested that all Cel-cest-1.2-dependent metabolites are derived from attachment of one or two of 16 different acyl moieties to the parent scaffolds (Figure 1f, Table S4), some of which we had previously shown to be incorporated into MOGLs.7 Metabolomic analysis of wildtype C. elegans supplemented with isotope labeled l-[U–13C6]-leucine and l-[3,3-D2]-tyrosine supported the assignment of isovaleryl as well as tyramine and octopamine moieties in the identified MOGLs (Figure S7, Table S4).7,22
Figure 2.
Characterization of Cel-cest-1.2-dependent metabolites. (a) Abundances of glucoside scaffolds in Cel-cest-1.2 mutants relative to wildtype C. elegans. (b) ESI− ion chromatograms for 2-O-acylated iglu#121 (25) and its 6-O-acylated isomer, iglu#12 (15), in Cel-cest-1.2 and wildtype C. elegans, showing abolishment specifically of the 2-O-acylated isomer. (c) Abundances of 2-O- (red) vs 6-O- (green) monoacylated MOGLs in Cel-cest-1.2 and Cbr-cest-2 mutants relative to wildtype C. elegans (N2) or wildtype C. briggsae (AF16), respectively. Data and error bars represent the mean of 4 biological replicates and standard deviation. (d) Synthetic scheme of 2-O-acylated MOGLs iglu#101 (26), iglu#121 (25), and iglu#401 (28) from iglu#1 (4).14,21
CEST-1.2 is Specifically Required for 2-O-Acylation.
On the basis of the previous examples, we proposed that Cel-cest-1.2-dependent MOGLs are 3-O-phosphorylated and feature 2-O- and/or 6-O acylation (Figures 1e and S4d).7,14 Importantly, almost all monoacylated MOGLs were represented by two isomers with near-identical MS/MS fragmentation patterns but distinct HPLC retention times. Of these, only the earlier eluting isomer was abolished in Cel-cest-1.2 mutants, whereas abundance of the later eluting isomers was generally unchanged or increased (Figures 2b,c and S8).
These results suggested that Cel-CEST-1.2 may be required for site-selective acylation of the parent glucoside scaffolds. To determine whether Cel-CEST-1.2 is responsible for 2- or 6-O-acylation, we selected the 2-O-acylated variants of three monoacylated MOGLs for total synthesis via established methods (Figure 2d).7,23 To selectively synthesize 2-O-acylated MOGLs, scaffold iglu#1 (4), was 4,6-di-O-protected using 1,3-dichloro-1,1,3,3-tetraisopropyldisiloxane. Esterification with different carboxylic acids gratuitously yielded primarily the 2-O-acylated derivative, which was 3-O-phosphorylated and subsequently deprotected to furnish the target MOGLs (Figure 2d). Synthetic samples of the 2-O-acylated iglu#121 (25), iglu#101 (26), and iglu#401 (28) matched HPLC retention times and MS/MS spectra of the corresponding natural compounds (Figures 2b and S8), confirming their structures. In all cases, these Cel-cest-1.2-dependent, 2-O-acylated glucosides have earlier HPLC retention time than their putative 6-O-acylated isomers, consistent with the previously reported retention time patterns of acylated uric acid glucosides.23 Since Cel-cest-1.2 mutants are defective specifically in the production of the earlier eluting isomer of monoacylated MOGLs, these observations indicate that Cel-CEST-1.2 is specifically required for 2-O-acylation of MOGLs (Table S4).
Cbr-CEST-2 is the Functional Ortholog of Cel-CEST-1.2.
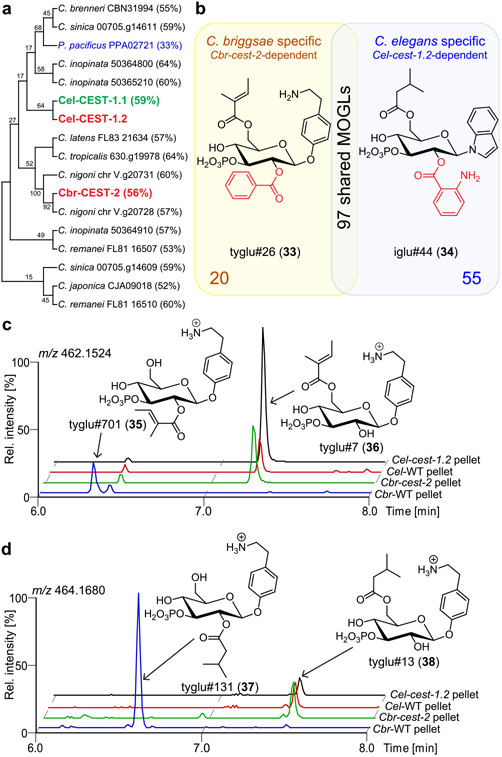
Cel-CEST-1.2 appears to be well conserved across the genus Caenorhabditis and possibly other nematode genera, e.g., Pristionchus (Figure 3a). We recently showed that MOGLs are also produced by C. briggsae, a species closely related to C. elegans, and that MOGL biosynthesis in C. briggsae also requires the LROs.7 Similar to C. elegans, the C. briggsae genome encodes a large family of carboxylesterase homologues, including Cbr-CEST-2, which has the highest sequence similarity to Cel-CEST-1.2 (Figure S9).24 Therefore, we hypothesized that the production of a subset of MOGLs, including any Cel-cest-1.2-dependent compounds also produced by C. briggsae, may require Cbr-CEST-2. Like Cel-CEST-1.2, Cbr-CEST-2 includes a C-terminal transmembrane domain and the conserved active site serine (Figures S1 and S2).
Figure 3.
(a) BLAST analysis dendrogram relating Cel-CEST-1.2 to homologous predicted proteins in other Caenorhabditis species and P. pacificus. Entries marked red were investigated in this study. Percentages represent percent identity with Cel-CEST-1.2. (b) Venn diagram showing representative MOGLs unique to either C. briggsae (yellow) or C. elegans (blue). (c, d) ESI+ ion chromatograms showing levels of C. briggsae specific, Cbr-CEST-2-dependent MOGLs, tyglu#701 (35, c) and tyglu#131 (37, d) in wildtype C. elegans, wildtype C. briggsae, Cel-cest-1.2, and Cbr-cest-2 mutants.
Using CRISPR/Cas9, we generated two Cbr-cest-2 null mutant strains and compared their endo- and exo-metabolomes with C. briggsae wildtype via HPLC-HRMS-based comparative metabolomics, as above. We found that Cbr-cest-2 mutants are defective in the production of >150 different MOGLs, including 97 MOGLs also produced by C. elegans, all of which are Cel-cest-1.2-dependent (Figure 3a,b, Table S4). These data suggest that, like Cel-CEST-1.2, Cbr-CEST-2 is specifically required for 2-O-acylation in MOGL biosynthesis (Figure 3b). We further detected several Cbr-cest-2-dependent MOGLs that are specific to C. briggsae. For example, Cbr-cest-2 mutants are defective in the biosynthesis of ascaroside-containing tyramine glucosides (e.g., tyglas#9, S7), which are not produced in C. elegans (Figure S10). Similarly, C. briggsae produce two isomers of tigloyl or isovaleroyl-modified tyglu glucosides, of which only the earlier eluting peak is Cbr-cest-2-dependent (tyglu#701 35, tyglu#131 37) (Figure 3c,d), whereas C. elegans only produce the later eluting isomer, which is Cel-cest-1.2-independent and thus likely represent the 6-O-acylated variant (Figure 3c,d). Taken together, these findings indicate that Cel-CEST-1.2 and Cbr-CEST-2 represent functional orthologs with highly similar substrate ranges and are required for 2-O-acylation of a range of scaffold glucosides.
Lifestage- and Starvation-Dependent Roles of Cel-CEST-1.2.
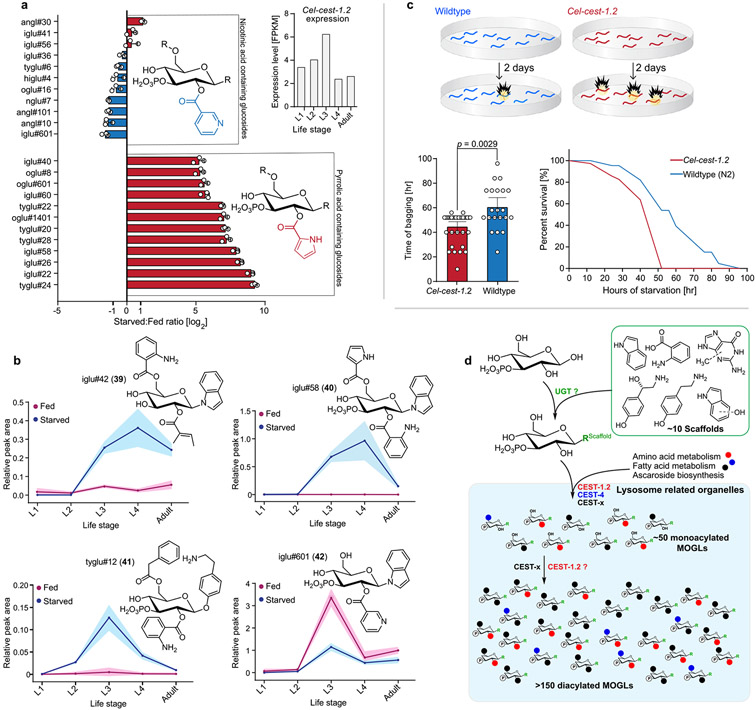
Biosynthesis of small molecules in C. elegans is often strongly dependent on developmental stage and nutritional state.25-27 Previous transcriptomic analysis showed that Cel-cest-1.2 expression peaks at the third larval stage (L3) and is induced by starvation (Figures 1c and 4a).17 To investigate the effect of developmental stage and starvation on the production of Cel-cest-1.2-dependent MOGLs, we obtained endo-metabolome samples from all four larval stages as well as gravid adults, under nutrient-replete conditions and after 24 h of starvation, followed by targeted analysis via HPLC-HRMS.
Figure 4.
Cel-cest-1.2-dependent MOGLs are induced by starvation and Cel-cest-1.2 is required for starvation survival. (a) Quantitation of nicotinic acid- and pyrrolic acid-containing MOGLs in starved relative to fed L3-stage larvae. Inset shows Cel-cest-1.2 expression levels during development. (b) Relative abundances of iglu#42 (39), iglu#58 (40), tyglu#12 (41), and iglu#601 (42) in fed and starved larvae during development. Data points represent the means, and shaded regions standard deviations. (c) Schematic for bioassay, using plates without food. Cel-cest-1.2 mutants exhibit reduced starvation survival due to bagging (a “bursting” of the worm bodies due to internal hatching of larvae). Average time of starvation survival (left) and fraction alive (right) of wildtype C. elegans and Cel-cest-1.2 mutants. (d) Model for MOGL biosynthesis. Scaffolds are glucosylated by putative glucuronosyltransferases (UGTs) and further modified in a combinatorial fashion via CEST homologues that attach diverse building blocks from amino acid and fatty acid metabolism (black, red, blue) within lysosome related organelles (LROs, shaded light blue).
Biosynthesis of most Cel-cest-1.2-dependent MOGLs was strongly induced by starvation. Pyrrolic acid-containing MOGLs were most strongly upregulated (e.g., iglu#58 (40)), whereas MOGLs incorporating nicotinic acid were not increased or even slightly downregulated (e.g., iglu#601 (42)) Figures 4a,b and S11-S13, Table S5). These trends were observed consistently across different glucose scaffolds (Figures 4a and S11-S13). In contrast, abundances of the unmodified scaffolds (e.g., iglu#2 (5)) were reduced during starvation, possibly due to lack of dietary input or because scaffold pools get depleted as a result of increased production of acylated MOGLs via Cel-CEST-1.2 and related CEST enzymes under these conditions (Figures S11-S13, Table S5). In addition, production of Cel-cest-1.2-dependent MOGLs was found to be strongly life stage-specific. Reflecting the expression pattern of Cel-cest-1.2 during development, Cel-cest-1.2-dependent metabolites were generally most abundant at the L3 larval stage; however, several compounds (e.g., iglu#42 (39) and iglu#58 (40)) showed alternate patterns with maximal production e.g. at the L4 larval stage (Figures 4b and S11-S13, Table S5). Production of most Cel-cest-1.2-dependent MOGLs was increased by starvation in most tested developmental stages, except the L1 larval stage, where starvation seemed to have little effect.
C. elegans is an important model for how starvation and dietary restriction affect lifespan in animals,28-31 and small molecules have been shown to play a major role in the underlying mechanisms.32 Because MOGL biosynthesis is strongly upregulated during starvation, we tested whether Cel-CEST-1.2 is required for starvation survival (Figure 4c). We found that lifespan of starved Cel-cest-1.2 adults was significantly reduced compared to wildtype (Figure 4c), whereas there were no significant differences in development or lifespan under food-replete conditions (Figure S14). Reduced lifespan during starvation of Cel-cest-1.2 animals was exclusively due to internal hatching of larvae, a matricide phenotype that results in bursting of the worm body, known as “bagging”.32-35
DISCUSSION
Our results demonstrate that Cel-CEST-1.2 and Cbr-CEST-2 are required for 2-O-acylation in the biosynthetic pathways of >150 different MOGLs. The product ranges in the two nematode species largely overlap, and differences may be due primarily to differences in available substrate pools. Despite the very large number of Cel-CEST-1.2/Cbr-CEST-2-dependent metabolites, their biosynthetic roles appear to be specific to 2-O-acylation, since every significant metabolic feature strongly downregulated or abolished in Cel-cest-1.2 or Cbr-cest-2 mutants, as detected in our comparative metabolomic analysis, could be assigned to a 2-O-acylated glucoside. Members of the α/β hydrolase family are known to exhibit broad substrate promiscuity,36 for example, the human Cel-CEST-1.2 homologue, carboxylesterase 2 (CES2) is capable of cleaving a diverse range of xenobiotics.37
In conjunction with the previous finding that Cel-cest-4 is specifically required for 6-O-attachment of anthranilate in indole glucosides (e.g., iglu#4 (11) in Figure 1b), our results for Cel-cest-1.2 or Cbr-cest-2 mutants allow proposing a combinatorial model for MOGL biosynthesis (Figure 4d). Following assembly of the glucoside scaffolds from indole, neurotransmitters (e.g., tyramine, octopamine) and other building blocks via UDP-glucuronosyltransferases, a wide range of acyl moieties are attached to the 2-position of glucose via Cel-cest-1.2 or the 6-position via Cel-cest-4 and additional homologues. Attachment of a second acyl moiety to produce diacylated MOGLs likely involves additional CEST-homologues. Whereas none of the abundant diacylated MOGLs are strictly cest-4-dependent,7 production of a large number of diacylated MOGLs is fully abolished in Cel-cest-1.2 mutants, suggesting that Cel-CEST-1.2 is primarily responsible for 2-O-acylation, whereas there must be additional homologues mediating 6-O-acylation, in addition to Cel-CEST-4, which compared to Cel-CEST-1.2, appears to have a much narrower substrate scope. Attempts to recapitulate the biosynthetic activities of CESTs in vitro have been unsuccessful so far, likely due to the presence of the C-terminal transmembrane domain which may cause improper folding under in vitro conditions.7,9,38
Our results further demonstrate that MOGL biosynthesis is highly regulated during development and depends on nutritional conditions. Different compound profiles at different life stages likely result in part from regulation of cest-expression, but may also reflect changes in substrate pools. For example, starvation is generally associated with increased protein turnover, which may result in an increase in amino acid degradation-derived building blocks, e.g., pyrrolic acid from proline or isovaleric and tiglic acid from leucine and isoleucine, respectively.39,40 Further, the relatives abundance of MOGLs may also depend on bacterial metabolism.22 For example. most bacteria occurring naturally with C. elegans produce much smaller amounts of indole than E. coli OP50.41 Correspondingly, we observed that C. elegans fed Providencia alcalifaciens JUb39, a bacterial species found with C. elegans in the wild, produce less indole-derived MOGLs compared to OP50-fed worms, whereas production of tyramine-derived MOGLs is increased, consistent with increased tyramine production in C. elegans fed Jub39 bacteria (Figure S15).22,42
Notably, MOGLs are mostly retained in the worm body and not excreted, suggesting that they serve specific intraorganismal function(s), paralleling the role of ascarosides in interorganismal signaling. Their highly context-specific production further supports the hypothesis that MOGLs may serve diverse biological functions. Our finding that Cel-cest-1.2 plays an important role for starvation survival and is conserved across other species provides a starting point for elucidating the role of MOGLs in C. elegans and other nematodes.
Supplementary Material
ACKNOWLEDGMENTS
We thank WormBase for sequences and life stage expression levels data, and D. Kiemle for assistance with NMR spectroscopy, Integrated DNA technologies for CRISPR reagents, and Tsui-Fen Chou for Cas9 protein.
Funding
This research was supported in part by the National Institutes of Health (R35GM131877 and U2CES030167 to F.C.S. and R24OD023041 to P.W.S.) and a National Science Foundation Graduate Research Fellowship (DGE 174301 to S.M.C.). F.C.S. is a Faculty Scholar of the Howard Hughes Medical Institute.
ABBREVIATIONS
- HPLC-HRMS
high performance liquid chromatography-high resolution mass spectrometry
- MOGL
modular glucoside
- MS/MS
tandem mass spectrometry
- LRO
lysosome related organelle
- UGT
uridine diphosphoglucuronosyltransferase
- UDP
uridine 5′-diphosphate
- CEST
carboxylesterase
- ESI−
electrospray ionization negative mode
- ESI+
electrospray ionization positive mode
- mCPBA
3-chloroperoxybenzoic acid
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c05908.
General methods, bioassay procedures, methods for metabolomics, synthetic procedures with NMR spectroscopic data, Figures S1-S15, and Tables S1-S3, NMR spectra (PDF)
Table S4 (XLSX)
Table S5 (XLSX)
Contributor Information
Chester J. J. Wrobel, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, United States
Jingfang Yu, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, United States.
Pedro R. Rodrigues, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, United States
Andreas H. Ludewig, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, United States
Brian J. Curtis, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, United States
Sarah M. Cohen, Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, California 91125, United States
Bennett W. Fox, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, United States
Michael P. O’Donnell, Department of Molecular, Cellular and Developmental Biology, New Haven, Connecticut 06511, United States
Paul W. Sternberg, Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, California 91125, United States
Frank C. Schroeder, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, United States.
REFERENCES
- (1).Schroeder FC Modular Assembly of Primary Metabolic Building Blocks: A Chemical Language in C. Elegans. Chem. Biol 2015, 22 (1), 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Butcher RA Decoding Chemical Communication in Nematodes. Nat. Prod. Rep 2017, 34 (5), 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Butcher RA; Fujita M; Schroeder FC; Clardy J Small-Molecule Pheromones That Control Dauer Development in Caenorhabditis Elegans. Nat. Chem. Biol 2007, 3 (7), 420–422. [DOI] [PubMed] [Google Scholar]
- (4).Jeong PY; Jung M; Yim YH; Kim H; Park M; Hong E; Lee W; Kim YH; Kim K; Paik YK Chemical Structure and Biological Activity of the Caenorhabditis Elegans Dauer-Inducing Pheromone. Nature 2005, 433 (7025), 541–545. [DOI] [PubMed] [Google Scholar]
- (5).Yu Y; Zhang YK; Manohar M; Artyukhin AB; Kumari A; Tenjo-Castano FJ; Nguyen H; Routray P; Choe A; Klessig DF; Schroeder FC Nematode Signaling Molecules Are Extensively Metabolized by Animals, Plants, and Microorganisms. ACS Chem. Biol 2021. 16 (6), 1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Manosalva P; Manohar M; von Reuss SH; Chen S; Koch A; Kaplan F; Choe A; Micikas RJ; Wang X; Kogel K-H; Sternberg PW; Williamson VM; Schroeder FC; Klessig DF Conserved Nematode Signalling Molecules Elicit Plant Defenses and Pathogen Resistance. Nat. Commun 2015, 6 (1), 7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Le HH; Wrobel CJJ; Cohen SM; Yu J; Park H; Helf MJ; Curtis BJ; Kruempel JC; Rodrigues PR; Hu PJ; Sternberg PW; Schroeder FC Modular Metabolite Assembly in Caenorhabditis Elegans Depends on Carboxylesterases and Formation of Lysosome-Related Organelles. eLife 2020, 9, 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Falcke JM; Bose N; Artyukhin AB; Rödelsperger C; Markov GV; Yim JJ; Grimm D; Claassen MH; Panda O; Baccile JA; Zhang YK; Le HH; Jolic D; Schroeder FC; Sommer RJ Linking Genomic and Metabolomic Natural Variation Uncovers Nematode Pheromone Biosynthesis. Cell Chem. Biol 2018, 25 (6), 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Faghih N; Bhar S; Zhou Y; Dar AR; Mai K; Bailey L; Basso KB; Butcher RA A Large Family of Enzymes Responsible for the Modular Architecture of Nematode Pheromones. J. Am. Chem. Soc 2020, 142 (32), 13645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chen AL; Lum KM; Lara-Gonzalez P; Ogasawara D; Cognetta AB; To A; Parsons WH; Simon GM; Desai A; Petrascheck M; Bar-Peled L; Cravatt BF Pharmacological Convergence Reveals a Lipid Pathway That Regulates C. Elegans Lifespan. Nat. Chem. Biol 2019, 15 (5), 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Long JZ; Cravatt BF The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease. Chem. Rev 2011, 111, 6022–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Hermann GJ; Schroeder LK; Hieb CA; Kershner AM; Rabbitts BM; Fonarev P; Grant BD; Priess JR Genetic Analysis of Lysosomal Trafficking in Caenorhabditis Elegans. Mol. Biol. Cell 2005, 16 (7), 3273–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).O’Donnell MP; Fox BW; Chao PH; Schroeder FC; Sengupta P A Neurotransmitter Produced by Gut Bacteria Modulates Host Sensory Behaviour. Nature 2020, 583 (7816), 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Stupp GS; von Reuss SH; Izrayelit Y; Ajredini R; Schroeder FC; Edison AS Chemical Detoxification of Small Molecules by Caenorhabditis Elegans. ACS Chem. Biol 2013, 8 (2), 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Hastings J; Mains A; Virk B; Rodriguez N; Murdoch S; Pearce J; Bergmann S; Le Novère N; Casanueva O Multi-Omics and Genome-Scale Modeling Reveal a Metabolic Shift During C. Elegans Aging. Front. Mol. Biosci 2019, 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Coburn C; Allman E; Mahanti P; Benedetto A; Cabreiro F; Pincus Z; Matthijssens F; Araiz C; Mandel A; Vlachos M; Edwards SA; Fischer G; Davidson A; Pryor RE; Stevens A; Slack FJ; Tavernarakis N; Braeckman BP; Schroeder FC; Nehrke K; Gems D Anthranilate Fluorescence Marks a Calcium-Propagated Necrotic Wave That Promotes Organismal Death in C. Elegans. PLoS Biol. 2013, 11 (7), e1001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Harris TW; Arnaboldi V; Cain S; Chan J; Chen WJ; Cho J; Davis P; Gao S; Grove CA; Kishore R; Lee RYN; Muller HM; Nakamura C; Nuin P; Paulini M; Raciti D; Rodgers FH; Russell M; Schindelman G; Auken KV; Wang Q; Williams G; Wright AJ; Yook K; Howe KL; Schedl T; Stein L; Sternberg PW WormBase: A Modern Model Organism Information Resource. Nucleic Acids Res. 2019, 48 (D1), D762–D767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Tautenhahn R; Patti GJ; Rinehart D; Siuzdak G XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal. Chem 2012, 84 (11), 5035–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Artyukhin AB; Zhang YK; Akagi AE; Panda O; Sternberg PW; Schroeder FC Metabolomic “Dark Matter” Dependent on Peroxisomal β-Oxidation in Caenorhabditis Elegans. J. Am. Chem. Soc 2018, 140 (8), 2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).von Reuss SH; Bose N; Srinivasan J; Yim JJ; Judkins JC; Sternberg PW; Schroeder FC Comparative Metabolomics Reveals Biogenesis of Ascarosides, a Modular Library of Small-Molecule Signals in C. Elegans. J. Am. Chem. Soc 2012, 134 (3), 1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Messaoudi S; Sancelme M; Polard-Housset V; Aboab B; Moreau P; Prudhomme M Synthesis and Biological Evaluation of Oxindoles and Benzimidazolinones Derivatives. Eur. J. Med. Chem 2004, 39 (5), 453–458. [DOI] [PubMed] [Google Scholar]
- (22).O’Donnell MP; Fox BW; Chao P-H; Schroeder FC; Sengupta P A Neurotransmitter Produced by Gut Bacteria Modulates Host Sensory Behaviour. Nature 2020, 583 (7816), 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Curtis BJ; Kim LJ; Wrobel CJJ; Eagan JM; Smith RA; Burch JE; Le HH; Artyukhin AB; Nelson HM; Schroeder FC Identification of Uric Acid Gluconucleoside-Ascaroside Conjugates in Caenorhabditis elegans by Combining Synthesis and MicroED. Org. Lett 2020, 22 (17), 6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kanzaki N; Tsai IJ; Tanaka R; Hunt VL; Liu D; Tsuyama K; Maeda Y; Namai S; Kumagai R; Tracey A; Holroyd N; Doyle SR; Woodruff GC; Murase K; Kitazume H; Chai C; Akagi A; Panda O; Ke HM; Schroeder FC; Wang J; Berriman M; Sternberg PW; Sugimoto A; Kikuchi T Biology and Genome of a Newly Discovered Sibling Species of Caenorhabditis Elegans. Nat. Commun 2018, 9 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Artyukhin AB; Yim JJ; Srinivasan J; Izrayelit Y; Bose N; von Reuss SH; Jo Y; Jordan JM; Baugh LR; Cheong M; Sternberg PW; Avery L; Schroeder FC Succinylated Octopamine Ascarosides and a New Pathway of Biogenic Amine Metabolism in Caenorhabditis Elegans. J. Biol. Chem 2013, 288 (26), 18778–18783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ludewig AH; Artyukhin AB; Aprison EZ; Rodrigues PR; Pulido DC; Burkhardt RN; Panda O; Zhang YK; Gudibanda P; Ruvinsky I; Schroeder FC An Excreted Small Molecule Promotes C. Elegans Reproductive Development and Aging. Nat. Chem. Biol 2019, 15 (8), 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hoki JS; Le HH; Mellott KE; Zhang YK; Fox BW; Rodrigues PR; Yu Y; Helf MJ; Baccile JA; Schroeder FC Deep Interrogation of Metabolism Using a Pathway-Targeted Click-Chemistry Approach. J. Am. Chem. Soc 2020, 142 (43), 18449–18459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lee GD; Wilson MA; Zhu M; Wolkow CA; De Cabo R; Ingram DK; Zou S Dietary Deprivation Extends Lifespan in Caenorhabditis Elegans. Aging Cell 2006, 5 (6), 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lakowski B; Hekimi S The Genetics of Caloric Restriction in Caenorhabditis Elegans. Proc. Natl. Acad. Sci. U. S. A 1998, 95 (22), 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Houthoofd K; Braeckman BP; Lenaerts I; Brys K; De Vreese A; Van Eygen S; Vanfleteren JR Axenic Growth Up-Regulates Mass-Specific Metabolic Rate, Stress Resistance, and Extends Life Span in Caenorhabditis Elegans. Exp. Gerontol 2002, 37 (12), 1371–1378. [DOI] [PubMed] [Google Scholar]
- (31).Houthoofd K; Braeckman BP; Johnson TE; Vanfleteren JR Life Extension via Dietary Restriction Is Independent of the Ins/IGF-1 Signalling Pathway in Caenorhabditis Elegans. Exp. Gerontol 2003, 38 (9), 947–954. [DOI] [PubMed] [Google Scholar]
- (32).Baugh LR; Hu PJ Starvation Responses throughout the Caenorhabditis elegans Life Cycle. Genetics 2020, 216 (4), 837–878, DOI: 10.1534/genetics.120.303565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chen J; Caswell-Chen EP Facultative Vivipary Is a Life-History Trait in Caenorhabditis Elegans. J. Nematol 2004, 36 (2), 107–113. [PMC free article] [PubMed] [Google Scholar]
- (34).Seidel HS; Kimble J The Oogenic Germline Starvation Response in c. Elegans. PLoS One 2011, 6 (12), e28074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Angelo G; Van Gilst MR Starvation Protects Germline Stem Cells and Extends Reproductive Longevity in C. Elegans. Science (Washington, DC, U. S.) 2009, 326 (5955), 954–958. [DOI] [PubMed] [Google Scholar]
- (36).Martínez-Martínez M; Coscolín C; Santiago G; Chow J; Stogios PJ; Bargiela R; Gertler C; Navarro-Fernández J; Bollinger A; Thies S; Méndez-García C; Popovic A; Brown G; Chernikova TN; García-Moyano A; Bjerga GEK; Pérez-García P; Hai T; Del Pozo MV; Stokke R; Steen IH; Cui H; Xu X; Nocek BP; Alcaide M; Distaso M; Mesa V; Peláez AI; Sánchez J; Buchholz PCF; Pleiss J; Fernández-Guerra A; Glöckner FO; Golyshina OV; Yakimov MM; Savchenko A; Jaeger KE; Yakunin AF; Streit WR; Golyshin PN; Guallar V; Ferrer M Determinants and Prediction of Esterase Substrate Promiscuity Patterns. ACS Chem. Biol 2018, 13 (1), 225–234. [DOI] [PubMed] [Google Scholar]
- (37).Imai T; Taketani M; Shii M; Hosokawa M; Chiba K Substrate Specificity of Carboxylesterase Isozymes and Their Contribution to Hydrolase Activity in Human Liver and Small Intestine. Drug Metab. Dispos 2006, 34 (10), 1734–1741. [DOI] [PubMed] [Google Scholar]
- (38).Falcke JM; Bose N; Artyukhin AB; Rödelsperger C; Markov GV; Yim JJ; Grimm D; Claassen MH; Panda O; Baccile JA; Zhang YK; Le HH; Jolic D; Schroeder FC; Sommer RJ Linking Genomic and Metabolomic Natural Variation Uncovers Nematode Pheromone Biosynthesis. Cell Chem. Biol 2018, 25 (6), 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Jones CB; Ott EM; Keener JM; Curtiss M; Sandrin V; Babst M Regulation of Membrane Protein Degradation by Starvation-Response Pathways. Traffic 2012, 13 (3), 468–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gretzmeier C; Eiselein S; Johnson GR; Engelke R; Nowag H; Zarei M; Küttner V; Becker AC; Rigbolt KTG; Høyer-Hansen M; Andersen JS; Münz C; Murphy RF; Dengjel J Degradation of Protein Translation Machinery by Amino Acid Starvation-Induced Macroautophagy. Autophagy 2017, 13 (6), 1064–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lee JH; Lee J Indole as an Intercellular Signal in Microbial Communities. FEMS Microbiology Reviews 2010, 34, 426. [DOI] [PubMed] [Google Scholar]
- (42).Samuel BS; Rowedder H; Braendle C; Félix M-A; Ruvkun G Caenorhabditis Elegans Responses to Bacteria from Its Natural Habitats. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (27), E3941–E3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.