Abstract
The expression of bromodomain-containing proteins that regulate chromatin structure and accessibility must be tightly controlled to ensure the appropriate regulation of gene expression. In the yeast S. cerevisiae, Bromodomain Factor 2 (BDF2) expression is extensively regulated post-transcriptionally during stress by RNase III-mediated decay (RMD), which is triggered by cleavage of the BDF2 mRNA in the nucleus by the RNase III homolog Rnt1p. Previous studies have shown that RMD-mediated down-regulation of BDF2 is hyperactivated in osmotic stress conditions, yet the mechanisms driving the enhanced nuclear cleavage of BDF2 RNA under these conditions remain unknown. Here, we show that RMD hyperactivation can be detected in multiple stress conditions that inhibit mRNA export, and that Rnt1p remains primarily localized in the nucleus during salt stress. We show that globally inhibiting mRNA nuclear export by anchoring away mRNA biogenesis or export factors out of the nucleus can recapitulate RMD hyperactivation in the absence of stress. RMD hyperactivation requires Rnt1p nuclear localization but does not depend on the BDF2 gene endogenous promoter, and its efficiency is affected by the structure of the stem–loop cleaved by Rnt1p. Because multiple stress conditions have been shown to mediate global inhibition of mRNA export, our results suggest that the hyperactivation of RMD is primarily the result of the increased nuclear retention of the BDF2 mRNA during stress.
Keywords: Rnt1p, RNase III, Bromodomain, mRNA export, stress
INTRODUCTION
The covalent modification of histones is a major epigenetic mechanism that regulates the accessibility of DNA for damage repair, transcriptional activation or repression and heterochromatin formation (Lawrence et al. 2016). Histones acetylation is a well-studied covalent modification that is generally associated with transcriptional activation (Kurdistani and Grunstein 2003). Bromodomain-containing proteins bind to acetylated histones and recruit various proteins to alter gene expression (Josling et al. 2012). As such, they are key players in transcriptional regulation, and bromodomains have recently emerged as therapeutic targets in oncogenesis and various pathological conditions (Morgado-Pascual et al. 2019; Gilan et al. 2020). Bromodomain factor 2 (Bdf2p) is one of the two bromodomain-containing proteins found in the yeast S. cerevisiae that recognizes acetylated lysines on histones. Bdf2p was found to establish heterochromatin boundaries and regulates the yeast salt stress response, although the specific mechanisms that govern these processes remain elusive (Fu et al. 2013). Bdf2p is not essential for growth in S. cerevisiae. However, strains lacking both Bdf2p and Bdf1p are inviable (Matangkasombut et al. 2000), suggesting a partial redundancy between the functions of the two proteins. Indeed, BDF2 overexpression can rescue the salt sensitivity and mitochondrial dysfunction of bdf1Δ mutants (Fu et al. 2013). This functional redundancy is further confirmed by the observation that Bdf2p can occupy sites normally bound by Bdf1p in mutants lacking Bdf1p (Durant and Pugh 2007). The absence of Bdf1p also increases the basal expression of BDF2 (Fu et al. 2013; Volanakis et al. 2013). Interestingly, Bdf2p has been found to interact with the general Pol.II transcription factor TFIID, implicating a possible broader role in regulating transcription (Matangkasombut et al. 2000; Fu et al. 2013). These observations show that a precise regulation of BDF2 is necessary to balance its expression relative to that of BDF1 and during stress.
BDF2 expression has been shown to be extensively regulated post-transcriptionally in the nucleus through two distinct pathways: spliceosome-mediated decay (SMD) (Volanakis et al. 2013) and RNase III-mediated decay (RMD) (Roy and Chanfreau 2014). During SMD, the BDF2 mRNA undergoes the first step of splicing at a 5′ splice site sequence (5′SS, Fig. 1A), but the subsequent intermediates are released and degraded, instead of proceeding through the second splicing step (Volanakis et al. 2013). RMD is initiated by cleavage of the BDF2 mRNA by Rnt1p, the sole representative of the RNase III family of double-stranded RNA endonucleases in S.cerevisiae (Elela et al. 1996; Roy and Chanfreau 2012). After cleavage in a stem–loop structure of BDF2 by Rnt1p (the Rnt1 cleavage site or RCS, Fig. 1A), the cleavage fragments are subsequently degraded by nuclear exoribonucleases (Roy and Chanfreau 2014). The major upstream cleavage product (CFm, Fig. 1A) can be detected upon partial inactivation of the nuclear exosome through a deletion of its nuclear component Rrp6p (Roy and Chanfreau 2014). RMD and SMD are not completely independent, as the 5′-exon released after SMD can undergo RMD (Roy and Chanfreau 2014), resulting in a shorter cleavage fragment shown as CFs on Figure 1A. Interestingly, the two major mechanisms of BDF2 nuclear decay are activated by different environmental stresses. During osmotic stress, RMD predominates over SMD while the opposite is true during DNA replication stress (Roy and Chanfreau 2014). The increase in the activity, or hyperactivation of RMD during salt stress results in a drastic decrease of the available pool of BDF2 transcripts and is also responsible for the extreme salt sensitivity of bdf1Δ mutants (Roy and Chanfreau 2014). However, the mechanism by which RMD is hyperactivated during stress remains unclear. In this study, we provide evidence that the increased cleavage of the BDF2 transcripts during specific stress conditions is primarily due to increased retention of the BDF2 mRNAs within the nucleus. These results show that RMD can act as an additional layer in regulating gene expression, where transcripts are retained within the nucleus and subsequently degraded by RMD.
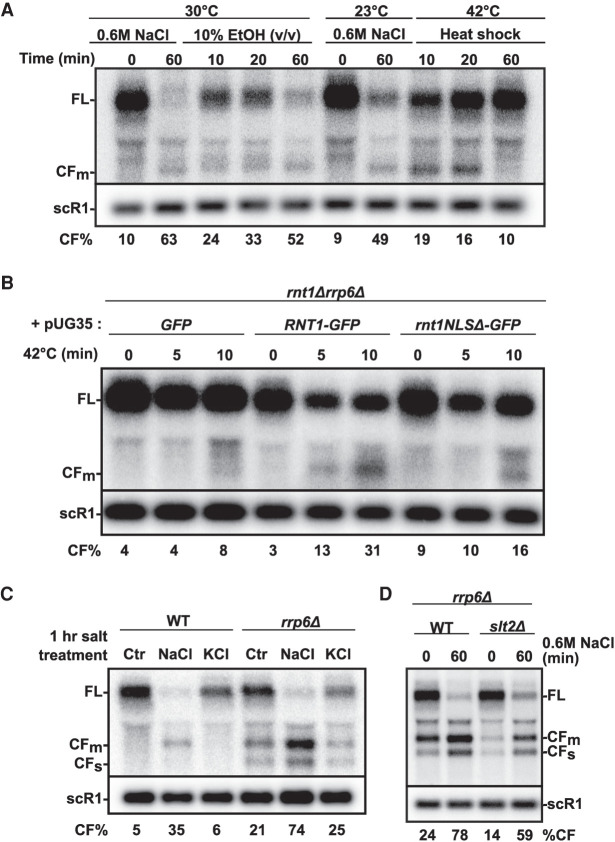
FIGURE 1.
Rnt1p protein levels and localization remain unchanged during after salt stress. (A) Schematic representation of the cleavage of the BDF2 mRNA by Rnt1p and by the spliceosome. Shown are the two 5′-cleavage products generated by Rnt1p-mediated decay (RMD; CFm) or by spliceosome mediated decay and RMD (CFs). These cleavage products are typically degraded by nuclear exonucleases but can be detected by inactivation of Rrp6p. The 3′ fragments are not represented. (B) Western blot analysis of Rnt1p harboring an HTP carboxy-terminal tag (Granneman et al. 2009). Cells were treated with or without 0.6 M NaCl for the indicated times. Tdh1p was used as a loading control. An untagged wild-type strain was used as a negative control. Numbers below the blot show the average of three independent biological replicates. (C) Northern blot analysis of BDF2 in the Rnt1-HTP tagged strain. Shown is a northern blot of BDF2 using a probe spanning the BDF2 initiation codon until the Rnt1p cleavage site (RCS; Fig. 1A) of RNAs extracted from a Rnt1-HTP tagged strain grown in normal medium or after a shift to NaCl (0.6 M, 1 h). The ncRNA scR1 was used as loading control. Numbers indicate the % of the BDF2 cleavage fragment CFm relative to all BDF2 transcripts. Numbers are the average of two independent experiments. (D) Localization of a Rnt1p-GFP tagged version before and after a 20 min shift to 1 M NaCl. Shown is the GFP signal from a strain expressing Rnt1–GFP, or GFP alone (bottom panel) in the indicated salt conditions, overlapped with the blue signal (DNA; Hoechst) or the red signal from the Cy3-labeled oligodT probe. (E) Northern blot analysis of BDF2 in a strain in which Rnt1p is exported to the nucleus using the anchor away technique. Shown is a northern blot of BDF2 showing the full length (FL) and cleavage fragment (CF) in a strain expressing Rnt1p tagged with a FKBP12-rapamycin binding domain (FRB) (Haruki et al. 2008). Strains were first treated with Rapamycin or control medium for 1 h, and then shifted to 0.6 M NaCl containing medium for 1 h or maintained in a similar medium for the control samples. Numbers indicate the levels of the full-length BDF2 relative to the scR1 control (average of three independent replicates).
RESULTS
Rnt1p protein levels remain stable in salt stress
Previous studies have shown that the BDF2 mRNA can be regulated by both spliceosome mediated decay (SMD) (Volanakis et al. 2013) as well as RNase III-mediated decay (RMD) (Roy and Chanfreau 2014). In the presence of high concentrations of NaCl (0.6–1 M), RMD is hyperactivated and promotes degradation of the BDF2 transcript (Roy and Chanfreau 2014). After treatment of cells with 0.6 to 1 M NaCl, the 5′-cleavage fragment generated by RMD becomes readily detectable by northern blot analysis, while the full-length BDF2 mRNA progressively disappears (Roy and Chanfreau 2014). To further investigate the mechanism responsible for RMD hyperactivation on the BDF2 transcript after exposure to high concentrations of NaCl, we first analyzed Rnt1p protein levels by western blot, as increased Rnt1p expression might be responsible for increased cleavage of BDF2. Western blot analysis of an HTP (His6-TEV-Protein A; Granneman et al. 2009)-tagged version of Rnt1p showed no significant changes in Rnt1p protein levels (paired t-tests P-values >0.1; quantifications, averages, standard deviations and statistical tests applied for all the quantitative data included in the article are shown in Supplemental Table S1) or electrophoretic mobility between samples grown in standard medium versus those treated with high salt (Fig. 1B). We verified that this HTP-tagged version of Rnt1p was functional for promoting BDF2 RMD (Fig. 1C). This suggests that the hyperactivation of RMD of the BDF2 transcript is not a consequence of an overall increase in Rnt1p protein levels.
Rnt1p remains localized in the nucleus during salt stress and its nuclear localization is necessary for BDF2 RMD
Rnt1p is primarily localized in the nucleoplasm and nucleolus in normal growth conditions (Catala et al. 2004; Henras et al. 2004). However, it is unknown if the subcellular localization of Rnt1p changes during stress or under different environmental conditions. We hypothesized that if the bulk of the BDF2 mRNA is cytoplasmic, an increase in Rnt1p cytoplasmic localization during osmotic stress may result in an increase in BDF2 RMD. In order to visualize the localization of Rnt1p within the cell, we utilized a strain expressing a GFP-tagged version of Rnt1p, which was previously shown to be functional (Henras et al. 2004) and which can promote BDF2 mRNA RMD during stress (see below). We did not observe a difference in the subcellular localization of Rnt1p in strains grown in normal conditions or treated with 1 M NaCl (Fig. 1D), as the major site of GFP accumulation was still detected in the nucleus, as previously shown (Henras et al. 2004). This localization was specific as it was not detected using a strain expressing a GFP control (Fig. 1C). The primary nuclear localization of Rnt1p during salt-induced stress suggests that Rnt1p-mediated cleavage of BDF2 transcripts during stress is likely to occur within the nucleus. Previous studies have shown that environmental stresses can cause global poly(A)+ mRNA retention within the nucleus (Piper 1995; Saavedra et al. 1996; Izawa et al. 2008). Thus, it is conceivable that salt stress may cause similar global nuclear retention of poly(A)+ mRNAs. In this scenario, nuclear retained transcripts would be more likely to undergo RMD. Using Cy3-labeled oligo d(T)50 to visualize polyadenylated mRNAs by fluorescence in situ hybridization (FISH), a distinct pattern of nuclear poly(A)+ mRNA aggregation was detected after exposure to 1 M NaCl salt stress, contrasting to the diffuse cytoplasmic pattern detected before stress (Fig. 1D). We tried to specifically detect the BDF2 mRNAs by FISH in these conditions, but we were unable to obtain consistent data using oligonucleotide probes complementary to the BDF2 sequence (not shown). Taken together, our data show that high salt stress does not induce an overall change in the nuclear localization of Rnt1p but results in an overall change in the localization of poly(A)+ mRNAs that may contribute to promoting RMD in the nucleus.
To further show that the nuclear localization of Rnt1p is necessary for BDF2 RMD, we depleted Rnt1p from the nucleus using the anchor away technique (Haruki et al. 2008) prior to salt stress exposure. We used a strain in which Rnt1p is tagged with a FKBP12-rapamycin binding domain (abbreviated as FRB), which can be used to promote the rapid export of nuclear proteins to the cytoplasm upon addition of rapamycin (Haruki et al. 2008). Anchoring away Rnt1p to the cytoplasm prior to 1 h of NaCl-induced stress was sufficient to prevent BDF2 RMD cleavage (Fig. 1E; lane 4 vs. lane 3 paired t-test P-value = 0.017), further supporting the hypothesis that RMD predominantly occurs within the nucleus. Altogether, these data indicate that Rnt1p localization remains unaltered in salt stress and that RMD is dependent on the nuclear localization of Rnt1p.
BDF2 RMD hyperactivation can be detected in a variety of stress conditions that are known to result in mRNA nuclear retention
The previous data showed that nuclear localization of Rnt1p is necessary for the cleavage of BDF2 transcripts, suggesting that an increase of BDF2 mRNA nuclear retention during salt stress may be a primary mechanism for RMD hyperactivation. To further test this hypothesis, we subjected S. cerevisiae to various stress conditions and analyzed BDF2 transcripts by northern blotting. RMD hyperactivation during these stress conditions was visualized either by estimating the amount of full-length mRNA remaining or by calculating the overall % of cleavage fragment (CF) over the overall BDF2 transcripts population [%CF = CF/(CF + FL) × 100]. Ethanol or heat shock stress are known to cause selective retention of bulk mRNAs within the nucleus and the rapid export of stress responsive transcripts (Piper et al. 1994; Saavedra et al. 1997; Izawa et al. 2008). Strikingly, we detected RMD hyperactivation of BDF2 in cells treated with either ethanol or heat shock (Fig. 2A) but the kinetics and final effect on full-length BDF2 levels varied. Ethanol treatment resulted in a global decrease of the BDF2 mRNA and an accumulation of the main cleavage fragment CFm after 1 h. However, during heat shock the cleavage fragment was detected at its peak within the first 20 min of treatment and the level of the BDF2 FL mRNA was not strongly affected. No cleavage fragment was detected after an hour of heat shock, as the cells may have recovered from the stress. This is consistent with RNA-seq analysis done after 45 min of heat shock (Wang et al. 2020), which showed that BDF2 full-length levels were similarly to control samples after prolonged heat-shock. Interestingly, a complete loss of the full length BDF2 transcript was not detected during salt treatment at 23°C, as compared to salt treatment at 30°C (compare FL between lanes 7 and 2, Fig. 2A). This suggests that the steady state growth temperature may influence the cleavage activity of Rnt1p as well. Overall, these data using heat shock and ethanol stress treatments suggest that the RMD hyperactivation seen on BDF2 mRNAs may be due to its nuclear retention, which has been shown to occur globally on mRNAs in these stress conditions (Piper et al. 1994; Saavedra et al. 1997; Izawa et al. 2008).
FIGURE 2.
Analysis of BDF2 RMD in various stress conditions. (A) Northern blot analysis of BDF2 following exposure to ethanol (10%), heat shock (42°C), or salt stress conditions (0.6 M NaCl). scR1 was used as a loading control. The migration of the full-length (FL) and of the main cleavage fragment (CFm) is indicated. Numbers indicate the % of the BDF2 cleavage fragments relative to all BDF2 transcripts. The numbers shown are the average of three independent biological replicates, except for lane 5 (60 min ethanol treatment) for which only two independent replicates were obtained. (B) Northern blot analysis of BDF2 in an rrp6Δrnt1Δ strain expressing GFP alone, Rnt1–GFP or Rnt1–GFP partially delocalized to the cytoplasm (NLSΔ) from pUG35 plasmids (Henras et al. 2004) in normal and heat-shock conditions. Numbers indicate the % of the BDF2 cleavage fragment relative to all BDF2 transcripts calculated from the northern blot shown on the picture which is the only biological replicate for this experiment. (C) Northern blot analysis of BDF2 in control medium or following a 1 h exposure to 0.6 M NaCl or 0.6 M KCl. Numbers indicate the % of the BDF2 cleavage fragment relative to all BDF2 transcripts (average of three independent replicates). (D) Northern blot analysis of BDF2 during NaCl-induced stress in the rrp6Δ and rrp6Δslt2Δ mutants. Legends as in Figure 2A. Numbers indicate the % of the BDF2 cleavage fragment relative to all BDF2 transcripts based on the average of three independent biological replicates.
To test the hypothesis that BDF2 RMD during heat-shock requires Rnt1p nuclear localization, we analyzed BDF2 during heat shock in a rnt1Δ rrp6Δ double deletion mutant strain expressing either GFP alone, the RNT1–GFP fusion used for the localization studies shown above, or a RNT1–GFP carrying a deletion of the nuclear localization signal of Rnt1p (NLSΔ), which results in a large fraction of Rnt1p delocalized to the cytoplasm (Henras et al. 2004). Cells expressing the Rnt1–GFP fusion exhibited a decrease of the FL form of BDF2 after heat-shock, with an increased accumulation of the BDF2 cleavage fragment compared to cells expressing GFP alone (Fig. 2B). This result shows that the Rnt1–GFP fusion is functional for RMD during heat shock. In contrast, cells expressing the Rnt1p–NLSΔ mutant showed a delay in the accumulation of the BDF2 cleavage fragment, with none detectable after 5 min of heat shock treatment, and a reduced accumulation after 10 min compared to the wild-type Rnt1–GFP (Fig. 2B). RMD was not detected in the strain expressing GFP alone, showing that the band detected upon heat-shock is the result of Rnt1p cleavage. Even though this experiment was only performed once, this result shows that a fully functional Rnt1p localization signal is required for optimal RMD of BDF2 during heat shock. These experiments also show that, as opposed to salt stress, heat shock results in a transient activation of RMD.
We further explored the type of ionic stress that can mediate RMD of BDF2. We found that the addition of high KCl concentrations (0.6 M) did not activate RMD, indicating that RMD hyperactivation is specific to the type of ionic stress (Fig. 2C). A similar result was obtained when nuclear exosome activity was impaired by the deletion of Rrp6p to increase detection of the CF (Fig. 2C). Altogether, these results demonstrate that BDF2 RMD can be hyperactivated in a variety of stress conditions that are known to specifically induce mRNA nuclear retention.
BDF2 RMD hyperactivation is reduced in the slt2Δ mutant but can be uncoupled from extracellular stress by anchoring away mRNA biogenesis or export factors
Specific stress conditions, including salt stress and heat stress can promote the nuclear retention of non-heat shock mRNAs as a mechanism to rapidly change gene expression (Carmody et al. 2010). Slt2p (also referred to as Mpk1p) is a key protein kinase involved in the stress response (Kim and Levin 2011), which is required for the retention of non-heat shock mRNAs during heat shock conditions (Carmody et al. 2010). We used a strain carrying a deletion of the SLT2 gene to determine if stress-induced hyperactivation of RMD is caused by BDF2 nuclear retention through the Slt2p pathway. We performed this experiment in an rrp6Δ background to maximize the detection of the BDF2 cleavage fragment. The absence of Slt2p significantly reduced BDF2 RMD activation during NaCl-induced stress (lane 4 vs. lane 2; paired t-test P-value = 0.008; Supplemental Table S1), as shown by the increased accumulation of the full-length BDF2 mRNA and the relative decrease of the cleavage product during stress compared to the wild-type control (Fig. 2D). However, the absence of Slt2p did not fully inhibit RMD activity on BDF2 during salt stress. This result suggests that Slt2p might contribute to the increased retention of BDF2 during stress, but that the BDF2 mRNA could also be retained through a different pathway not defined by Slt2p. For example, specific stress signals may impact the mRNP formation, processing and export of several transcripts differently (Izawa et al. 2008).
To further test the hypothesis that the hyperactivation of RMD on BDF2 is primarily due to its nuclear retention during stress, we tested the effect of globally inhibiting mRNA export to the cytoplasm independently from any extracellular stress. We used the anchor-away technique (Haruki et al. 2008) to rapidly deplete from the nucleus several proteins that are involved directly or indirectly in the export of mRNAs to the cytoplasm. We first focused on the poly(A) binding protein Nab2p and on the 3′-end processing factor Nab4p (Hrp1p), as defects in poly(A) tail binding or formation have been shown to inhibit mRNA export (Hammell et al. 2002). As a control, salt treatment of strains in which Nab2p and Nab4p were FRB-tagged (but without adding rapamycin which promotes the anchor away process) resulted in a reduction of the full length BDF2 transcripts and in the appearance of the cleavage product CFm (Fig. 3A). Strikingly, cleavage of BDF2 transcripts was detected in the absence of any salt stress after anchoring away Nab2p or Nab4p (Fig. 3A). The size of the fragment detected upon anchoring away Nab2p or Nab4p in the absence of stress was identical to the size of the fragment detected during salt stress (Fig. 3A), which showed that RMD was activated upon anchoring away Nab2p or Nab4p. The extent of BDF2 cleavage differed, with Nab4p resulting in a stronger accumulation of the CFm and in a very strong reduction in the levels of the FL form of BDF2; however, this reduction was not solely due to RMD, as long extended forms could also be detected due to transcription termination defects arising from defective 3′-end formation when Nab4p is inactivated (Minvielle-Sebastia et al. 1998). Nevertheless, these results suggested that the inhibition of mRNA export triggered by defects in poly(A) formation or binding is sufficient to recapitulate the salt stress-induced RMD hyperactivation of BDF2. However, anchoring away Nab2p or Nab4p did not result in an activation of RMD to the same extent as that detected during salt stress, and a further loss of the full-length transcript was observed when cells were exposed to salt stress prior to Rapamycin treatment (Fig. 3A). This additive effect may either indicate a synergy between the retention of unprocessed RNAs and the retention of mature RNAs during stress for RMD activation, or that the anchoring away of either of these two factors may not completely abolish mRNA export.
FIGURE 3.
Anchoring away mRNA biogenesis or mRNA export factors triggers RMD hyperactivation of BDF2 in the absence of stress. (A) Northern blot analysis of BDF2 in strains expressing FRB-tagged versions of Nab2p, and Nab4p in salt stress conditions and/or in conditions triggering nuclear export of these factors upon addition of Rapamycin. Cells grown to log phase were split in half, with one kept in normal medium and the other half treated with 0.6 M salt. After an hour of salt treatment, each culture was split in half again and treated with either 1 µg/mL of rapamycin or its vehicle solvent (90% ethanol and 10% Tween-20) for an additional hour. scR1 was used as a loading control. The BDF2 full-length (FL) and cleavage fragments (CFm) are shown, with the scR1 loading control. Numbers indicating the % of the BDF2 cleavage fragment relative to all BDF2 transcripts are the average of three independent biological replicates. (B) Northern blot analysis of BDF2 in strains expressing FRB-tagged versions of Rrp6p and of the cleavage and polyadenylation factors Yth1p or Ysh1p, in salt stress conditions and/or upon addition of Rapamycin. Legends are as in Figure 3A. Numbers indicating the % of the BDF2 cleavage fragment relative to all BDF2 transcripts are the average of three independent biological replicates, except for lanes 1, 4, 7, and 10 (two independent replicates), and 2 and 3 (one replicate). (C) Northern blot analysis of BDF2 in a strain expressing FRB-tagged versions of Rrp6p and of the mRNA export factor Mex67p in salt stress and/or upon addition of Rapamycin. Legends are as in Figure 3A. Numbers indicating the % of the BDF2 cleavage fragment relative to all BDF2 transcripts are the average of three independent biological replicates. (D) Northern blot analysis of the BDF2 and HSP12 mRNAs in a wild-type strain and in a strain expressing BDF2 from the HSP12 promoter in normal and in salt stress conditions. Legends as in Figure 3A. Numbers indicating the % of the BDF2 cleavage fragment relative to all BDF2 transcripts are the average of ten (lanes 1 and 2) or three (lanes 3 and 4) independent biological replicates.
To further explore the hypothesis that inhibiting 3′-end processing can result in BDF2 RMD hyperactivation, we constructed strains in which the core cleavage and polyadenylation (CPA) factors Yth1p and Ysh1p can be anchored away from the nucleus, concomitantly with Rrp6p (Fig. 3B). This double anchor away strategy was used to allow a better detection of the BDF2 cleavage product as anchoring away Rrp6p strongly stabilizes nuclear exosome targets (Roy et al. 2016), possibly because other subunits of the exosome might be anchored away concomitantly. Strikingly, anchoring away both Rrp6p and Ysh1p or Yth1p resulted in a strong accumulation of the BDF2 cleavage product (Fig. 3B) in the absence of any extracellular stress. The fragment detected upon anchoring away Ysh1p or Yth1p and Rrp6p in the absence of stress comigrated with the fragment detected during salt stress treatment alone (Fig. 3A), which showed that this fragment was generated by Rnt1p cleavage. The accumulation of the BDF2 cleavage fragment and the decrease of the full-length mRNA were exacerbated when strains were exposed to salt stress prior to anchoring away these CPA factors. Similar to what was observed with Nab4p, anchoring away Ysh1p and Yth1p also resulted in the appearance of extended species of BDF2, likely because of the transcription termination defects resulting from CPA inactivation. These results show that inactivation of different CPA factors can promote the hyperactivation of BDF2 RMD.
To formally demonstrate that inhibition of mRNA nuclear export can trigger RMD hyperactivation, we performed nuclear depletion of the essential mRNA export factor, Mex67p concomitantly with Rrp6p. Mex67p is a core member of the mRNA export complex (Hurt et al. 2000) and its nuclear depletion through the anchor-away technique completely abolishes poly(A)+ mRNA export (Haruki et al. 2008). Strikingly, co-nuclear depletion of Mex67p and Rrp6p fully reproduced BDF2 RMD hyperactivation (Fig. 3C) in the absence of extracellular stress. Nuclear depletion of Mex67p resulted in a complete loss of the full length BDF2 transcripts, a phenotype stronger than the nuclear depletion of Nab2p or of the 3′-end processing factors Nab4p, Yth1p, or Ysh1p. The complete cleavage of the BDF2 transcripts was further confirmed by the observation that no additional cleavage product accumulated when the anchored-away strains were treated with salt stress (Fig. 3C). Overall, these results indicate that the subcellular localization of BDF2 mRNAs plays a pivotal role in determining their degradation fate through RMD, and that inhibition of mRNA nuclear export by triggering export of key mRNA biogenesis or export factors can fully recapitulate RMD hyperactivation of BDF2 in the absence of extracellular stress.
Promoter identity is not required to promote BDF2 RMD during stress
Elements within gene promoters can affect the stability of mRNAs expressed from these promoters (Trcek et al. 2011; Catala and Abou Elela 2019). Promoter-dependent regulation of mRNA stability is controlled through the cotranscriptional recruitment of several proteins that initiate mRNA decay. Rnt1p itself can be recruited to the promoter before recognizing the Rnt1p cleavage site (RCS) within the ORF (Catala and Abou Elela 2019). To determine if BDF2 RMD relies on the recruitment of Rnt1p to the BDF2 promoter, we swapped the endogenous BDF2 promoter with that of HSP12. The HSP12 mRNA does not undergo RMD, and we hypothesized that this promoter would be unable to recruit Rnt1p despite containing many stress response elements (STRE) (Varela et al. 1995). Furthermore, the HSP12 promoter is strongly activated under salt stress, allowing us to investigate the RMD-mediated regulation of the BDF2 transcript generated from this promoter (pHSP12-BDF2). The BDF2 mRNA generated from pHSP12- was still targeted by Rnt1p for rapid cleavage under salt stress (Fig. 3D). In fact, the strong induction of the HSP12 promoter under salt stress generated more transcripts to be targeted by RMD, resulting in an increase of accumulation of the cleavage fragment compared to wild type post-salt stress. RMD was however not sufficient to completely eliminate the highly expressed pHSP12-BDF2 transcript, possibly due to the known ability of STRE to promote rapid export in heat shock through recruitment of the mRNA export factor Mex67p by Hsf1p (Zander et al. 2016). We conclude that the RMD hyperactivation on the BDF2 transcript does not require recruitment of Rnt1p to the endogenous BDF2 promoter.
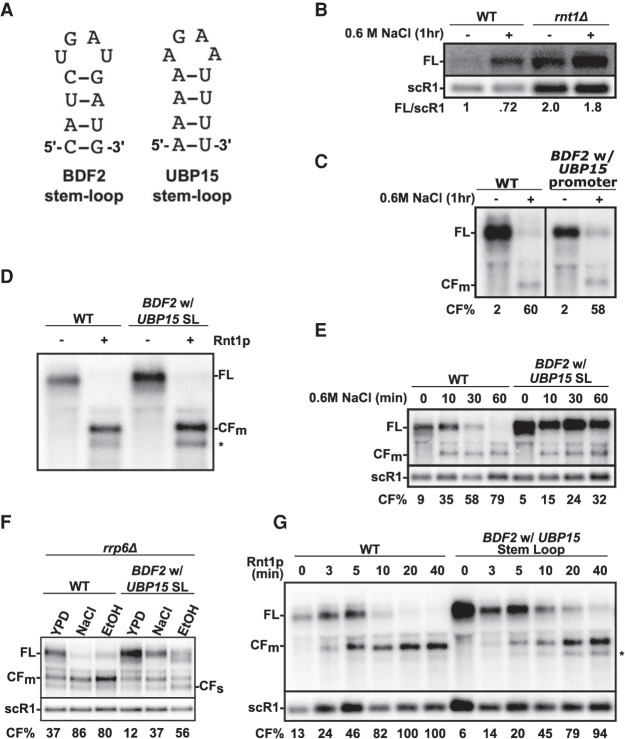
The identity of the stem–loop cleaved by Rnt1p influences RMD efficiency during stress
Previous studies have demonstrated that specific structural features of the stem–loop recognized by Rnt1p may affect cleavage efficiency (Comeau et al. 2016). To further investigate how the stem–loop present in BDF2 may affect RMD hyperactivation, we replaced the endogenous BDF2 RCS with that of UBP15 (Fig. 4A). The UBP15 mRNA does not undergo RMD hyperactivation in salt stress (Fig. 4B) and replacing the BDF2 promoter by the UBP15 promoter does not abrogate RMD (Fig. 4C), which showed that the lack of cleavage of UBP15 in vivo during stress is not due to its endogenous promoter, which could have promoted its export from the nucleus as shown for other stress induced genes (Zander et al. 2016). However, the BDF2 mRNA containing the stem–loop (SL) present in UBP15 was fully cleaved when incubated for 1 h with recombinant Rnt1p in vitro (Fig. 4D). Incubation of total RNAs containing the BDF2 hybrid transcripts containing the UBP15 SL with recombinant Rnt1p resulted in detection of a cleavage product with a migration identical to that detected upon in vitro cleavage of the wild-type BDF2 (Fig. 4D). This indicates that the hybrid BDF2 transcripts can be indeed targeted and cleaved by Rnt1p in the same stem–loop structure. We note that the UBP15 terminal loop also induced the cleavage at an additional, minor site (asterisk on Fig. 4D). Based on these in vitro cleavage data, it is unclear whether the absence of RMD activity on UBP15 in vivo is due to the identity of its RCS or to other factors that might prohibit cleavage in vivo. To further investigate the impact of the RCS identity on RMD, we analyzed the levels of wild-type BDF2 and of the mutant containing the UBP15 SL during stress. The BDF2 hybrid transcript containing the UBP15 SL was less efficiently down-regulated by RMD during a kinetic of salt exposure (Fig. 4E), with significant decreases of RMD efficiency compared to the WT based on the %CF (all time points paired t-tests P-values <0.05; Supplemental Table S1). To better characterize the impact of the UBP15 stem–loop on BDF2, we introduced this mutation in the context of the rrp6Δ mutant, and we analyzed BDF2 levels in control medium (YPD) or after 1 h of 0.6 M NaCl or 10% thanol treatment (Fig. 4F). In rich medium, the presence of the UBP15 SL resulted in a substantial increase of the FL form and a decrease of both CFm and CFs (Fig. 4F), reflecting a significant decrease in Rnt1p cleavage activity in normal growth conditions (lane 1 vs. 4; t-test P-value = 0.005; Supplemental Table S1). The UBP15 stem–loop also decreased the hyperactivation of BDF2 RMD during stress, based on the %CF ratios indicated and as shown by a substantial amount of the full length BDF2 hybrid transcript remaining after an hour of salt or ethanol treatment compared to the wild-type BDF2 (paired t-test P-values: NaCl treatment = 0.001; ethanol treatment = 0.05). To further characterize the impact of the UBP15 stem–loop on Rnt1p cleavage activity, a kinetic analysis of in vitro cleavage was performed using purified Rnt1p and total RNAs as a source of substrate RNA. As shown in Figure 4G the rate of in vitro cleavage of the hybrid transcript was slower than that observed for the wild-type BDF2 transcript (Fig. 4G). Although this time course experiment was performed only once, the decreased cleavage efficiency is consistent with the decreased cleavage activity detected in vivo in Figure 4E,F. Inefficient cleavage of this substrate compared to the WT could be due to the fact that the UBP15 stem–loop is rich in A–U base pairs near the terminal tetraloop, which is unusual for Rnt1p substrates (Chanfreau 2003). Taken together, our data demonstrate that specific features of the RCS can affect RMD activity during normal and stress conditions and that optimal degradation of BDF2 during stress requires both its specific stem–loop structure and inhibition of RNA export, but is not dependent on expression of BDF2 from its natural promoter.
FIGURE 4.
The identity of the Rnt1p cleavage site (RCS) influences RMD hyperactivation in stress conditions. (A) Predicted secondary structures of the terminal stem–loops present at the top of the RCS of BDF2 and UBP15 using Mfold (Zuker 2003). (B) Analysis of UBP15 mRNA expression in WT and rnt1Δ cells in normal growth conditions and upon salt stress treatment. Shown is a northern blot analysis of UBP15 in the corresponding strains. scR1 was used as a loading control. This experiment was performed once and the ratios indicate the level of full-length UBP15 relative to scR1 based on the northern blot shown. (C) Northern blot analysis of BDF2 in normal or NaCl-stress conditions for the endogenous BDF2 gene or the gene expressing BDF2 from the UBP15 promoter. The FL and main cleavage fragments are indicated. This experiment was performed once and the %CF was calculated from the northern blot shown on this figure. (D) In vitro cleavage assay of BDF2 transcripts containing the RCS of UBP15. Shown is a northern blot of total RNAs from the corresponding strains after incubation in buffer or with recombinant Rnt1p for 1 h. The location of the main cleavage fragment (CFm) is indicated. The asterisk indicates a secondary, minor cleavage event detected for the BDF2–UBP15 SL substrate. (E) Northern blot of wild-type BDF2 and BDF2 transcripts harboring the RCS from UBP15 after exposure to 0.6 M NaCl stress. Legends as in Figure 2. Numbers indicating the % of the BDF2 cleavage fragment relative to all BDF2 transcripts are the average of three independent biological replicates. (F) Northern blot of BDF2 in an rrp6Δ mutant expressing the wild-type BDF2 or a BDF2 mutant with the UBP15 stem–loop in normal medium, or after exposure to 0.6 M NaCl or 10% ethanol for 1 h. Numbers indicating the % of the BDF2 cleavage fragment relative to all BDF2 transcripts are the average of three independent biological replicates. (G) Time course of in vitro cleavage of the wild-type BDF2 and BDF2 RNA with the UBP15 terminal stem–loop by recombinant Rnt1p. Total RNAs extracted from the wild-type strain or from a strain expressing BDF2 with the UBP15 terminal stem–loop were incubated with recombinant Rnt1p for the indicated times. The reactions were stopped, the RNAs repurified and analyzed by northern blot. The numbers indicate the % of BDF2 cleavage [(CF/CF + FL) × 100] relative to time zero for each substrate for the time course experiment shown on this figure.
DISCUSSION
Bdf2p is a bromodomain protein involved in multiple aspects of epigenetic control through its ability to recognize acetylated histones. We previously showed that the BDF2 transcript is subject to both RMD and SMD pathways which limit and regulate its expression (Roy and Chanfreau 2014). This regulation is of particular importance during stress conditions, where a rewiring of transcription and translation is necessary to promote cellular fitness during stress. In high salt stress, an increase in activity of RMD on BDF2 causes its transcript to become completely undetectable. Repression of BDF2 in stress conditions is necessary for the robust expression of the stress responsive gene GPH1, and potentially other stress responsive genes as well (Roy and Chanfreau 2014). Despite the significance of regulating the expression of BDF2, the mechanism that triggers RMD hyperactivation during salt stress was not clearly understood. In this study, we demonstrate that the increase of RMD activity on BDF2 during stress does not arise from a direct change in the expression of Rnt1p or its localization within the cell. Rather, our study suggests that the nuclear retention of BDF2 transcripts during stress causes RMD hyperactivation. A global block of nuclear export of mRNAs induced by stress conditions such as heat or ethanol shock, or by nuclear depletion of specific mRNA biogenesis or export factors, can reproduce salt stress-induced RMD hyperactivation on BDF2. We also show that inactivation of the key stress response factor Slt2p reduces the efficiency of BDF2 RMD during stress. We interpreted this reduction in RMD in the slt2Δ mutant as a consequence of the role of Slt2p in mediating stress-induced mRNA nuclear retention (Carmody et al. 2010). However, we do not know if the reduction of RMD efficiency detected in this mutant for RMD is dependent on its kinase and signaling activity, as Slt2p has kinase-independent roles during stress (Kim and Levin 2011). Taken together, these results show that the primary mechanism underlying the hyperactivation of RMD for BDF2 is the inhibition of mRNA export and the retention of transcripts in the nucleus. RMD hyperactivation is independent of the identity of the BDF2 transcript promoter region, as switching its promoter to that of HSP12 or UBP15 does not prevent RMD. This suggests that promoter-dependent recruitment of Rnt1p, which is required for RMD of other transcripts (Catala and Abou Elela 2019) does not play a role in the hyperactivation of RMD of BDF2 observed in stress conditions.
We further provide evidence that the identity of the Rnt1p cleavage stem–loop (RCS) within BDF2 further influences its susceptibility to cleavage, as replacing the endogenous BDF2 terminal stem–loop by that of UBP15 decreases BDF2 RMD in vivo, and Rnt1p cleavage efficiency in vitro. This supports previous evidence that the nucleotide base-pairing of the product termini can determine the Rnt1p substrate reactivity (Comeau et al. 2016). The effect of the UBP15 stem–loop on Rnt1p cleavage might be linked to its stretch of A–U base pairs near the terminal tetraloop (Fig. 4A), which is not usually found in natural Rnt1p substrates (Chanfreau 2003). It is unclear whether certain stress conditions, such as heat shock or salt stress, may alter the structure of the RCS in vivo and influence its cleavage efficiency by Rnt1p. High temperatures or specific salt conditions may contribute to the stabilization or destabilization of the BDF2 RCS and thereby modulate cleavage efficiency. This may contribute to explain some of the differences in BDF2 RMD efficiency detected between heat shock treatment, ethanol or salt stress. Alternatively, it is possible that these different stress conditions affect mRNA export out of the nucleus differently, or that they could impact degradation pathways which facilitate the detection of the BDF2 cleavage products.
Altogether, our results indicate that Rnt1p may help regulate expression of specific genes through the cleavage and degradation of specific substrates based on their localization and stem–loop structures. The nuclear export of many mRNAs is inhibited during stress, which provides the opportunity for these mRNAs to be targeted by Rnt1p for degradation in these conditions to differing extents, depending on substrate reactivity. Previous studies have shown that blocking the nuclear export of mRNAs by nuclear depletion of Mex67p results in the rapid degradation of newly synthesized RNAs (Tudek et al. 2018). A similar effect was observed for polyadenylated RNAs when the poly(A) binding protein Nab2p was depleted from the nucleus (Schmid et al. 2015). Strikingly, anchoring away both of these factors is sufficient to promote BDF2 RMD in the absence of stress (Fig. 3). Therefore, it is possible that Rnt1p may play a role in the general degradation mechanism previously described by the Jensen group upon nuclear depletion of these factors, and that Rnt1p-mediated cleavage may contribute to the elimination of specific mRNAs containing compatible cleavage sites during stress-induced nuclear retention of mRNAs. However, not all mRNAs sequestered in the nucleus during stress or after nuclear depletion of CPA or export factors contain stem–loop structures compatible with Rnt1p cleavage, and it is likely that the sequence content of mRNAs has evolved to either promote RMD of mRNAs that are not required or perhaps detrimental during stress conditions.
MATERIALS AND METHODS
Yeast strains and plasmids
All strains used in this study were derived from BMA64, HHY168 or as described below:
- Strains previously described or from external sources:
- BMA64 (Chanfreau et al. 1998): MATa; ura3-1; trp1Δ2; leu2-3;112; his3-11,15; ade2-1; can1-100
- rnt1Δ: BMA64, rnt1Δ::TRP (Chanfreau et al. 1998)
- BY4742: MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (Open Biosystems)
- HHY168 (Haruki et al. 2008) MATalpha tor1-1 fpr1::NAT RPL13A-2×FKBP1 2::TRP1 ade 2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL psi+ (Haruki et al. 2008)
- yCW2: MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RNT1-HTTP::KIURA3 (gift of D. Tollervey).
- yCW6: BY4742, rrp6Δ:: hphMX6 (Wang et al. 2020)
- yCW17: yCW6, slt2Δ::kanMX6 (Wang et al. 2020)
- Strains generated in this study:
- yCW4: MATalpha tor1-1 fpr1::hphMX6 RPL13A-2×FKBP12::TRP1 ade 2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL psi+
- yCW5: yCW4 with RNT1-FRB::kanMX6
- yCW7: yCW4 with NAB2-FRB::kanMX6
- yCW8: yCW4 with NAB4-FRB::kanMX6
- yCW9: yCW4 with YTH1-FRB::kanMX6 RRP6-FRB::his3MX6
- yCW10: yCW4 with YSH1-FRB::kanMX6 RRP6-FRB::his3MX6
- yCW11: yCW4 with MEX67-FRB::kanMX6 RRP6-FRB::his3MX6
- yCW12: BMA64 with pHSP12-BDF2
- yCW13: BMA64 with BDF2 with UPB15 SL
- yCW14: BMA64 with BDF2 with UPB15 SL rrp6Δ::hphMX6
- yCW18: BMA64 rnt1Δ::TRP, rrp6Δ::hphMX6
- yCW19: BMA64 with BDF2 with UBP15 promoter
Strains were constructed using a high efficiency transformation method (Gietz and Schiestl 2007). Mutations within the BDF2 transcripts were constructed through the delitto perfetto approach (Storici and Resnick 2006). The Rnt1p target stem–loop within the BDF2 transcript (ChrIV:332367-80) was replaced with the CORE integration cassette, consisting of the URA3 and KanMX6 genes. Successful transformants were selected through their resistance to G418, and further confirmed through PCR. Afterwards, the CORE integration cassette was replaced with various Rnt1p target stem–loops using the transformation protocol as described above. Successful transformants were initially selected on the basis for their ability to grow on 5-Fluoroorotic acid (5-FOA) due to the loss of URA3, inability to grow on G418 due to the loss of kanMX6, and then further confirmed through PCR and Sanger sequencing (Laragen, Inc.).
Anchor away strains were created in a modified HHY168 (Haruki et al. 2008) background where the natMX6 marker was replaced with the hphMX4 marker amplified from pAG32 (Goldstein and McCusker 1999). The genes of interest were carboxy-terminally tagged with the rapamycin binding domain (FRB) using the transformation method as described. Plasmids expressing Rnt1–GFP and Rnt1–GFP with a deletion of the nuclear localization sequence are described in Henras et al. (2004).
Oligonucleotides used for mutagenesis (F and R stand for forward and reverse primers)
BDF2 Promoter Core oligonucleotides:
F:AGAGGCGAAAAAAGAGTGCAACGTCAACAACGCTAAAAGAGAGCTCGTTTTCGACACTGG
R:AGCCGCCGAGGTTTATTTCGCTCAATCTGTTTGTTTCAGTTCCTTACCATTAAGTTGATC
HSP12::BDF2 promoter swap:
F:AATGGAGTGAAGCAGGCAGGGTGACCCTCTAGCTAAAAAA GATCCCACTAACGGCCCAGC; R:CAGAATGTGCGTGTCTTGTATCCATGTTAGTACGAGACAT TGTTGTATTTAGTTTTTTTT
UBP15::BDF2 promoter swap:
F:AGAGGCGAAAAAAGAGTGCAACGTCAACAACGCTAAAAGA CAAGAGAGCAGTAGTAAGAG
R:CAGAATGTGCGTGTCTTGTATCCATGTTAGTACGAGACAT TGTTTGTTTGAAGAGACTAA
BDF2 Rnt1 cleavage site Core insertion:
F:GGATTCAGATCTTGAAGAGGATAACTATTCTTCTTCATATGAGCTCGTTTTCGACACTGG
R:TAGTTATATCGTTTTCATTTATGTCTTCATCATCATATTCTCCTTACCATTAAGTTGATC
UBP15 stem–loop swap:
R:ATATTTTGAATGAAATTGAAACGGATTCAGATCTTGAAGAGGATGCAAAATTTGGTCTCGGACAAAAAGAATTTTCAAAG
F:TTTGTTCCAAATATTGAATAGCCGGATTAGTTATATCGTTTTCATTACTTAATTTGATCTTTGAAAATTCTTTTTGTCCG
Yeast media and growth conditions
All strains were grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) or minimal media (0.67% w/v yeast nitrogen base, 2% w/v dextrose, and 0.2% w/v amino acid mixture) at 30°C unless noted otherwise. For protein and RNA extractions, 50 mL of culture were harvested at OD600 0.4–0.6 by centrifugation at 4000 rpm (Sigma Rotor 11030) for 1.5 min and transferred to 2 mL screw capped Eppendorf tubes. The cells were pelleted and flash frozen in liquid nitrogen. For experiments involving treatment involving anchor away and salt stress conditions, specific conditions are described in the results or legends of each figure. For heat shift experiments, cells were grown to exponential phase at 23°C before equal volumes of 61°C preheated YPD were added to bring the temperature to 42°C. The cultures were immediately harvested at the indicated times. For ethanol treatment, cells were grown to exponential phase before equal volumes of YPD or YPD containing 20% ethanol (v/v) were added, and then harvested at the indicated times.
Yeast RNA extraction and northern blot analysis
For most experiments, northern blot analysis was performed in triplicate (unless indicated otherwise) on purified total RNAs as described previously (Roy and Chanfreau 2014). Briefly, RNAs were extracted using phenol-chloroform, precipitated with ethanol and sodium acetate, and resuspended in water. A total of 5 µg of glyoxylated RNAs were run on a 1.8% agarose gel and transferred to nylon membranes for probing.
Riboprobe and oligonucleotide probe synthesis for northern blotting analysis
The radiolabeled riboprobes were transcribed in vitro using T3 RNA polymerase (Promega) as described previously (Roy and Chanfreau 2014). The BDF2 riboprobe hybridizes from the beginning of the open-reading frame until the Rnt1p cleavage site.
Templates used were synthesized by PCR using the primers: BDF2 F: GCACATTCTGCTTTACTGGCAGC and BDF2 T3R: GGCTAAATTAACCCTCACTAAAGG TTCAAGATCTGAATCCGTTT.
The SCR1 Oligonucleotide used for the loading control: ATCCCGGCCGCCTCCATCAC was radiolabeled using γ-32P-ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs) according to the manufacturer's protocol.
In vitro Rnt1p cleavage assay
Recombinant Rnt1p was purified as described previously (Henras et al. 2005). In vitro cleavage reactions were performed in 50 µL reactions consisting of 50 µg of total yeast RNA, 10 pmol of purified recombinant Rnt1p in Rnt1p cleavage buffer (30 mM Tris pH 7.5, 150 mM KCl, 5 mM spermidine, 200 mM MgCl2, 0.1 mM DTT, 0.1 mM EDTA). The reactions were incubated at 30°C and halted by the addition of 150 µL of RNA buffer at the times indicated. The reactions were then purified through using phenol-chloroform. Briefly, 200 µL of phenol: chloroform: isoamyl alcohol were added to the samples. The samples were vortexed for 1 min and spun down for 2 min at 15,000 rpm. The top aqueous layer was added to a fresh Eppendorf tube containing 1 mL ethanol, 40 µL 3 M sodium acetate pH 5.2, and 1 µL of GlycoBlue (Ambion). Precipitation of the RNA was facilitated through incubating the samples at −80°C for 30 min and then pelleted by centrifugation at 15,000 rpm for 10 min. The pellets were washed with 200 µL of 70% ethanol and resuspended in 15 µL of nuclease-free water.
Protein extraction and western blot analysis
Western blot analysis was performed using protein extracts from an Rnt1p-HTP strain kindly provided by D. Tollervey (U. Edinburgh). The HTP carboxy-terminal tag (Granneman et al. 2009) consists of a 6-His sequence, followed by a TEV protease cleavage site and a Protein A sequence (PMID: 19482942). Total protein was prepared from the Rnt1p-HTP strain and an untagged control strain grown to mid-log phase in 50 mL YPD liquid medium and treated with 0.6 M NaCl for 0, 15, and 60 min. The culture was harvested by centrifugation for 5 min at 3500 rpm, washed with ddH2O, and resuspended in lysis buffer (200 mM Tris-HCl pH 8.0; 320 mM Ammonium sulfate; 5 mM MgCl2; 10 mM EGTA pH 8.0; 20 mM EDTA pH 8.0; 1 mMDTT; 20% glycerol; 1 mM PMSF; 2 mM benzamidine HCl and protease inhibitor cocktail). The sample was vortexed with glass beads (roughly equivalent volume to size of cell pellet) for 8 min at 4°C. Supernatant was collected by centrifugation at max speed (13.2k rpm) for 5 min at 4°C. Total protein concentration was measured using the Bradford method with the Bio-Rad Protein Assay (#500-0006) according to the manufacturer's protocol. A total of 10 µg total protein from crude extracts was analyzed by 8% SDS-PAGE and transferred to PVDF membranes for western blot analysis. Rnt1p-HTP was detected with anti-protein A antibody conjugated to horseradish peroxidase (HRP) (Invitrogen PA1-26853) diluted 1:8000 in blocking solution (1× PBS-T, 5% milk) with the Pierce ECL Western Blotting Substrate (Thermo Fisher #32209) according to the manufacturer's protocol.
Microscopy
Strains expressing GFP-tagged Rnt1p (Henras et al. 2004) were grown to mid-log phase in standard growth conditions and treated with 0.6 M or 0.1 M NaCl for 20 min before being prepared for imaging via the Stellaris RNA FISH protocol for S. cerevisiae with the following modifications: All centrifugation was performed at 850g for 6 min instead of 400g for 5 min, and Hoescht stain was used instead of DAPI for nuclear staining. The FISH probe used for visualization of mRNA localization was an oligo-d(T)25 labeled with Cy3 (MilliporeSigma). Microscopy was performed on a Leica DMI4000B Confocal microscope. All images were taken with identical settings and with a z-stack of six images with 1 µm steps, from which one z-slice was chosen from each stack for further processing. Image processing was then performed and equally applied to all images with the ImageJ software.
Reproducibility and statistics
In general, most experiments were performed in triplicate with the exception of the experiments presented in Figures 2B, 4B–D,G which included only one replicate, and some specific lanes of some figures (see figure legends). The quantification of the western blot and northern blot signals for experiments shown in all figures and for all replicates is included in Supplemental Table S1, with the calculation of the average and the standard deviations. Paired t-test P-values are included in Supplemental Table S1 for the comparisons of averages corresponding to specific lanes for each experiment.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joseph Ong for help with the confocal microscope and Dr. Anne Hong-Hermesdorf for providing us access to the microscope. This work was supported by the National Institute of General Medical Sciences (R35 GM130370 to G.F.C.) and a National Research Service Award Training Grant (GM007185 to C.W. and K.B.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078880.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Carmody SR, Tran EJ, Apponi LH, Corbett AH, Wente SR. 2010. The mitogen-activated protein kinase Slt2 regulates nuclear retention of non-heat shock mRNAs during heat shock-induced stress. Mol Cell Biol 30: 5168–5179. 10.1128/MCB.00735-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala M, Abou Elela S. 2019. Promoter-dependent nuclear RNA degradation ensures cell cycle-specific gene expression. Commun Biol 2: 211. 10.1038/s42003-019-0441-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala M, Lamontagne B, Larose S, Ghazal G, Elela SA. 2004. Cell cycle-dependent nuclear localization of yeast RNase III is required for efficient cell division. Mol Biol Cell 15: 3015–3030. 10.1091/mbc.e04-03-0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G. 2003. Conservation of RNase III processing pathways and specificity in hemiascomycetes. Eukaryot Cell 2: 901–909. 10.1128/EC.2.5.901-909.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Rotondo G, Legrain P, Jacquier A. 1998. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J 17: 3726–3737. 10.1093/emboj/17.13.3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau M-A, Lafontaine DA, Abou Elela S. 2016. The catalytic efficiency of yeast ribonuclease III depends on substrate specific product release rate. Nucleic Acids Res 44: 7911–7921. 10.1093/nar/gkw507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant M, Pugh BF. 2007. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol Cell Biol 27: 5327–5335. 10.1128/MCB.00468-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elela SA, Igel H, Ares M Jr. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85: 115–124. 10.1016/S0092-8674(00)81087-9 [DOI] [PubMed] [Google Scholar]
- Fu J, Hou J, Liu L, Chen L, Wang M, Shen Y, Zhang Z, Bao X. 2013. Interplay between BDF1 and BDF2 and their roles in regulating the yeast salt stress response. FEBS J 280: 1991–2001. 10.1111/febs.12219 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2: 31–34. 10.1038/nprot.2007.13 [DOI] [PubMed] [Google Scholar]
- Gilan O, Rioja I, Knezevic K, Bell MJ, Yeung MM, Harker NR, Lam EYN, Chung C, Bamborough P, Petretich M, et al. 2020. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immuno-inflammation. Science 368: eaaz8455. 10.1126/science.aaz8455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Granneman S, Kudla G, Petfalski E, Tollervey D. 2009. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci 106: 9613–9618. 10.1073/pnas.0901997106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Gross S, Zenklusen D, Heath C V, Stutz F, Moore C, Cole CN. 2002. Coupling of termination, 3′ processing, and mRNA export. Mol Cell Biol 22: 6441–6457. 10.1128/MCB.22.18.6441-6457.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31: 925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Henras AK, Bertrand E, Chanfreau G. 2004. A cotranscriptional model for 3′-end processing of the Saccharomyces cerevisiae pre-ribosomal RNA precursor. RNA 10: 1572–1585. 10.1261/rna.7750804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Sam M, Hiley SL, Wu H, Hughes TR, Feigon J, Chanfreau GF. 2005. Biochemical and genomic analysis of substrate recognition by the double-stranded RNA binding domain of yeast RNase III. RNA (New York, NY) 11: 1225–1237. 10.1261/rna.2760705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Strasser K, Segref A, Bailer S, Schlaich N, Presutti C, Tollervey D, Jansen R. 2000. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J Biol Chem 275: 8361–8368. 10.1074/jbc.275.12.8361 [DOI] [PubMed] [Google Scholar]
- Izawa S, Kita T, Ikeda K, Inoue Y, Piper P, Tani T, Derby RJ, Hiraoka Y, Spector DL, Saavedra C, et al. 2008. Heat shock and ethanol stress provoke distinctly different responses in 3′-processing and nuclear export of HSP mRNA in Saccharomyces cerevisiae. Biochem J 414: 111–119. 10.1042/BJ20071567 [DOI] [PubMed] [Google Scholar]
- Josling GA, Selvarajah SA, Petter M, Duffy MF. 2012. The role of bromodomain proteins in regulating gene expression. Genes (Basel) 3: 320–343. 10.3390/genes3020320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Levin DE. 2011. Mpk1 MAPK association with the paf1 complex blocks sen1-mediated premature transcription termination. Cell 144: 745–756. 10.1016/j.cell.2011.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M. 2003. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol 4: 276–284. 10.1038/nrm1075 [DOI] [PubMed] [Google Scholar]
- Lawrence M, Daujat S, Schneider R. 2016. Lateral thinking: how histone modifications regulate gene expression. Trends Genet 32: 42–56. 10.1016/j.tig.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev 14: 951–962. [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Beyer K, Krecic AM, Hector RE, Swanson MS, Keller W. 1998. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J 17: 7454–7468. 10.1093/emboj/17.24.7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgado-Pascual JL, Rayego-Mateos S, Tejedor L, Suarez-Alvarez B, Ruiz-Ortega M. 2019. Bromodomain and extraterminal proteins as novel epigenetic targets for renal diseases. Front Pharmacol 10: 1315. 10.3389/fphar.2019.01315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper PW. 1995. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol Lett 134: 121–127. 10.1111/j.1574-6968.1995.tb07925.x [DOI] [PubMed] [Google Scholar]
- Piper PW, Talreja K, Panaretou B, Moradas-Ferreira P, Byrne K, Praekelt UM, Meacock P, Recnacq M, Boucherie H. 1994. Induction of major heat-shock proteins of Saccharomyces cerevisiae, including plasma membrane Hsp30, by ethanol levels above a critical threshold. Microbiology 140: 3031–3038. 10.1099/13500872-140-11-3031 [DOI] [PubMed] [Google Scholar]
- Roy K, Chanfreau GF. 2012. The diverse functions of fungal RNase III enzymes in RNA metabolism. Enzymes 31: 213–235. 10.1016/B978-0-12-404740-2.00010-0 [DOI] [PubMed] [Google Scholar]
- Roy K, Chanfreau G. 2014. Stress-induced nuclear RNA degradation pathways regulate yeast bromodomain factor 2 to promote cell survival. PLoS Genet 10: e1004661. 10.1371/journal.pgen.1004661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Gabunilas J, Gillespie A, Ngo D, Chanfreau GF. 2016. Common genomic elements promote transcriptional and DNA replication roadblocks. Genome Res 26: 1363–1375. 10.1101/gr.204776.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra C, Tuug KS, Amberg DC, Hopper AK, Cole CN. 1996. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev 10: 1608–1620. 10.1101/gad.10.13.1608 [DOI] [PubMed] [Google Scholar]
- Saavedra CA, Hammell CM, Heath C V, Cole CN. 1997. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev 11: 2845–2856. 10.1101/gad.11.21.2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Olszewski P, Pelechano V, Gupta I, Steinmetz LM, Jensen TH. 2015. The nuclear polyA-binding protein Nab2p is essential for mRNA production. Cell Rep 12: 128–139. 10.1016/j.celrep.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Storici F, Resnick MA. 2006. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol 409: 329–345. 10.1016/S0076-6879(05)09019-1 [DOI] [PubMed] [Google Scholar]
- Trcek T, Larson DR, Moldón A, Query CC, Singer RH. 2011. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147: 1484–1497. 10.1016/j.cell.2011.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudek A, Schmid M, Makaras M, Barrass JD, Beggs JD, Jensen TH. 2018. A nuclear export block triggers the decay of newly synthesized polyadenylated RNA. Cell Rep 24: 2457–2467. 10.1016/j.celrep.2018.07.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela JOCS, Praekelt UM, Meacock PA, Planta RJ, Mager WH. 1995. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol Cell Biol 15: 6232–6245. 10.1128/MCB.15.11.6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volanakis A, Passoni M, Hector RD, Shah S, Kilchert C, Granneman S, Vasiljeva L. 2013. Spliceosome-mediated decay (SMD) regulates expression of nonintronic genes in budding yeast. Genes Dev 27: 2025–2038. 10.1101/gad.221960.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Y, DeMario SM, Mandric I, Gonzalez-Figueroa C, Chanfreau GF. 2020. Rrp6 moonlights in an RNA exosome-independent manner to promote cell survival and gene expression during stress. Cell Rep 31: 107754. 10.1016/j.celrep.2020.107754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander G, Hackmann A, Bender L, Becker D, Lingner T, Salinas G, Krebber H. 2016. MRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature 540: 593–596. 10.1038/nature20572 [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.