Significance
Retrotransposons are noninfectious, mobile genetic elements that proliferate in host genomes via an RNA intermediate that is copied into DNA by a reverse transcriptase (RT) enzyme. RTs are important for biotechnological applications involving information capture from RNA since RNA is first converted into complementary DNA for detection or sequencing. Here, we biochemically characterized RTs from two retroelements and uncovered several activities that allowed us to design a streamlined, efficient workflow for determining the inventory of RNA sequences in processed RNA pools. The unique properties of nonretroviral RT activities obviate many technical issues associated with current methods of RNA sequence analysis, with wide applications in research, biotechnology, and diagnostics.
Keywords: non-LTR retroelement reverse transcriptase, RNA sequencing, miRNA, tRNA, noncoding RNA
Abstract
Selfish, non-long terminal repeat (non-LTR) retroelements and mobile group II introns encode reverse transcriptases (RTs) that can initiate DNA synthesis without substantial base pairing of primer and template. Biochemical characterization of these enzymes has been limited by recombinant expression challenges, hampering understanding of their properties and the possible exploitation of their properties for research and biotechnology. We investigated the activities of representative RTs using a modified non-LTR RT from Bombyx mori and a group II intron RT from Eubacterium rectale. Only the non-LTR RT supported robust and serial template jumping, producing one complementary DNA (cDNA) from several templates each copied end to end. We also discovered an unexpected terminal deoxynucleotidyl transferase activity of the RTs that adds nucleotide(s) of choice to 3′ ends of single- and/or double-stranded RNA or DNA. Combining these two types of activity with additional insights about nontemplated nucleotide additions to duplexed cDNA product, we developed a streamlined protocol for fusion of next-generation sequencing adaptors to both cDNA ends in a single RT reaction. When benchmarked using a reference pool of microRNAs (miRNAs), library production by Ordered Two-Template Relay (OTTR) using recombinant non-LTR retroelement RT outperformed all commercially available kits and rivaled the low bias of technically demanding home-brew protocols. We applied OTTR to inventory RNAs purified from extracellular vesicles, identifying miRNAs as well as myriad other noncoding RNAs (ncRNAs) and ncRNA fragments. Our results establish the utility of OTTR for automation-friendly, low-bias, end-to-end RNA sequence inventories of complex ncRNA samples.
Retroelements are mobile genome segments that use an RNA intermediate to template the synthesis of DNA inserted at a new genome location. This group of selfishly replicating DNAs includes eukaryotic long terminal repeat (LTR) and non-LTR retrotransposons, as well as prokaryotic mobile introns also found in eukaryotic organelles. Genome sequencing projects have revealed evolutionary episodes of dramatic retroelement proliferation, for example, the spread of human non-LTR long interspersed nuclear element 1 to constitute about 20% of our genome (1). Many non-LTR retroelements in the genomes of living organisms retain the ancestral eukaryotic retroelement architecture (2) with a single open reading frame (ORF) between unique 5′ and 3′ untranslated regions (UTRs). Not surprisingly, the element-encoded protein multitasks in its interactions with RNA and DNA, reverse transcriptase (RT) activity, and often DNA-nickase activity. Some of these retroelements show site-specific insertion, which would limit their copy number, decrease their toxicity, and increase their potential for long-term evolutionary persistence (3).
The only detailed biochemical characterization of a purified RT from the ancestral single-ORF non-LTR retroelement families is of the R2 element protein from the silk moth Bombyx mori in pioneering work by the Eickbush laboratory (4). R2 and other R-elements insert exclusively into a precise sequence of the ribosomal RNA (rRNA) precursor gene transcribed by RNA polymerase I (5). The R2 ORF encodes a protein comprised of N-terminal DNA-binding motifs (zinc-finger and Myb domains), a central RT domain, a C-terminal restriction-like endonuclease (EN) domain, and other regions of unknown function (SI Appendix, Fig. S1A). After binding the target site and introducing a nick to create a DNA primer 3′ end, the protein then switches to reverse transcription of a bound template RNA to produce complementary DNA (cDNA). This process is termed target-primed reverse transcription (TPRT) (4). To complete non-LTR retroelement insertion, second-strand DNA nicking and synthesis must also occur, the latter of which could in theory be performed by the retroelement protein and/or a cellular DNA polymerase (4).
Recombinant B. mori R2 protein produced in bacteria, combined with target DNA duplex and an RNA containing the retroelement 3′ UTR, is sufficient to reconstitute site-specific TPRT in vitro (6). The RT initiates cDNA synthesis by “template jumping,” which we define as engaging the 3′ end of an RNA to template cDNA synthesis without base pairing or with just one to two base pairs that would only be stable within the enzyme active site. Group II intron RTs are known to template jump in vitro, but in cells, intron insertion begins by reverse-splicing of the catalytic RNA followed by cDNA synthesis on contiguous DNA-RNA template (7). Retroviral RTs have template-jumping activity in vitro exploited for cDNA library 3′ adaptor addition (8), but the required amount of base pairing between cDNA and 3′ adaptor template (AT) is uncertain (9). Template jumping differs from retroviral RT “template switching,” which we define as occurring by cDNA product release from one template and reannealing to a new template anywhere within a transcript. This template switching activity is essential to complete the synthesis of LTRs (10).
In principle, the use of template jumping to make cDNA libraries that capture template sequences end to end, flanked by 5′ and 3′ adaptors, could be a boon for research and biotechnological applications. New methods of RNA sequencing (RNA-seq) have illuminated an ever-increasing diversity of RNA types, but challenges associated with library generation from noncoding RNA (ncRNA) limit what is known about specifically processed forms of ncRNA in cells and in extracellular vesicles (EVs) (11–14). Toward the goal of comprehensive, unbiased, end-to-end ncRNA-seq, we sought to use retroelement RT(s) for serial template jumping to add distinct 5′ and 3′ adaptors during cDNA library synthesis. We first compared RTs from group II introns and non-LTR retroelements for maximal template-jumping activity, and then we engineered the rampant template jumping of a truncated, modified R2 RT to perform two template jumps in specific order. We describe streamlined, automation-friendly, single-tube cDNA library production for next-generation sequencing (NGS), with adaptor indexing either in the initial cDNA library synthesis or by low-cycle PCR. We benchmarked the technology by sequencing cDNA libraries produced from a commercial reference standard of 962 microRNAs (miRNAs). Next, to gain biological insight, we used the technology to sequence small RNAs (sRNAs) in EVs secreted by human cell lines, with results that have implications for models of EV biogenesis and function.
Results
Comparison of Template Jumping by Nonretroviral RTs.
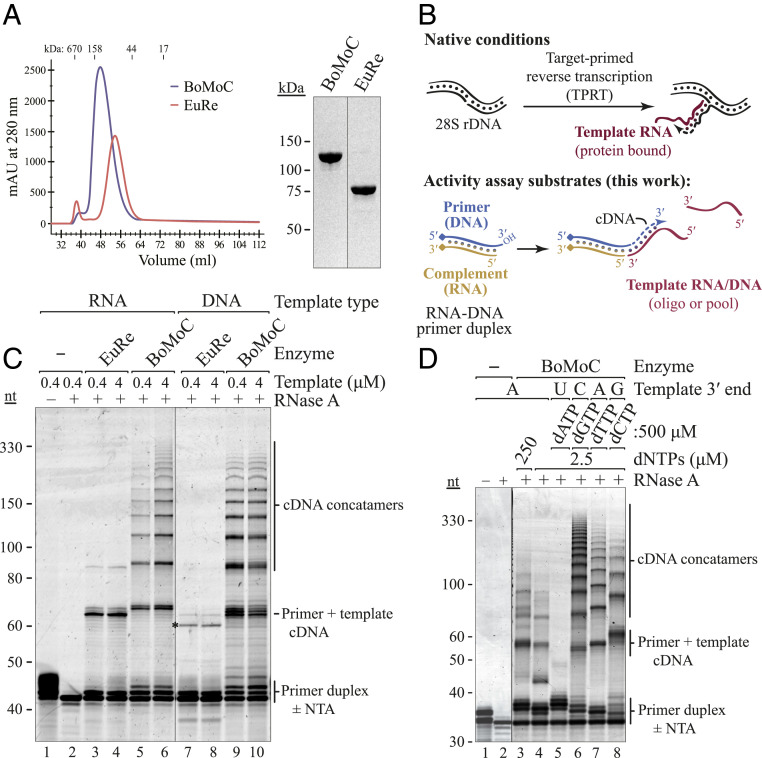
We evaluated the ability of nonretroviral RTs to use physically separate, discontinuous template molecules for continuous cDNA synthesis (primer and template sequences are listed in SI Appendix, Table S1). We screened recombinant versions of bacterial intron and eukaryotic non-LTR RTs for robust expression, purification, and serial template jumping. Proteins were expressed as N-terminal maltose binding protein (MBP) fusions with a C-terminal 6-histidine (6xHis) tag. Among the group II intron RTs tested under pilot screen conditions, the top candidate was from Eubacterium rectale (EuRe; Fig. 1 and SI Appendix, Fig. S1A). In papers published subsequent to our initial screening, this same enzyme, differing in tag configuration, was shown in studies from the Pyle laboratory to support remarkably processive cDNA synthesis (15, 16). Group II intron RTs synthesize cDNA across base- and sugar-modified templates with high tolerance for RNA structure (17), contributing to their processivity. In our assays of serial template jumping, the best performing enzyme was an N-terminally truncated B. mori R2 RT termed BoMoC (Fig. 1 and SI Appendix, Fig. S1A). Because removal of the C-terminal EN domain of BoMoC was unfavorable for enzyme stability, we introduced an EN active-site mutation previously characterized in the full-length protein (18) to produce an EN-crippled version of BoMoC. This modified, truncated R2 RT enzyme was used for all of our subsequent experiments.
Fig. 1.
Nonretroviral RTs differ in template jumping processivity. (A) SEC (Left) and SDS-PAGE (Right) of purified BoMoC and EuRe RTs. (B) Schematic of non-LTR retrotransposon cDNA synthesis by TPRT (Top) and cDNA library synthesis by OTTR (Bottom). Covalently linked blocking group indicated by diamond. (C) SYBR Gold-stained denaturing PAGE gel of BoMoC and EuRe RT products using +1T primer duplex (400 nM) and a single-stranded DNA or RNA oligonucleotide template harboring a 3′A (400 nM or 4 μM). To encourage template jumping, RT assays contained 2.5 μM mixed dNTP supplemented with 500 μM dTTP. Size differences between RNA-templated cDNA products of BoMoC and EuRe RNA are due to the more robust BoMoC cDNA extension by nontemplated NTA. An asterisk indicates the mobility of slightly shorter than expected cDNA products observed for EuRe synthesis on the DNA oligonucleotide template, possibly due to premature termination. The first two lanes are plus or minus RNase A to show removal of the primer duplex RNA strand. (D) SYBR Gold-stained denaturing PAGE gel of BoMoC RT products using blunt-end primer duplex with dNTPs and RNA templates indicated.
Recombinant EuRe and BoMoC RTs were purified extensively to remove nucleic acid and nuclease contamination. Purification involved binding to and elution from nickel agarose and heparin agarose, with a final size-exclusion chromatography (SEC) step, a strategy also used in previous EuRe purifications (16, 19). Very high ionic strength buffers were essential to release bound nucleic acids and decrease aggregation. Both EuRe and BoMoC fractionated as monomers by SEC and migrated in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) consistent with predicted fusion proteins of 91 and 137 kDa, respectively (Fig. 1A). Measurement of the absorbance ratio at 260 and 280 nm suggested a lack of nucleic acid copurifying with the RT proteins. MBP tag removal from bacterially produced BoMoC reduced its stability; therefore, the RTs were used as fusion proteins for all assays.
We assayed for template jumping by using an oligonucleotide RNA-DNA primer duplex and single-stranded RNA or DNA template, partially mimicking the non-LTR retroelement RT process of TPRT (Fig. 1B). We designed primer duplexes that had a 3′ hydroxyl (OH) DNA strand with or without a 3′T overhang, which gives the primer potential to form a single base pair with a 3′A RNA template. Oligonucleotide 3′ ends other than that on the DNA primer were blocked such that only the intended primer could be elongated (SI Appendix, Table S1). BoMoC synthesized cDNA products by primer extension across at least 10 template molecules, a high apparent processivity of template jumping similar in reactions with RNA or DNA templates (Fig. 1C). In comparison to BoMoC, EuRe showed more limited template jumping and a stronger preference for using RNA as template (Fig. 1C). Additional differences in the cDNA extension products of BoMoC and EuRe arise from distinct preferences of the RTs for adding untemplated nucleotides (nt) to duplex cDNA 3′ ends (SI Appendix, Fig. S1B, and discussed in additional detail in text below).
To investigate the influence of different DNA primer 3′ overhangs on template jumping, we used primer duplexes that differed only in length of the DNA 3′ overhang. More than 2 nt of overhang was strongly inhibitory for cDNA synthesis by BoMoC, even if templates had a 3′ sequence that was fully complementary to the primer 3′ overhang (SI Appendix, Fig. S1C). We next tested how overhang sequence affected template choice by comparing use of +1C and +1T overhang primers for jumping to templates with 3′A and 3′G. The +1T primer supported cDNA synthesis from a template with 3′A but not 3′G, whereas the +1C primer supported by cDNA synthesis from 3'G and with low efficiency 3'A templates (SI Appendix, Fig. S1D). However, products from the +1C primer using a template with 3′A were slightly shorter than products from reactions using the +1T primer and the same template, indicative of the +1C overhang pairing with the template G 1 nt internal to the template 3′ end. The ability of a G-C base pair to allow for initiation slightly internal to the template 3′ end was previously noted for full-length B. mori R2 RT protein under conditions when the enzyme cannot make an appropriate base pairing of primer overhang and template 3′ end (20). From these results, we conclude that use of a primer containing a +1 overhang at least partially suppresses the use of templates with a noncomplementary 3′ end. Also, primer 3′ overhangs of >2 nt are inhibitory for template jumping. Under standard assay conditions, BoMoC preferred a 1 nt versus 2 nt overhang, which is opposite the preference of a retroviral RT (21).
Relationship between Nontemplated Nucleotide Addition and Template Jumping.
DNA-templated DNA polymerases, especially those without a 3′-5′ proofreading exonuclease activity, tend to dissociate after adding a single-nt overhang to a cDNA duplex (22–25). BoMoC, as an RNA- or DNA-templated polymerase, adds several nontemplated nt to a fully duplexed primer or product 3′ end. This can be clearly visualized in the primer-extension products 1 to 5 nt longer than the starting primer (Fig. 1 C and D and SI Appendix, Fig. S1 B and E). The addition of a typically 3 to 4 nt 3′ overhang, with some product having a 5 nt 3′ overhang, suggests that BoMoC may have even more robust nontemplated nucleotide addition (NTA) than the full-length R2 RT protein shown to add a 2 to 3 nt 3′ overhang (20). This difference could be inherent to the protein sequences or, more likely, a result of differences in enzyme purification, storage, and reaction conditions.
NTA could facilitate template jumping by creating a cDNA 3′ overhang that base pairs to a template 3′ end. On the other hand, NTA could be the consequence of aborted template jumping rather than a stimulus for it. For retroviral RTs, different studies come to different conclusions about the dNTP preference and number of nt added by NTA as well as the role of NTA in template jumping (9, 21, 26–29). For BoMoC, we first compared NTA and template-jumping activities in the presence of a 200-fold excess of each single dNTP over the other dNTPs using a blunt-end primer duplex and templates with a 3′ nt complementary to the dNTP in excess (Fig. 1D). Reactions with a high dATP concentration promoted maximal NTA, as noted with full-length R2 RT (20). Template jumping in this high dATP reaction was dramatically suppressed, yielding almost only products corresponding to primer extended by +3 and +4 NTA (Fig. 1D, lane 5). In comparison, reactions with excess dGTP allowed 2 to 3 nt of NTA and maximal template jumping (Fig. 1D, lane 6). Reactions with excess dCTP or dTTP generated products with typically 2 nt of NTA and some template jumping but less than attained in reactions with excess dGTP (Fig. 1D, lanes 7 to 8). Together, these assays do not point to a simple relationship between efficiency of NTA and template jumping. We suggest that the +2 to +5 NTA products that accumulate are inhibitory to template jumping, whereas the low level of +1 NTA product in part reflects its use for additional cDNA synthesis. From this perspective, NTA is stimulatory for template jumping, but only under conditions that slow extension of a +1 nt overhang to +2 or more nt and that favor the addition of a +1 nt overhang complementary to an intended template 3′ end.
Curiously, in reactions using primers with a +1T 3′ overhang, we observed very little NTA to extend the +1 overhang even if the same dNTP concentration induced up to 5 nt of NTA on a blunt-end primer duplex (SI Appendix, Fig. S1E; compare NTA products of the primer). The reduction of NTA using a +1T primer appeared to promote the initial template jump, particularly in reactions with equal concentrations of each dNTP (SI Appendix, Fig. S1E; note that only the first template jump would be influenced by a primer 3′ overhang). This led us to develop a strategy for ordered serial template jumping dependent on a +1T RNA-DNA duplex to capture the first template (see Ordered Two-Template Relay for cDNA Library Synthesis below).
Terminal Transferase Activity in the Presence of Manganese Ions.
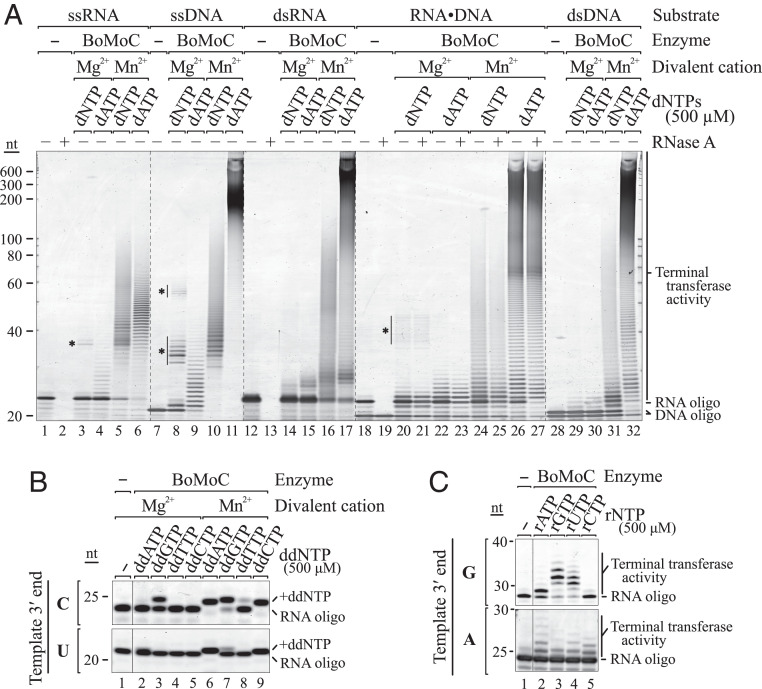
Polymerases require divalent cations for catalysis. Typically, Mg2+ functions as the cofactor under physiological conditions, but other divalent ions, including Mn2+, can support some level of DNA synthesis. To determine how BoMoC activity is influenced by use of Mn2+, we substituted Mn2+ for Mg2+ in template-jump reactions. Expected cDNA products were not detected; instead, a smear of variable-length product was observed. Surprisingly, in reactions with Mn2+, BoMoC added nontemplated nt(s) to the 3′ end of single-stranded RNAs or DNAs and also to double-stranded RNA-RNA, DNA-DNA, or RNA-DNA substrates (Fig. 2A). This type of activity is often described as terminal transferase or “tailing” activity (30, 31). Assays of EuRe for Mn2+-dependent terminal transferase activity showed it to have less tailing activity than BoMoC on double-stranded substrates relative to single-stranded substrates (SI Appendix, Fig. S2A). Previous studies have shown that full-length B. mori R2 RT can use single-stranded RNA to prime synthesis across another noncomplementary oligonucleotide or transcript (32). We observed some products in Mg2+ reactions with BoMoC and EuRe that likely arise from this nonselective priming (Fig. 2A and SI Appendix, Fig. S2A, asterisks). Cross-priming of single-stranded RNA or DNA molecules intended to be template molecules compromises the template pool by depletion and synthesis of artifact chimeric products.
Fig. 2.
BoMoC acts as a terminal transferase in the presence of Mn2+. SYBR Gold-stained denaturing PAGE gels of the terminal transferase reaction products of BoMoC are shown. (A) Input single-stranded (ss) RNA and DNA and double-stranded (ds) blunt-ended RNA, DNA, and RNA-DNA duplex were assayed in Mg2+ and Mn2+ reaction conditions in the presence of 500 μM of each dNTP or dATP alone. Products marked with asterisks indicate template copying primed by a noncomplementary oligonucleotide. (B) RNA oligonucleotide with 3′C or 3′U was assayed for extension by a single ddNTP in Mg2+ and Mn2+ conditions in the presence of 500 μM of an individual ddNTP. Single NTA in Mg2+ results from cDNA synthesis priming by a noncomplementary oligonucleotide. (C) RNA oligonucleotide with 3′G or 3′A was assayed for extension under Mn2+ conditions in the presence of 500 μM of an individual rNTP.
In Mn2+ reactions with a single dNTP, BoMoC added a dNTP-dependent length of homopolymer tract to single-stranded RNA (SI Appendix, Fig. S2B) or DNA (SI Appendix, Fig. S2C). Tailing of single-stranded RNA or DNA by incorporation of dATP was especially rampant compared to tailing with mixed dNTPs or other individual dNTPs (Fig. 2A and SI Appendix, Fig. S2 B and C). Studies of the Tf1 Schizosaccharomyces pombe LTR retroelement RT demonstrated tailing in Mn2+ specifically with dATP (33), although this activity seems much less robust than tailing by BoMoC. In single-stranded RNA or DNA tailing reactions, BoMoC had substrate preference related to the primer 3′ nt. For example, a substrate ending with 3′G generally showed compromised tailing with dCTP, whereas a template with 3′A generally showed compromised tailing with dTTP (SI Appendix, Fig. S2 B and C). We suggest that in Mn2+ reaction conditions, a single-stranded nucleic acid will preferentially bind as a template in the active site in the presence of a 3′-end complementary dNTP, whereas with a noncomplementary dNTP, it will more readily bind as primer to enable its extension by tailing.
To investigate whether BoMoC could give template nucleic acids a single shared 3′ nt, we assayed incorporation of ddNTPs in tailing reactions. Each ddNTP could be incorporated to some extent (Fig. 2B). However, as observed for tailing reactions with dNTPs, tailing with a ddNTP was influenced by both the ddNTP base and the template 3′ nt. The most efficient and general labeling was observed in reactions with ddATP and was improved for difficult substrates by lower reaction temperatures (30 °C compared to 37 °C), the presence of a crowding agent (polyethylene glycol [PEG]-8000), and increased reaction time (SI Appendix, Fig. S2D). A limited extent of tailing by ribonucleotide addition was also observed (Fig. 2C), even without an active-site mutation to remove steric hindrance on the ribose 2′ hydroxyl (34).
Ordered Two-Template Relay for cDNA Library Synthesis.
NGS libraries from sRNA are commonly prepared by sequential ligation of adaptors to input RNA 3′ and 5′ ends, often with a gel purification step after each ligation step, followed by cDNA synthesis and PCR. Ligase bias in miRNA capture is reduced by the use of degenerate sequence (“4N”) at adaptor ends (35). However, 4N protocols are time consuming in manual effort, technically challenging, and require high-input RNA amount due to many steps with product loss. We sought to exploit the serial template jumping ability of BoMoC as the basis of a ligation-independent method for end-to-end sRNA sequence capture into NGS libraries.
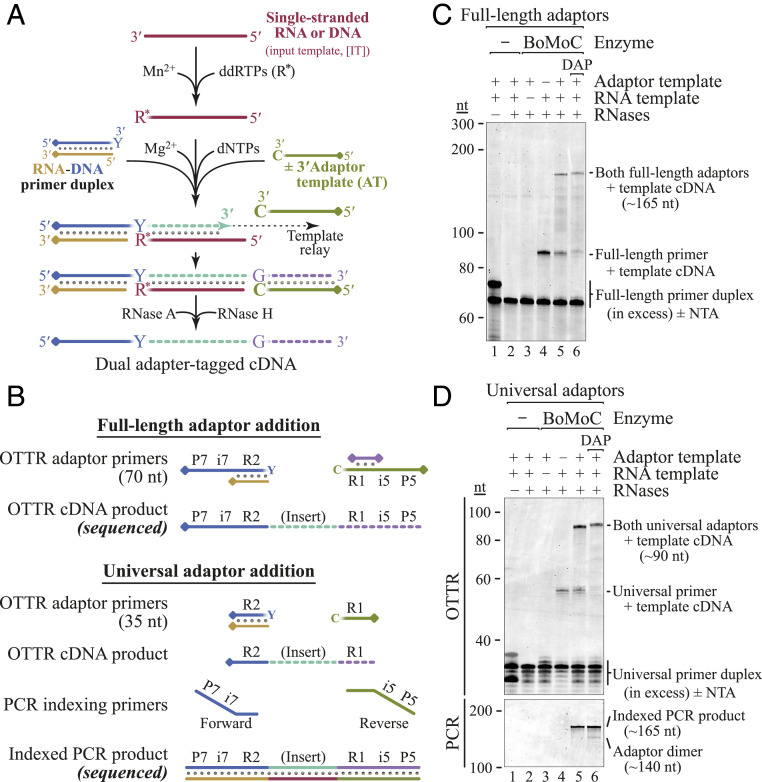
Ultimately, we developed Ordered Two-Template Relay (OTTR): a single-tube reverse transcription reaction for dual-end adaptor-tagged cDNA library synthesis (Fig. 3A and SI Appendix, Fig. S3). First, we used the terminal transferase activity of BoMoC to add a single ddRTP (ddATP and/or ddGTP) to input template (IT) RNA 3′ ends (Fig. 3A, maroon line). By utilizing primer duplex(es) with a +1Y (+1T and/or +1C) overhang (Fig. 3A, blue/tan duplex), all IT molecules could form a single base pair between template and primer 3′ ends. This strategy exploits our observation that a primer +1Y overhang is particularly resistant to additional NTA that would inactivate the primer for template jumping. In the same RT reaction, we added a template for synthesis of a single copy of cDNA 3′ adaptor. To disfavor use of this cDNA 3′ AT until after cDNA synthesis across an IT, the AT has a 3′C (Fig. 3A, green line). By manipulating dNTP concentrations and adding a dNTP analog to the reaction, we encouraged extension of the IT cDNA by a single NTA of dGTP. This gives the AT, but not IT, an ability to form a single base pair with the intermediate-stage cDNA 3′ overhang, recruiting the 3′C AT for the second template jump to complete library synthesis. If the AT has a 5′ block to additional template jumping, the desired cDNA library is produced. Adaptor dimer formation is limited by the mismatch between primer +1Y and AT 3′C. Copying of more than one molecule of input sRNA is strongly suppressed by the extremely poor use of a dYTP for NTA to the intermediate-stage cDNA and by poor elongation of a mismatched cDNA 3′G by template jumping to an IT with a 3′ ddR. Importantly, 3′ tailing of input sRNA with a nonextendable ddNTP prevents artifact generation by BoMoC use of template RNA to prime cDNA synthesis on another template RNA (36, 37), which would deplete the template pool and generate nonnative fusions.
Fig. 3.
OTTR for NGS cDNA library generation. (A) Optimized workflow for single-tube synthesis of cDNA libraries. A pool of RNA and/or DNA input molecules (maroon) is first labeled by BoMoC with 3′ ddRTP. Buffer conditions are then toggled from Mn2+ to Mg2+ and any free ddRTPs are inactivated. Next, dNTPs, oligonucleotides, and BoMoC are added to initiate cDNA synthesis from the RNA-DNA primer duplex across the IT (maroon), ending after copying the AT (green). If desired, products can then be treated with RNase A and RNase H to remove RNA, yielding the desired cDNA. The illustrated blocking groups are detailed in SI Appendix, Table S1. (B) Schematic of primers involved in Illumina Full-length (Top) or Universal (Bottom) adaptor addition and their respective cDNA library products. DNA primers were the complement of P7-i7-R2 or R2, while ATs were P5-i5-R1 or R1. In the Full-length adaptor strategy, only cDNA products elongated by copying the AT can bind to the flow cell. The covalently linked blocking group is indicated by a diamond. (C and D) Proof of principle for OTTR library generation using an RNA oligonucleotide template with Full-length (C) or Universal (D) adaptors. All reactions contained primer duplex. Only reactions containing primer duplex, RNA template, AT, and BoMoC (lanes 5 and 6) generate properly sized cDNA library product. Universal adaptor RT reactions required PCR amplification for P5 and P7 sequence fusion and indexing (D, Bottom). DAP, 2-amino-2′-deoxyadenosine triphosphate.
In the OTTR protocol, 5′ and 3′ cDNA adaptor sequences can be varied as desired. We confirmed cDNA library synthesis using both the Illumina NGS “Universal” read 1 (R1) and read 2 (R2) adaptor sequences and the “Full-length” P5-i5-R1 and P7-i7-R2 adaptor sequences (Fig. 3 and SI Appendix, Table S1). The Universal adaptor cDNA libraries were indexed by low-cycle PCR (4 to 8 cycles) with P5-i5 and P7-i7 primers, whereas the Full-length adaptor cDNA libraries were indexed by inclusion of different i5 and i7 bar codes in the 5′ cDNA primer and 3′ cDNA AT included in the RT reaction (Fig. 3B).
We optimized OTTR using RNA oligonucleotide templates. The intended dual-adaptor–flanked cDNAs were generated when all reaction components were present (Fig. 3 C and D). In our initial workflows, the yield of complete cDNA library product was diminished by accumulation of single-template-jump products (Fig. 3 C and D, lane 5), which accumulated with +2 or more nt of NTA. We found that replacement of most of the dATP in the reaction with 2-amino-2′-deoxyadenosine triphosphate (DAP) nearly eliminated the accumulation of intermediate-length cDNA products (Fig. 3 C and D, lane 6).
OTTR Outperforms Commercial Kits for miRNA Library Generation.
To evaluate OTTR for NGS library preparation, we first compared libraries generated from the miRXplore miRNA reference standard. This reference material contains a reported 963 distinct synthetic miRNAs at equimolar ratio (in fact, 962, as 2 sequences are identical). Each RNA oligonucleotide has a 5′ monophosphate and 3′ hydroxyl group, like native miRNA. Many commercially available cDNA library production kits have been evaluated using the miRXplore reference standard, enabling us to sample independently obtained data to benchmark OTTR against commercial kits (35, 38–40, SI Appendix, Table S2).
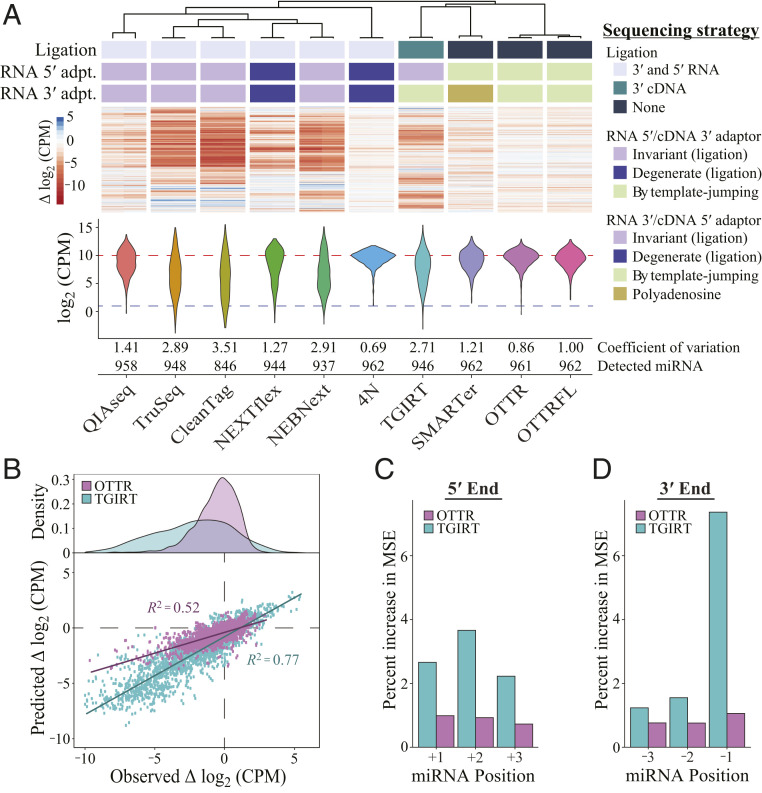
Quantitative evaluation of miRNA capture bias in the cDNA libraries of miRXplore standard was done by calculating the coefficient of variation (CV), defined as the ratio of SD to mean read counts totaled across all input sequences in a sample. If individual miRNAs have read counts far from the expected mean, the CV increases; therefore, the lower the CV, the better the library. The same read-count information can be visualized as a violin plot of miRNA read counts, with each miRNA adding to violin width or height on the vertical axis of read counts (Fig. 4A). A good library has a short and wide violin, indicating that most miRNAs had read counts close to expected. In addition to CV, read-count violin plots, and the number of miRNAs detected per fixed number of sampled sequencing reads, we evaluated bias by clustering libraries according to similarities of bias for each individual miRNA (Fig. 4A).
Fig. 4.
OTTR outperforms library generation protocols from commercially available kits. Sequencing reads other than from OTTR were sampled by downloading published datasets (35, 38–40). (A) Unsupervised hierarchical clustering of ∆log2CPM of miRNA read counts from libraries made using different protocols, with side-by-side paired technical replicates for each. ∆log2CPM = log2 (CPMexpected/CPMobserved), where CPMexpected is 962−1 × 1,000,000. The dendrogram indicates the relatedness of miRNA read-count bias. Annotations below the dendrogram indicate protocol distinctions in ligase and polymerase usage. Ligated adaptors were considered “degenerate” or “invariant” based on whether the adapter sequence had mixed-base positions. “Polyadenosine” indicates tailing of the input RNA by polyA polymerase necessary for binding of an oligothymidine RT primer. DESeq2 was used to normalize read counts for each set of replicates before conversion to log2CPM, and the distributions for combined replicates are presented as violin plots. Across the violins, the red dashed line defines the expected mean log2CPM of equimolar representation, and the blue dashed line defines the detection cutoff, which was CPM > 2. (B) Evaluation of random forest models’ predicted ∆log2CPM and observed ∆log2CPM for each miRNA based on the 5′-most (+1, +2, and +3) or 3′-most (−3, −2, −1) bases. (C and D) Percent increase in mean squared error (MSE) or relative importance for each variable of the random forest model trained on OTTR and TGIRT datasets (C: +1, +2, and +3 for the three 5′-most bases where +1 is the exact 5′ end; D: −3, −2, and −1 for the three 3′-most bases where −1 is the exact 3′ end). Variables with a higher-percent MSE are considered more important in the random forest model when predicting the log2CPM.
We randomly sampled a matched number of deposited reads from Illumina NGS libraries of the miRXplore standard and reads from two types of OTTR libraries: one with Universal cDNA adaptors and indexing by PCR (“OTTR”) and one with Full-length NGS cDNA adaptors and no PCR (“OTTRFL”). The CVs of OTTR libraries were lower than all commercial kits, and the violin plots showed more miRNAs with read counts near the expected log2-scale count per million of 10 (Fig. 4 A, Bottom). The lower quality of libraries using OTTRFL versus OTTR with PCR appears related to longer single-stranded adaptor sequence regions because OTTRFL library quality improved when some of the single-stranded region was made double-stranded by annealing oligonucleotides (Fig. 3B). Continued improvements of OTTRFL will be worthwhile for applications that benefit from PCR-free NGS library production, for example, cell-free DNA sequencing for noninvasive prenatal testing, which has nanogram amounts of input DNA and high value gained from minimal time between sample collection and sequence analysis.
The lowest CV among the sampled cDNA libraries was observed for an extensively optimized, ligation-based 4N protocol with a technically challenging workflow (35). Multiple laboratories have developed variations of the 4N protocol, but only one noncommercial 4N protocol has a published dataset using the miRXplore standard (35). The noncommercial 4N and commercial ligation-based protocols clustered in their profiles of miRNA read-count deviation from equal representation (Fig. 4 A, Top), as did OTTRFL and OTTR with PCR.
We were particularly interested in comparison of miRNA capture bias between OTTR and other protocols that use template jumping for one step of adaptor addition. TGIRT-seq (41, 42) uses a bacterial thermostable intron RT, TGIRT, for an intended single template jump to initiate cDNA synthesis (i.e., to jump “on” an IT RNA). SMARTer-seq (43) uses a modified retroviral RT for an inefficient single template jump to extend the initial cDNA by synthesis across an AT (i.e., to jump “off” the duplex of IT and its cDNA). Only OTTR exploits serial ordered template jumping to add both adaptors in a single step. The bias of TGIRT-based library generation is substantially higher than that of OTTR (Fig. 4A). Comparison of TGIRT-seq to OTTR using a scatter plot of expected versus observed individual miRNA read counts offers a granular visualization of their difference (Fig. 4B). TGIRT-seq bias appears to derive predominantly from the identity of the template 3′ nt that would engage the +1N primer overhang to support template jumping, with less bias from the template 5′ end (Fig. 4 C and D). Although the SMARTer protocol has the best CV among commercial kits, it is also outperformed by OTTR (Fig. 4A) and is compromised in utility by the loss of sRNA 3′ end information due to polyadenosine tailing prior to dT-primed cDNA synthesis.
OTTR for EV RNA-Seq.
Many categories of sRNA, including miRNA, piwi-interacting RNA, transfer RNA (tRNA)/tRNA fragments (tRFs), and Y RNAs, play important roles in the regulation of gene expression (44, 45). The profile of these RNAs in the bloodstream and other biofluids holds promise as an approach for diagnostic monitoring of human disease (46, 47). Extracellular RNAs with more than a fleeting half-life are contained within EVs, a vesicle population that includes low-density EVs released by budding from the plasma membrane and higher-density EVs released upon fusion of cytoplasmic multivesicular bodies with the plasma membrane (48).
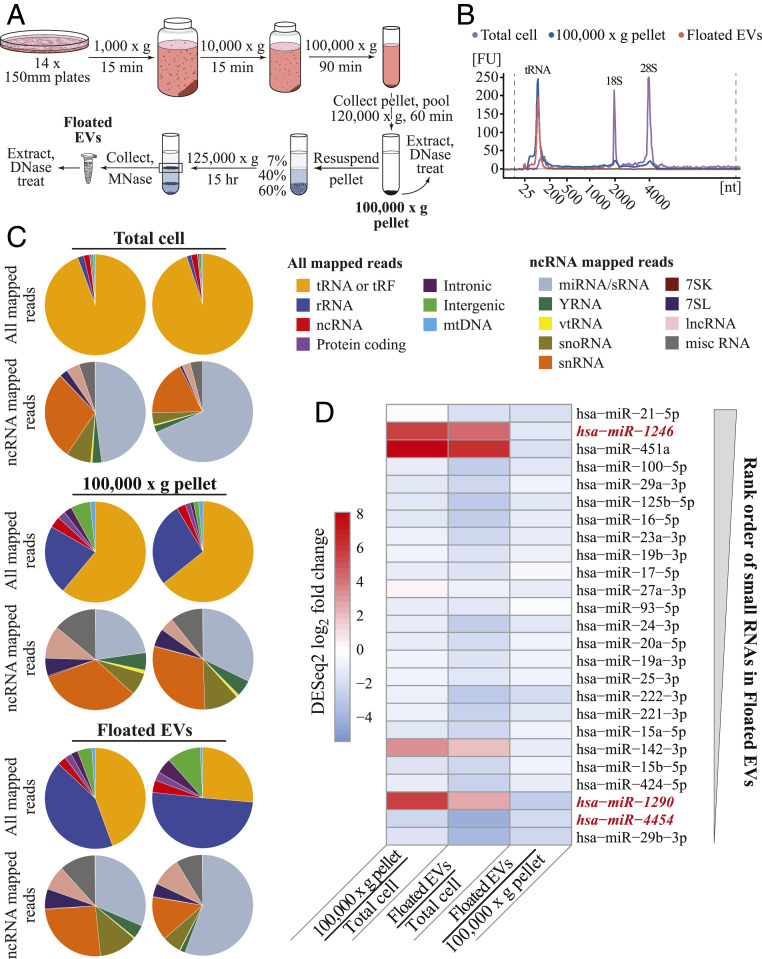
To improve community knowledge of EV sRNA inventories, we generated and sequenced OTTR cDNA libraries from EV populations (Fig. 5A). From the breast cancer–derived MDA-MB-231 cell line, EVs were sampled as crude EV preparations from conditioned medium (100,000 × g pellet containing vesicular and nonvesicular sedimentable material) and as highly purified vesicles floated in a sucrose density step gradient (Floated EVs) and treated with micrococcal nuclease before detergent lysis to remove nucleic acids not enclosed within the vesicles (42). The length profile of total cellular RNA included major peaks for 18S and 28S rRNAs and tRNAs, whereas bulk or highly purified EV RNAs had lengths predominantly of tRNA size or smaller (Fig. 5B). For sequencing comparison we used filtration to enrich total cellular sRNA of less than 200 nt prior to library generation. We also used a different human cell line, HEK 293T, to generate similarly size-enriched total cellular sRNA and floated vesicles for comparison (SI Appendix, Fig. S4).
Fig. 5.
OTTR RNA-seq inventorying of EV sRNA. (A) Schematic of EV purification. (B) Agilent Bioanalyzer RNA traces for cellular RNA (purple), the 100,000 × g pellet (blue), and Floated EVs (peach). Peaks corresponding to tRNA and 18S and 28S rRNA are indicated. (C) Pie charts of mapped read assignments of MDA-MB-231 sRNA libraries from two biological replicates. tRAX and miRDeep2 were used to map tRNA and miRNA reads, respectively. rRNA, ncRNA, and protein-coding reads aligned to annotated transcripts or genomic ncRNA loci. Intronic, intergenic, and mitochondrial (mt) DNA reads mapped in corresponding locations. Among ncRNA reads, vt is vault and miscRNA includes all ncRNA not split out into other pie slices. (D) EV miRNA enrichment in MDA-MB-231 based on DESeq2 log2 fold change estimates between, as pairwise combinations, 100,000 × g pellet and Total cell, Floated EVs and Total cell, and Floated EVs and 100,000 × g pellet. The 25 miRBase sRNAs included in the panel are the most abundant in read count in MDA-MB-231 Floated EVs, with most abundant of the 25 at top as schematized by the thicker end of the gray wedge. The sequences not considered bona fide miRNA based on miRGeneDB are in red italic. The sequence of miRBase hsa-mir-142 corresponds to miRGeneDB hsa-mir-142-v3.
Isolated RNA pools were used directly, without gel purification, for OTTR cDNA library synthesis and sequencing. Unsurprisingly, total cellular sRNA and EV library reads were dominated by tRNAs or tRFs and rRNA fragments, as evident from pie charts comparing RNA species across all mapped reads and EV-enriched populations (Fig. 5C and SI Appendix, Fig. S4A and Table S3). Reads from full-length tRNAs as well as tRFs had genome-mismatched nt at expected positions of posttranscriptional modification (SI Appendix, Fig. S5).
To evaluate ncRNA representation in more detail, we split the non-tRNA, non-rRNA part of ncRNA into its own set of pie slices (Fig. 5C and SI Appendix, Fig. S4A and Table S3). Among ncRNA categories, RNAs annotated in miRBase were readily detectable in all samples (gray slices of ncRNA read pies, Fig. 5C and SI Appendix, Fig. S4A). The miRBase inventory includes some RNAs that do not appear to be authentic miRNAs (49). These RNAs, together with the more-confidently identified miRNAs, we classify as sRNA. In general, from rank-ordered abundance of floated EV sequence read counts (Fig. 5D and SI Appendix, Fig. S4 B and C), relative read-count abundance of individual sRNA differed across cell types but generally EV enrichment of a particular sRNA did not. EV samples also contained a sizeable read count from fragments of small nuclear RNA (snRNA), small nucleolar RNA, long ncRNA, and 7SL RNA (ncRNA read pies, Fig. 5C and SI Appendix, Fig. S4A and Table S3).
Most of the relatively abundant EV RNA with sequences mapped to miRBase are as well or better represented in total cellular sRNA than in EVs (Fig. 5D and SI Appendix, Fig. S4 B and C; see SI Appendix, Fig. S6 for read mapping of representative miR-16_5p). We detected EV enrichment of some validated miRNA in both cell types, such as miR-142-3p (Fig. 5D and SI Appendix, Figs. S4B and S6), consistent with previous studies (42). However, the two most abundant EV-enriched sRNAs for both cell types, miR-1246 and miR-451a (Fig. 5D and SI Appendix, Fig. S4 B and C), have noncanonical biogenesis pathways: miR-451a precursor is cleaved by Drosha, then Ago2, then exonuclease(s) (50), and miR-1246 is produced at high levels in cancer cells by degradation of U2 snRNA in an unknown, Drosha-independent, and Dicer-independent manner (51). Mapped reads for these two sRNA have relatively dispersed 5′ and/or 3′ end positions (SI Appendix, Fig. S6). Both miR-451a and miR-1246 are candidate cancer biomarkers (52). Overall, we suggest that NGS libraries that oblige defined end-to-end sequence capture of entire sRNA pools will be useful for appreciating differences in the precision of RNA processing and for EV RNA diagnostics.
Discussion
Here, we describe an approach of controlled serial template jumping to synthesize a cDNA library that captures input sRNA sequences end to end. Our approach uses cDNA synthesis to fuse 5′ and 3′ adaptor sequences. Discontinuous templates produce a continuous cDNA with three segments in specific order: the 5′ adaptor primer, the complement of a single template from the input RNA pool, and the 3′ adaptor. To enforce this order of template copying, the primer has a 3′ pyrimidine single-nt overhang, the input pool of templates has a 3′ purine nt, and the 3′ AT has a 3′ cytidine (Fig. 3A). The comprehensiveness of sRNA sequence capture by OTTR relies on an unanticipated ability of nonretroviral RTs to act as robust terminal transferase enzymes in reactions with Mn2+ as the divalent ion. BoMoC terminal transferase activity may lend itself to the development of standalone protocols for 3′-end labeling of single- and double-stranded RNA and DNA.
In addition to the simplicity and low bias of OTTR for NGS library production, the OTTR strategy has additional benefits. First, ddNTP labeling of the input pool 3′ ends precludes templates from the self-priming and cross-priming that create aberrant cDNA fusions (36, 37). Second, the use of serial template jumps to add flanking adaptors obliges end-to-end copying of ITs. B. mori R2 RT can template jump only when synthesis reaches the 5′ end of an engaged template, as enzyme paused midtemplate does not support template jumping (20), so partial cDNAs would drop out of the library due to lack of a 3′ adaptor. Third, OTTR requires much less input than used in other protocols of precise end-to-end RNA capture, for example, in previous TGIRT-seq of similar preparations of EV RNA (42). Adaptation of this protocol to labor-intensive sRNA sequencing workflows, like those for ribosome profiling, could motivate more researchers to attempt RNA-seq library production.
It is general consensus in the EV field that better RNA-seq is needed, despite hundreds of millions of dollars invested in EV RNA surveys to date (53–55). The gold-standard 4N method has relatively low bias. We note that the full diversity of noncommercial 4N protocols is not evaluated in this work, and some 4N methods could have lower bias than the one benchmarked with miRXplore. However, home-brew 4N methods are challenging for nonexperts to perform, labor intensive, and not efficient in conversion of input RNA to cDNA. OTTR overcomes all of these barriers, and the simplicity of the protocol makes it amenable to future automation. Even minor changes in enzyme storage buffer tested subsequent to this work further decrease the bias of OTTR libraries (SI Appendix, Fig. S3). Sequence information gained from our OTTR EV RNA libraries adds weight to previous conclusions that EVs contain distinctly processed fragments of larger RNAs (14, 54, 56), here established to be preexisting fragments by the need for template jumping to add 5′ and 3′ adaptors. The relative simplicity and low input requirement of the OTTR protocol, combined with future automation, can provide a necessary research and clinical tool for distinguishing sRNA differences in normal and cancer cells and applying the information gained to high-throughput diagnostics using liquid biopsies.
Materials and Methods
Recombinant Protein Expression and Purification.
Codon-optimized ORFs for EuRe and BoMoC RT proteins were ordered from GenScript. Proteins were produced in Escherichia coli by expression from MacroLab vector 2bct with a C-terminal 6xHis tag (https://qb3.berkeley.edu/facility/qb3-macrolab/) modified to include an N-terminal MBP tag. Cells were grown in 2xYT medium using Rosetta2(DE3)pLysS cells and induced at OD600 0.9 at 16 °C overnight with 0.5 mM isopropylthio-β-galactoside (IPTG). Lysis of the induced cell pellet took place in 20 mM Tris⋅HCl pH 7.4, 1 M NaCl, 10% glycerol, 1 mM MgCl2, 1 mM β-mercaptoethanol (BME), and protease inhibitors by sonication on ice for 3.5 min (10 s on, 10 s off). Three-step purification was initiated by column binding 6xHis-tagged proteins on nickel agarose. Following binding, the column was washed with 5 volumes of 20 mM Tris⋅HCl pH 7.4, 1 M KCl, 20 mM imidazole, 10% glycerol, and 1 mM BME. Elution from the resin proceeded with 20 mM Tris⋅HCl pH 7.4, 1 M KCl, 400 mM imidazole, 10% glycerol, and 1 mM BME. Protein eluted from the Ni column was desalted into heparin buffer A (5 mM Hepes-KOH pH 7.4, 400 mM KCl, 2% glycerol, 0.2 mM dithiothreitol [DTT]) and then bound to heparin-Sepharose. The column was washed with 5 column volumes heparin buffer A and then ramped up by gradient over 15 column volumes to 20 mM Hepes-KOH pH 7.4, 2 M KCl, 10% glycerol, and 1 mM DTT. Eluted protein peak was pooled and diluted back to 20 mM Hepes-KOH pH 7.4, 400 mM KCl, 10% glycerol, and 1 mM DTT. Protein was size-fractionated using a HiPrep 16/60 Sephacryl S-200HR in 25 mM Hepes-KOH pH 7.4, 0.8 M KCl, 10% glycerol, and 1 mM DTT. Purified protein concentration was determined by ultraviolet absorbance using the calculated extinction coefficient (EuRe: 1.325 M−1/cm−1, BoMoC: 1.350 M−1/cm−1) and validated as homogeneous full-length RT fusion protein by SDS-PAGE. Final protein concentrations stored at −80 °C were 4.1 mg/mL for EuRe and 8.0 mg/mL for BoMoC. Working stock of protein was diluted ∼5 fold in 25 mM Hepes-KOH, 0.8 M KCl, 50% glycerol, and 1 mM DTT then moved to −20 °C.
RT Assay Conditions.
Templated cDNA synthesis and NTA assays were carried out under the following conditions: 25 mM Tris⋅HCl pH 7.5, 75 to 150 mM KCl, 1 to 5 mM MgCl2, 5 mM DTT, 2.5% glycerol, 90 to 400 nM RNA-DNA duplex, 45 nM to 4 μM RNA or DNA template, 2.5 nM to 500 μM dNTP, and 0.5 to 2 μM enzyme with or without 2% PEG-6000. Samples were incubated for 30 min to 2 h at 37 °C, heat inactivated at 65 °C for 5 min, nuclease treated with 0.5 μg/μL RNase A (Sigma, R6513), and 0.5 units of thermostable RNase H (NEB, M0523S), then stopped with 10 mM Tris pH 8.0, 0.5 mM ethylenediaminetetraacetic acid, and 0.1% SDS. Products were extracted with phenol:chloroform:isoamyl alcohol (PCI; 25:24:1), ethanol precipitated using 10 μg glycogen as a carrier, and air dried for 5 min prior to resuspending in 5 μL H2O. Products were separated by 7.5% to 15% denaturing urea-PAGE gel then stained using SYBR Gold and imaged by Typhoon Trio.
Terminal Transferase Assay Conditions.
Terminal transferase activity assays were performed under conditions similar to those described previously except for the presence of MnCl2 rather than MgCl2. Briefly, assays contained 25 mM Tris⋅HCl pH 7.5, 75 to 150 mM KCl, 5 mM MnCl2, 5 mM DTT, 2.5% glycerol, 400 nM RNA-RNA, RNA-DNA, or DNA-DNA duplex or single-stranded RNA or DNA template, 500 μM dNTP/NTP/ddNTP, and 0.5 μM enzyme with or without 5% PEG-8000. Samples were incubated for 30 min at 30 to 37 °C, heat inactivated at 65 °C for 5 min, and processed as discussed previously with or without the post-RT nuclease treatment step.
OTTR Library Generation and Sequencing.
Input RNA (10 ng) from the miRXplore Universal Reference Standard (Miltenyi Biotech, 130-094-407), EV sRNA, or mirVana size-selected total cellular RNA was diluted into 20 mM Tris⋅HCl pH 7.5, 150 mM KCl, 0.5 mM DTT, 5% PEG-8000, 2 mM MnCl2, 250 μM ddATP (±250 μM ddGTP), and 0.7 μM BoMoC and then incubated for 1.5 to 2 h at 30 °C. For ddGTP chase of initial labeling with ddATP, the reaction was allowed to proceed for 1.5 h at 30 °C with ddATP only and then chased with 250 μM ddGTP and incubated for another 30 min at 30 °C. The reaction was stopped by incubating at 65 °C for 5 min followed by addition of 5 mM MgCl2 and 0.5 units of shrimp alkaline phosphatase (NEB, M0371S). The phosphatase reaction was incubated at 37 °C for 15 min, stopped by addition of 5 mM EGTA, then incubated at 65 °C for 5 min. Subsequently, buffers were added to give an additional 0.5 mM MgCl2 and 45 mM KCl plus 2% PEG-6000, 200 μM dGTP, 40 μM dTTP and dCTP, 2 μM dATP, 150 μM DAP, 90 nM RNA-DNA primer duplex with +1T and +1C overhangs, 180 nM terminating AT, and 0.5 μM BoMoC. Samples were processed as previously discussed and then resuspended in 10 μL H2O. If Universal adaptors were used in the RT reaction, libraries were generated using 5 μL of the purified RT reaction product and 4 to 8 cycles of PCR with Q5 high fidelity polymerase (NEB, M0491S). PCR products were column purified and separated by 7.5% urea-PAGE to remove adaptor dimer, and the desired product was isolated by diffusion overnight at 37 °C followed by PCI extraction, ethanol precipitation, and resuspension in 10 μL H2O. If Full-length adaptors were used, the RT reaction was treated with RNase A and RNase H prior to cleanup, and no PCR amplification was performed. Quantification of libraries prior to sequencing used qPCR with primers specific to the Illumina P5 and P7 adaptor sequences and standards from the NEBNext Library Quant Kit (NEB, E7630S). Sequencing of prepared libraries was performed using an Illumina MiniSeq with the 75-cycle high-output kit. Library yield using Universal adaptors ranged from 20 to 30 nM following four cycles of PCR for the miRXplore reference standard and from 6 to 10 nM with more complex input pools (total cellular sRNA and EV RNA) following four cycles of PCR. Yield using Full-length adaptors with the miRXplore reference standard was ∼1 nM. To recover 10 M reads for total cellular and EV RNA-seq, ∼1.5% of each library was taken to produce the starting 1 nM sample pool.
The SI Appendix, Supplemental Methods provides full details of read mapping and bioinformatics analysis as well as EV purification. Sequence reads can be accessed with the BioProject ID PRJNA726257.
Acknowledgments
H.E.U., L.F., S.C.P., and K.C. were supported by NIH Grants R35 GM130315 and DP1 HL156819, as well as the Bakar Fellows Program (to K.C.) and NIH Grant T32 GM007232 (to L.F.). M.M.T.-D., X.-M.L., and R.S. were supported by the Howard Hughes Medical Institute.
Footnotes
Competing interest statement: H.E.U., L.F., S.C.P., and K.C. are named inventors on University of California, Berkeley patent applications describing OTTR technology. H.E.U. and K.C. are founders of Karnateq, Inc., which licensed this technology.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107900118/-/DCSupplemental.
Data Availability
RNA sequence data have been deposited in GenBank as BioProject ID PRJNA726257.
References
- 1.Lander E. S.et al.; International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Eickbush T. H., Jamburuthugoda V. K., The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 134, 221–234 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sultana T., Zamborlini A., Cristofari G., Lesage P., Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 18, 292–308 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Eickbush T. H., Eickbush D. G., Integration, regulation, and long-term stability of R2 retrotransposons. Microbiol. Spect. 3, MDNA3-0011-2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiwara H., Site-specific non-LTR retrotransposons. Microbiol. Spect. 3, MDNA3-0001-2014 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Luan D. D., Eickbush T. H., RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 15, 3882–3891 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambowitz A. M., Belfort M., Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbiol. Spect. 3, MDNA3-0050-2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y. Y., Machleder E. M., Chenchik A., Li R., Siebert P. D., Reverse transcriptase template switching: A SMART approach for full-length cDNA library construction. Biotechniques 30, 892–897 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Wulf M. G., et al., Non-templated addition and template switching by Moloney murine leukemia virus (MMLV)-based reverse transcriptases co-occur and compete with each other. J. Biol. Chem. 294, 18220–18231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Negroni M., Buc H., Retroviral recombination: What drives the switch? Nat. Rev. Mol. Cell Biol. 2, 151–155 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Boivin V., et al., Reducing the structure bias of RNA-seq reveals a large number of non-annotated non-coding RNA. Nucleic Acids Res. 48, 2271–2286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozsolak F., Milos P. M., RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stark R., Grzelak M., Hadfield J., RNA sequencing: The teenage years. Nat. Rev. Genet. 20, 631–656 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Abramowicz A., Story M. D., The long and short of it: The emerging roles of non-coding RNA in small extracellular vesicles. Cancers (Basel) 12, 1445 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo L. T., et al., Sequencing and structure probing of long RNAs using MarathonRT: A next-generation reverse transcriptase. J. Mol. Biol. 432, 3338–3352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao C., Liu F., Pyle A. M., An ultraprocessive, accurate reverse transcriptase encoded by a metazoan group II intron. RNA 24, 183–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katibah G. E., et al., Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc. Natl. Acad. Sci. U.S.A. 111, 12025–12030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Malik H. S., Eickbush T. H., Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. U.S.A. 96, 7847–7852 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao C., Pyle A. M., Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat. Struct. Mol. Biol. 23, 558–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibillo A., Eickbush T. H., End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J. Biol. Chem. 279, 14945–14953 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Oz-Gleenberg I., Herschhorn A., Hizi A., Reverse transcriptases can clamp together nucleic acids strands with two complementary bases at their 3′-termini for initiating DNA synthesis. Nucleic Acids Res. 39, 1042–1053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark J. M., Joyce C. M., Beardsley G. P., Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J. Mol. Biol. 198, 123–127 (1987). [DOI] [PubMed] [Google Scholar]
- 23.Holton T. A., Graham M. W., A simple and efficient method for direct cloning of PCR products using ddT-tailed vectors. Nucleic Acids Res. 19, 1156 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchuk D., Drumm M., Saulino A., Collins F. S., Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 19, 1154–1154 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark J. M., Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 16, 9677–9686 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golinelli M.-P., Hughes S. H., Nontemplated base addition by HIV-1 RT can induce nonspecific strand transfer in vitro. Virology 294, 122–134 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Patel P. H., Preston B. D., Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc. Natl. Acad. Sci. U.S.A. 91, 549–553 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohtsubo Y., Nagata Y., Tsuda M., Efficient N-tailing of blunt DNA ends by Moloney murine leukemia virus reverse transcriptase. Sci. Rep. 7, 41769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luczkowiak J., Matamoros T., Menéndez-Arias L., Template-primer binding affinity and RNase H cleavage specificity contribute to the strand transfer efficiency of HIV-1 reverse transcriptase. J. Biol. Chem. 293, 13351–13363 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin G., Keller W., RNA-specific ribonucleotidyl transferases. RNA 13, 1834–1849 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motea E. A., Berdis A. J., Terminal deoxynucleotidyl transferase: The story of a misguided DNA polymerase. Biochim. Biophys. Acta 1804, 1151–1166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bibiłło A., Eickbush T. H., The reverse transcriptase of the R2 non-LTR retrotransposon: Continuous synthesis of cDNA on non-continuous RNA templates. J. Mol. Biol. 316, 459–473 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Oz-Gleenberg I., Herzig E., Hizi A., Template-independent DNA synthesis activity associated with the reverse transcriptase of the long terminal repeat retrotransposon Tf1. FEBS J. 279, 142–153 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Brown J. A., Suo Z., Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry 50, 1135–1142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giraldez M. D., et al., Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 36, 746–757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaushik N., et al., Role of glutamine 151 of human immunodeficiency virus type-1 reverse transcriptase in substrate selection as assessed by site-directed mutagenesis. Biochemistry 39, 2912–2920 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Bibillo A., Eickbush T. H., High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J. Biol. Chem. 277, 34836–34845 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Coenen-Stass A. M. L., et al., Evaluation of methodologies for microRNA biomarker detection by next generation sequencing. RNA Biol. 15, 1133–1145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shore S., et al., Small RNA library preparation method for next-generation sequencing using chemical modifications to prevent adapter dimer formation. PLoS One 11, e0167009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H., Yao J., Wu D. C., Lambowitz A. M., Improved TGIRT-seq methods for comprehensive transcriptome profiling with decreased adapter dimer formation and bias correction. Sci. Rep. 9, 7953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohr S., et al., Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA 19, 958–970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Temoche-Diaz M. M., et al., Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. eLife 8, e47544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Everaert C., et al., Performance assessment of total RNA sequencing of human biofluids and extracellular vesicles. Sci. Rep. 9, 17574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris K. V., Mattick J. S., The rise of regulatory RNA. Nat. Rev. Genet. 15, 423–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishna S., Raghavan S., DasGupta R., Palakodeti D., tRNA-derived fragments (tRFs): Establishing their turf in post-transcriptional gene regulation. Cell. Mol. Life Sci. 78, 2607–2619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin C.-P., He L., Noncoding RNAs in cancer development. Annu. Rev. Cancer Biol. 1, 163–184 (2017). [Google Scholar]
- 47.Hulstaert E., et al., Charting extracellular transcriptomes in the human biofluid RNA atlas. Cell Rep. 33, 108552 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Shurtleff M. J., Temoche-Diaz M. M., Schekman R., Extracellular vesicles and cancer: Caveat lector. Annu. Rev. Cancer Biol. 2, 395–411 (2018). [Google Scholar]
- 49.Fromm B., et al., MirGeneDB 2.0: The metazoan microRNA complement. Nucleic Acids Res. 48, D132–D141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kretov D. A., et al., Ago2-dependent processing allows miR-451 to evade the global microRNA turnover elicited during erythropoiesis. Mol. Cell 78, 317–328.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y.-F., Hannafon B. N., Khatri U., Gin A., Ding W.-Q., The origin of exosomal miR-1246 in human cancer cells. RNA Biol. 16, 770–784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J., Guo H., Yang Y., Zhang Y., Liu H., A meta-analysis on the prognosis of exosomal miRNAs in all solid tumor patients. Medicine (Baltimore) 98, e15335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veziroglu E. M., Mias G. I., Characterizing extracellular vesicles and their diverse RNA contents. Front. Genet. 11, 700 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan S., et al., Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell 177, 446–462.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tosar J. P., Cayota A., Extracellular tRNAs and tRNA-derived fragments. RNA Biol. 17, 1149–1167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidal M., Exosomes: Revisiting their role as “garbage bags”. Traffic 20, 815–828 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequence data have been deposited in GenBank as BioProject ID PRJNA726257.