Significance
Application scope, underlying mechanisms, and potential limitation of AAV/CRISPR-mediated hepatic gene editing remain unexplored. Here we report that a synthetic enhancer/promoter (P2) incorporation empowered AAV/CRISPR to restore FIX-encoding capacity of a severe F9 defect involving the regulatory region. Systemic analyses revealed a critical role of host cell heterogenicity in determining the therapeutic benefit of this regimen, wherein a subclinical inflammation posttreatment regulated the P2 activity and the retention of the edited F9 alleles in a hepatocytic subset–dependent manner. Moreover, preeminent hepatic presence of hematopoietic cells implicated their involvement in fueling and restricting this hepatic inflammation. Collectively, these results characterize the critical factors determining the efficacy and durability of AAV/CRIPSR-mediated gene editing in dealing with complicated hepatic monogenic disorders.
Keywords: gene therapy, hemophilia B, AAV/CRISPR, liver immunity
Abstract
Infusing CRISPR/donor-loaded adeno-associated viral vectors (AAV/CRISPR) could enable in vivo hepatic gene editing to remedy hemophilia B (HB) with inherited deficiency of clotting factor IX (FIX). Yet, current regimens focus on correcting HB with simple mutations in the coding region of the F9, overlooking those carrying complicated mutations involving the regulatory region. Moreover, a possible adverse effect of treatment-related inflammation remains unaddressed. Here we report that a single DNA cutting-mediated long-range replacement restored the FIX-encoding function of a mutant F9 (mF9) carrying both regulatory and coding defects in a severe mouse HB model, wherein incorporation of a synthetic Alb enhancer/promoter-mimic (P2) ensured FIX elevation to clinically meaningful levels. Through single-cell RNA sequencing (scRNA-seq) of liver tissues, we revealed that a subclinical hepatic inflammation post-AAV/CRISPR administration regulated the vulnerability of the edited mF9-harboring host cells to cytotoxic T lymphocytes (CTLs) and the P2 activity in a hepatocytic subset–dependent manner via modulating specific sets of liver-enriched transcription factors (LETFs). Collectively, our study establishes an AAV/CRISPR-mediated gene-editing protocol applicable to complicated monogenetic disorders, underscoring the potentiality of improving therapeutic benefits through managing inflammation.
Various types of genetic defects occurring in the F9 locus in the X chromosome cause the failure of hepatocytes to produce coagulation factor IX (FIX), which leads to hemophilia B (HB). HB manifests crippling bleeding diathesis that severely affects the development and life quality of patients and is a representative monogenic disorder being intensively tested for gene therapy (1, 2). Current clinical trials favor a gene augmentation strategy in which an episomal FIX-expressing cassette is delivered into hepatocytes through recombinant adeno-associated virus vector (AAV). The general structure of the vector includes a piece of particular sequence, designated as the inverted terminal repeats (ITRs), that flanks the FIX-expressing cassette at both ends. Discrete serotypes of AAV have different tissue-specific tropisms, mainly determined by their capsids. Through AAV2 or AAV8 serotype of vectors targeting liver tissue, a large number of AAV genomes containing a promoter-F9 cDNA cassette were delivered into individual hepatocytes wherein they produce F9 mRNA (3, 4). Nevertheless, due to an inherent instability of these episomal AAVs in host cells, their expression might decline toward subtherapeutic levels (5–7).
Alternatively, a gene-editing regimen in which the encoding function of the mutant F9 (mF9) of hepatocytes is restored through in situ genomic editing is expected to produce FIX in a more durable manner. In this regard, CRISPR-Cas9 represents an effective sequence-specific genomic editing tool that comprises a target sequence-pairing simple guide RNA (sgRNA) and a DNA-cutting nuclease Cas9. The resultant DNA double strand breaks (DSBs) would stimulate the endogenous DNA repair mechanisms including nonhomologous end joining (NHEJ), homology-directed recombination (HDR), or homology-independent targeted insertion (HITI) (8, 9). NHEJ, in which the two ends of DSBs are rejoined with random insertion or deletion of a few bases (indel), is often used for knocking out certain target genes, whereas HDR or HITI, with the availability of exogenous donor template containing two flanking sequences homologous to the genomic sequences surrounding the DSBs, namely, left homology arm (LHA) and right homology arm (RHA), underlies the targeted genomic replacement or insertion usually in a much lower efficiency than the NHEJ. Employing a mouse HB caused by a mF9 harboring a 12-bp deletion within exon 8, previous studies demonstrated the feasibility of the AAV/CRISPR strategy codelivering an intron1-cutting SaCas9 (Staphylococcus aureus Cas9)/sgRNA and a donor containing exons 2 to 8 of F9 cDNA in ameliorating the associated coagulopathy (8, 9). DNA sequencing indicated an expected precise insertion of exons 2 to 8 of F9 cDNA into intron 1 of mF9 via HITI and HDR (8, 9).
Nevertheless, several issues remain to be addressed, which could potentially hinder the therapeutic application of AAV/CRISPR-mediated in vivo gene editing. Firstly, the genetic defects of mF9 in HB patients present as diverse forms that scatter across its exons 1 to 8 and the 5′ or 3′ regulatory region (1, 10), and it is unknown whether the AAV/CRISPR delivery would efficiently rectify the HB coagulopathy caused by more complicated genetic defects involving enhancer/promoters. Secondly, the exact major liver cell type(s) and DNA recombination mechanisms responsible for producing the functional FIX mRNA after gene editing are unclear. Thirdly, although AAV itself and transgene expression in hepatocytes have long been regarded as nonpathogenic and low immunogenic (11–14), growing evidence accumulated that therapeutic AAV infusion into even unprimed recipients, with or without CRISPR codelivery, would provoke hepatic inflammation (15–17). Whether this inflammation would undermine the therapeutic benefits is largely unexplored. In this study, we demonstrated the efficacy and safety of a specific AAV/CRISPR protocol employing synthetic enhancer/promoter integration and HDR-mediated replacement of large DNA sequences in dealing with a severe HB model carrying a complex F9 defect. Moreover, a systemic analysis based on single-cell RNA sequencing (scRNA-seq) of liver tissues characterized previously unappreciated negative effects of the AAV/CRISPR infusion-caused subclinical inflammation on the expression and retention of the edited mF9 alleles in a hepatocyte subset–dependent manner.
Results
HDR-Edited P2 Integration in a Severe HB Model Ensures a Therapeutic FIX Elevation.
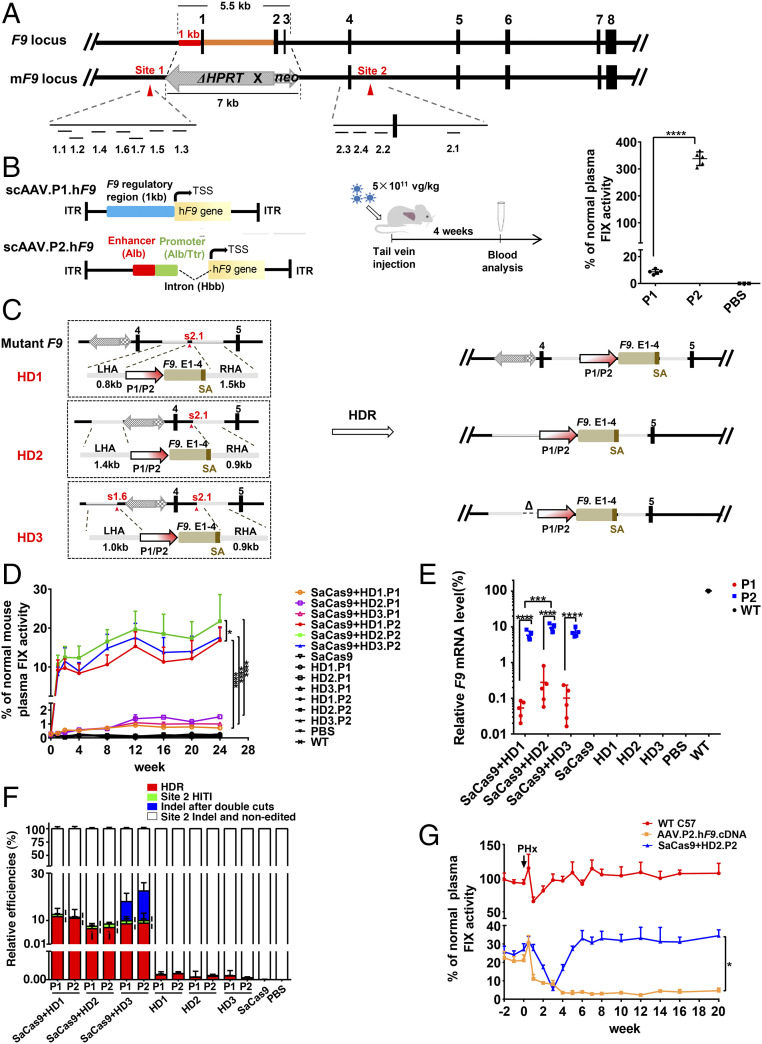
To screen out a gene-editing strategy for the HB patients carrying complicated mutations, we choose a severe HB mouse model, in which the genomic region covering a 1-kb regulatory region upstream of the transcription start site (TSS) and ∼4.5-kb 5′ coding region stretching into intron 3 of wild-type (WT) F9 was replaced by a ∼7-kb foreign neoΔHPRT as the selection marker in the creation of this disease model (Fig. 1A). Firstly, the regions upstream (site 1) and downstream (site 2) of neoΔHPRT were sought for the potential sgRNA targets (Fig. 1A). Initial T7EI enzymatic cutting analysis of cells in vitro showed that five site 1–targeting sgRNAs and four site 2–targeting sgRNAs were able to induce an indel rate of over 35% (SI Appendix, Fig. S1A). Further indel rate analysis of liver in vivo pointed to sgRNAs 1.6, and sgRNA 2.1 as the potentially effective ones (SI Appendix, Fig. S1 B–D).
Fig. 1.
HDR-mediated P2 integration and neoΔHPRT removal ensure a stably therapeutic FIX elevation in a severe HB model. (A) Genetic structures of WT F9 and mF9. The dashes depict the locations of 11 candidate sgRNAs of target sites. (B) Structures of AAV.P1.hF9 cDNA and AAV.P2.hF9 cDNA vectors. Middle shows the flowchart of experiments (5.0 ×1011 vg/kg). The plasma FIX levels in P1, P2, and PBS groups are shown in the Right graph (P1 and P2 groups, n = 5; PBS group, n = 3). (C) The Left part of the diagram depicts three protocols of HDR (HD1–HD3). The Right part illustrates the expected edited forms of mF9 post-HDR. (D) Plasma FIX activity (n = 6) was monitored over 24 wk after AAV infusion (donor-sgRNA: 2.0 × 1014 vg/kg; SaCas9: 4.0 × 1013 vg/kg). (E) The relative F9 mRNA levels in each group as indicated were evaluated by RT-qPCR (n = 5). (F) The primer locations that discriminate four groups of mF9 alleles from each other using qPCR (n = 5). P values (Ì) for differences between HD1.P1/2 and HD2.P1/2 HDR rates were <0.01, and the P values (¦) for the differences between the HDR rates and the site 2 HITI rates in the same groups were <0.001. (G) HB mice were intravenously injected with AAV.P2.hF9.cDNA alone (1.0 ×1011 vg/kg) or with dual AAVs with HD2.P2 (HD2.P2: 2.0 × 1014 vg/kg, SaCas9: 4.0 × 1013 vg/kg). After 32 wk of observation of plasma FIX activity, a PHx was performed on these two groups of HB mice and a group of age-matched WT mice (n = 3). The plasma FIX activity was monitored at time points as indicated. Data are presented as mean ± SEM, *P < 0.05, ***P < 0.001, ****P < 0.0001. vg/kg, vector genomes per kilogram.
The missing native sequence in this mF9 contained not only the 1 kb of regulatory sequence at its 5′ end (namely P1) but also an intron 1–embedded enhancer at its 3′ end following exon 1 (18), whose whole size is too large to be packaged by an AAV vector. To restore its FIX-encoding function, we considered substituting these two missing regulatory elements with a compact (∼0.6 kb) synthetic enhancer/promoter (namely, P2) comprising several regulatory elements extracted from the genes being highly expressed in liver, such as an albumin (Alb)-like enhancer element, a chimeric Alb/transthyretin (Ttr) promoter, and a hemoglobin subunit beta (Hbb) intron (Fig. 1B) (19). To test the transcriptional efficiency of P2, we infused hepatocyte-tropism AAV8 particles packed with either the P1-directed hF9 cDNA or the P2-directed hF9 cDNA expressional cassette into HB mice. The plasma FIX levels measured 4 wk later in the P2-F9 group were ∼30-fold higher than those detected in the P1-F9 group (Fig. 1B), thus verifying a robust transcription-activating ability of the P2 within hepatocytes.
Considering that the residual defective fragment may affect the transcription of the edited mF9 if the genetic repair was done simply through a targeted insertion (Fig. 1C, HD1) (8, 9), we also designed a strategy of genetic replacement (Fig. 1C, HD2). Since a previous study showed that double DNA cuttings might facilitate the HDR-mediated replacement of large fragments (20), a parallel protocol based on double DNA cuttings was included (Fig. 1C, HD3). Thus, with s2.1 and s1.6 as the chosen sgRNA(s), three types of donors (HD1 to 3) were designed in which a P1- or P2-coupled F9 exons 1 to 4 (P1-E1-4 or P2-E1-4) was flanked by three pairs of RHAs and LHAs (Fig. 1C). With a single s2.1 cutting, the HD1 simply mediated a targeted insertion of the P1-E1-4 or P2-E1-4 within intron 4 and the foreign neoΔHPRT remained postedition. The HD2 was expected to mediate a replacement of the ∼17.5-kb neoΔHPRT-containing genomic region by the P1-E1-4 or P2-E1-4. In the case of HD3, employment of double gRNA-mediated double DNA cuttings would result in a replacement slightly different from that made in HD2, in which a 0.4-kb native regulatory region immediately upstream of P1 was lost in the edited mF9 (Fig. 1C).
Then we intravenously injected the dual AAVs that harbored both SaCas9 and sgRNA/donor, or the single AAV controls or phosphate-buffered saline (PBS) into HB mice. The FIX plasma activity increased with increasing AAV dosage (SI Appendix, Fig. S1E), indicating a dose-dependent mF9-editing effect within a certain range. Notably, the FIX activity elevated to >10% of normal value was only seen in the dual AAV groups that harbored the P2 (Fig. 1D), which was confirmed via measuring activated partial thromboplastin time (APTT), thromboelastogram (TEG), and hemostasis function (SI Appendix, Fig. S1 F–H). Likewise, as measured by semiquantitative reverse transcription nested PCR (RT-qPCR), the P2 incorporation greatly increased the F9 mRNA levels (4.38 to 12.45%), up to 10- to 20-fold of those detected in the corresponding P1 groups (0.02 to 0.81%) (Fig. 1E and SI Appendix, Fig. S1I). Consistently, immunochemical FIX staining on liver tissue sections confirmed these findings (SI Appendix, Fig. S1J). These results highlighted an important role of P2 integration in correcting coagulopathy.
Theoretically, diverse forms of mF9 alleles existed postedition, which included the HDR and 1/2 of the HITIs at site 2 (forward) that would produce functional F9 mRNA as well as two other groups that did not (SI Appendix, Fig. S2A). Unlike what was shown by a previous study (9), 3′ semiquantitative nested PCR (qPCR) showed that in all six dual AAV groups, the HDR rates were nearly 10-fold higher than the rates of site 2 forward HITI (6.5 to 11.6% versus 0.5 to 1.5%) (Fig. 1F and SI Appendix, Fig. S2 B and C). Noticeably, FIX plasma activity and F9 mRNA levels were considerably higher in the dual AAV group with HD2.P2 than in those with HD1.P2 (Fig. 1 D and E), although the HDR rates employing HD2 donors were ∼2-fold less than the HDR rates employing HD1 donors (6.5% versus 11.6% on average) (Fig. 1F). These results indicated that neoΔHPRT removal significantly facilitated the expression of the functionally edited mF9. Unexpectedly, the HD3 donor employing double genomic cuttings did not significantly increase HDR rates compared to HD2 donor employing a single genomic cutting (Fig. 1F). Thus, the integration of the HD2.P2 conferred a robust rate on the transcription of edited mF9.
As expected, qPCR assay revealed that the episomal AAV copy numbers within liver declined over time after having enabled a genomic editing (SI Appendix, Fig. S2D). Nevertheless, after a partial hepatoectomy (PHx), a restoration to the original plasma FIX level was seen in the HB mice treated with the AAV/CRISPR group but not in the HB mice treated with the single AAV group harboring the episomal P2-F9 (Fig. 1G). In agreement, the episomal AAVs were almost lost while a substantial proportion of HDR-edited mF9 alleles remained detectable 20 wk after the PHx (SI Appendix, Fig. S2 E and F).
Characterization of Adverse Effects of AAV/CRISPR-Mediated Gene Editing.
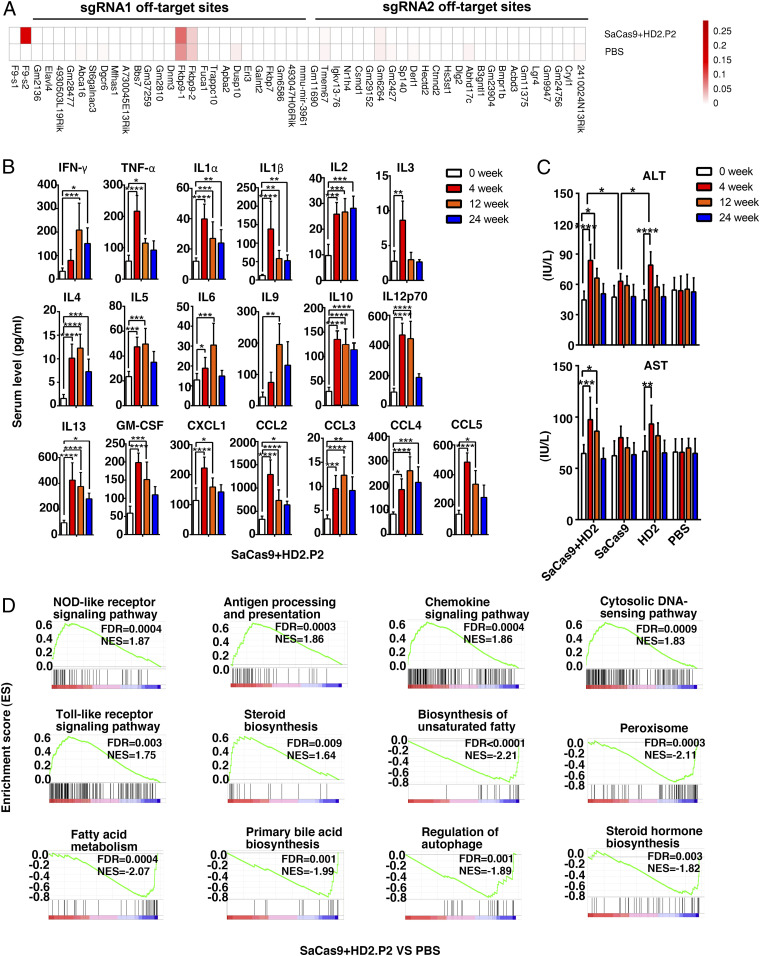
The off-target effect caused by SaCas9/sgRNA is still a major obstacle that precludes its clinical application. Through measuring the AAV copy number and targeted indel rate of multiple organs or tissues, we confirmed that the gene editing events occurred selectively in the liver (SI Appendix, Fig. S3 A–C). Whole genome sequencing of hepatic DNA samples detected two to four types of point mutations or 2-nt deletions that were clustered in a few genes, including Gm6264, Gm2427, and Pomt2. Nonetheless, their rates did not significantly differ between PBS-treated and dual AAV/CRISPR-treated HB mice (SI Appendix, Fig. S3D). Moreover, we also picked up the top 25 potential off-target sites of sgRNA 1.6 and sgRNA 2.1 to perform amplicon deep sequencing. Again, no increased mutation rate by sgRNA 1.6/SaCas9 or sgRNA 2.1/SaCas9 was detected (Fig. 2A). The apparent mutation rates observed at two potential sgRNA1 off-target sites within Fkbp9 in the PBS group might be caused by PCR amplification, as many repetitive DNA sequences were the same as the PCR primer used (SI Appendix, Fig. S3E).
Fig. 2.
Characterization of genomic off-target effects and subclinical hepatic inflammation post-AAV/CRISPR delivery. (A) Top 25 potential off-target sites of the s2.1 sgRNA- or s1.6 sgRNA-guided SaCas9 genomic cuttings were predicted through CCTop-CRISPR/Cas9 software, and examined by deep sequencing (1,000×, n = 3) in PBS and dual AAV with HD2.P2 groups. Extended off-target sites and relevant analyses are provided in Datasets S1 and S2. (B) Serum levels of 19 inflammatory cytokines in HB mice were assayed by the Luminex Liquidchip system at indicated time points after dual AAV infusion with HD2.P2 (n = 5). (C) Serum ALT and AST levels were assayed in HB mice at indicated time points after the treatment with PBS, single AAV groups, or dual AAVs (n = 6). (D) GSEA of RNA-seq data of HB hepatic tissues extracted at 24 wk after treatment with PBS or dual AAVs. Data are presented as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. NES, normalized enrichment score; FDR, false discovery rate.
The inflammation and FIX-neutralizing antibody resulting from the immune responses against AAV/CRISPR and FIX protein are also major concerns in HB gene therapy. We used the Luminex Liquidchip system to monitor serum levels of multiple inflammatory cytokines. As exemplified in the dual AAV group with HD2.P2, the levels of 19 out of 23 inflammatory cytokines such as IFN-γ, TNF-α, IL1β, IL6, and CCL5 increased post-AAV infusion compared to the PBS group, and these elevations peaked in 4 wk after treatment and declined over time (Fig. 2B). Similarly, the serum levels of hepatic alanine transaminase (ALT) and aspartate transaminase (AST) moderately peaked in 4 wk after treatment and then declined (Fig. 2C), thus suggesting a causal connection between AAV/CRISPR delivery–caused inflammation and a possibly mild hepatic injury. Interestingly, ALT elevations in the dual AAV group and single HD2.P2-carrying AAV group were similarly higher than that detected in the single SaCas9-carrying AAV group (Fig. 2C). Since five- to sixfold more AAVs were administered in the two former groups, the hepatic inflammation/injury was probably mostly attributed to AAV dosage rather than SaCas9 or/and FIX expression. Besides, FIX-neutralizing antibody was not detected in HB mice receiving dual AAV infusion in our study (SI Appendix, Fig. S3F).
Next, we performed bulk RNA sequencing analysis of liver samples from PBS and dual AAV groups. Notably, 904 out of 34,745 examined genes were still up-regulated and 213 of the genes down-regulated (fold changes >2) in the dual AAV group compared to the PBS group in 24 wk after AAV infusion (SI Appendix, Fig. S3G). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis and gene set enrichment analysis (GSEA) highly suggested that the dysregulated expressions were predominantly involved in inflammation and hepatic functionality (Fig. 2D and SI Appendix, Fig. S3H), verifying a hepatic inflammation after gene editing. Nonetheless, HB mice receiving dual AAVs manifested no obvious weight loss and activity reduction compared to those in the PBS-treated group, indicating a type of subclinical manifestation of inflammation.
Antiinflammation Promotes P2 Activity and Retention of the Hepatocytes Carrying Functionally Edited mF9 Alleles.
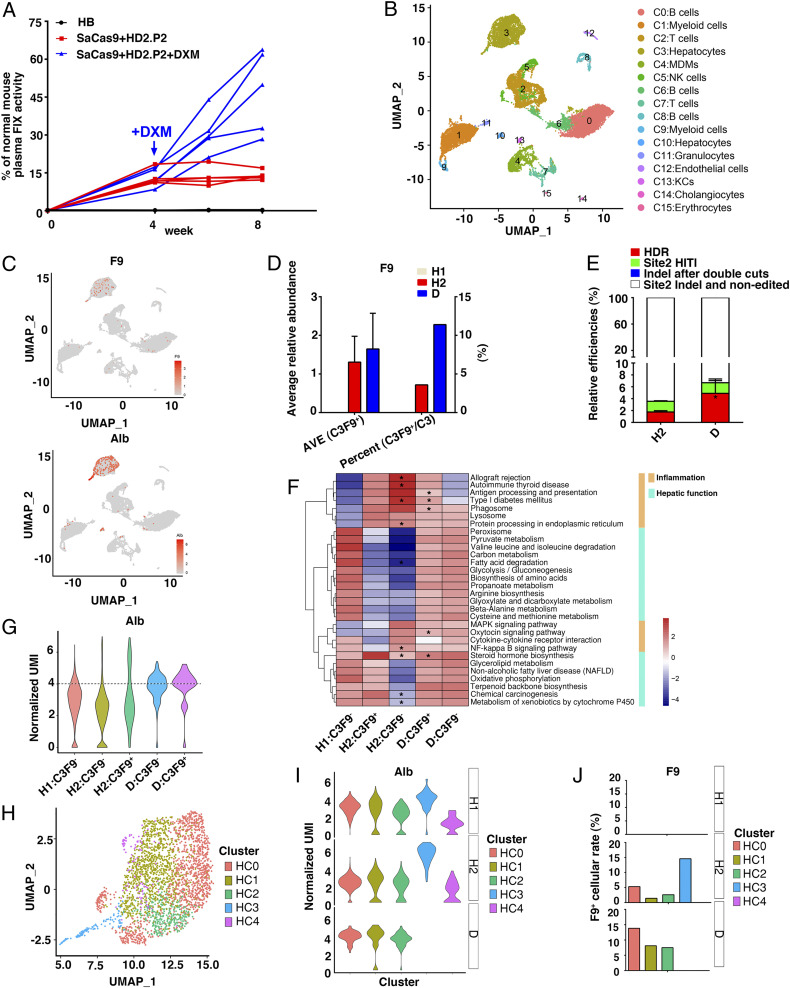
To test whether this inflammation influenced the therapeutic outcomes, we administered dexamethasone (DXM), a drug widely accepted to control AAV infusion–caused inflammation (21, 22), starting from week 5 after dual AAV infusion. Remarkably, DXM administration for 4 wk substantially increased the plasma FIX activity by three- to fourfold, resulting in elevating effects comparable to those achieved through increasing AAV dosage by >2.5-fold (Fig. 3A and SI Appendix, Fig. S1E).
Fig. 3.
DXM promotes expressional opening and retention of the functionally edited mF9 alleles within the major subsets of hepatocytes. (A) HB mice were intravenously injected with PBS (n = 5) or with dual AAVs with HD2.P2 (HD2.P2: 1.0 × 1014 vg/kg, SaCas9: 2.0 × 1013 vg/kg, n = 10). After 4 wk, HB mice infused with dual AAVs were divided into two groups (n = 5) and treated with or without a daily DXM (0.2 mg/d/mouse) for 4 wk. (B) UMAP visualization of mouse liver cell clusters pooled from HB mice injected with PBS (H1 group, 4,753 single cellular events, 64,885 reads per cell), dual AAVs without DMX treatment (H2 group, 12,793 single cellular events, 16,596 reads per cell), or dual AAVs + DXM (D group, 2,806 single cellular events, 60,555 reads per cell) (n = 3 mice for each group). (C) The F9 and Alb projection among different clusters visualized by UMAP map. The color bar indicates log2 normalized expression. The points with high Alb expression (UMIs >4) are shown. (D) The average relative abundance of F9 mRNA level in C3F9+ hepatocytes and the F9+ cellular rates among C3 hepatocytes in H1, H2, and D groups. (E) The relative rates of four groups of mF9 alleles as indicated were evaluated by qPCR in hepatic tissues of the H2 group and the D group (n = 3). (F) GSEA analyses of the pathways as indicated among H1:C3F9−, H2:C3F9−, H2:C3F9+, D:C3F9−, and D:C3F9+ groups. *P < 0.05 individually H1:C3F9− subset versus H2:C3F9+ subset, and D:C3F9+ subset versus H2:C3F9+ subset of each pathway. (G) Violin plots showing overall average expression (log transformed) of Alb gene among H1:C3F9−, H2:C3F9−, H2:C3F9+, D:C3F9−, and D:C3F9+ groups. The cellular events above the dotted line are regarded as Albhi. (H) UMAP plot comprising scRNA-seq data of C3 hepatocytes (HC0, HC1, HC2, HC3, and HC4). Clusters and associated cell types are indicated by name and color. (I) Violin plots showing overall average expression (log transformed) of Alb among HC0–HC4 cellular subsets under the H1, H2, and D conditions. (J) F9+ cellular rates among HC0–HC4 subsets under the H1, H2, and D conditions.
To understand the underlying mechanisms, scRNA-seq was performed on pooled liver cells from the HB mice injected with PBS (H1 group), the dual AAVs without DXM treatment (H2 group), or the dual AAVs treated with DXM (D group) (n = 3 mice for each group) (23). Comparing the scRNA-seq data with the bulk RNA-seq data (SI Appendix, Fig. S3 G and H) showed a significant correlation in the H1 group (r = 0.64) and the H2 group (r = 0.66) (SI Appendix, Fig. S4A). Uniform manifold approximation and projection (UMAP) visualization identified 16 cellular clusters (C0–C15) from all of the single cells through classifying the differentially expressed genes, including two hepatocyte subsets (a major C3 and a minor C10) and multiple types of hematopoietic lineages (Fig. 3B and SI Appendix, Fig. S4B). The C3 proportions within liver cells were similar under the H1 (18%) and H2 (14%) conditions, but declined under the D (7.3%) condition (SI Appendix, Fig. S4C).
The F9 projection among different clusters visualized by the UMAP map showed that the F9 mRNA-positive cells were located in the C3 hepatocyte subset (Fig. 3C). The F9 mRNA-positive C3 cells (C3F9+) rate among the C3 population was expectedly increased in the D group as compared to the H2 one (11.39% versus 3.59%), although an increase in the average F9 mRNA level per C3F9+ cell was not statistically significant (Fig. 3D). Meanwhile, the average frequency of functionally edited mF9 copies (HDR + 1/2 site 2 HITI) among liver cells was increased (2.65 to 5.79%) (Fig. 3E), indicating a protective role of DXM on the retention of the hepatocytes that harbored a functionally edited mF9 (C3FEs). Given that the C3 hepatocytes accounted for only a fraction of liver cells (SI Appendix, Fig. S4C), the average rates of C3FEs could be much higher than the actual C3F9+ rates (SI Appendix, Fig. S4D); thus a portion of C3FEs might display as the C3F9− phenotype probably due to repressed P2 activity.
As expected, AAV infusion enhanced inflammatory status of the C3 hepatocytes while compromising hepatic functionality (SI Appendix, Fig. S4E). Paradoxically, the C3F9+ fraction displayed a weaker inflammatory feature but better functionality than the C3F9− hepatocytes under the H2 condition (Fig. 3F). Consistently, C3F9+ cells were highly overlapped with the Albhi C3 hepatocytes (the value of normalized unique molecular identifers [UMIs] >4) indicative of a high hepatic functionality (Fig. 3 C and G and SI Appendix, Fig. S4F). These observations suggested that the P2 activity among a portion of C3FEs was switched off by a higher inflammatory status of the host cells, which was reversed by DXM. Similarly, compared to the untreated H1:C3F9− cells, the H2:C3F9− cells but not H2:C3F9+ cells exhibited enhanced P53 signaling and apoptosis pathways, which was ameliorated by DXM (SI Appendix, Fig. S4G). Given that hepatocytes were highly heterogeneous in terms of functionality and inflammatory status (24, 25), these observations indicated that inflammatory status of the C3FEs negatively affected their survival and the P2 activity.
To explore how an inherent hepatic heterogeneity regulates the P2-driven F9 expression (24, 25), we further divided the C3 hepatocytes into five subgroups (HC0–HC4) by UMAP at higher resolution (Fig. 3H and SI Appendix, Fig. S5A). Pseudotime analysis implicated that the HC0, HC1, and HC2 subsets were more closely related in development (SI Appendix, Fig. S5B), which constituted the major fractions of C3 hepatocytes under the H1, H2, and D conditions (SI Appendix, Fig. S5C). Virtually, the minor HC3 and HC4 subsets under the H1 or H2 condition (slightly expanded) were hardly detectable under the D condition (SI Appendix, Fig. S5C). Interestingly, under the H2 and D conditions the F9+ cellular rates among individual HC0–HC3 subsets paralleled with the cellular rates of Albhi (Fig. 3 I and J) (24, 25), confirming a tight association between P2 activity and Alb mRNA levels. Moreover, after injection of AAV/CRISPR, Alb expression was down-regulated in the HC0 to HC2 subsets but up-regulated in the HC3 subset (Fig. 3I). Therefore, inflammation up-regulated or down-regulated the P2 activity or Alb expression depending on host cell types.
Inflammation Differentially Regulates P2 Activity via Two Distinct Sets of Liver-Enriched Transcription Factors.
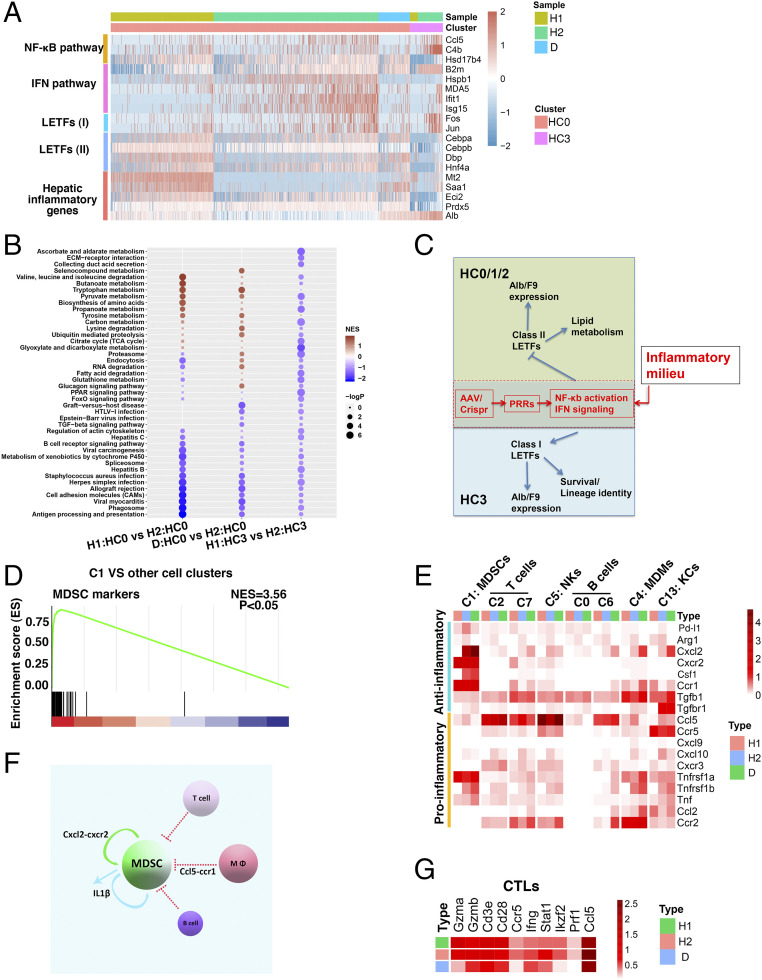
Next, we employed ingenuity pathway analysis (IPA) and scenic analysis to explore the underlying molecular mechanisms, which revealed that F9 and/or Alb were potentially regulated by two sets of coregulated liver-enriched transcription factors (LETFs) containing class I members Fos, Junb, and AP1 (comprising Jun and Fos) in the HC3 to HC4 subsets and class II members Dpb, Hnf4a, Cebpα, and Cebpβ in the HC0 to HC2 subsets, respectively (SI Appendix, Fig. S6 A–C and Dataset S3). To characterize how inflammation regulated the hepatic function in a subset-dependent manner via the LETFs, we firstly compared the inflammation-disfavoring HC0 and inflammation-favoring HC3 subsets (Fig. 4A and SI Appendix, Fig. S5B). In the HC0 subset, the mRNA levels of the IFN signaling signature genes, including Ifih1, B2m, Hspb1, Isg15, and Ifit1, were increased while the expression of class II LETFs, including Dpb, Hnf4a, Cebpα, and Cebpβ, was decreased after injection of AAV/CRISPR. In parallel to this, the hepatic functional genes, including Mt2, Saa1, Eci2, Prdx5, and Alb, were inhibited after injection of AAV/CRISPR but then were at least partially restored by DXM. In line with the previous findings that LETFs were distinctly regulated by hepatic inflammatory cues and that AP1 (Jun and Fos) was presumably activated by the NF-κB pathway (26, 27), in the HC3 subset the specific up-regulation of the NF-κB pathway target genes Hsd17b4, Ccl5, and C4b was evident after injection of AAV/CRISPR, which was accompanied by the up-regulated expressions of the class I LETF members Jun and Fos and hepatic functional genes such as Alb (Fig. 4A). Two similar alteration patterns were seen in the HC1–HC2 subsets and the HC4 subset, respectively (SI Appendix, Fig. S6D). As also supported by GSEA analysis of the HC0 and the HC3 subsets (Fig. 4B), these results indicated that the expressions of F9 and Alb as well as other genes indicative of hepatocytic functionality in two kinds of C3 hepatocytes were regulated in opposite ways through distinct LETFs when inflammatory signaling pathways including the NF-κB and IFN pathways were activated inside hepatocytes (Fig. 4C). In corroboration of this, after injection of AAV/CRISPR, the expression levels of fibrinogen, F2, F5, F10, F11, and F12 paralleled those alterations occurring in the Alb/F9 in different hepatocyte subsets (SI Appendix, Fig. S6E). Nevertheless, after DXM treatment, not all expressional alterations of individual clotting factors in the HC0-HC2 were consistent with the elevation of F9/Alb (SI Appendix, Fig. S6E), indicating an extra and complex clotting factor regulatory activity of DXM in addition to its ameliorating AAV/CRISPR-caused inflammation.
Fig. 4.
Hepatic inflammation differentially regulates F9 and Alb expressions via two specific sets of LETFs. (A) Heatmap showing expressions of the selected gene modules as indicated in HC0 and HC3 cells under the H1, H2, and D conditions. The color bar indicates log2-normalized expression. (B) GSEA analyses of H1:HC0 versus H2:HC0, D:HC0 versus H2:HC0, and H1:HC3 versus H2:HC3. (C) Hypothesis: Alb and F9 expressions indicative of hepatocyte functionality among discrete hepatocytes are driven by distinct sets of LETFs whose expressions and activities are modulated by inflammatory signaling in opposite ways. (D) GSEA analysis of C1 versus the other 15 cellular clusters in terms of MDSC signature genes. (E) Average heatmap depicting the expressional alterations of major cytokines/ligands in major hematopoietic clusters including C1:MDSCs, C2/7:T cells, C5:NK, C0/6:B cells, C4:MDMs, and C13:KCs under the H1, H2, and D conditions. (F) Diagram depicting the C1 pool regulation by two major cytokine–receptor communications pathways under the H2 or D condition. (G) Average heatmap depicting the expressional alterations of CTL effector molecules as indicated under the H1, H2, and D conditions.
Hematopoietic Cells Are Actively Involved in the Regulation of Inflammation.
Then we sought to investigate whether the preeminent presence and dynamic alterations of myeloid clusters (C1, C4, and C13), NK/T cell clusters (C5, C2, and C7), and others under the H1, H2, and D conditions could play a role in regulating hepatic inflammation (SI Appendix, Fig. S4C). Of the myeloid clusters, apart from previously reported C4 monocyte-derived macrophage (C4:MDMs, Ccr2+/loCD11b+/loF4/80loMafb+/−Csf1r+CD74+/loH2-A/E+/lo) and C13 Kupffer cells (C13:KCs, Ccr2−CD11b−/loF4/80hiApoe+) that infiltrated liver (SI Appendix, Fig. S7A) (28), the C1 myeloid cells generally displayed Lyz2+CD11b+Ly6c+/loLy6glo/−Pu.1+Cebpβ+Cebpα−Irf8− and expressed the mMono3 cells’ signature genes (SI Appendix, Fig. S7 B and C) (29). Meanwhile, specific expressions of Arg2 and Il1b but not MHC class II antigens (H2-A/E) along with a marked expressional correlation to myeloid-derived suppressor cells (MDSCs) pointed to the C1 as immunosuppressive cells (Fig. 4D and SI Appendix, Fig. S7D) (30). In accordance, C1 proportion was greatly decreased after injection of AAV/CRISPR but increased to a profound level by DXM treatment (SI Appendix, Fig. S4C), thus indicating a critical role of C1:MDSCs in controlling hepatic inflammation. Besides, at higher T distributed stochastic neighbor embedding (t-SNE) resolution of natural killer (NK) and T cells a TC4:CD8+ T effector (cytotoxic T lymphocyte [CTL]) subset and a TC10:Treg subset were also identified (SI Appendix, Fig. S7 E and F).
Then we explored the molecular mechanism of the interaction between these cells. mRNAs of numerous pattern recognition receptors (PRRs), cytokines, and receptors were detected in discrete liver cells (SI Appendix, Fig. S8A). Next, employing the CellPhoneDB v2.0 software, we integrated our scRNA-seq data (including 114 receptors and 129 ligands whose mRNA levels with UMI values were >1) into the construction of intercellular interactive maps. As expected, a close cytokine-receptor network between liver-resident hematopoietic immune cells and the inflammation-favoring HC3-HC4 subsets existed under the H1 condition (SI Appendix, Fig. S8B). After injection of AAV/CRISPR, the molecular signaling among hematopoietic immune cells intensified, thus forming an active proinflammatory hub wherein a Ccl5-Ccr1 axis, the NK, T cells, MDMs, and KCs but not the C1:MDSCs developed a closer relationship with the HC0 subset (SI Appendix, Figs. S8B and 4F). Expectedly, this proinflammatory hub was largely dissolved by DXM (SI Appendix, Fig. S8B), implicating its involvement in the development of AAV/CRISPR-caused hepatic inflammation.
Previous work implicated PRRs, such as TLR2, TLR9, and RLRs, may participate in AAV-triggered hepatic inflammation (31–33). Nevertheless, the up-regulations of Ifi204, MDA5, and Rig-I but not others were evident in hepatocytes and multiple hematopoietic cells, including the C1:MDSCs after injection of AAV/CRISPR, which was at least partially reversed by DXM (SI Appendix, Fig. S8C). Particularly, since the PRR activation led to inflammasome activation that in turn processed the premature Il1b for maturation and release, and the expressions of the Il1 receptor subunits, including Il1r, Il1rap, and antagonist Il1rn were mostly detected in C1:MDSCs (SI Appendix, Fig. S8A), a paracrine and/or an autocrine IL1β signaling may contribute to the hepatic inflammation development and regulation of C1:MDSCs themselves (Fig. 4F) (30). Moreover, MDSC inducer Cxcl2 was up-regulated in the C1:MDSCs and other cells under both H2 and D conditions (34, 35), which might promote C1:MDSC activation directly through Cxcr2 prominently expressed in C1:MDSCs (Fig. 4 E and F). On the other hand, while the Ccl5 up-regulation might exert a moderate activating effect on NKs whose Ccr5 expression was decreased under the H2 condition, it also helped to suppress C1:MDSCs via signaling Ccr1 selectively expressed on C1:MDSCs (Fig. 4 E and F) (36, 37). In this regard, Ccl5 up-regulation was evident in the C4 and C13 macrophages after injection of AAV/CRISPR, which was diminished by DXM (Fig. 4 E and F). Moreover, Ccl5 was also generated by T cells, NK cells, and the bulk C6 B cells (Fig. 4E and SI Appendix, Fig. S8A). Notably, the TC4:CTL proportion increased after injection of AAV/CRISPR, but was decreased by DXM (SI Appendix, Fig. S8D), with parallel up- and down-regulations of major CTL effector molecules, including granzyme, Ifng, and perforin (Fig. 4G). In light of the data mentioned above (SI Appendix, Fig. S4 D and G) (38), these CTLs were probably involved in the clearance of certain C3FEs wherein the inflammatory signalings were more highly activated.
Discussion
In this study, we characterized three AAV/CRISPR-mediated HDR strategies in dealing with a massive mF9 defect involving multiple interposed regulatory and encoding genomic regions, and thus identified a simple, safe, and effective protocol that employed a long-range DNA replacement strategy to be integrated with the synthetic P2 enhancer/promoter composite (19). Moreover, we identified the actual hepatocyte types that produced FIX after gene edition. In this regard, we have revealed a previously unappreciated effect of a subclinical and likely transient inflammation after AAV/CRISPR-mediated mF9 editing that attenuated the expression strength and durability of the edited F9 through affecting distinct sets of LETFs. We also uncovered previously unappreciated roles of multiple types of liver-resident hematopoietic lineages, including a particular MDSC-enriched myeloid subset in determining the inflammatory status. These results highlight a notion that the early inflammation management even in those AAV-unprimed recipients should greatly increase the efficacy and durability of AAV/CRISPR-mediated hepatic gene editing.
A TNF-α–related inflammatory reaction was shown to inhibit the transgenic FIX expression from episomal AAVs in hepatocytes (39). Investigating the mechanisms underlying the inhibitory effect of the subclinical inflammation on the expression and/or retention of the functionally edited mF9 alleles, we found that P2-driven F9 expression was largely predetermined by the host cell type. The hepatic functionality and the P2 activity alike were inhibited in the major hepatocytes (HC0–HC2) while being stimulated in the minor hepatocytes (HC3), which was executed by NF-κB and IFN signaling pathways via modulating distinct LETFs—class I and class II LETFs. Relevantly, the most likely targets of CTL clearance specifically under the H2 condition were those AAV-transduced C3 hepatocytes being at a proinflammatory state (40, 41). This systemic mechanistic analysis sets up a basis for developing a hepatocyte subtype-dependent AAV/CRISPR-mediated hepatic gene editing strategy that may improve its therapeutic efficacy and durability.
The activation of antigen-specific Tregs was implicated in imparting immunotolerance to AAV infusion in mice (42). In our study, scRNA-seq analysis helped identify a particularly large pool of liver-resident C1:MDSCs. Their predominance over Tregs in pool size under physiological conditions or following AAV/CRISPR infusion with or without DXM treatment suggested a likely pivotal role of this population in restraining the hepatic inflammation. On the other hand, the evaluation of serum Ccl5, its expression in multiple hepatic proinflammatory hematopoietic cells, and a central location with the envisioned proinflammatory hub indicated it as a prominent proinflammatory factor under the H2 condition. Further exploration into how C1:MDSCs and Ccl5-mediated signalings regulate the inflammation process after AAV/CRISPR delivery might bring about specific inflammation managing strategies other than DXM administration (36, 37).
Materials and Methods
Detailed descriptions regarding vector construction, AAV preparation, experimental treatment and monitoring of HB mice, liver cell isolation, single-cell RNA sequencing and analysis, inflammatory cytokine assay, histology analysis, and next generation sequencing are described here.
All high-throughput sequencing data can be viewed in NODE (https://www.biosino.org/node) by pasting the accession ID (DNA data: OEP001209; RNA data: OEP001206) into the text search box.
Animal Model.
HB mice (factor IX promoter and exons 1 to 3 knockout [F9-KO]) on a C57BL/6 background were obtained from The Jackson Laboratory (43). The strain was B6.129P2-F9tm1Dws/J. The genotypes of mice were identified by PCR according to the method recommended by The Jackson Laboratory. The C57BL/6 wild-type and HB mice were bred and experimentally treated in a specific-pathogen-free environment in the Animal Institute of Ruijin Hospital in accordance with the institutional guidelines, as provided by the Laboratory Animal Resource Center of Shanghai Jiao-Tong University School of Medicine and the Chinese Association for Laboratory Animal Sciences. Adult male mice aged 8 to 12 wk were randomly selected as experimental animals. The animals were housed under room temperature 18 to 22 °C, humidity 30 to 70%, and a 12-h light/dark cycle controlled automatically. A pelleted standard rodent diet and fresh drinking water were provided ad libitum. The mice were anesthetized by intraperitoneal administration of 1% pentobarbital sodium (Cat. No. P11011, Merck) for tail bleeding test. A PHx was performed under deep surgical anesthesia induced by isoflurane (Cat. No. 100150, Yuyan).
HB Mice Treatment and Monitoring.
AAV8 vectors formulated in 200 μL of PBS were injected into male HB mice through the tail vein. The blood was withdrawn from the animals for serum biochemistry at 1- to 4-wk intervals, and also for measuring hepatic enzyme and inflammatory cytokine levels. The treated animals were also killed for liver immunohistochemistry analysis and the extraction of liver DNA and mRNA.
Vector Construction.
The candidate single-guide RNAs (sgRNAs) were designed using an online design tool (https://zlab.bio/guide-design-resources) to minimize potential off-target effects. The candidate sgRNAs were cloned into the lentiCRISPRv2 vector coexpressing SpCas9 (Cat. No. 52961, Addgene plasmid repository) or the pUC57 vector containing AAV ITR-flanked donors (Genscript). The right or left homology arms that flank the P1- or P2-coupled F9 exons 1 to 4 fusion cDNA was amplified from 0.8- to 1.5-kb genomic regions flanking the target sites using the genomic DNA from the HB mouse. The DNA templates for calibration of site 2 indels, nonedited mF9 alleles, or indels after double cuts were constructed by deleting the promoter and F9 exons 1 to 4 cDNA segments from donor HD2 and HD3. The plasmid for site 2 HITI-positive control contained two tandem RHA segments, which were separated by one ITR.
Virus Production and Titration.
For in-house viral production, lentiviral vectors coexpressing SpCas9 and sgRNA were packaged in 293T cells (human embryonic kidney cell line, HEK-293; The Chinese Academy of Sciences Cell Bank, National Collection of Authenticated Cell Cultures) transduced with triple plasmid transfection methods (long terminal repeat–structured viral vector expression plasmid, lentivirus capsid protein expression plasmid, and helper plasmid). Forty-eight hours after changing the culture medium, cell supernatant was collected and concentrated by high-speed centrifugation.
AAV8 vector expressing SaCas9 or harboring donors was produced by triple transfection methods and purified by CsCl density gradient centrifugation as described in previous works (44). Briefly described, AAV-293 cells (hHEK-293; The Chinese Academy of Sciences Cell Bank, National Collection of Authenticated Cell Cultures) were cotransfected with the plasmid with ITR structure, AAV8 packaging plasmid, and helper plasmid. Seventy-two hours later, cells and cell supernatant containing AAV vectors were collected. Cells were then ruptured, and the lysis supernatant was precipitated together with cell supernatant mentioned above. The precipitate was centrifuged with a two-step CsCl density gradient centrifugation to remove the empty capsid and impurities. These viral vectors were titrated by real-time qPCR using a customized Taq-Man probe against the transgene (Cat. No. 4440040, ABI).
Chromogenic Assay.
The activity of plasma FIX was tested by chromogenic assay (Cat. No. 221806, HYPHEN BioMed). The plasma was isolated from the centrifugation of 3.8% sodium citrate–added blood. The pooled standard plasma sample of normal wild-type C57BL/6 mice was used in 1:50, 1:100, 1:200, 1:400, 1:800, 1:2,000, 1:4,000, and 1:10,000 dilutions. Samples from treated HB mice were diluted as 1:100. Fifty-microliter samples per well were applied for the assay. After the termination of the reaction, the absorbance at the wavelength of 405 nm was read by a microplate reader.
aPTT.
aPTT was carried out by mixing a 50-μL plasma sample with 50 μL of SynthASil aPTT reagent (Cat. No. 0020006300, HemosIL; Werfen). Clot formation was initiated by addition of 50 μL 20 mM calcium chloride (Cat. No. 0019741910, HemosIL; Werfen) according to the manufacturer’s instructions and the aPTT was tested with a Stago coagulation analyzer.
TEG R Time.
TEG was applied to evaluate the blood coagulation function through R time. Four hundred microliters of whole blood, added with 3.8% sodium citrate solution at 1:9 ratio, was mixed with 16 μL of kaolin activator. Fifteen minutes postactivation, 340 μL of activated whole blood was used for the assay after adding 20 μL of 0.2 M calcium chloride following the manufacturer’s instructions (TEG 5000 Thrombelastograph Hemostasis Analyzer, Haemoscope Corporation).
Tail Bleeding Test.
After narcotizing mice, we preheated the tail for 10 min in 37 °C saline. Bleeding was initiated by a scalpel-made 2.5-mm cut of the tail tip and continued by submerging the cut tail into 2 mL of saline. The bleeding time and blood loss were monitored for 10 min, at which point the bleeding was forcedly terminated. The blood loss was determined by measuring the hemoglobin concentration in the saline. One hundred microliters of a blood/saline mixture was diluted with 2 mL of hemoglobin reagent (hemiglobincyanide colorimetry) (Cat. No. C021-1, Jiancheng), and placed at room temperature for 5 min before absorbance at 540 nm was measured on a Nanodrop 2000 photometer (Thermo Fisher Scientific). The absorbance was converted to a hemoglobin concentration by standard curve provided in the hemoglobin reagent kit.
ALT and AST Analysis.
ALT and AST from serum were measured via Reitman–Frankel colorimetric assay according to the manufacturer’s instructions for the alanine aminotransferase assay kit and aspartate aminotransferase assay kit (Cat. No. AST: C010-2, ALT: C009-2, Jiancheng).
Inflammatory Cytokine Measurement.
Luminex assay was utilized for measuring 23 inflammatory factors in the serum of whole blood with anticoagulant. The serum protein levels of inflammatory cytokines were quantified by ELISA according to the manufacturer’s instructions (R&D Systems).
Histology Analysis.
Immunohistochemistry (IHC) staining was performed on the sections of 4% paraformaldehyde-fixed liver with a polyclonal rabbit antibody against the mouse FIX as the primary antibody (Cat. No. ab175075, Abcam) with 1:200 dilution (15 μg/mL). Goat anti-rabbit IgG-linked horseradish peroxidase (HRP) (Cat. No. GB23303, Servicebio) was used as the secondary antibody with 1:200 dilution (10 μg/mL). Finally, diaminobenzidine tetrahydrochloride (DAB) (Cat. No. K5007, Dako Agilent) chromogen reagent was applied for chromogenic reaction.
Indel Rate Determination by T7EI Cleavage Assay.
NIH 3T3 cells (NIH Swiss mouse embryonic fibroblasts cell line; The Chinese Academy of Sciences Cell Bank, National Collection of Authenticated Cell Cultures) were seeded into 24-well plates 1 d before infection with lentiviral vectors at designated multiplicities of infection and harvested at 72 h posttransduction for DNA extraction. HB 8- to 12-wk-old mice were injected with AAV8 vectors at 2 × 1012 vg/kg (vector genomes per kilogram) via tail vein, and the hepatic DNA was extracted at 1-wk postinjection using a DNA Extraction kit (Cat. No. 69504, Qiagen) following the manufacturer’s protocol. Then the standard protocol of T7EI assay (Cat. No. M0302L, NEB) was used to determine the frequency of the small insertions and deletions (indels) characteristic of DSB repair by NHEJ. The indel rate was analyzed by semiquantitative ImageJ software. ƒcut = (Cleavage Band1 + Cleavage Band2)/(Uncleaved Band + Cleavage Band1 + Cleavage Band2); % of NHEJ = 100 × [1 − (1 − ƒcut)1/2]. The primers for T7EI assays of candidate sgRNAs are listed in Dataset S4. The primers covering the potential target regions of SaCas9/sgRNA2.1 were used for indel efficiencies analysis through T7EI assays in liver and other tissues.
qPCR.
qPCR (Cat. No. 4440040, ABI) was performed for efficiencies analysis of HDR, site 2 HITI, indel after double cuts, site 2 indel, and the nonedited mF9. To exclude possible interference of episomal vectors, we chose ∼2 kb of a region beyond the RHA as the template of qPCR. To reduce the interference among three groups from differences in PCR primers and amplification lengths, the same reverse primer (R1) was used and the targeted sites of forward primers F1–F4 had the same distance from R1. PCR was performed using the KOD-Plus kit (Cat. No. KOD-201, Toyobo). Then the nested qPCR with the same set of primers F5/R2 (that were outside of RHA) and Taq-Man probe was performed. The four kinds of PCR were calibrated by plasmids using the same PCR program. The primers for PCR and qPCR are listed in Dataset S5.1.
The primers of qPCR for relative DNA levels of SaCas9 and HD2.P2 in liver and other tissues are listed in Dataset S5.2. The primers of qPCR for relative DNA level of AAV.P2.hF9 in liver are listed in Dataset S5.3.
RT-qPCR Assay on mRNA Levels of F9 and Other Genes.
Total RNA was isolated from liver or other tissues using the RNeasy Mini kit (Cat. No. 74104, Qiagen), followed by reverse transcription with the M-MLV Reverse Transcriptase kit (Cat. No. 28025013, Invitrogen). The hepatic mRNA samples in treated mice were then amplified by RT-PCR and RT-qPCR to analyze the transcription of F9. The primers for qPCR and PCR are listed in Dataset S5.4.
PHx.
The partial hepatectomy was performed as previously described (45). Starting 2 wk prior to the operation, the blood was withdrawn from treated mice every week. The activity of plasma FIX was tested by chromogenic assay.
Next Generation Sequencing and Whole Genome Sequencing for Off-Target Analysis.
Off-target sites were predicted by the online tool CCTop-CRISPR/Cas9 target online predictor: https://cctop.cos.uni-heidelberg.de:8043/ (up to 8-bp mismatch). The potential off-target sites and the primers for off-target PCR are listed in Datasets S1 and S2. The genomic regions centered at the target sites were PCR amplified and the multiplexed libraries of amplicons were generated via the 48.48 Access Array microfluidic platform according to the manufacturer’s protocol provided by Fluidigm. Deep sequencing was performed on the MiSeq platform provided by Illumina according to established Illumina protocols.
For whole genome sequencing, DNA was extracted from liver samples of treated HB mice using a DNeasy Blood & Tissue kit (Cat. No. 69504, Qiagen). Libraries were generated by the KAPA protocol provided from Roche using 100 ng DNA and sequenced on an Illumina NovaSeq6000 (150-bp paired end) machines. Data were processed according to standard Illumina sequencing analysis procedures.
Bulk RNA Sequencing and Analyses.
Total RNA was isolated from liver samples using an RNeasy Mini kit (Cat. No. 74104, Qiagen) following the manufacturer’s instructions, and RNA quantity was further assessed by RNA integrity number (RIN) value of 9.5 using an Agilent 2100 Bioanalyzer. cDNA libraries were constructed using a mRNA-Seq Sample Preparation kit (Illumina) following the manufacturer’s instructions. The quality and size of libraries were measured by the Agilent 2100, according to the reagent kit guide. Then, the samples were sequenced on an Illumina X Ten. Significant differentially expressed genes (DEGs) were identified as those with a false discovery rate (FDR) value above the threshold (Q < 0.05) and fold change >2 using edgeR software. GO (http://geneontology.org/) enrichment and KEGG (https://www.kegg.jp/) pathway analyses of DEGs were performed using DAVID software. GSEA with a C2.KEGG gene set was analyzed by GSEA 3.0 software. The P value <0.05 was taken as the threshold for both the functional and pathway enrichment analyses.
Intraperitoneal Injection of Dexamethasone.
Ten male HB 10- to 12-wk-old mice were injected with AAV.SaCas9 (2 × 1013 vg/kg) and AAV.HD2.P2 (1 × 1014 vg/kg) dual vectors formulated in 200 μL of PBS by tail vein, and five male HB mice were injected with 200 μL PBS through tail vein in parallel. After 4 wk post-injection, the plasma samples of these treated HB mice were withdrawn for FIX activity test. At the same time, half of the dual AAV-treated mice were treated with 0.2 mg/d/mouse dexamethasone by intraperitoneal injection.
Mouse Liver Cell Isolation.
A 24-G needle was inserted into the inferior vena cava of killed mice and secured with a clamp, and chelating solution (0.05 M Hepes pH 7.2, 10 mM ethylene glycol tetraacetic acid in Hank’s balanced salt solution without CaCl2 and MgCl2) was run at a low speed (1.5 to 2 mL/min). The portal vein was then cut and perfusion speed was increased to a flow rate of 7 mL/min. After that, the liver was transported in a sterile culture dish with 10 mL 1× Dulbecco’s phosphate-buffered saline (DPBS) (Cat. No. 14190144, Gibco) on ice to remove the residual tissue storage solution, and then minced on ice. We used 0.25% trypsin (Cat. No. 25200072, Gibco) and 10 μg/mL DNase I (Cat. No. 11284932001, Roche) in PBS (Cat. No. 10010049, Gibco) with 5% fetal bovine serum (FBS) (Cat. No. 10099141, Gibco) to digest the tissues. Liver tissues were dissociated at 37 °C with a shaking speed of 50 rpm for 40 min. Cell suspensions were filtered using a 40-μm nylon cell strainer and red blood cells were removed by 1× Red Blood Cell Lysis Solution. Dissociated cells were washed with 1× DPBS containing 2% FBS. Cells were stained with 0.4% Trypan blue (Cat. No. 15250061, Gibco) to check the viability on a Countess II Automated Cell Counter.
Single-Cell RNA Sequencing and Analysis.
The single liver cells were subjected to scRNA-seq analysis using 10× Genomics Chromium Single-Cell 3′ according to the manufacturer’s instructions. Library generation was performed following the Chromium Single Cell 3′ Reagents kits version 3 user guide (CG000206 Rev D). The libraries were sequenced using Illumina NovaSeq6000 platforms to achieve an average of 47,335 reads per cell. Approximately 55.0% of the sequence reads can be confidently mapped to the mouse transcriptome. Alignment of 3′ end counting libraries from scRNA-seq analyses was completed using 10× Genomics Cell Ranger 2.1.0. Each library was aligned to an indexed mm10 genome using the Cell Ranger Count.
The Seurat pipeline (version 2.3.0) was used for cluster identification in scRNA-seq datasets. Unique sequencing reads for each gene were normalized to total UMIs in each cell to obtain normalized UMI values. Using Seurat’s FindIntegrationAnchors and IntegrateData, liver cells were integrated together into a single analysis. Specific markers for each cluster identified by Seurat were determined using the “find all markers” function. For cell type subset analyses, clusters with high expression of cell type markers in their analyses were subset out, and standard Seurat workflow was applied on each cluster (46). The UMAP plots, t-SNE plots, violin plots, bar plots, circular plot, bubble plots, feature plots, and heatmaps were generated by R and Java TreeView. Using the R package Monocle (version 2.8.0), differentiation hierarchies among hepatocyte subsets were reconstructed. We used high dispersion genes identified by Monocle to sort cells in pseudotime order. IPA analysis was used to determine pathways or gene networks that may be altered across clusters based on scRNA-seq results. Via Qiagen Digital Insights (https://www.qiagenbioinformatics.com), we uploaded each cluster marker gene to IPA, and used the “core analysis” function to analyze upstream transcriptional regulators and gene networks. GSEA functions of R packages clusterProfiler (47) were applied to identify a priori–defined gene sets that showed statistically significant differences between the given clusters. The gene sets of apoptosis and the p53 signaling pathway in SI Appendix, Fig. S6C was from the Hallmark dataset of the MSigDB database. For correlation analysis between C0–C15 clusters and MDSCs in SI Appendix, Fig. S8B, the 105 of the 200 top marker genes’ overlapped between gene signatures for G- and M-MDSCs in a previous study (30) were applied. SCENIC analysis was run as described in a previous study (48). SCENIC code and tutorials are available at https://scenic.aertslab.org. The UMI count matrix was inputted and the SCENIC analysis step was run with default parameters. As a result, we found 13 additional regulons. CellPhoneDB analysis was used to infer cell to cell communication networks from scRNA-seq data. The analysis was performed with CellPhoneDB v2.0; the procedures and a practical step-by-step guide to help implement the protocol were present in a previous study (49).
Statistical Analysis.
Data were analyzed by GraphPad 6.0 software, Student’s t test compared to controls, one-way ANOVA with Tukey’s post hoc analysis or two-way ANOVA with Tukey’s, and Sidak’s multiple comparisons test at the proper places. Statistical P values <0.05 were considered significant.
Acknowledgments
We thank Ms. Ruihong Zhang, Ms. Zheng Ruan, Ms. Xiangqin Weng, and Ms. Yan Sheng of the Shanghai Institute of Hematology for their technical support; and Hongjuan Deng, Xiaotao Cheng (Shanghai Biotechnology Corporation), and Li Zhang (School of Statistics, East China Normal University) for single-cell RNA-seq statistical consultation (Xiaotao Cheng for supporting Ingenuity Pathway Analysis). We thank Dr. Ernest Johann Helwig (Tongji Medical College, Huazhong University of Science and Technology) for language editing. This research was supported by the Double First-Class Project (WF510162602) from the Ministry of Education, State Key Laboratory of Medical Genomics, the Shanghai Shenkang Hospital Development Center (SHDC2), Innovative Research Team of High-Level Local Universities in Shanghai, the National Major Scientific and Technological Special Project (2018ZX09101001), Shanghai Collaborative Innovation Program on Regenerative Medicine and Stem Cell Research (2019CXJQ01), the National Key Research and Development Program of China (2018YFA0107802), 111 Project (B17029), the Chinese National Key Basic Research Project 973 (2013CB966800), the National Natural Science Foundation of China (81470012 and 81861148030), Shanghai Major Project for Clinical Medicine (2017ZZ01002), Novo Nordisk Hemophilia Research Fund in China, and PhD Innovation Fund of Shanghai Jiao-Tong University School of Medicine (BXJ201616).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110887118/-/DCSupplemental.
Data Availability
All high-throughput sequencing data can be viewed in NODE (http://www.biosino.org/node) by pasting the accession ID (DNA data: OEP001209`OEP001206) into the text search box.
References
- 1.Rallapalli P. M., Kemball-Cook G., Tuddenham E. G., Gomez K., Perkins S. J., An interactive mutation database for human coagulation factor IX provides novel insights into the phenotypes and genetics of hemophilia B. J. Thromb. Haemost. 11, 1329–1340 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Pierce G. F., Gene therapy for hemophilia: Anticipating the unexpected. Blood Adv. 4, 3788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.George L. A., et al., Hemophilia B gene therapy with a high-specific-activity factor IX variant. N. Engl. J. Med. 377, 2215–2227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miesbach W., et al., Gene therapy with adeno-associated virus vector 5-human factor IX in adults with hemophilia B. Blood 131, 1022–1031 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakai H., et al., Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 75, 6969–6976 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manno C. S., et al., Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 12, 342–347 (2006). Correction in: Nat. Med. 12, 592 (2006). [DOI] [PubMed] [Google Scholar]
- 7.George L. A., et al., Long-term follow-up of the first in human intravascular delivery of AAV for gene transfer: AAV2-hFIX16 for severe hemophilia B. Mol. Ther. 28, 2073–2082 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., et al., CRISPR/Cas9-mediated in vivo gene targeting corrects hemostasis in newborn and adult factor IX-knockout mice. Blood 133, 2745–2752 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Ohmori T., et al., CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci. Rep. 7, 4159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funnell A. P., Crossley M., Hemophilia B Leyden and once mysterious cis-regulatory mutations. Trends Genet. 30, 18–23 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Guilbaud M., et al., Five years of successful inducible transgene expression following locoregional adeno-associated virus delivery in nonhuman primates with no detectable immunity. Hum. Gene Ther. 30, 802–813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martino A. T., Markusic D. M., Immune response mechanisms against AAV vectors in animal models. Mol. Ther. Methods Clin. Dev. 17, 198–208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaiss A. K., et al., Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J. Virol. 76, 4580–4590 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mingozzi F., et al., Induction of immune tolerance to coagulation factor IX antigen by in vivo hepatic gene transfer. J. Clin. Invest. 111, 1347–1356 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandamme C., Adjali O., Mingozzi F., Unraveling the complex story of immune responses to AAV vectors trial after trial. Hum. Gene Ther. 28, 1061–1074 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdera H. C., Kuranda K., Mingozzi F., AAV vector immunogenicity in humans: A long journey to successful gene transfer. Mol. Ther. 28, 723–746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson C. E., et al., Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 25, 427–432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao C. H., et al., Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro. Mol. Ther. 1, 522–532 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Zhang R., Wang Q., Zhang L., Chen S., Optimized human factor IX expression cassettes for hepatic-directed gene therapy of hemophilia B. Front. Med. 9, 90–99 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Byrne S. M., Ortiz L., Mali P., Aach J., Church G. M., Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 43, e21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nathwani A. C., et al., Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 371, 1994–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nathwani A. C., et al., Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 365, 2357–2365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W., et al., Sample preparation method for isolation of single-cell types from mouse liver for proteomic studies. Proteomics 11, 3556–3564 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Aizarani N., et al., A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacParland S. A., et al., Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruminy P., et al., Gene transcription in hepatocytes during the acute phase of a systemic inflammation: From transcription factors to target genes. Inflamm. Res. 50, 383–390 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Lu X., et al., NF-κB increased expression of 17β-hydroxysteroid dehydrogenase 4 promotes HepG2 proliferation via inactivating estradiol. Mol. Cell. Endocrinol. 401, 1–11 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Krenkel O., Tacke F., Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 17, 306–321 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Zilionis R., et al., Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alshetaiwi H., et al., Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci. Immunol. 5, eaay6017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hösel M., et al., Toll-like receptor 2-mediated innate immune response in human nonparenchymal liver cells toward adeno-associated viral vectors. Hepatology 55, 287–297 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Zhu J., Huang X., Yang Y., The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice. J. Clin. Invest. 119, 2388–2398 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao W., et al., Double-stranded RNA innate immune response activation from long-term adeno-associated virus vector transduction. JCI Insight 3, e120474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neamah W. H., et al., AhR activation leads to massive mobilization of myeloid-derived suppressor cells with immunosuppressive activity through regulation of CXCR2 and microRNA miR-150-5p and miR-543-3p that target anti-inflammatory genes. J. Immunol. 203, 1830–1844 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han X., et al., CXCR2 expression on granulocyte and macrophage progenitors under tumor conditions contributes to mo-MDSC generation via SAP18/ERK/STAT3. Cell Death Dis. 10, 598 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M., et al., CCL5 deficiency promotes liver repair by improving inflammation resolution and liver regeneration through M2 macrophage polarization. Cell. Mol. Immunol. 17, 753–764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno C., et al., CCR5 deficiency exacerbates T-cell-mediated hepatitis in mice. Hepatology 42, 854–862 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Mingozzi F., et al., CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat. Med. 13, 419–422 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Breous E., Somanathan S., Bell P., Wilson J. M., Inflammation promotes the loss of adeno-associated virus-mediated transgene expression in mouse liver. Gastroenterology 141, 348–357, 357.e1–357.e3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S. R. P., Hoffman B. E., Terhorst C., de Jong Y. P., Herzog R. W., The balance between CD8+ T cell-mediated clearance of AAV-encoded antigen in the liver and tolerance is dependent on the vector dose. Mol. Ther. 25, 880–891 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charlesworth C. T., et al., Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 25, 249–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meliani A., et al., Antigen-selective modulation of AAV immunogenicity with tolerogenic rapamycin nanoparticles enables successful vector re-administration. Nat. Commun. 9, 4098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin H. F., Maeda N., Smithies O., Straight D. L., Stafford D. W., A coagulation factor IX-deficient mouse model for human hemophilia B. Blood 90, 3962–3966 (1997). [PubMed] [Google Scholar]
- 44.Ayuso E., et al., High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 17, 503–510 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Mitchell C., Willenbring H., A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 3, 1167–1170 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Giladi A., et al., Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 20, 836–846 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aibar S., et al., SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Efremova M., Vento-Tormo M., Teichmann S. A., Vento-Tormo R., CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All high-throughput sequencing data can be viewed in NODE (http://www.biosino.org/node) by pasting the accession ID (DNA data: OEP001209`OEP001206) into the text search box.