Abstract
In the ribosomal DNA of Saccharomyces cerevisiae, sequences in the nontranscribed spacer 3′ of the 35S ribosomal RNA gene are important to the polar arrest of replication forks at a site called the replication fork barrier (RFB) and also to the cis-acting, mitotic hyperrecombination site called HOT1. We have found that the RFB and HOT1 activity share some but not all of their essential sequences. Many of the mutations that reduce HOT1 recombination also decrease or eliminate fork arrest at one of two closely spaced RFB sites, RFB1 and RFB2. A simple model for the juxtaposition of RFB and HOT1 sequences is that the breakage of strands in replication forks arrested at RFB stimulates recombination. Contrary to this model, we show here that HOT1-stimulated recombination does not require the arrest of forks at the RFB. Therefore, while HOT1 activity is independent of replication fork arrest, HOT1 and RFB require some common sequences, suggesting the existence of a common trans-acting factor(s).
The ribosomal DNA (rDNA) locus in the yeast Saccharomyces cerevisiae consists of 9.1-kb tandem repeats with the 35S rRNA gene, the much smaller 5S rRNA gene, and two nontranscribed spacer (NTS) regions (see Fig. 1) (see references 29 and 22 for reviews of sequence elements in the NTS). NTS2, located between the 5′ ends of the two genes, contains the promoter for the 35S rRNA gene, a weak origin of replication named the rDNA ARS, and sequences essential for the cis-acting mitotic recombination hot spot HOT1. The 35S RNA polymerase I transcriptional enhancer lies in NTS1 near the 3′ end of the 35S gene. NTS1 also contains sequences important for the polar arrest of replication forks (replication fork barrier [RFB]) and HOT1. The extent of sequence overlap and the interdependence of these two events in DNA metabolism are unknown.
FIG. 1.
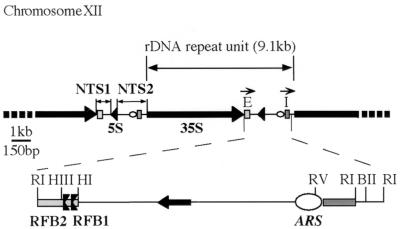
Features of the NTS region of S. cerevisiae. (Top) Repetitive nature of the rDNA array, where 100 to 200 rDNA repeat units (9.1 kb) are found in tandem on chromosome XII. The NTS lies between 35S genes and is separated into NTS1 and NTS2 by the intervening 5S gene. Fragments containing the 35S rDNA transcriptional enhancer and initiator (called E and I, respectively) are essential to HOT1-stimulated recombination (35) and are labeled with the arrows oriented in the direction of RNA polymerase I transcription of the 35S gene. (Bottom) A 2.68-kb EcoRI (RI) fragment that spans most of the NTS region. Only relevant restriction sites are shown. The positions of RFB1 and RFB2 within the 129-bp HindIII-HpaI (HIII-HI) fragment of the E fragment are delineated in this report. RFB1 and RFB2 block replication forks polarly, inhibiting forks traveling in the direction opposite of 35S gene transcription (2). The rDNA ARS, near the EcoRV (RV) site, is also noted. BII, BglII.
The rDNA RFB was first identified in S. cerevisiae, when high-resolution two-dimensional (2D) gel electrophoresis revealed two closely spaced sites where forks arrest (2), herein called RFB1 and RFB2. RFBs appear to be a highly conserved feature of rDNAs, with barriers being found at the 3′ end of the rRNA genes in a number of other organisms (9, 21, 23, 32, 36, 38). The yeast RFBs efficiently block replication forks traveling in the direction opposite to 35S transcription, together impeding ∼90% of encountered forks (2). Fork arrest is not a consequence of transcription per se, since replication forks still arrest at the RFB in cells lacking functional RNA polymerase I (2). The RFB sequences are also not inherently difficult to replicate (2), and thus fork arrest is thought to result from the binding of proteins at the RFB sequences. A protein-mediated mechanism of fork arrest in the rDNA RFB has also been implicated in peas and Tetrahymena thermophila (24, 37) and reported to involve the transcription-terminating factor TTF-I in mice and humans (8, 23).
HOT1 sequences from the rDNA, when assayed at ectopic sites in the genome, stimulate mitotic homologous recombination between intra- and interchromosomal repeats (14). Subcloning analysis showed that the sequences necessary for HOT1 recombination are localized to two noncontiguous regions of the rDNA NTS (35); the E fragment contains the enhancer for 35S transcription, and the I fragment contains the 35S promoter and initiation site (see Fig. 1). When the HOT1 sequences E and I are inserted next to a construct consisting of direct repeats of his4 sequences on chromosome III (see Fig. 2A), recombination can be elevated more than 350-fold (12). Through studies of recombination at this ectopic site, HOT1 activity has been shown to require RNA polymerase I transcription of the repeat elements involved in recombination (12, 35). Mutations in four genes, HRM1 through HRM4, reduce HOT1-stimulated recombination (19). HRM1 was later found to be identical to FOB1 (3), a gene that was identified in a search for mutants defective for both HOT1 and RFB activities (17). Studies on FOB1 indicate that the protein is important for the expansion and contraction of the rDNA array (15) and plays a role in regulating life span (3). The FOB1 protein is a candidate for creating the physical fork barrier at the RFB, but it is not yet known whether the protein functions by directly binding to DNA.
FIG. 2.
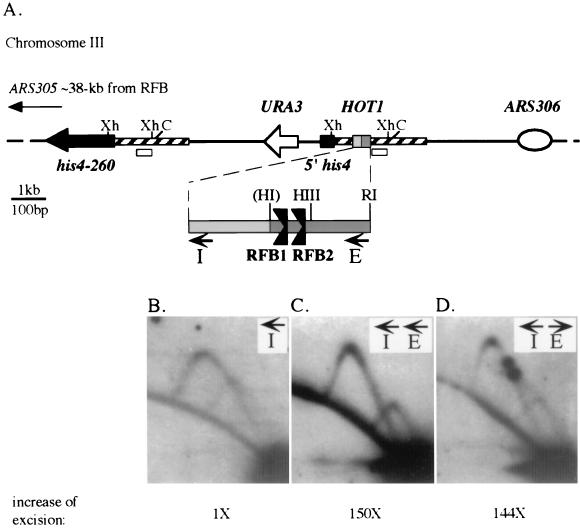
HOT1-stimulated recombination independent of RFB fork arrest. (A) Construct for HOT1 quantitative intrachromosomal recombination assay. The sequences that lie between the ClaI (C) sites were integrated into the his4 locus on chromosome III of RLK 88-3C as previously described (12). The E-I HOT1 sequences are inserted to the 5′ side of the repeated his4 sequences to stimulate homologous recombination between these sequences. Intramolecular recombination between repeated sequences of the 5′ end of his4 or the flanking chromosomal DNA, indicated by the striped area, can result in excision of the intervening URA3 marker. The two flanking origins are indicated: ARS305 and ARS306 are located ∼38 and ∼6 kb, respectively, from the E-I region. The bottom map displays the orientation of the 255-bp I and 320-bp E fragments and the RFBs. The arrows for E and I and restriction sites in E are indicated as in Fig. 1. The HpaI (HI) site is not present in this construct. Xh, XhoI. (B, C, and D) Autoradiograms of high-resolution 2D gel of three HOT1 constructs at the his4 locus on chromosome III. For each gel, the XhoI fragment was probed with chromosomal sequences (open rectangles in panel A). The XhoI fragment of interest is a 1.7-kb fragment for B and a 2-kb fragment for C and D. Due to the duplicated chromosomal and his4 sequences in the HOT1 cassette, the probe hybridized to a second XhoI fragment near the his4-260 gene. Hybridization to this smaller fragment, 1.4 kb, is observed as a second simple Y arc in the lower right corner of the 2D gels. The number beneath each gel is the fold stimulation of excision by HOT1, taken from Voelkel-Meiman et al. (35). These data were reconfirmed in the present study (data not shown). The strains used were M51 (B), M39 (C), and M78 (D) (35).
Evidence from Escherichia coli that the arrest of replication forks at sequence-specific sites may be recombinogenic (1, 11, 11a; reviewed in references 18 and 31) has led to the hypothesis that forks blocked at the RFB contribute substantially to HOT1 recombination (15, 17). However, the apparent difference between the transcription requirements for fork arrest at the RFB and for HOT1-stimulated recombination and the requirement of the I fragment for only the latter event might suggest that these activities are independent. We report here that fork arrest is not required for HOT1 recombination. However, we show that RFB activity and HOT1 recombination share some common cis-acting sequences in the rDNA NTS1 region.
MATERIALS AND METHODS
Construction of RFB plasmids.
Plasmid pBB3NTS (see Fig. 3A) was provided by Katherine Friedman and was constructed as follows. Vector pBB3 was constructed by ligating the 967-bp NdeI-SmaI URA3 fragment to the 2.435-kb NdeI-SmaI pUC18 vector. Yeast RM14-3a DNA prepared by glass bead lysis (10) was cleaved with EcoRI. Fragments were separated by gel electrophoresis, and a visible rDNA band of approximately 2.5 kb was excised and electroeluted. This fragment was cloned into the unique EcoRI site of vector pBB3. The resulting plasmid was partially digested with EcoRV, and a 425-bp NheI-HindIII fragment containing ARS1 was blunt-ended with Klenow enzyme (Boehringer) and cloned into the EcoRV site of the rDNA. ARS1 was oriented so that its HindIII site is closest to the RFB. The HindIII-HpaI fragment in plasmid pBB3NTSΔHH was deleted by digesting plasmid pBB3NTS with HindIII and HpaI, filling in the HindIII site with Klenow polymerase, and ligating the resulting ends.
FIG. 3.
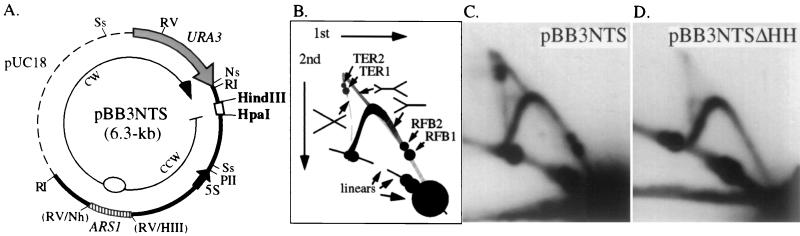
Deletion of the HindIII-HpaI region eliminates RFB1 and RFB2. (A) Map of the 6.3-kb plasmid pBB3NTS. The dashed line is vector sequence from pUC18. Only relevant restriction sites are noted. The thicker line between the EcoRI (RI) sites corresponds to the 2.46-kb EcoRI NTS region (a subfragment from the restriction map in Fig. 1) from the rDNA of RM14-3a. The locations of ARS1 and URA3 are indicated. A 425-bp NheI-HindIII (Nh-H3) fragment containing ARS1 was blunt-ended and inserted into the EcoRV (RV) site near the rDNA ARS (Fig. 1) to improve the efficiency of extrachromosomal maintenance of the plasmid. Bidirectional replication initiating from ARS1 creates a CCW fork that is blocked by the RFBs before meeting the CW fork. Ss, SspI; Ns, NsiI; Sp, SphI; PII, PvuII. (B) Schematic diagram of the migration of different replication intermediates in 2D gels shown in C and D. Accumulation of arrested forks results in the two intense spots of hybridization (RFB1 and RFB2) along the arc of Y intermediates. The nearly vertical dashed line represents the pattern of hybridization seen for X-shaped, or terminating, molecules. Termination at RFB1 and RFB2 results in the accumulation of X-shaped molecules TER1 and TER2, respectively. The thicker diagonal gray line corresponds to the hybridization pattern for double-Y replication intermediates. (C and D) High-resolution 2D gels of the 2.2-kb SspI (Ss in panel A) fragment from pBB3NTS and pBB3NTSΔHH, respectively, probed with URA3 sequences.
YEp24 plasmids containing the HindIII-HpaI RFB fragment were constructed in two steps following the procedure used by Kobayashi et al. (16). The HindIII site of the HindIII-HpaI fragment from pBB3NTS was blunted with Klenow polymerase, and the fragment was ligated into the HincII site of the polylinker of pUC18. Plasmids were screened for orientation of the insert by sequencing across the plasmid with the M13 sequencing primer 1211 (New England Biolabs). For each orientation, the SphI-BamHI fragment of the pUC18 derivatives was cloned between the SphI and BamHI sites of YEp24 to create plasmids YEp24HH+ and YEp24HH−. YEp24HH+ (see Fig. 5A, top) contains the HindIII-HpaI fragment in the orientation expected to block replication forks coming from the 2μm origin of replication.
FIG. 5.
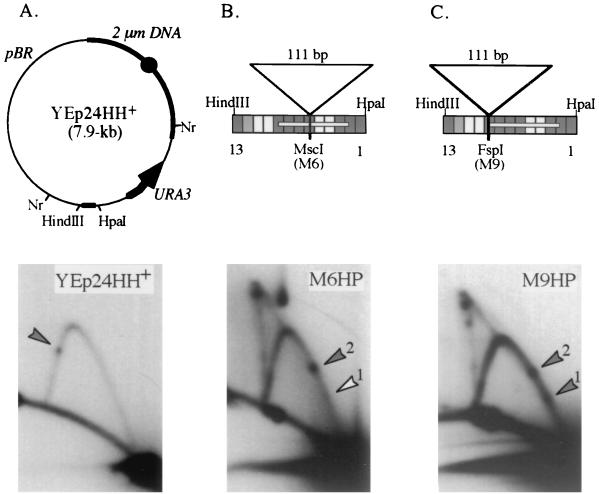
HindIII-HpaI sequences sufficient only for RFB1. (A) Map of the 7.9-kb plasmid YEp24HH+ and a high-resolution 2D gel of the 2.4-kb NruI (Nr) fragment. The 2μm origin (solid circle) and the HindIII-HpaI insert are labeled. Also shown are the locations of the URA3 gene and pBR322 (pBR) sequences. (B and C) Maps of 111-bp insertion mutations between RFB1 and RFB2. Disruptions in the HindIII-HpaI region were tested for RFB function in plasmid pBB3NTS (see Fig. 3A for map). The 2.3-kb SspI fragment was probed with URA3 sequences. The map above each gel is similar to the diagram in Fig. 4A, and mutations 1 and 13 are noted for orientation. The white bar within the chart shows the location of the 69-bp minimum RFB sequence determined by Kobayashi et al. (16). Mutation M6HP has an 111-bp insertion (from pUC18) in the MscI site created by the M6 mutation. Mutation M9HP contains the same 111-bp fragment inserted just outside of the 69-bp sequence in the FspI site created by the M9 mutation. The spot located above the double-Y line of replication intermediates in B is background hybridization. See the legend to Fig. 4 for other details.
Two plasmids were constructed for in vitro mutagenesis. The first construct, used to make mutations M2 through M11, in which blocks of DNA in the region required for RFB activity were replaced, was made by inserting the 837-bp NTS1 EcoRI-PvuII fragment from pBB3NTS into the EcoRI and PvuII sites of the polylinker of pUC118 to create pUC118RFB. Mutated HindIII-HpaI fragments were excised from pUC118RFB and ligated between the HindIII and HpaI sites of pBB3NTS in place of the wild-type fragment. All mutations were confirmed by sequencing and then tested for RFB function. The second mutagenesis construct, used to make mutations M1, M12, and M13, consisted of an insertion of the NsiI-PvuII fragment of pBB3NTS in place of the small NsiI-PvuII fragment of a modified YIp5 vector (pMUTBIAS, provided by Katherine Kolor, has a mutation in the ampicillin resistance gene created by filling in a PstI site). The resulting plasmid, pMUTBIASRFB, was ampicillin sensitive. For sequencing and testing of RFB function, the NsiI-SphI fragment of a mutated pMUTBIASRFB was ligated between the SphI and NsiI sites of pBB3NTS in place of the wild-type fragment.
Site-directed mutagenesis.
Site-directed mutations were made in the HindIII-HpaI fragment by oligonucleotide-directed mutagenesis (4). The annealing reaction mixture consisted of 70 ng of vector, 25 pmol of each kinase-treated oligonucleotide, 2 μl of solution TN (0.2 M Tris-HCl [pH 7.5], 0.5 M NaCl), and 2 μl of 0.1 M MgCl2 in a total volume of 20 μl. This reaction mixture was incubated at 100°C for 3 min and then chilled for 5 min on ice. The synthesis reaction mixture consisted of the 20-μl annealing reaction, 3 μl of solution TDD (5 mM deoxynucleoside triphosphates, 0.1 M Tris-HCl [pH 7.5], 20 mM dithiothreitol), 1 μl (3 U) of T4 DNA polymerase (New England Biolabs), and 1 μl (400 U) of T4 DNA ligase (New England Biolabs) in a total volume of 30 μl. This reaction mix was incubated at 37°C for 90 min. The synthesis reaction was stopped by the addition of 3 μl of solution SE (0.25% sodium dodecyl sulfate, 5 mM EDTA) and a 5-min incubation at 65°C.
For mutagenesis of plasmid pUC118RFB, 5 μl of the synthesis reaction mixture was used to transform the mismatch repair-defective E. coli strain DSM3 (33). Primary transformants were cultured in 10 ml of Luria-Bertani medium with 10 μg of ampicillin per ml overnight. Plasmids were recovered with a Qiagen Midi Column procedure. Plasmid DNA (1 μg) was digested with ScaI in a 20-μl reaction mix to select against the parental plasmid which had not incorporated the selection oligonucleotide (CTGTGACTGGTGACGCGTCAACCAAGTC). Then 5 μl of the digest was transformed into E. coli DH5α. Plasmids which had lost the ScaI site were then screened for the presence of the new restriction site indicative of a mutation in the HindIII-HpaI fragment. Approximately 75% of plasmids that had incorporated the selection oligonucleotide had also incorporated the mutagenesis oligonucleotide.
For mutagenesis of plasmid pMUTBIASRFB, 5 μl of the synthesis reaction mix was used to transform the mismatch repair-defective E. coli strain DSM3, and after a 2-h incubation at 37°C, transformants were spread on plates containing ampicillin (10 μg/ml). Incorporation of the selection oligonucleotide (CACCACGATGCCTGCAGCAATTGGCAAC) restores the PstI site in the ampicillin resistance gene so that cells containing the plasmid are ampicillin resistant. Ampicillin-resistant colonies were screened by restriction digest to determine if the plasmid had incorporated the mutagenesis oligonucleotide. The wild-type and mutant sequences for each HindIII-HpaI mutation (M1 to M13) are shown in Table 1.
TABLE 1.
Mutations in the HindIII-HpaI region
| Mutation | Wild-type sequence | Mutant sequence | New restriction site |
|---|---|---|---|
| M1 | TTTCCTATAGTT | GGGATCCGCTGG | BamHI |
| M2 | GAAAAGCTCA | AGGCCTAGAG | StuI |
| M3 | AGAGAATTGA | CTCTCCGGAC | BspEI |
| M4 | GTATAAGTTT | TGCGCCCGGG | SmaI |
| M5 | ATGAGTGCTT | CGTCCCATGG | NcoI |
| M6 | AGCGGCAAAC | CTATTGGCCA | MscI |
| M7 | GCACCATCAG | TACGTCGACT | SalI |
| M8 | AGTTTTTTCC | CCGGGGGGAA | SmaI |
| M9 | TTCATGGAGC | GCACGTTCTA | FspI |
| M10 | GACAGTTTGC | TCACTGGGAA | EcoRI |
| M11 | GATTTGCCCG | ACGCGTAAAT | MluI |
| M12 | AGCGTGAAAG | CTATGTTCTA | XbaI |
| M13 | AAGCTTCCCG | CCATGGAAAT | NcoI |
Construction of plasmids containing HOT1 mutations.
The C20 single-base-pair mutation (12) was reconstructed in the RFB test plasmid by two in vitro mutagenesis steps. First, a 2-bp mutation which created an MluI site and included the C20 single-base-pair conversion was produced by using the pMUTBIASRFB construct as explained above. Second, the NsiI-PvuII fragment that included the 2-bp mutation was moved into a fresh pMUTBIAS vector, mutagenized to the C20 single-base-pair mutation, and screened for the loss of the MluI site. The C20 mutation was cloned into pBB3NTS for sequencing and testing for RFB function as explained above.
To house the other HOT1 mutations for 2D gel analysis, pBB3NTS was modified to create plasmid L3520 by replacing the HpaI site with an XbaI linker and deleting the EcoRI site distal to the enhancer. The 320-bp EcoRI-XbaI enhancer-containing fragment of L3520 was deleted and replaced with an EcoRI-HindIII-XbaI polylinker to create L3520ΔE. The HOT1 mutants G182, G188, G190, and N35 (mutant sequences are described in reference 12) were recovered as a 320-bp EcoRI-XbaI fragment and ligated into the polylinker site of L3520ΔE for assay by 2D gel.
2D agarose gel conditions.
DNA for 2D gels was isolated from asynchronous, log-phase cultures as described previously (7). The yeast strain RM14-3a (MATa cdc7-1 bar1 ura3-53 trp1-289 leu2-3,112 his6) was used to construct the strains for all 2D gel experiments. From 1 to 4 μg of DNA was used for each 2D gel. The conditions for the first dimension of the gels to test the mutant plasmids and integrated constructs were 0.5% agarose and 1 V/cm for approximately 22 h at 23°C. Conditions for the second dimension were 1.5% agarose, 0.3 μg of ethidium bromide per ml, and 4 to 5 V/cm for 5 to 6 h at 4°C. Probes were labeled by the hexanucleotide priming method (5).
HOT1 quantitative intrachromosomal recombination assay.
Mutations M4, M5, M6, M10, and M11 were isolated from the pBB3NTS clones as a 320-bp EcoRI-HpaI fragment. The EcoRI and HpaI ends of the fragment were converted to BamHI and XbaI, respectively, by the addition of linkers. The resulting fragment replaced the corresponding region of the plasmid G141 (12). All plasmids were digested at the unique ClaI site and targeted to the his4 locus of chromosome III in RLK 88-3C (35). Mutant chromosomal constructs were confirmed by Southern blot hybridization, and the HOT1 activity of these mutants was assayed as previously described (12).
RESULTS
Does replication fork arrest contribute to HOT1 recombination?
An appealing hypothesis to explain HOT1 mitotic hyperrecombination is that replication forks stalled at the RFB are fragile (17, 31). DNA strand breakage at the RFB may generate a lesion that is repaired by homologous recombination. This possibility led us to test whether the two phenotypes are related mechanistically. That is, is the level of HOT1-stimulated recombination detected at the chromosome III site dependent on the efficiency of the RFB at this site? The ultimate test of this hypothesis is to determine whether forks are actually being blocked at the chromosome III HOT1 assay site and whether the presence or absence of barriers correlates with the functional status of HOT1.
The E and I fragments of the rDNA NTS (Fig. 1) that are required for HOT1 activity were assayed for their ability to stimulate recombination at an ectopic site on chromosome III (Fig. 2A) (12, 35). Previous work revealed that both orientations of the E element conferred similar levels of HOT1 recombination (35). If fork arrest at the polar RFB does play a role in HOT1 activity, then replication forks must proceed through the E fragment equally in both directions within different cells in a population to establish a similar number of blocked forks. The E fragment is located between ARS305 and ARS306, which fire at similar times early in S phase (30) and initiate very efficiently (28). However, the asymmetrical location of E, closer to ARS306 than to ARS305, predicts that E would be replicated by forks from ARS306 (Fig. 2A). The normal test orientation of the E fragment in the HOT1 chromosome III assay is such that replication forks reaching this locus from ARS306 travel in the direction where they do not confront the barrier activity in E. Consistent with this expectation, a test for barriers in the XhoI fragment containing HOT1 sequences gave no evidence for the RFB (Fig. 2C). The 2D gel pattern was comparable to that of the construct that was missing the 320-bp EcoRI-HpaI E fragment (Fig. 2B). These data suggest that this region is replicated by forks traveling from ARS306. To confirm this proposal, we performed 2D gel analysis of the direction of replication fork movement (7) in these constructs. We observed that >95% of the replication forks arrive at the HOT1-RFB locus from ARS306 (data not shown). Therefore, >95% of the forks replicate through the E region in the permissive direction.
The possibility that the RFB is not active at this chromosome III location was tested by inverting the E element so that it would now be expected to arrest forks arriving from ARS306. High-resolution 2D gel analysis shows that both arrest sites in the HOT1 XhoI fragment, RFB1 and RFB2, are functional (Fig. 2D). This observation was confirmed by examining two other restriction fragments in which the RFB was located at different positions (data not shown). From these results, we conclude that a replication fork reaches the E region from ARS306 and that, if oriented properly, the RFB is capable of efficient fork arrest. Therefore, the two constructs that differ in the orientation of E have significant differences in fork blocking at the RFB. If arresting forks did contribute to HOT1 activity, we would expect a significant increase in levels of HOT1-stimulated recombination from the construct that impeded forks. However, as observed earlier (35) and reconfirmed for the present study, the two constructs were indistinguishable in the level of excisive recombination (bottom of Fig. 2C and D and data not shown). These findings clearly demonstrate that the stimulation of recombination associated with the HOT1 sequences is not dependent upon stalled replication forks.
Role of the 129-bp HindIII-HpaI fragment in RFB1 and RFB2.
Although HOT1 recombination does not require replication fork arrest, it is possible that common cis-acting sequences play a role in these two activities. To test this possibility, it was first necessary to better define the sequences involved in the rDNA RFB activity.
Previous mapping of the barrier in the yeast rDNA NTS by 2D gel analysis demonstrated that forks initiating at the rDNA ARS in NTS2 arrest between the HindIII and HpaI sites (Fig. 1) (2, 16). High-resolution 2D gel analysis of the chromosomal RFB revealed two discrete arrest sites of unequal strength in this region, RFB1 and RFB2 (2). Because the barriers continue to function when transplanted to plasmids (2, 16), it was possible to perform deletion analysis to identify the sequences important for RFB activity. Kobayashi et al. (16) determined that a 69-bp region within the 129-bp HindIII-HpaI fragment was sufficient to generate reduced RFB activity on a plasmid. However, their 2D gel conditions failed to resolve the two arrest sites, and it is not known whether the 69-bp region or the 129-bp HindIII-HpaI fragment containing it is sufficient for arrest at both RFB1 and RFB2.
To determine if sequences for both RFB1 and RFB2 activity are present in the 129-bp HindIII-HpaI fragment, we analyzed replication of an NTS region from which this fragment was deleted. First, a plasmid was constructed to include the 2.46-kb EcoRI fragment that spans most of the NTS region of the rDNA (Fig. 1). The rDNA ARS that lies within this EcoRI fragment proved to be inefficient in episome maintenance, with the plasmid often integrating into chromosomal rDNA. Therefore, the efficient ARS1 origin was inserted at the EcoRV site near the rDNA ARS (Fig. 1), creating plasmid pBB3NTS (Fig. 3A). The location and orientation of the RFB with respect to the origin in pBB3NTS ensured that the replication fork proceeding counterclockwise (CCW) from ARS1 will encounter the RFB before the fork moving clockwise (CW) (Fig. 3A). Therefore, replication intermediates in which the CCW fork arrests at the RFB until it is met by the CW fork will accumulate. To detect the accumulation of these branched molecules, we examined the 2.2-kb SspI fragment from pBB3NTS under 2D gel conditions in which the two arrest sites, RFB1 and RFB2, were revealed (Fig. 3C). Although the plasmid RFB generated less intense spots relative to chromosomal barriers (2), the plasmid RFB blocks replication forks long enough for site-specific termination events to occur. Accumulation of the two expected replication termination structures, TER1 and TER2, that correspond with the arrest at RFB1 and RFB2, respectively, are observed along the hybridization line of X-shaped molecules (Fig. 3B and C).
Next, the role of the HindIII-HpaI fragment in RFB1 and RFB2 was assessed by deleting it from pBB3NTS to create pBB3NTSΔHH. Under high-resolution 2D gel analysis, the replication intermediates from pBB3NTSΔHH lacked any trace of either arrest site and either specific termination site (Fig. 3D). These results indicate that the HindIII-HpaI fragment is necessary for the function of both RFB1 and RFB2 on a plasmid. They also demonstrate that no other regions within the EcoRI fragment, which spans most of the NTS region, were sufficient to significantly arrest the progress of replication forks.
Sequences within the HindIII-HpaI fragment responsible for RFB1 and RFB2.
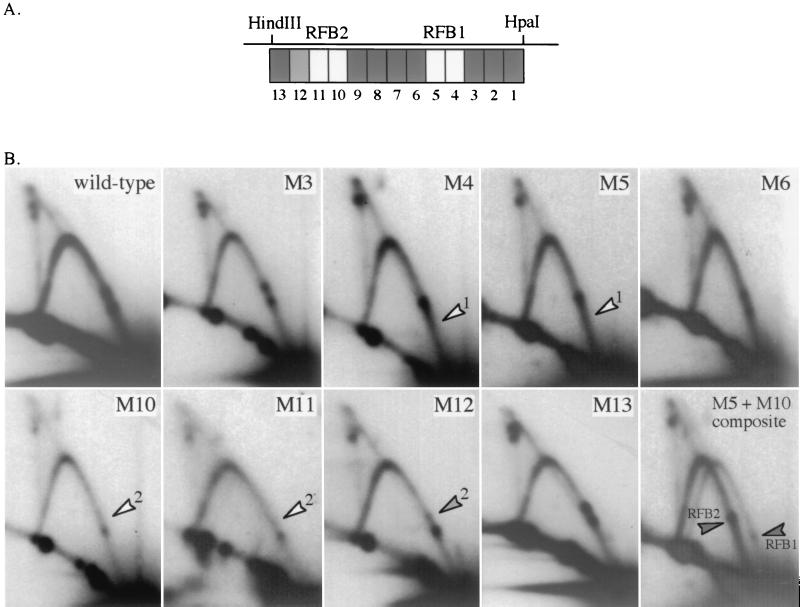
Since it appeared that the HindIII-HpaI fragment is necessary to generate RFB1 and RFB2 at an ectopic site on a plasmid, we subjected the 129-bp HindIII-HpaI region to systematic mutagenesis of consecutive 10-bp regions (12 bp in the case of M1). Mutations in the HindIII-HpaI region were generated in a mutagenesis plasmid (see Materials and Methods) and then transferred to pBB3NTS, in which RFB function could be tested in the context of almost the entire NTS region. Each base pair within the sequence targeted for mutagenesis was replaced with another base pair. An effort was made to create the least conservative changes possible: adenines were replaced with cytosines, guanines were replaced by thymines, and vice versa. However, for ease of screening, each mutation created a new restriction site that sometimes made it impossible to make the least conservative change in the region. In no case did any base within the mutated sequence remain the same as it was in the wild-type sequence (Table 1). All mutated HindIII-HpaI regions were sequenced to confirm that no unintended base pair changes were created.
High-resolution 2D gel analyses of SspI fragments from each of the 13 block mutations were performed. Figure 4A summarizes all of the 2D results obtained, and the autoradiograms of wild-type plasmid and some mutant plasmids are shown in Fig. 4B. Eight of the 13 mutations do not differ significantly from wild-type RFB behavior (Fig. 4A; M3, M6, and M13 in Fig. 4B). All exhibit two intense spots along the simple Y arc, corresponding in location to RFB1 and RFB2 and the two prominent termination signals. Mutations M4 and M5 displayed only one barrier, located at the position of RFB2 and a single termination species at the position of TER2 (Fig. 4B). This result indicates that the sequences within the 20 bp covered by mutations M4 and M5 are required for RFB1 but not for RFB2 when the NTS region is on a plasmid.
FIG. 4.
Scanning mutagenesis of the 129-bp HindIII-HpaI region. (A) Diagram showing the locations of the 12 10-bp (M13 to M2) and 1 12-bp (M1) block mutations within the HindIII-HpaI fragment. The mutant and the wild-type sequences are listed in Table 1. The raw data in panel B are summarized by the shading of the boxes in A: black, similar to wild type; gray, reduced fork arrest; white, absence of fork arrest. (B) High-resolution 2D gel analysis of the block mutations in the HindIII-HpaI region. Mutations were tested for RFB function in plasmid pBB3NTS (see Fig. 3A for map). The 2.2-kb SspI fragment was probed with URA3 sequences. A 2D gel of pBB3NTS containing the wild-type HindIII-HpaI sequence is shown for comparison. Open arrowheads point to loss of an RFB, and gray arrowheads point to a significantly decreased RFB. The M5 + M10 composite is an overlay of the M5 and M10 autoradiograms shifted laterally to display the relative and easily distinguishable positions of RFB1 and RFB2, indicated by solid arrowheads.
Similar analysis of mutations M10, M11, and M12 uncovered the importance of a nearby 30-bp stretch in causing the arrest of forks at RFB2. M10 and M11 completely abolished arrest at RFB2 (Fig. 4B). These changes in an RFB activity are again reflected in the shift of terminating structures, in this case to TER1. M12 produced a somewhat reduced accumulation of arrested forks at RFB2 and termination structures at TER2. Overlapping autoradiograms of M5 and M10, offset horizontally by 4 mm, allow the unambiguous assignment of RFB1 and RFB2 in the two mutations (last panel of Fig. 4B).
To test whether the plasmid results are valid in the context of the chromosomal rDNA locus, plasmids containing the mutation at either M5 or M11 were integrated into the rDNA on chromosome XII. The plasmid used for chromosomal integration was similar to pBB3NTS but lacked the ARS1 sequence and contained a fragment of λ DNA to serve as a unique hybridization probe. Transplacement of just the NTS region was attempted; however, when 10 transformant cultures of each mutant type were screened for the presence of the unique restriction site created by the mutation, all were found to have lost the restriction site, indicating that the mutations had been repaired through gene conversion to wild-type sequence. A lower rate of gene conversion was seen when the entire plasmid was integrated into the rDNA in the BglII site at the 5′ end of the 35S gene (Fig. 1); 25 to 50% of the transformants screened retained the restriction site indicative of the mutated constructs. High-resolution 2D gel analysis of the chromosomal M5 mutation showed the complete loss of RFB1 at the rDNA locus (data not shown), a result which is consistent with the plasmid analysis. Similarly, the chromosomal M11 mutation abolished RFB2 activity (data not shown). The results for the rDNA integrants of mutations M5 and M11 support the validity of using the plasmid model to investigate RFB function.
Sequences sufficient for RFB1 and RFB2 activity.
Kobayashi et al. (16) found that the 129-bp HindIII-HpaI fragment alone had RFB activity, although it seemed reduced. We recreated their minimal HindIII-HpaI plasmid (YEp24HH+; Fig. 5A, top) to test whether their minimal construct was sufficient for both RFBs or only one. Analysis by high-resolution 2D gels showed the presence of only a single RFB species (Fig. 5A, bottom). Further deletion of the 129-bp fragment by Kobayashi et al. reduced the sequences sufficient for RFB activity to a 69-bp fragment (white bar in Fig. 5B and C, top). Since these sequences include M4 and M5, which are required for RFB1, but do not include those sequences necessary for RFB2 (M10 to M12), we can conclude that the 69-bp region is sufficient for RFB1.
The 69-bp sequence sufficient for RFB1 activity cannot be further trimmed by deletion without complete loss of RFB activity (16), yet our mutagenesis data suggest that the sequences within these deletions, about 35 of these 69 bp (M6 to mid-M9), may not be necessary for RFB1 (white bar inside map in Fig. 5B and C, top). One possibility is that the M6 to M9 region contains functionally redundant sequence elements that contribute, together with sequences in M4 and M5, to RFB1 activity. If so, moving the M4-M5 and M6-M9 regions apart might affect RFB1 function. As a test of this idea, we inserted a 111-bp fragment of pUC18 DNA into the restriction site of the M6 mutant sequence (Fig. 5B, top). The insertion mutation in the HindIII-HpaI region was tested for RFB function in plasmid pBB3NTS (Fig. 3A). High-resolution 2D gel analysis shows that RFB1 activity is eliminated (Fig. 5B, bottom). As a control, the same 111-bp fragment was inserted outside of the 69-bp region, in the restriction site of the mutant M9 sequence (Fig. 5C, top). This construct retains both RFB1 and RFB2 activity (Fig. 5C, bottom), with the distance between the two spots increased compared with the barrier spacing of the wild-type HindIII-HpaI fragment (compare with Fig. 3C). These findings support the functionally redundant sequence hypothesis and indicate that the spacing between these functionally redundant sequences and M4-M5 is crucial for RFB1 fork arrest.
From these data, we conclude that the HindIII-HpaI NTS fragment contains discrete sequences that uniquely contribute to the specification of RFB1 and RFB2. It should be noted that the identified sequences required for RFB1 and RFB2 function do not necessarily coincide with the sequences at which the nascent strands are arrested. Sequences essential for RFB1 map to the 20-bp region defined by mutations M4 and M5 and unspecified, redundant sequences in the region between M6 and M9. RFB2 depends absolutely on the sequences defined by M10 and M11 and to a lesser extent on sequences in the region defined by M12. However, these 30 bp are not by themselves sufficient for RFB2; on low-resolution 2D gels, Brewer et al. (2) determined that sequences that lie 35S gene-proximal to the HindIII site contributed to RFB activity. In addition to the HindIII-HpaI fragment, sequences within the 188-bp EcoRI-HindIII fragment most likely comprise the region sufficient for full RFB2 activity.
Correlation between RFB and HOT1 sequences.
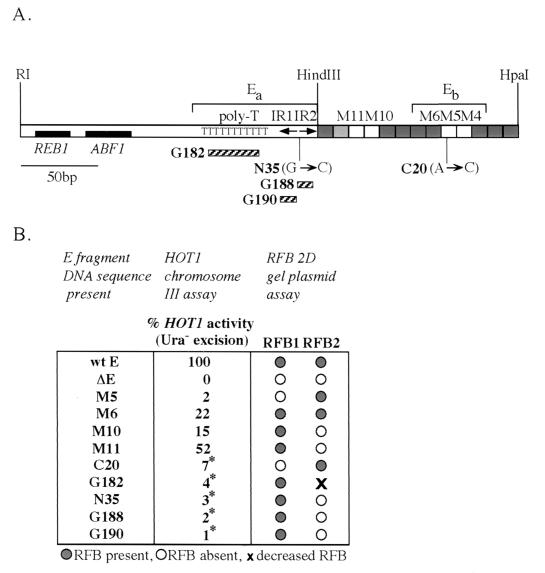
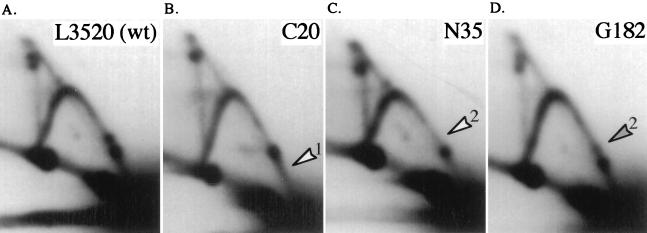
Mutations in the E fragment that abolished HOT1 activity were identified by Huang and Keil (12) in a screen using the chromosome III HOT1 recombination assay (Fig. 2A). Those mutations were scattered in the right halves of both the HindIII-HpaI (Eb) and EcoRI-HindIII (Ea) fragments of E (see Fig. 7A). Five of these mutations (G182, N35, G188, G190, and C20) were moved into the RFB test plasmid pBB3NTS, and the presence of RFB activity was determined by high-resolution 2D gels. Results for the wild-type construct (L3520) and three of the mutations are shown in Fig. 6. C20, a single-base-pair mutation in the M5 region, cleanly abolished RFB1 (Fig. 6B). Within the inverted repeat region of Ea, the point mutation N35 and the scrambled sequence block mutations G188 and G190 eliminated RFB2 (Fig. 6C and data not shown). G182, a block mutation in the poly(T) region and the most distal mutation from the HindIII-HpaI fragment tested, had a more modest effect on RFB2, showing a reduced efficiency of fork arrest at this site (Fig. 6D). As would be predicted, deleting the entire E fragment results in the elimination of both barriers (data not shown). All of these mutations also result in the drastic reduction of HOT1 recombination (Fig. 7B) (12). These data show that sequences required for RFB1 and RFB2 overlap sequences essential to HOT1-stimulated recombination.
FIG. 7.
Summary of the effect of HOT1 and RFB activity on E fragment mutations. (A) Map of the 320-bp EcoRI-HpaI E fragment located from the rDNA NTS1 (Fig. 1). The HindIII-HpaI fragment is represented as in Fig. 4A. Only the mutants that were tested for HOT1 activity are noted. Ea and Eb are the two regions within the E fragment that were identified by linker insertion mutagenesis to be required for HOT1 activity (34). The striped bars underneath the map represent the HOT1 block mutations tested in the RFB plasmid assay: G182 in the poly(T) region and G190 and G188 in inverted repeats IR1 and IR2, respectively. Mutant and wild-type sequences for the HOT1 block mutations are given in Huang and Keil (12). The base pair changes and locations of the point mutations N35 and C20 are indicated. The locations of two identified 35S enhancer-binding proteins, REB1 and ABF1, are noted (13, 27). (B) Summary chart of all the mutant sequences tested. wt E indicates the presence of wild-type E sequences; ΔE signifies that the E fragment is absent. HOT1 recombination was measured as the relative excision rate of the URA3 marker in the chromosome III construct (see Fig. 2A) as previously described (12). *, data from reference 12.
FIG. 6.
E fragment mutations that affect HOT1 and RFB activity. (A) 2D gel of the 2.2-kb SspI fragment of wild-type (wt) L3520. L3520 differs from pBB3NTS (see Fig. 3A) in two restriction sites (detailed in Materials and Methods). L3520 was used to clone the N35 and G182 mutations, which are shown in the other panels, while C20 was cloned into pBB3NTS. (B, C, and D) 2D gel analysis of the 2.2-kb SspI fragment of C20, N35, and G182. C20 and N35 are point mutations (see Fig. 7A for wild-type and mutant sequences), and G182 is a sequence-scrambled block mutation (see reference 12 for wild-type and mutant sequences). The locations of these mutations within the E fragment are shown in Fig. 7A. See the legend to Fig. 4 for other details.
Since the screen for HOT1 mutations may not have been a saturating screen, the failure to find HOT1 mutations that mapped to the M10 and M11 region may not be meaningful. To test the role of the M10-M11 region in HOT1 recombination and to test additional 10-bp linker mutations, both normal and mutant for RFB function, five of the mutations were cloned into the E fragment of the HOT1 assay cassette on chromosome III (Fig. 2A) and their effects on HOT1 recombination were assessed (Fig. 7B). M5, as expected from the previous C20 result, completely eliminated HOT1 activity. However, the adjacent M6 mutation, which had no effect on either RFB, reduced HOT1 activity substantially (about fivefold). Of the two mutations that eliminate RFB2, M10 significantly reduced HOT1 activity (sixfold), whereas M11 decreased HOT1 by only twofold. The results with M6 and M11 indicate that either HOT1 or RFB activity can be dramatically decreased or eliminated by mutation without great reduction in the other one. Therefore, we find that the cis-acting sequences for RFB and HOT1 are not entirely coincident.
Mutation M4 behaved anomalously, eliminating RFB1 but producing variable effects on HOT1 recombination. The recombinational excision rates for 14 independent M4 transformants ranged from less than 1% to more than 100% of the wild-type value (data not shown). Most of the isolates showed a very modest (twofold) decrease in recombination. Sequence analysis showed that the M4 mutation was still present in all 14 transformants examined. The reason for the unique variability of mutation M4 on HOT1-stimulated recombination is unknown at this time.
DISCUSSION
HOT1-stimulated recombination is independent of blocked replication forks.
Sequences necessary for both RFB activity and HOT1 recombination reside at the 3′ end of the 35S gene. An attractive model to explain this colocalization is that replication forks arrested at the RFB are prone to strand breaks that can stimulate homologous recombination (17). If this model were correct, then, because the arrest is polar, the direction of replication of the HOT1 sequences should determine the level of HOT1-stimulated recombination. Earlier work showing that the E element functions in an orientation-independent manner in HOT1 recombination (35) cast doubt on this model. Here we have demonstrated directly, both by fork direction analysis and by 2D gel visualization of fork arrests, that RFB arrested forks are not required for HOT1-stimulated recombination. These findings are surprising, considering the current model that proposes that RFB arrested forks stimulate rDNA recombination by initiating a breakage event (3, 15, 30a). Our studies do not support the paradigm that stalled forks are fragile sites at which recombinational repair is induced. Instead, we favor the idea that proteins that stimulate HOT1 recombination may, as a consequence of DNA binding, have the ability to arrest replication forks.
While there is evidence in E. coli that the arrest of replication forks can lead to double-strand breaks (1a, 11, 26), there is no physical evidence that normal fork arrest at the yeast RFB causes breaks in vivo. Indeed, two observations suggest that replication forks arrested at the yeast RFB may have less single-stranded character than moving forks and thus may be more stable. Linskens and Huberman (20) observed that RFB arrested forks behave on BND-cellulose chromatography as if they possessed more double-stranded regions than moving forks. Consistent with this interpretation, Lucchini and Sogo (25) noted that the DNA immediately behind the stalled RFB fork appeared to be mostly double stranded when visualized by electron microscopy after psoralen cross-linking. The lagging strands at forks arrested at the RFB site thus appeared to have completed Okazaki fragment replication and ligation. Therefore, the RFB arrested forks may be less fragile, hence less susceptible to breakage and recombinational repair, than moving forks.
Sequences responsible for RFB1 and RFB2 are distinct and independent.
Earlier work revealed that the S. cerevisiae rDNA RFB consisted of two discrete arrest sites (2), herein named RFB1 and RFB2. Using high-resolution 2D gel analysis of plasmid replication intermediates, we have now further defined the sequences required for arrest at these two sites. While the 129-bp HindIII-HpaI region in NTS1 (Fig. 1) is necessary for fork arrest at both RFB1 and RFB2, it is sufficient for a barrier only at RFB1. Sequences located in the adjacent, 35S gene-proximal 188-bp EcoRI-HindIII fragment (the 35S enhancer) most likely contain the additional sequences sufficient for fork arrest at RFB2 (2). No other regions within the EcoRI fragment that spans most of the NTS region are sufficient to impede progression of a replication fork.
Essential sequences for RFB1 and RFB2 were localized to two distinct regions, 20 and 30 bp in length and separated by 40 bp. The 20-bp region covered by mutations M4 and M5, which abolish RFB1 activity, shows no sequence similarity to the 30-bp region covered by mutations M10, M11, and M12, which affect fork arrest at RFB2. Interestingly, the M4 and M5 region includes 10 matches to a 12-bp stretch of the pea RFB (CTTGTATAAGTT) that Hernandez et al. uncovered in a search for RFB homology between pea and S. cerevisiae (9). A shorter 7-bp match to this 12-bp pea sequence was also found within the yeast 188-bp EcoRI-HindIII fragment (9), neighboring the right end of the REB1 binding site (Fig. 7A). While this additional restriction fragment appears to be important to RFB2, it is not yet known if this specific 7-bp sequence is needed for RFB2 arrest.
Although the molecular mechanism that results in replication fork arrest at the S. cerevisiae rDNA RFB has not yet been determined, the binding of proteins is likely to be involved (2). Our results from mutating different sequences show that fork arrests at RFB1 and RFB2 are eliminated independently from one another. We also demonstrate that the distance between the barriers can be increased by 111 bp without reduction in fork arrest activity at these sites (Fig. 5C, bottom). These data make it unlikely that one protein molecule binds simultaneously at RFB1 and RFB2. Thus, the RFB1 and RFB2 essential sequences most likely correspond to binding sites for either two different proteins or one protein with two independent binding sites.
Sequences essential for HOT1 recombination and the rDNA RFB overlap.
The fact that sequences necessary for HOT1 and RFB activity colocalize within NTS1 posed the possibility that the two activities may require the same cis-acting sequences. While there is some sequence sharing, it is not complete: mutations C20 and N35 (see Fig. 7A for location) reduce HOT1 activity to less than 6% of the wild-type level while simultaneously eliminating one of the barriers (RFB1 and RFB2, respectively); however, mutation M6 displays normal barrier activities while reducing HOT1 activity to 22% of the wild-type level, and mutation M11 abolishes the barrier at RFB2, where it has only a modest effect on HOT1 (Fig. 7B). These results should not be surprising, since HOT1 and RFB also respond differently to mutations in trans-acting factors. For example, HOT1 recombination is dependent upon 35S rRNA gene transcription by RNA polymerase I (12), while the RFBs function independently of this process (2).
The abundant nuclear transcription factors REB1 and ABF1 (REB2) have long been known to bind within the E fragment (13, 27). While their sites of binding are in close proximity to the HOT1 and RFB essential sequences (see Fig. 7A for binding sites), they clearly do not overlap. Deletion of both sites resulted in only a modest decrease in HOT1 activity (12). Neither protein is required for RFB1 function, considering that their sites are not included in sequences sufficient for RFB1. The REB1 protein is most likely not involved in RFB2, since both barriers are normal in strains with a temperature-sensitive allele of REB1 grown at the restrictive temperature for several hours (6). However, the effect of ABF1 on RFB2 function is not yet known because scanning mutagenesis of the EcoRI-HindIII region has not been done.
Using chromatin immunoprecipitation, two yeast helicases, Pif1p and Rrm3p, were recently found to preferentially associate with rDNA chromatin, including the RFB region (12a). By measuring the generation of extrachromosomal rDNA circles in null mutants, Ivessa et al. (12a) showed that Rrm3p suppressed and Pif1p promoted rDNA recombination. The authors also performed 2D gel analyses on the direction of replication fork movement downstream of the RFB block and showed that a greater number of forks bypassed the RFB in a pif1 mutant than in the wild type. This finding suggests that Pif1p plays a role in maintaining an efficient fork arrest at the RFB. Further evidence also suggests that Rrm3p is an important factor in resolving forks terminating at the RFB. Neither Pif1p nor Rrm3p is essential to RFB activity, since forks are still arrested at the RFB site in the null mutants. However, it seems that Pif1p and Rrm3p are two newly identified trans-acting factors that appear to be involved in both RFB activity and rDNA recombination.
The FOB1 protein is localized to the nucleolus and is essential both for arresting replication forks at the RFB and for HOT1-stimulated recombination (3, 17). It plays a role in the maintenance of rDNA repeat number (15) and in the accumulation of rDNA extrachromosomal circles, which appear to influence life span (3). Fob1p does not affect rDNA transcription (R. Prusty, unpublished data) and does not appear to confer any significant growth defects (15). Based on the properties of a fob1 mutant (17) and on the results reported here, Fob1p must influence protein-DNA interactions at several sites, those determining replication fork arrest at RFB1 and at RFB2 and those mediating HOT1-stimulated recombination. It seems likely that the FOB1 protein facilitates the binding of different proteins to these different sites. Further studies will be needed to clarify the protein-DNA interactions and their functional consequences at this complex locus in the rDNA.
ACKNOWLEDGMENTS
We thank Katherine Kolor for plasmid pMUTBIAS and Katherine Friedman for plasmid pBB3NTS. We thank Heather McCune for comments on the manuscript and M. K. Raghuramen for help with manuscript preparation.
This work was supported by National Institute of General Medical Sciences grant 18926 to B.J.B. and W.L.F. and by U.S. Public Health Service grant R01 GM36422 to R.L.K. T.R.W. was supported during part of this work by U.S. Public Health Service training grant T32 GM07735.
Theresa R. Ward, Margaret L. Hoang, and Reeta Prusty contributed equally to this work.
REFERENCES
- 1.Bierne H, Ehrlich S D, Michel B. The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. EMBO J. 1991;10:2699–2705. doi: 10.1002/j.1460-2075.1991.tb07814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Bierne H, Ehrlich S D, Michel B. Deletions at stalled replication forks occur by two different pathways. EMBO J. 1997;16:3332–3340. doi: 10.1093/emboj/16.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brewer B J, Lockshon D, Fangman W L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 3.Defossez P-A, Prusty R, Kaeberlein M, Lin S-J, Ferrigno P, Silver P A, Keil R L, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 4.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 6.Friedman K L. Determinants of late origin activation in yeast. Ph.D. thesis. Seattle: University of Washington; 1996. [Google Scholar]
- 7.Friedman K L, Brewer B J. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 8.Gerber J-K, Goegel E, Berger C, Wallisch M, Muller F, Grummt I, Grummt F. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell. 1997;90:559–567. doi: 10.1016/s0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez P, Martin P L, Martinez R M L, Schvartzman J B. Conserved features in the mode of replication of eukaryotic ribosomal RNA genes. EMBO J. 1993;12:1475–1485. doi: 10.1002/j.1460-2075.1993.tb05791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 11.Horiuchi T, Fujimura Y. Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J Bacteriol. 1995;177:783–791. doi: 10.1128/jb.177.3.783-791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M J. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J Bacteriol. 1994;176:4656–4663. doi: 10.1128/jb.176.15.4656-4663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang G S, Keil R L. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics. 1995;141:845–855. doi: 10.1093/genetics/141.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Ivessa A S, Zhou J-Q, Zakian V A. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 13.Kang J J, Yokoi T J, Holland M J. Binding sites for abundant nuclear factors modulate RNA polymerase I-dependent enhancer function in Saccharomyces cerevisiae. J Biol Chem. 1995;270:28723–28732. doi: 10.1074/jbc.270.48.28723. [DOI] [PubMed] [Google Scholar]
- 14.Keil R L, Roeder G S. cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, Heck D J, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi T, Hidaka M, Nishizawa M, Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet. 1992;233:355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 18.Kuzminov A. Instability of inhibited replication forks in E. coli. Bioessays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y-H, Keil R L. Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics. 1991;127:31–38. doi: 10.1093/genetics/127.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linskens M H, Huberman J A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little R D, Platt T H, Schildkraut C L. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Estrano C, Schvartzman J B, Hernandez P. The replication of ribosomal RNA genes in eukaryotes. Chromosomes Today. 1997;12:149–169. [Google Scholar]
- 23.Lopez-Estrano C, Schvartzman J B, Krimer D B, Hernandez P. Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J Mol Biol. 1998;277:249–256. doi: 10.1006/jmbi.1997.1607. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Estrano C, Schvartzman J B, Krimer D B, Hernandez P. Characterization of the pea rDNA replication fork barrier: putative cis-acting and trans-acting factors. Plant Mol Biol. 1999;40:99–110. doi: 10.1023/a:1026405311132. [DOI] [PubMed] [Google Scholar]
- 25.Lucchini R, Sogo J M. Chromatin structure and transcriptional activity around the replication forks arrested at the 3′ end of the yeast rRNA genes. Mol Cell Biol. 1994;14:318–326. doi: 10.1128/mcb.14.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrow B E, Johnson S P, Warner J R. Proteins that bind to the yeast rDNA enhancer. J Biol Chem. 1989;264:9061–9068. [PubMed] [Google Scholar]
- 28.Newlon C S, Collins I, Dershowitz A, Deshpande A M, Greenfeder S A, Ong L Y, Theis J F. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- 29.Reeder R H. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293–327. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds A E, McCarroll R M, Newlon C S, Fangman W L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Rothstein R, Gangloff S. The shuffling of a mortal coil. Nat Genet. 1999;22:4–6. doi: 10.1038/8705. [DOI] [PubMed] [Google Scholar]
- 31.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination, or “gimme a break.”. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 32.Sanchez J A, Kim S M, Huberman J A. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp Cell Res. 1998;238:220–230. doi: 10.1006/excr.1997.3835. [DOI] [PubMed] [Google Scholar]
- 33.Seligman L, Manoil C. An amphipathic sequence determinant of membrane protein topology. J Biol Chem. 1994;269:19888–19896. [PubMed] [Google Scholar]
- 34.Stewart S E, Roeder G S. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voelkel-Meiman K, Keil R L, Roeder G S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 36.Wiesendanger B, Lucchini R, Koller T, Sogo J M. Replication fork barriers in the Xenopus rDNA. Nucleic Acids Res. 1994;22:5038–5046. doi: 10.1093/nar/22.23.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue M, Reischmann K P, Kapler G M. Conserved cis- and trans-acting determinants for replication initiation and regulation of replication fork movement in tetrahymenid species. Nucleic Acids Res. 1998;26:4635–4644. doi: 10.1093/nar/26.20.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Macalpine D M, Kapler G M. Developmental regulation of DNA replication: replication fork barriers and programmed gene amplification in Tetrahymena thermophila. Mol Cell Biol. 1997;17:6147–6156. doi: 10.1128/mcb.17.10.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]