Abstract
The 2′-5′ oligoadenylate (2-5A)/RNase L pathway is one of the enzymatic pathways induced by interferon. RNase L is a latent endoribonuclease which is activated by 2-5A and inhibited by a specific protein known as RLI (RNase L inhibitor). This system has an important role in regulating viral infection. Additionally, variations in RNase L activity have been observed during cell growth and differentiation but the significance of the 2-5A/RNase L/RLI pathway in these latter processes is not known. To determine the roles of RNase L and RLI in muscle differentiation, C2 mouse myoblasts were transfected with sense and antisense RLI cDNA constructs. Importantly, the overexpression of RLI in C2 cells was associated with diminished RNase L activity, an increased level of MyoD mRNA, and accelerated kinetics of muscle differentiation. Inversely, transfection of the RLI antisense construct was associated with increased RNase L activity, a diminished level of MyoD mRNA, and delayed differentiation. In agreement with these data, MyoD mRNA levels were also decreased in C2 cells transfected with an inducible RNase L construct. The effect of RNase L activity on MyoD mRNA levels was relatively specific because expression of several other mRNAs was not altered in C2 transfectants. Therefore, RNase L is directly involved in myoblast differentiation, probably through its role in regulating MyoD stability. This is the first identification of a potential mRNA target for RNase L.
The 2′-5′ oligoadenylate (2-5A)/RNase L system is an interferon (IFN)-inducible RNA degradation pathway which is responsible for many of the antiviral and antiproliferative effects of IFNs (37, 41).
The 2-5A pathway is composed of at least three types of enzymatic activities: 2-5A-synthetase, 2-5A-degrading enzymes, and RNase L. 2-5A, an oligoadenylate with 2′-5′ phosphodiester bonds, activates RNase L (53), a latent endoribonuclease. Upon activation, RNase L cleaves mRNAs 3′ of UpNp sequences, thus leading to the inhibition of protein synthesis (14, 21).
The activity of RNase L was originally thought to be modulated solely by the concentration of the 2-5A activator (11, 21). Moreover, we have previously established that RNase L activity can also be regulated by RLI (RNase L inhibitor), a protein inhibitor (5). Overexpression of the RLI cDNA in HeLa cells results in the inhibition of the IFN-activated 2-5A pathway. RLI is induced by viruses such as encephalomyocarditis virus (EMCV) and human immunodeficiency virus (HIV), causing an inhibition of the 2-5A/RNase L system (27, 28). The role of the 2-5A/RNase L pathway in the selective reduction of viral mRNA during EMCV and HIV infection has been demonstrated elsewhere (16, 25, 27).
Variations in intracellular 2-5A and 2-5A-synthetase levels have been observed during cell growth and differentiation even in the absence of exogenous IFN treatment. Indeed, expression of IFN-inducible proteins, such as 2-5A-synthetase, double-standed RNA-activated protein kinase (PKR), and p202 (a member of the “200 family” of murine proteins) have been observed during myoblast differentiation (4, 9, 38). Recently, Kronfeld et al. (23) established the involvement of PKR in the regulation of myogenesis using PKR transfected C2C12 cells. Moreover, transfection studies have provided direct proof of the role played by 2-5A-synthetase in the proliferation of human glioblastoma (35) and NIH 3T3 cells (51), as well as in myeloid differentiation (36).
To more precisely assess the role of the RNase L and RLI in cell behavior, we studied their effects on muscle cell differentiation using C2 myoblasts. These cells can be grown in proliferative medium without activation of the myogenic program, but muscle differentiation is induced upon reduction of growth factors in the cell culture media. Induction of muscle-specific genes and cell fusion, resulting in the formation of mature muscle cells known as myotubes, occurs following the exit of these cells from the cell cycle (33, 52). These different muscle cell events are controlled by muscle-specific transcription factors belonging to the MyoD family (8, 30, 40, 47). The MyoD family is composed of four myogenic basic helix-loop-helix (bHLH) transcription factors: MyoD, myogenin, Myf5, and MRF4. At least in vitro, MyoD appears to be the master gene of muscle skeletal differentiation, orchestrating the onset of skeletal muscle differentiation. Indeed, even in the presence of Myf5, inactivation of MyoD expression by a MyoD antisense strategy leads to an inhibition of differentiation (29).
Here we show that the timing and myogenic differentiation of C2 cells is modulated by the level of RNase L activity. In C2 cells transfected with an RLI antisense cDNA construction (VAS), RNase L activity is increased. In these clones, MyoD and myogenin levels are low and the expression of α actin and troponin T, two genes expressed at the differentiation stage, is delayed compared to control C2 cells transfected with an empty vector (VV). In contrast, C2 cells overexpressing an RLI sense cDNA construct (VS) differentiate more rapidly, as attested by the earlier kinetic expression of α-actin and troponin T. We also find that this increased differentiation capacity is associated with high levels of MyoD and myogenin. This is likely due to an increased stability of MyoD mRNA in these transfectants. Importantly, this effect appears to be specific since the half-life of other mRNAs was not altered. Finally, in C2 cells which express RNase L after induction by IPTG (isopropyl-β-d-thiogalactopyranoside), MyoD mRNA levels are greatly reduced. Therefore, our work identifies the muscle-regulatory MyoD as a potential cellular mRNA target of RNase L and explains how RNase L and RLI might control skeletal muscle cell differentiation.
MATERIALS AND METHODS
Cells and cell extracts.
Permissive C2 myoblasts (clone C2.7) (a generous gift of A. Bonnieu, INRA, Montpellier, France) have been previously described (33, 52). Proliferating myoblasts were routinely maintained in Dulbecco modified Eagle medium (DMEM) (Gibco-BRL) supplemented with 10% (vol/vol) fetal calf serum (growth medium, GM) at low cell density (300 cells/cm2) and subcultured twice a week. For differentiation experiments, cells were plated at high density (104 cells/cm2 in GM), and after 2 days the medium was changed to DMEM–2% fetal calf serum (differentiating medium, DM). Cells were collected 24 h after seeding (day 0) and at 24-h intervals thereafter for preparation of cell extracts. At the indicated times, cells were resuspended in 2 volumes of radioimmunoprecipitation assay buffer (50 mM HEPES, pH 7.5; 400 mM NaCl; 1% [vol/vol] NP-40; 1 mM phenylmethylsulfonyl fluoride; 10 μg of aprotinin per ml; 150 μg of leupeptin per ml), disrupted in a Dounce homogenizer, and centrifuged at 10,000 × g (S10). The protein concentration in the supernatant (S10) was determined by spectrophotometry (48).
Expression vectors and transfections.
The coding sequence of the human RLI cDNA (5) was directionally subcloned in pcDNA3neo (Invitrogen) by standard procedures (39). RLI antisense-pcDNA3neo (7 μg), RLI sense-pcDNA3neo (7 μg), or the empty vector pcDNA3neo (7 μg) was transfected into C2 cells by calcium phosphate coprecipitation (39). Stable transfectants were selected by culturing the cells in the presence of 1 mg of G418 (Gibco-BRL) per ml. The yield of transfection was approximately 80%, so we did not isolate individual clones. The polyclonal cell population expressing the transfected antisense RLI cDNA was named VAS, the polyclonal cell population expressing the transfected sense RLI cDNA was named VS, and the polyclonal cell population transfected with the empty vector was named VV.
For conditional expression of RNase L, the LacSwich II inductible mammalian expression system (Stratagene) was used. First the pCMVLacI repressor vector (0.5 μg of DNA) was transfected into C2 cells with Fugene (2 μl) according to the manufacturer's instructions (Boehringer). After selection of stable transfectants by culturing the cells in the presence of hygromycin (200 U/ml), the expression of the Lac repressor expression was tested by Northern blotting (39). After transfer, the nylon sheets were incubated with a [32P]cDNA probe corresponding to the XbaI-StuI fragment of the LacI repressor cDNA radiolabeled using the multiprime procedure (Gibco-BRL). After autoradiography on a PhosphorImager 445-SI (Molecular Dynamics), the mRNAs were quantified by image analysis with the ImageQuant program (Molecular Dynamics). The clone expressing the greatest quantity of Lac repressor was then transfected by calcium phosphate coprecipitation (39) with the pOPRSVI vector harboring the coding sequence of the human RNase L cDNA (7 μg) (53). Stable transfectants were selected by culturing the cells in the presence of 1 mg of G418 (Gibco-BRL) per ml. For analysis, clones were treated with 5 mM IPTG for 8 h. Cells were then collected, and each clone was analyzed for RNase L expression by the 2-5A radiocovalent binding assay (see below). One clone (RNase L3) was selected and used in further experiments.
Western blot assay.
Proteins (100 μg) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrophoretically to a nitrocellulose membrane according to the method of Towbin et al. (45). The nitrocellulose membranes were incubated in phosphate-buffered saline (PBS; 140 mM NaCl, 2 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.4]) supplemented with 5% (wt/vol) skimmed milk for 30 min and then soaked overnight at 4°C in the same buffer with one of the following antibodies: anti-Myf-5 (C-20), anti-MyoD (M-318), anti-myogenin (M-225), or anti-p21 (C-19) (all rabbit polyclonal antibodies from Santa Cruz Biotechnology) at 0.5 μg/ml; mouse monoclonal anti-α-sarcomeric actin (1/500 dilution), rabbit polyclonal antiactin (1/100 dilution), mouse monoclonal antitropomyosin (1/400 dilution), mouse monoclonal anti-troponin T (1/1,000 dilution), all from Sigma Immuno Chemicals; rabbit polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (1/3,000 dilution), a generous gift of G. Cathala (IGMM, Montpellier, France); and rabbit polyclonal anti-RLI (1/500 dilution) (28). The filters were washed with PBS supplemented with 0.05% (vol/vol) Tween 20 and incubated for 1 h at room temperature with donkey anti-rabbit or sheep anti-mouse immunoglobulin antibody conjugated to horseradish peroxidase (Amersham). Specific proteins were visualized using a chemiluminescence kit (NEN). The gels were scanned, and protein bands were quantified by image analysis with the Intelligent Quantifier program (Bio Image Systems Corp.). The experiments were done in triplicate.
Immunofluorescence.
Each polyclonal population of transfected C2 myoblasts and myotubes were fixed for 5 min in 3.7% (vol/vol) formalin-PBS, followed by a 30-s extraction in −20°C acetone and rehydratation in PBS containing 0.5% (vol/vol) bovine serum albumin (BSA). Expression of MyoD was analyzed using the 5.8A mouse monoclonal antibody against MyoD diluted 1/5 (a gift of P. Dias and P. J. Houghton [10]). Primary antibody diluted in PBS-BSA was incubated for 1 h at 37°C and then washed in PBS, followed by a 30-min incubation with biotinylated anti-mouse antibody (1/200 dilution; Amersham). Biotinylated antibodies were finally revealed after a 30-min incubation with streptavidin-Texas red (1/200 dilution; Amersham). DNA was stained with a Hoechst stain (0.1 mg/ml; Sigma). The experiments were done in triplicate.
Radiobinding and radiocovalent binding assay: affinity labeling of RNase L with 2-5ApCp.
Transfected C2 cells were harvested at various time points before and after differentiation, as indicated in the legends to the figures. For radiobinding (22), cell extracts (600 μg of protein) of VV-, VAS-, and VS-expressing cells were incubated with 20,000 cpm of 2-5A4-3′-[32P]pCp (2-5ApCp; 3,000 Ci/mmol) on ice for 15 min as previously described (6). The radiolabeled RNase L was then precipitated at −20°C for 5 min by using 300 μl of polyethylene glycol 6000 (25% [wt/vol]) after addition of 150 μl of bovine serum as a carrier. After centrifugation (10,000 × g, 10 min), the radioactivity of the pellet containing the 2-5ApCp bound to RNase L was measured. The experiments were done in triplicate and the standard deviation is indicated on the plots.
For the radiocovalent binding assay (50), the radiolabeled 2-5A4-3′-[32P]pCp probe was oxidized at the 3′ end as previously described (2). Cell extracts (600 μg of protein) from the RNase L3 clone were incubated with the oxidized 2-5A4-3′-[32P]pCp probe for 30 min in ice and for a further 20 min at room temperature in the presence of sodium cyanoborohydride (20 mM, final concentration). Proteins were analyzed by SDS-PAGE in a 10% (vol/vol) polyacrylamide gel (24), and the labeled proteins were visualized after autoradiography. The gels were scanned, and the protein bands were quantified by image analysis with the Intelligent Quantifier program (Bio Image Systems Corp.). The experiments were done in triplicate, and the standard deviation is indicated on the plots.
Analysis of mRNA stability.
VV, VAS, and VS cells were plated at 104 cells/cm2 in GM and, after 2 days, the medium was changed to DM. At day 0 (24 h after seeding, before differentiation) or at day 4 (after the induction of differentiation) cells were treated with actinomycin D (5 μg/ml) for 0, 30, 60, 120, or 240 min; mRNA was then prepared and analyzed by Northern blotting (39). After transfer, the nylon sheets were incubated with different [32P]cDNA probes (as indicated in the figure legends) synthesized by the multiprime procedure (Gibco-BRL). After autoradiography on a PhosphorImager 445-SI (Molecular Dynamics), the mRNAs were quantified by image analysis with the ImageQuant program (Molecular Dynamics). Each lane was normalized with the S26 probe (46). The experiments were done in triplicate, and the standard deviation is indicated on the plots.
Nuclear run-on transcription assay.
Preparation of nuclei and elongation of nascent transcripts were performed as described by Greenberg and Ziff (15), except that heparin was added to the incubation mixture (final concentration, 1 mg/ml) (7). GAPDH, MyoD, Myf5, myogenin, and α-actin plasmid DNA were spotted onto nitrocellulose. Prehybridation and hybridization were performed in 4× STE (600 mM NaCl; 80 mM Tris-HCl, pH 7.5; 4 mM EDTA), 0.2% (wt/vol) SDS, 0.1% (wt/vol) PPi, 2 mg of heparin per ml, and 50% (vol/vol) formamide. Filters were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS (wt/vol) for 10 min at 20°C and then in 0.2 × SSC at 65°C for 30 min. Filters were then RNase A treated in 2× SSC for 15 min at 37°C. After autoradiography on a PhosphorImager 445-SI (Molecular Dynamics), the mRNAs were quantified by image analysis with the ImageQuant program (Molecular Dynamics). The experiments were performed in triplicate, and the standard deviation is indicated on the plots.
rRNA cleavage.
Cells were resuspended in 1 volume of buffer (5 mM Tris-HCl, pH 7.6; 1.25% [vol/vol] glycerol; 20 mM KCl; 1.25 mM magnesium acetate), vortexed for 1 min, and kept on ice for 10 min. The cell-buffer suspension was passed through a 1-ml tuberculin syringe and centrifuged at 15,000 × g 2 min. The protein concentration of the supernatant was determined by spectrophotometry (48). Cell extracts (100 μg of protein) were incubated for 1 h at 30°C in the presence or absence of 2-5A4 (1 μM, final concentration). The total RNA was extracted, denatured, and analyzed by electrophoresis on 1.8% agarose gel. The gels were stained with ethidium bromide, and the RNA bands were visualized under UV light (18–20, 49). The experiments were done in triplicate.
RESULTS
RNase L and RLI are sequentially induced during C2 differentiation.
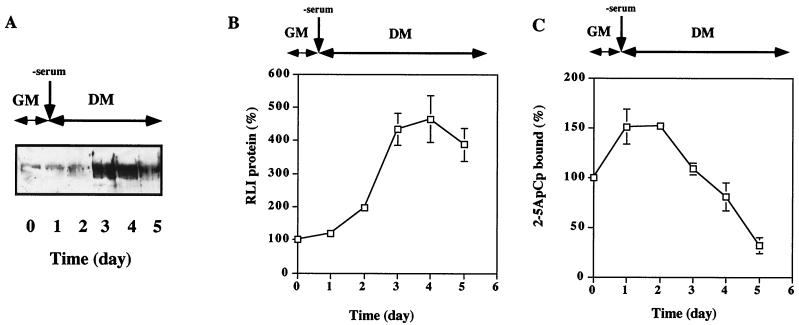
We first studied the behavior of RLI and RNase L during the induction of differentiation in the myogenic C2 cell line. The cells were cultivated in either GM or growth-factor-deprived medium (DM), and cell extracts were prepared at different times. RLI was identified by immunoblot analysis using a previously described RLI-specific polyclonal antibody (28). Because the specificity and the affinity of RNase L for its activator, 2-5A, is very high, and the nuclease activity of RNase L is strictly dependent on its activation by 2-5A (12, 53), the measurement of the binding of 2-5A to RNase L is a good indicator of the presence of activatable RNase L. Thus, RNase L levels were followed by monitoring its 2-5A binding activity. RLI levels did not change during the first day in DM compared to its basal level in GM. Moreover, it was induced after 2 days in DM, with a maximal level observed at day three (Fig. 1A and B). In contrast, RNase L 2-5A binding activity increased immediately upon culture of C2 cells in DM but decreased by day 3 when RLI was highest (Fig. 1C).
FIG. 1.
Increases in the RLI and RNase L proteins during the differentiation of C2 myoblasts into myotubes. (A) C2 cells were grown in GM and then shifted to DM on day 1. At the indicated times, cells were harvested and lysed as described in Materials and Methods. Total protein samples (100 μg) were analyzed for RLI protein by Western blotting with a polyclonal RLI antiserum. (B) Densitometric analysis of the gel shown in panel A. A value of 100% corresponds to the amount of RLI protein in proliferating myoblasts at day 0. Error bars refer to the standard deviation obtained in three independent experiments. (C) C2 cells were grown in GM and then shifted to DM on day 1. At the indicated times, cells were harvested and lysed. Proteins (600 μg) were incubated with radiolabeled 2-5A4-3′-[32P]pCp (2-5ApCp) in a radiobinding assay; 100% corresponds to the amount of 2-5ApCp bound to RNase L in proliferating myoblasts. Error bars refer to the standard deviation obtained in three independent experiments.
Alterations in RLI levels in C2 cells modulates their differentiation potential.
RNase L 2-5A binding activity was induced transiently at the beginning of the differentiation process. Concomitant with the decrease in RNase L 2-5A binding activity, RLI was induced. The ratio between these two proteins appeared to be important in the differentiation of C2 cells. To show the direct involvement of RLI and RNase L in the differentiation process, C2 cells were transfected with the empty pcDNA3 vector (as control cells, we used VV cells), RLI sense-pcDNA3 (VS cells), or RLI antisense-pcDNA3 (VAS cells).
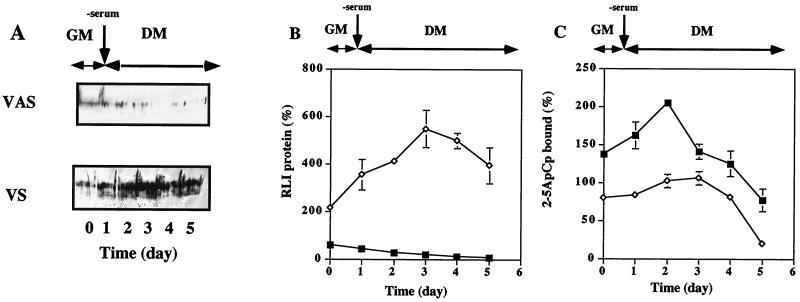
RLI and RNase L 2-5A binding activity were monitored during differentiation as described above. The results obtained are presented in Fig. 2. RLI and RNase L 2-5A binding activity were equivalent in the control (VV) and untransfected C2 cells (data not shown).
FIG. 2.
Increases in RLI and RNase L proteins during the differentiation of transfected C2 cells. (A) Pooled C2 cells stably transfected with an RLI cDNA sense construct (VS) and an RLI cDNA antisense construction (VAS) were grown in GM and then shifted to DM on day 1. At the indicated times, cells were lysed, and RLI expression was assessed in protein samples (100 μg) using an RLI-specific antiserum. (B) Densitometric analysis of the gel shown in panel A. A value of 100% corresponds to the amount of RLI protein at day 0 in proliferating C2 control myoblasts. Error bars refer to the standard deviation obtained in three independent experiments. Symbols: ◊, VS cells; ■, VAS cells. (C) The cells described in panel A were lysed, and proteins (600 μg) were incubated with radiolabeled 2-5A4-3′-[32P]pCp (2-5ApCp) in a radiobinding assay; 100% corresponds to the amount of 2-5ApCp bound to RNase L in proliferating C2 control myoblasts. Error bars refer to the standard deviation obtained in three independent experiments. Symbols: ◊, VS cells; ■, VAS cells.
The basal level of RLI in the polyclonal RLI antisense (VAS) cell population was 40% lower than in C2 and control cells (Fig. 2A and B), and RNase L 2-5A binding activity was correspondingly increased by nearly 40% (Fig. 2C). Under differentiation conditions, no RLI induction was observed in these VAS transfectants, and RNase L 2-5A binding activity was twofold higher than that observed in C2 and control cells (Fig. 2C).
The basal level of RLI in the polyclonal RLI sense (VS) cell population was twofold higher than that detected in C2 and control cells, and during differentiation RLI levels remained increased (Fig. 2A and B). Consequently, although there was an increase in RNase L 2-5A binding activity during differentiation, this level was lower than that observed in the control and RLI antisense (VAS) cells (Fig. 2C).
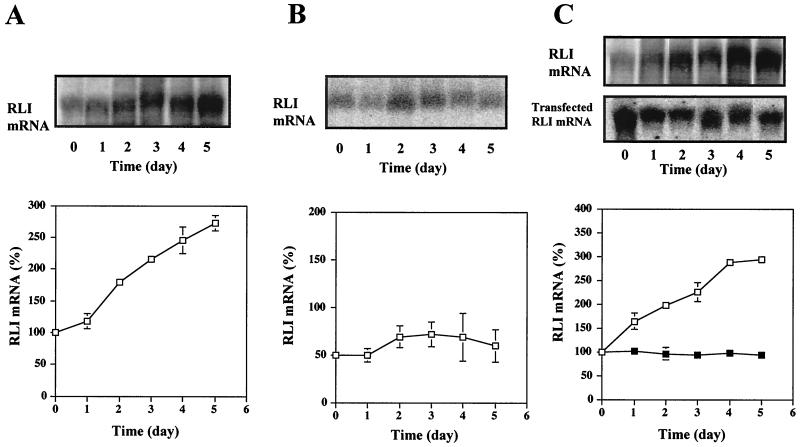
We checked the RLI mRNA level in these different transfectants. In control (VV) cells, RLI mRNA was induced during C2 differentiation (Fig. 3A). In contrast, the level of RLI mRNA was significantly lower in the polyclonal RLI antisense (VAS) cell population, indicating that the transfected antisense cDNA exercised its inhibitory effect at the mRNA level (Fig. 3B). In the polyclonal RLI sense (VS) cell population, expression of the endogenous RLI mRNA (3.5 kb) (3) and the transfected RLI cDNA containing only the coding sequence of RLI (1.8 kb) were observed (Fig. 3C). In these latter cells, expression of the endogenous RLI mRNA was induced during C2 differentiation in a manner similar to that observed in control cells. In contrast, expression of the transfected RLI cDNA was stable during differentiation.
FIG. 3.
RLI mRNA levels increase during the differentiation of transfected C2 cells. Pooled stable control C2 cells transfected by an empty pcDNA3 vector (VV) (A), pooled stable C2 cells transfected with an RLI cDNA antisense construction (VAS) (B), and pooled stable C2 cells transfected with an RLI cDNA sense construction (VS) (C) were grown in GM and then shifted to DM on day 1. At the indicated times, cells were harvested, and mRNAs were extracted and analyzed by Northern blot using the murine RLI [32P]cDNA probe (VV, VAS, and VS [□]). The VS samples were also probed with a human RLI [32P]cDNA fragment (■) as indicated. Densitometric analysis of the gels are presented. A value of 100% corresponds to the amount of RLI mRNA in proliferating C2 control myoblasts at day 0. Error bars refer to standard deviation obtained in three independent experiments.
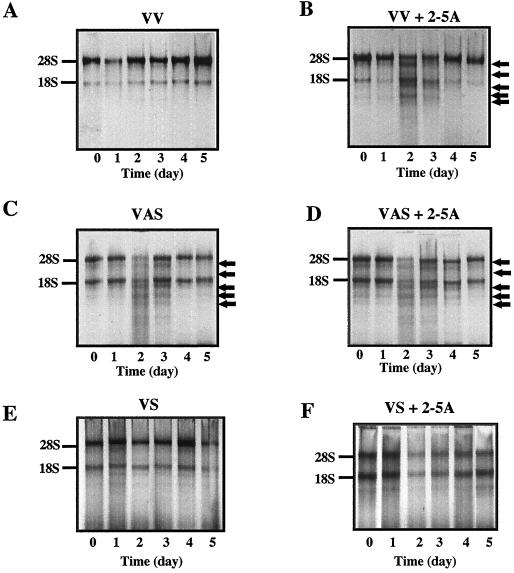
These different levels of RLI mRNA and protein resulted in a wide range of RNase L 2-5A binding activities, as expected (Fig. 2). To determine whether the nuclease activity of RNase L was also modified in these different transfectants, the nuclease activity of RNase L was monitored in vitro by studying the cleavage of rRNA. Activation of RNase L by 2-5A gives rise to a specific pattern of degradation of rRNA (42, 49). As illustrated in Fig. 4A and B, the cleavage of rRNA in control (VV) cells, after activation of RNase L, was visible at days 2 and 3. The nuclease activity of RNase L was visible in the RLI antisense (VAS) cell population even in the absence of exogenous 2-5A (Fig. 4C and D). It is likely that because the level of RLI was so low in these cells, the basal level of 2-5A in the cell was sufficient to activate RNase L. In contrast, in the polyclonal RLI sense (VS) cell population, no cleavage of rRNA was observed even after the addition of exogenous 2-5A (Fig. 4E and F). Thus, the levels of RLI and RNase L activity in these polyclonal cell populations varied greatly both before and during differentiation.
FIG. 4.
Variations in RNase L activity following transfection with RLI sense and antisense constructs. Pooled stable control C2 cells transfected with an empty pcDNA3 vector (VV, panels A and B), the RLI antisense cDNA (VAS, panels C and D) and the RLI sense cDNA (VS, panels E and F) were seeded in GM and then shifted to DM (day 1). Cells were harvested at the indicated times. Proteins (100 μg) were incubated for 60 min at 30°C in the absence (A, C, and E) or presence (B, D, and F) of 2-5A4 (1 μM, final concentration). rRNAs were analyzed on 1.8% (wt/vol) agarose gels. Intact 28S and 18S rRNA migration is indicated at the left of each gel. Major rRNA degradation products are indicated by arrows.
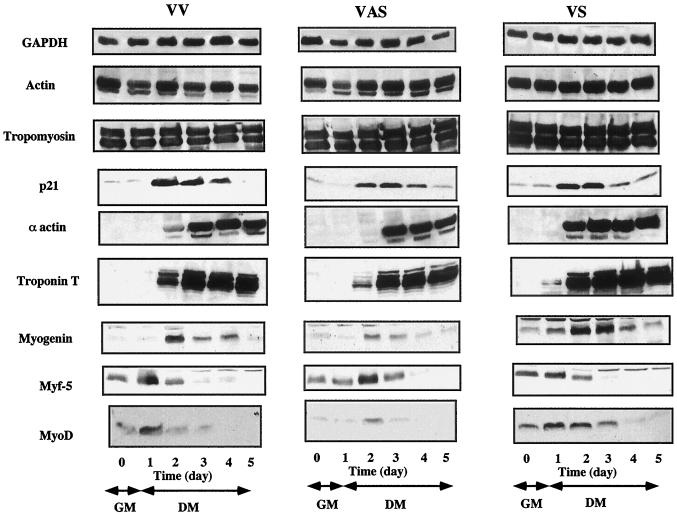
To establish whether variations in the levels of RLI and RNase L activity affect myogenic differentiation, the appearance of muscle-specific proteins and muscle-specific transcription factors was studied in these three polyclonal cell populations (VV, VAS, and VS). Initially, the differentiation process was monitored by studying the expression of housekeeping and myogenesis-specific proteins by Western blot. As illustrated in Fig. 5, no differences were observed for constitutively expressed ubiquitous proteins such as GAPDH, actin, and tropomyosin in the three C2 transfected populations. The same result was observed for p21, a protein which is not myoblast specific but whose expression is regulated during the cell cycle. This protein was strongly induced to the same level in the three transfected populations at day 2 (Fig. 5). In contrast, when we studied the structural proteins, there was clearly a difference in the kinetics of expression of proteins whose expression is regulated during myogenesis. α-Actin expression was observed 1 day later in RLI antisense (VAS) cells compared to control (VV) cells, while its level was maximal at an earlier time point in RLI sense (VS) cells (Fig. 5). Troponin T levels were 3.5 times lower in RLI antisense (VAS) cells compared to control (VV) cells at day 2. Notably, its expression was noted as early as 1 day following differentiation in RLI sense (VS) cells (Fig. 5). Nevertheless, while the kinetics of expression of α-actin and troponin T differed in these populations, their maximal levels of expression were equivalent.
FIG. 5.
Kinetics of protein expression in differentiation C2 cells. Pooled stable control C2 cells transfected with an empty pcDNA3 vector (VV), the RLI sense cDNA (VS), and the RLI antisense cDNA (VAS) were seeded in GM and then shifted to DM (day 1). Cells were harvested at the indicated times. Proteins (100 μg) were analyzed by Western blotting with the different antisera as indicated.
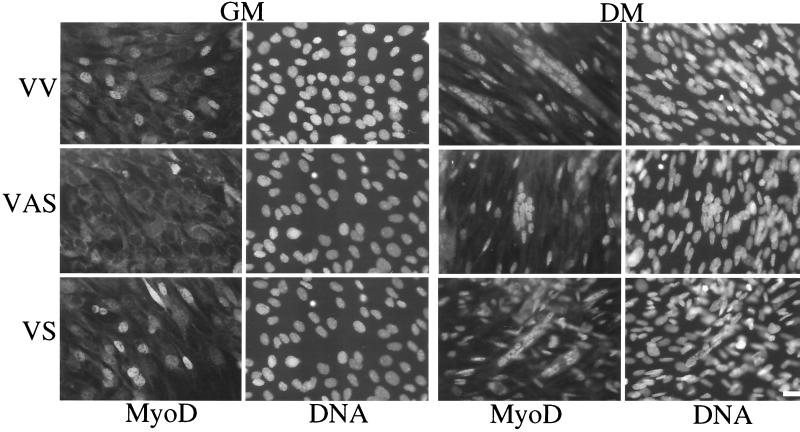
Regarding the bHLH factors, Myf5 levels were comparable in RLI sense (VS) cells, control (VV) cells, and RLI antisense (VAS) cells in GM. After the shift to DM, Myf5 induction was delayed by 1 day in RLI antisense (VAS) cells. A more striking difference was observed for MyoD and myogenin. The level of MyoD was greatly increased in RLI sense (VS) cells compared to control (VV) cells. In RLI antisense (VAS) cells, MyoD protein was almost undetectable even after the shift to DM, but it was observed at day 2, 1 day after maximal levels were detected in control (VV) and RLI sense (VS) cells. This difference in MyoD protein expression was confirmed by immunofluoresence using another antibody (Fig. 6). The MyoD protein was overexpressed in RLI sense (VS) cells cultured either in GM or in DM. Additionally, MyoD protein was practically undetectable in RLI antisense (VAS) cells cultured in GM.
FIG. 6.
Decreased expression of MyoD in VAS cells and overexpression of MyoD in VS cells. Each polyclonal population of transfected C2 (VV, VAS, and VS) myoblasts grown in GM and myotubes differentiated in DM were fixed as described in Materials and Methods. Expression of MyoD was analyzed using the 5.8A mouse monoclonal antibody against MyoD. DNA was stained with Hoechst stain (0.1 mg/ml).
Similary, the myogenin level was high in RLI sense (VS) cells. Myogenin was already present in RLI sense (VS) cells before differentiation (days 0 and 1) and reached five- and twofold-higher levels than that detected in RLI antisense (VAS) and control (VV) cells, respectively. In RLI antisense (VAS) cells, the myogenin level was very low and remained low even after the beginning of differentiation (Fig. 5). Therefore, changes in MyoD and myogenin levels, as well as a modulation of the differentiation capacity, were major consequences of varying RLI levels in C2 cells.
MyoD mRNA stability is dependent on the modification of RNase L activity.
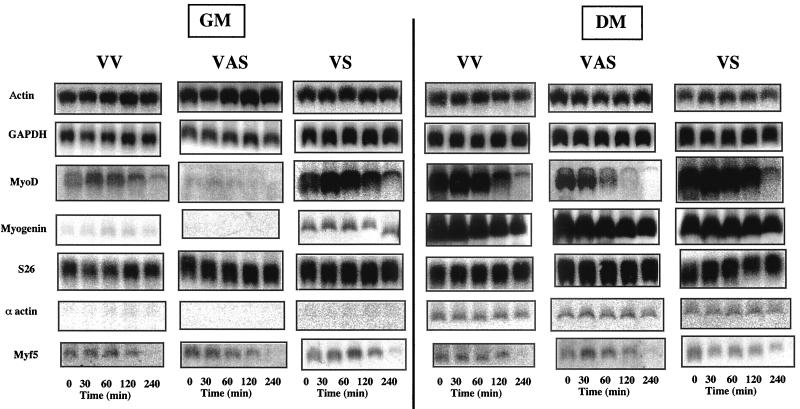
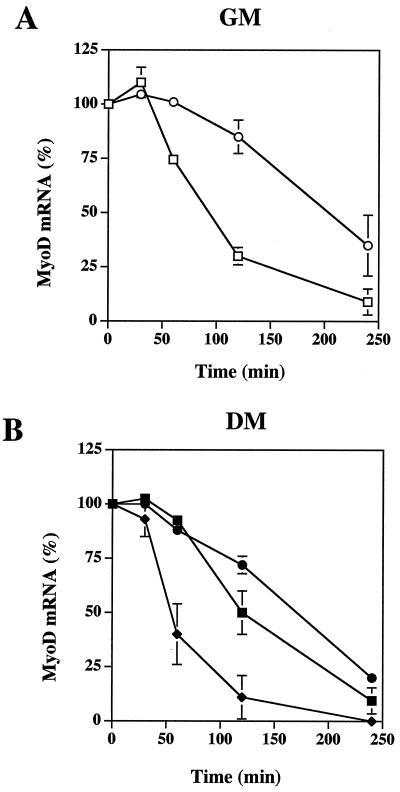
The 2-5A/RNase L system is an RNA degradation pathway. Variations in MyoD and myogenin proteins could be due to a degradation of their mRNAs by RNase L. To test this hypothesis, we measured the stability of different mRNAs in transfected C2 cells. At day 0, under GM conditions, and at day 4, after the shift to DM when RNase L activity decreases, all transfected C2 cells were treated with actinomycin D in order to measure the half-life of the mRNAs. Cells were collected at different times after the addition of actinomycin D, and mRNAs were extracted and analyzed by Northern blotting (Fig. 7). We found that while the half-life of actin, GAPDH, S26, α-actin, Myf5, and myogenin mRNAs were equivalent in all three polyclonal populations, the half-life of the MyoD mRNA was modified. In GM conditions, the half-life of the MyoD mRNA in RLI antisense (VAS) cells could not be determined because the mRNA was not detectable. In control (VV) cells, the half-life of MyoD mRNA was approximatively 90 min, a result similar to the results reported in P2 cells by Thayer et al. (43). In RLI sense (VS) cells, MyoD mRNA was stabilized, with a half-life of approximately 200 min (Fig. 8A). After the decrease in RNase L activity, at day 4, MyoD mRNA was detectable in RLI antisense (VAS) cells, and its half-life was approximately 50 min (Fig. 8B). Collectively, these data demonstrate that MyoD mRNA stability was associated with RNase L activity.
FIG. 7.
MyoD mRNA stability is altered by the expression of RLI. Each polyclonal population of transfected C2 (VV, VAS, and VS) myoblasts grown in GM and myotubes grown in DM were treated with actinomycin D (5 μg/ml). At the indicated times, cells were harvested and the mRNAs were extracted and analyzed by Northern blotting with the different [32P]cDNA probes. mRNAs were visualized on a PhosphorImager 445-SI (Molecular Dynamics).
FIG. 8.
The half-life of the MyoD mRNA is altered by the expression of RLI. Each polyclonal population of transfected C2 (VV, VAS, and VS) myoblasts (GM) and myotubes (DM) were treated with actinomycin D (5 μg/ml), and the mRNAs were analyzed as described in Fig. 7. After visualization on a PhosphorImager 445-SI (Molecular Dynamics), MyoD mRNAs were quantified by image analysis with the ImageQuant program (Molecular Dynamics). Each lane was normalized with the S26 probe. Densitometric analysis of the gel is presented. A value of 100% corresponds to the amount of MyoD mRNA at time zero in each polyclonal population of transfected C2 (VV, VAS, and VS) myoblasts (GM) (A) (□, control VV cells; ○, RLI sense VS cells) or myotubes (DM) (B) (■, control VV cells; ●, RLI sense VS cells; ⧫, RLI antisense cells). Error bars refer to the standard deviation obtained in three independent experiments.
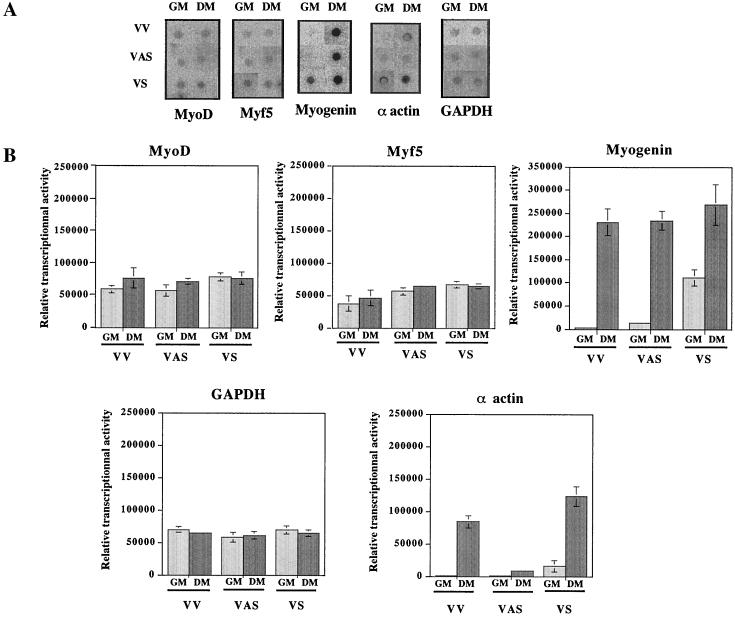
Given that only the half-life of the MyoD mRNA appeared to be affected, it was important to assure that the difference observed in MyoD mRNA levels were really due to posttranscriptional regulation. We therefore performed nuclear run-on experiments to assess the transcription rate of these genes in the different transfectants (Fig. 9). Importantly, transcription levels of MyoD mRNA were comparable in GM and DM and were roughly equal in all three populations. The transcription of GAPDH mRNA was approximately equivalent in the three populations of cells both before and after differentiation. There was a small increase in Myf5 mRNA transcription in RLI sense (VS) cells and RLI antisense (VAS) cells in GM and DM compared to control (VV) cells. α-Actin and myogenin mRNA transcription were already detectable in RLI sense (VS) cells before differentiation and increased during differentiation.
FIG. 9.
Nuclear run-on transcription analysis in RLI-transfected C2 cells. (A) Each polyclonal population of transfected C2 (VV, VAS, and VS) myoblasts grown in GM and myotubes grown in DM was harvested, and the nuclei were extracted as described in Materials and Methods. In vitro-elongated RNAs were hybridized to a blot of DNAs corresponding to MyoD, Myf5, myogenin, α-actin, and GAPDH. (B) After visualization on a PhosphorImager 445-SI (Molecular Dynamics), the mRNAs were quantified by image analysis with the ImageQuant program (Molecular Dynamics). A densitometric analysis of the autoradiography shown in panel A is presented. Error bars refer to the standard deviation obtained in three independent experiments.
Therefore, the difference observed in the level of MyoD mRNA was due to a modification of its mRNA stability, whereas the difference observed in myogenin mRNA levels was likely due to a modification of its transcription.
Decreased level of MyoD mRNA in C2 cells overexpressing RNase L.
To more precisely study the role played by RNase L in MyoD expression, we modified RNase L activity in C2 cells using another method. C2 cells were transfected with a vector from which RNase L was expressed upon induction with IPTG. First, C2 cells were transfected with a vector allowing constitutive expression of the Lac repressor. Clonal populations of these cells were tested for expression of the Lac repressor and then transfected with a vector wherein RNase L expression was under the control of the Lac operator. Under these conditions, the Lac repressor binds to the Lac operator, blocking transcription of RNase L. Synthetic inducers such as IPTG, which bind to the Lac repressor, effectively decrease the affinity of the repressor for the operator, resulting in the transcription of RNase L.
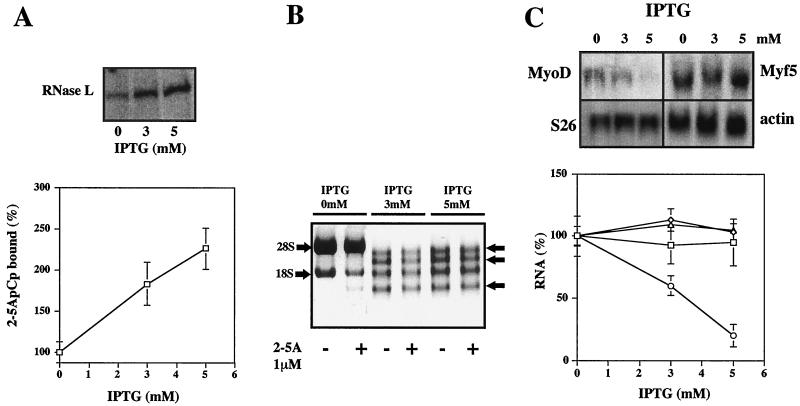
In a selected C2 clone (RNase L3), RNase L expression was induced with different concentrations of IPTG (3 and 5 mM), and the RNase L 2-5A binding activity was studied (Fig. 10). IPTG induced RNase L expression, as assessed by an induction in 2-5A binding activity and nuclease activity (Fig. 10A and B). As in the VAS clone, cleavage of rRNAs was observed even in the absence of 2-5A. In this clone, the level of MyoD mRNA was greatly decreased (Fig. 10C), whereas Myf5, S26, and actin mRNA levels were not affected. Moreover, in this clone, there was a direct correlation between the level of RNase L and the level of MyoD mRNA.
FIG. 10.
MyoD mRNA levels are decreased in a C2 clone overexpressing RNase L. (A) The RNase L3 clone was treated with increasing concentrations of IPTG during 8 h. Cells were then harvested and analyzed for RNase L 2-5A binding activity with the 2-5A radiocovalent binding assay. A densitometric analysis of the gel is shown. A value of 100% corresponds to the amount of 2-5A binding activity in proliferating RNase L3 clone myoblasts in the absence of IPTG. Error bars refer to the standard deviation obtained in three independent experiments. (B) RNase L rRNAs cleavage activity was assessed in the absence or presence of 2-5A (1 μM) as described in Fig. 4. (C) mRNAs were analyzed by Northern blotting using the indicated [32P]cDNA probes. Bands were visualized on a PhosphorImager 445-SI (Molecular Dynamics). A densitometric analysis of the gels is shown (◊, actin mRNA; □, Myf5 mRNA; ▵, S26 mRNA; ○, MyoD mRNA). A value of 100% corresponds to the amount of each mRNA in proliferating RNase L3 clone myoblasts in the absence of IPTG. Error bars refer to the standard deviation obtained in three independent experiments.
DISCUSSION
The role performed by RLI in the control of RNase L activity during the differentiation of cultured C2 myoblasts to myotubes is essential. To more precisely study the role of RLI and RNase L during muscle differentiation, the level of expression of these two proteins was modulated. Upon expression of RLI antisense and sense cDNA constructs in C2 cells, RNase L activity was increased (VAS cells) and decreased (VS cells), respectively. RNase L activity was also increased upon conditional expression of RNase L.
The data presented here indicate that RNase L activity increases during the differentiation of cultured C2 myoblasts to myotubes. RNase L levels were maximal at day 2 following differentiation induction and then decreased concomitantly with the expression of its inhibitor RLI (Fig. 1, 3, and 4). Nevertheless, the decrease in RNase L activity could not be due solely to the induction of RLI since RNase L activity also decreased in cells where RLI expression was severely diminished by an RLI antisense construct (Fig. 2, 3, and 4). We were unable to determine whether RNase L is regulated at the transcriptional level because we failed to detect RNase L mRNA in C2 cells. This is in agreement with prior studies showing that the level of RNase L mRNA is very low in the absence of IFN treatment (53).
The over- or underexpression of RLI resulted in a modification of the kinetics of appearance of myotube-specific proteins such as α-actin and troponin T (Fig. 5). The expression of these two proteins was delayed in RLI antisense (VAS) cells (conditions where RNase L activity was increased), whereas they were expressed earlier in RLI sense (VS) cells (RLI was overexpressed, and RNase L activity was decreased). The expression of such specific proteins is a consequence of an entire differentiation program which is based on the expression of muscle-specific transcription factors. The most striking variation between the different transfectants was the expression of two of these specific transcription factors: MyoD and myogenin (Fig. 5, 6, and 7).
Myogenin mRNA and protein were detectable even under nondifferentiating conditions (GM) in RLI sense (VS) cells. In these cells, the increase in myogenin mRNA was due to augmented transcription since the half-life of the mRNA was not changed (Fig. 7 and 9). In contrast, the stability of MyoD mRNA was significantly altered in cells with modified RNase L activity (Fig. 7, 8, 9, and 10). MyoD is a muscle-specific transcription factor that can activate downstream myogenic structural genes and myogenic conversion in many different cell types (32). MyoD also activates its own transcription, as well as the transcription of myogenin. This activation is direct and does not involve any other intermediates (17, 43). Therefore, the modification of MyoD expression by posttranscriptional regulation affects the level of myogenin mRNA transcripts. In cells with a high level of MyoD (VS cells), we observed a small increase in MyoD transcription and an important increase in myogenin transcription, whereas in cells where the level of MyoD was low (VAS cells), MyoD transcription was slightly decreased and myogenin transcription was very low (Fig. 9). Consequently, the expression of other structural genes such as α-actin and troponin T which are controlled, even indirectly, by MyoD and myogenin was also modified. Finally, it is interesting to note that all three populations of transfectants (VV, VAS, and VS), as well as the inductible RNase L3 clone, expressed comparable levels of Myf5, providing additional evidence that Myf5 and MyoD are not fully interchangeable. Previously, Montarras et al. showed that MyoD but not Myf5 is essential for the differentiation of C2 cells (29).
Although we observed a direct correlation between the level of RNase L activity and MyoD expression, we could not completely exclude the possibility that MyoD is not a direct target of RNase L but rather is indirectly regulated by another RNase which is controlled by RNase L. Likewise, although we have not found that RLI changes the activity on other known RNases (5), it is possible that RLI regulates also the activity of an as-yet-undefined RNase.
RNase L is not the only IFN-induced protein that is increased during myoblasts differentiation. 2-5A-synthetase is induced in rat and mouse myoblasts (4). PKR is induced after the shift of mouse C2C12 myoblasts to differentiating conditions. Transfection of PKR in C2C12 cells results in the increased expression of MyoD and myogenin, and the differentiation process is induced (23). Another protein, p202, is induced later in the differentiation of C2C12 cells. In this case, the overexpression of p202 results in a reduced level of MyoD by affecting its expression at the transcriptional level and inhibits MyoD and myogenin transcriptional activity (9). These proteins, all activated during muscle differentiation, operate at different levels and by different pathways, but they all seem to regulate MyoD. PKR induces MyoD and myogenin by an unknown mechanism, whereas p202 and RNase L act at the transcriptional and posttranscriptional levels, respectively. The activation of these proteins seems to be sequential and may explain why previous studies, assessing the role of IFN during differentiation, appear to be conflicting. Some studies describe an inhibitory effect of IFN on myogenesis (26, 31), whereas others report a positive effect (1, 13, 44).
Here we have demonstrated that RNase L and RLI are directly involved in muscle cell regulation and have established MyoD as a potential target of the RNase L pathway. It now remains to be determined how RNase L affects MyoD mRNA stability in the presence of different cellular mRNAs. It is highly unlikely that MyoD is the only mRNA which is regulated by RNase L.
ACKNOWLEDGMENTS
We thank A. Bonnieu (INRA, Montpellier, France) for the gift of C2 cells and MyoD, Myf5, myogenin, α-actin plasmids; G. Cathala (IGMM, Montpellier, France) for polyclonal antibody against GAPDH and fruitful discussions; and P. Dias and P. Houghton for the gift of 5.8A mouse monoclonal antibody against MyoD. We are also very grateful to N. Taylor (IGMM, Montpellier, France) for critical reading of the manuscript.
This work was done in B. Lebleu's laboratory and was supported by the Association pour la Recherche Contre le Cancer (ARC) research grants (9492 and 9298). F. Le Roy is a recipient of a doctoral fellowship from the ARC, and L. Milligan is a recipient of a doctoral fellowship from the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Andre P, Braun S, Passaquin A, Coupin G, Bartholeyns J, Warter J, Poindron P. Rat interferon enhances the expression of acetylcholine receptors in rat myotubes in culture. J Neurosci Res. 1988;19:297–302. doi: 10.1002/jnr.490190304. [DOI] [PubMed] [Google Scholar]
- 2.Bayard B, Bisbal C, Silhol M, Cnockaert J, Huez G, Lebleu B. Increased stability and antiviral activity of 2′-O-phosphoglyceryl derivatives of (2′-5′)oligo(adenylate) Eur J Biochem. 1984;142:291–298. doi: 10.1111/j.1432-1033.1984.tb08284.x. [DOI] [PubMed] [Google Scholar]
- 3.Benoit De Coignac A, Bisbal C, Lebleu B, Salehzada T. cDNA cloning and expression analysis of the murine ribonuclease L inhibitor. Gene. 1998;209:149–156. doi: 10.1016/s0378-1119(98)00040-7. [DOI] [PubMed] [Google Scholar]
- 4.Birnbaum M, Trink B, Shainberg A, Salzberg S. Activation of the interferon system during myogenesis in vitro. Differentiation. 1990;45:138–145. doi: 10.1111/j.1432-0436.1990.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 5.Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of an RNase L inhibitor. A new component of the interferon-regulated 2-5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 6.Bisbal C, Salehzada T, Lebleu B, Bayard B. Characterization of two murine (2′-5′)(A)n-dependent endonucleases of different molecular mass. Eur J Biochem. 1989;179:595–602. doi: 10.1111/j.1432-1033.1989.tb14588.x. [DOI] [PubMed] [Google Scholar]
- 7.Bonnieu A, Rech J, Jeanteur P, Fort P. Requirements for c-fos mRNA downregulation in growth stimulated murine cells. Oncogene. 1989;4:881–888. [PubMed] [Google Scholar]
- 8.Braun T, Bober E, Buschhausen D G, Kohtz S, Grzeschik K H, Arnold H H, Kotz S. Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J. 1989;8:3617–3625. doi: 10.1002/j.1460-2075.1989.tb08535.x. . (Erratum, 8:4358.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta B, Min W, Burma S, Lengyel P. Increase in p202 expression during skeletal muscle differentiation: inhibition of MyoD protein expression and activity by p202. Mol Cell Biol. 1998;18:1074–1083. doi: 10.1128/mcb.18.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dias P, Parham D M, Shapiro D N, Tapscott S J, Houghton P J. Monoclonal antibodies to the myogenic regulatory protein MyoD1: epitope mapping and diagnostic utility. Cancer Res. 1992;52:6431–6439. [PubMed] [Google Scholar]
- 11.Dong B, Silverman R H. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 12.Dong B, Xu L, Zhou A, Hassel B A, Lee X, Torrence P F, Silverman R H. Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J Biol Chem. 1994;269:14153–14158. [PubMed] [Google Scholar]
- 13.Fisher P B, Miranda A F, Babiss L E, Pestka S, Weinstein I B. Opposing effects of interferon produced in bacteria and of tumor promoters on myogenesis in human myoblast cultures. Proc Natl Acad Sci USA. 1983;80:2961–2965. doi: 10.1073/pnas.80.10.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg M E, Ziff E B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 16.Gribaudo G, Lembo D, Cavallo G, Landolfo S, Lengyel P. Interferon action: binding of viral RNA to the 40-kilodalton 2′-5′-oligoadenylate synthetase in interferon-treated HeLa cells infected with encephalomyocarditis virus. J Virol. 1991;65:1748–1757. doi: 10.1128/jvi.65.4.1748-1757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollenberg S M, Cheng P F, Weintraub H. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc Natl Acad Sci USA. 1993;90:8028–8032. doi: 10.1073/pnas.90.17.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kariko K, Li S W, Sobol R J, Suhadolnik R J, Charubala R, Pfleiderer W. Phosphorothioate analogues of 2′,5′-oligoadenylate: activation of 2′,5′-oligoadenylate-dependent endoribonuclease by 2′,5′-phosphorothioate cores and 5′-monophosphates. Biochemistry. 1987;26:7136–7142. doi: 10.1021/bi00396a040. [DOI] [PubMed] [Google Scholar]
- 19.Kariko K, Ludwig J. n-Decyl-NHpppA2′p5′A2′p5′A: a phosphatase-resistant, active pppA2′p5′A2′p5′A analog. Biochem Biophys Res Commun. 1985;128:695–698. doi: 10.1016/0006-291x(85)90102-0. [DOI] [PubMed] [Google Scholar]
- 20.Kariko K, Sobol R J, Suhadolnik L, Li S W, Reichenbach N L, Suhadolnik R J, Charubala R, Pfleiderer W. Phosphorothioate analogues of 2′,5′-oligoadenylate. Enzymatically synthesized 2′,5′-phosphorothioate dimer and trimer: unequivocal structural assignment and activation of 2′,5′-oligoadenylate-dependent endoribonuclease. Biochemistry. 1987;26:7127–7135. doi: 10.1021/bi00396a039. [DOI] [PubMed] [Google Scholar]
- 21.Kerr I M, Brown R E. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight M, Cayley P J, Silverman R H, Wreschner D H, Gilbert C S, Brown R E, Kerr I M. Radioimmune, radiobinding and HPLC analysis of 2-5A and related oligonucleotides from intact cells. Nature. 1980;288:189–192. doi: 10.1038/288189a0. [DOI] [PubMed] [Google Scholar]
- 23.Kronfeld K Y, Vilchik S, Hyman T, Leibkowicz F, Salzberg S. Involvement of PKR in the regulation of myogenesis. Cell Growth Differ. 1999;10:201–212. [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Li X L, Blackford J A, Hassel B A. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J Virol. 1998;72:2752–2759. doi: 10.1128/jvi.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lough J, Keay S, Sabran J L, Grossberg S E. Inhibition of chicken myogenesis in vitro by partially purified interferon. Biochem Biophys Res Commun. 1982;109:92–99. doi: 10.1016/0006-291x(82)91570-4. [DOI] [PubMed] [Google Scholar]
- 27.Martinand C, Montavon C, Salehzada T, Silhol M, Lebleu B, Bisbal C. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and downregulates the 2-5A/RNase L pathway in human T cells. J Virol. 1999;73:290–296. doi: 10.1128/jvi.73.1.290-296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinand C, Salehzada T, Silhol M, Lebleu B, Bisbal C. RNase L inhibitor (RLI) antisense constructions block partially the downregulation of the 2-5A/RNase L pathway in encephalomyocarditis virus (EMCV)-infected cells. Eur J Biochem. 1998;254:238–247. doi: 10.1046/j.1432-1327.1998.2540248.x. [DOI] [PubMed] [Google Scholar]
- 29.Montarras D, Aurade F, Johnson T, IIan J, Gros F, Pinset C. Autonomous differentiation in the mouse myogenic cell line, C2, involves a mutual positive control between insulin-like growth factor II and MyoD, operating as early as at the myoblast stage. J Cell Sci. 1996;109:551–560. doi: 10.1242/jcs.109.3.551. [DOI] [PubMed] [Google Scholar]
- 30.Montarras D, Chelly J, Bober E, Arnold H, Ott M O, Gros F, Pinset C. Developmental patterns in the expression of Myf5, MyoD, myogenin, and MRF4 during myogenesis. New Biol. 1991;3:592–600. [PubMed] [Google Scholar]
- 31.Multhauf C, Lough J. Interferon-mediated inhibition of differentiation in a murine myoblast cell line. J Cell Physiol. 1986;126:211–215. doi: 10.1002/jcp.1041260209. [DOI] [PubMed] [Google Scholar]
- 32.Olson E N. MyoD family: a paradigm for development? Genes Dev. 1990;4:1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- 33.Pinset C, Montarras D, Chenevert J, Minty A, Barton P, Laurent C, Gros F. Control of myogenesis in the mouse myogenic C2 cell line by medium composition and by insulin: characterization of permissive and inducible C2 myoblasts. Differentiation. 1988;38:28–34. doi: 10.1111/j.1432-0436.1988.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 34.Romeo G, Affarbis E, Battistini A, Rossi G. Cell and animal effects of IFNs: natural occurrence and differentiation effects and mechanisms. In: Baron S, Coppenhaver D H, Dianzani F, Fleischmann R, Hughes T K, Klimpel G R, Niesel D W, Stanton G J, Tyring S K, editors. Interferon: principles and medical applications. Galveston, Tex: The University of Texas Medical Branch at Galveston Department of Microbiology; 1992. pp. 271–287. [Google Scholar]
- 35.Rysiecki G, Gewert D R, Williams B R. Constitutive expression of a 2′,5′-oligoadenylate synthetase cDNA results in increased antiviral activity and growth suppression. J Interferon Res. 1989;9:649–657. doi: 10.1089/jir.1989.9.649. [DOI] [PubMed] [Google Scholar]
- 36.Salzberg S, Hyman T, Turm H, Kinar Y, Schwartz Y, Nir U, Lejbkowicz F, Huberman E. Ectopic expression of 2-5A synthetase in myeloid cells induces growth arrest and facilitates the appearance of a myeloid differentiation marker. Cancer Res. 1997;57:2732–2740. [PubMed] [Google Scholar]
- 37.Salzberg S, Lanciano F, Hacohen D. Reversibility of the antiproliferative effect of interferon. Nat Immun Cell Growth Regul. 1990;9:191–202. [PubMed] [Google Scholar]
- 38.Salzberg S, Mandelboim M, Zalcberg M, Shainberg A, Mandelbaum M. Interruption of myogenesis by transforming growth factor beta 1 or EGTA inhibits expression and activity of the myogenic-associated (2′-5′)oligoadenylate synthetase and PKR. Exp Cell Res. 1995;219:223–232. doi: 10.1006/excr.1995.1222. . (Erratum, 220:509.) [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 40.Sassoon D A. Myogenic regulatory factors: dissecting their role and regulation during vertebrate embryogenesis. Dev Biol. 1993;156:11–23. doi: 10.1006/dbio.1993.1055. [DOI] [PubMed] [Google Scholar]
- 41.Sen G C, Lengyel P. The interferon system: a bird's eye view of its biochemistry. J Biol Chem. 1992;267:5017–5020. [PubMed] [Google Scholar]
- 42.Silverman R H, Skehel J J, James T C, Wreschner D H, Kerr I M. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol. 1983;46:1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thayer M J, Tapscott S J, Davis R L, Wright W E, Lassar A B, Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989;58:241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- 44.Tomita Y, Hasegawa S. Multiple effects of interferon on myogenesis in chicken myoblast cultures. Biochim Biophys Acta. 1984;804:370–376. doi: 10.1016/0167-4889(84)90141-1. [DOI] [PubMed] [Google Scholar]
- 45.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent S, Marty L, Fort P. S26 ribosomal protein RNA: an invariant control for gene regulation experiments in eucaryotic cells and tissues. Nucleic Acids Res. 1993;21:1498. doi: 10.1093/nar/21.6.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 48.Whitaker J R, Granum P E. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal Biochem. 1980;109:156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]
- 49.Wreschner D H, James T C, Silverman R H, Kerr I M. Ribosomal RNA cleavage, nuclease activation and 2-5A(ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res. 1981;9:1571–1581. doi: 10.1093/nar/9.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wreschner D H, Silverman R H, James T C, Gilbert C S, Kerr I M. Affinity labelling and characterization of the ppp(A2′p)nA-dependent endoribonuclease from different mammalian sources. Eur J Biochem. 1982;124:261–268. doi: 10.1111/j.1432-1033.1982.tb06586.x. [DOI] [PubMed] [Google Scholar]
- 51.Yaffe A, Schwarz Y, Hacohen D, Kinar Y, Nir U, Salzberg S. Inhibition of 2-5A synthetase expression by antisense RNA interferes with interferon-mediated antiviral and antiproliferative effects and induces anchorage-independent cell growth. Cell Growth Differ. 1996;7:969–978. [PubMed] [Google Scholar]
- 52.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 53.Zhou A, Hassel B A, Silverman R H. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]