ABSTRACT
Streptococcus pneumoniae colonizes the nasopharynx of children and the elderly but also kills millions worldwide yearly. The secondary bile acid metabolite deoxycholic acid (DoC) affects the viability of human pathogens but also plays multiple roles in host physiology. We assessed in vitro the antimicrobial activity of DoC and investigated its potential to eradicate S. pneumoniae colonization using a model of human nasopharyngeal colonization and an in vivo mouse model of colonization. At a physiological concentration, DoC (0.5 mg/ml; 1.27 mM) killed all tested S. pneumoniae strains (n = 48) 2 h postinoculation. The model of nasopharyngeal colonization showed that DoC eradicated colonization by S. pneumoniae strains as soon as 10 min postexposure. The mechanism of action did not involve activation of autolysis, since the autolysis-defective double mutants ΔlytAΔlytC and ΔspxBΔlctO were as susceptible to DoC as was the wild type (WT). Oral streptococcal species (n = 20), however, were not susceptible to DoC (0.5 mg/ml). Unlike trimethoprim, whose spontaneous resistance frequency (srF) for TIGR4 or EF3030 was ≥1 × 10−9, no spontaneous resistance was observed with DoC (srF, ≥1 × 10–12). Finally, the efficacy of DoC to eradicate S. pneumoniae colonization was assessed in vivo using a topical route via intranasal (i.n.) administration and as a prophylactic treatment. Mice challenged with S. pneumoniae EF3030 carried a median of 4.05 × 105 CFU/ml 4 days postinoculation compared to 6.67 × 104 CFU/ml for mice treated with DoC. Mice in the prophylactic group had an ∼99% reduction of the pneumococcal density (median, 2.61 × 103 CFU/ml). Thus, DoC, an endogenous human bile salt, has therapeutic potential against S. pneumoniae.
KEYWORDS: Streptococcus pneumoniae, bile acids, colonization, deoxycholate, eradication
INTRODUCTION
Streptococcus pneumoniae colonizes millions of people worldwide yearly, with a colonization prevalence particularly high in children and the elderly (1–3). In children, the prevalence of pneumococcal carriage can be as high as 90% in those from low- to middle-income countries and between 25 and 40% in children from high-income countries (1, 4–8). Along the same lines, carriage of S. pneumoniae in adults 18 to 49.9 years of age can be as high as 50% (8, 9), while pneumococcal carriage in a more vulnerable population, those older than 60, is similar (10). Besides the carriage prevalence, recent studies have demonstrated that a high nasopharyngeal colonization density of S. pneumoniae is associated with cases of severe pneumonia as well as other lower respiratory tract infections in children (11–13). An increased pneumococcal density has also been implicated with transmission in animal models (14) and in children with influenza-like illness (15).
The frequency of pneumococcal colonization is also increased in individuals infected with the influenza virus (5, 16, 17). Recent evidence also demonstrated that pneumococcal colonization density increased in those individuals with asymptomatic infection with the influenza virus (18). Although it is too early to draw conclusions, the frequency of pneumococcal carriage increased in populations with a positive swab for SARS-CoV-2, whereby we may experience a surge of pneumococcal disease (PD) cases in the next few years (19, 20).
Pneumococcal carriage is a risk for developing PD, and therefore it is considered an immediate and necessary precursor of PD (1, 2, 21, 22). An important intervention with a demonstrated positive impact on S. pneumoniae carriage has been vaccination (23). Pneumococcal conjugated vaccines (PCV) have been introduced in many parts of the world since 2001, when PCV7 was licensed in the United States (24, 25). The introduction of these vaccines reduced the burden of PD caused by vaccine serotypes on a global scale and has also decreased nasopharyngeal carriage of pneumococcal vaccine types in vaccinated populations (23, 26). The overall carriage prevalence has not changed, because of a phenomenon called “serotype replacement”; i.e., vaccine-escape strains have replaced vaccine type (VT) strains in the nasopharynx, resulting in pneumococcal carriage rates similar to those observed prior to the introduction of vaccines (27–29). However, we have observed that the S. pneumoniae carriage density in individuals colonized with a non-vaccine type increases postvaccination with PCV7 (92). Therefore, additional interventions, or prophylactic strategies, are needed to reduce pneumococcal carriage, carriage density, and the burden of pneumococcal disease.
Pathogenic and commensal bacteria are susceptible to bile from different mammals (30–32). Recent studies have demonstrated that the lack of primary and secondary bile acid metabolites is implicated in the development of intestinal infectious disease (30–32). Bile consists of ∼95% water in which are dissolved a number of endogenous solid constituents, including bile salts, bilirubin phospholipid, cholesterol, amino acids, steroids, enzymes, porphyrins, vitamins, and heavy metals (33). Bile salts are the major organic solutes in bile and normally function to emulsify dietary fats and facilitate their intestinal absorption. The two main primary bile salts that are synthesized in the mammalian liver are cholic acid, and chenodeoxycholic acid (33, 34). Intestinal bacteria then produce secondary bile acids through enzymatic reactions, with the addition of two hydroxyl groups to cholic acid producing one of the most abundant secondary metabolites, deoxycholic acid (DoC) (33, 35). Endogenously produced, secondary bile acids have widespread effects on the host and resident microbiota and have therefore been used to treat different diseases (35). Cholic acid is used to treat patients with genetic deficiencies in the synthesis of bile acids due to single enzyme deficiencies; the typical dose is 10 to 15 mg/kg of body weight once daily (36). DoC has been utilized in humans at a concentration of 15 mg/kg/day to decrease plasma high-density lipoprotein (HDL) cholesterol and low-density lipoprotein (LDL) cholesterol (36), and it is also FDA-approved to reduce fat deposits (37, 38).
In addition to their physiological role in digesting lipids, primary and secondary bile acids are important regulators of intestinal homeostasis since either reducing the intestinal pool of these secondary metabolites or supplying them from an exogenous source has proven important for the susceptibility of the host to infections (34). For example, in germfree mice or mice treated with antibiotics to deplete intestinal bacteria that produce secondary bile acids, the lack of these secondary metabolites caused deficient TLR7-MyD88 signaling, resulting in an increased susceptibility to systemic chikungunya infection (30). Studies using another secondary bile acid, ursodeoxycholic acid (UDA), demonstrated that UDA contributes to colonization resistance in a mouse model of Clostridiodes difficile infection by inhibiting in vitro toxin production, growth, and spore germination of strains (39). Similarly, an in vitro study demonstrated that reducing levels of DoC produced by intestinal bacteria allowed an increased density of C. difficile, indicating that DoC is an important defense against intestinal colonization by this pathogen (40). Moreover, oral administration to mice of DoC reduced Campylobacter jejuni-induced colitis (31). The growth of another intestinal pathogen, Clostridium perfringens, was inhibited in vitro with as little as 50 μM DoC. Using an animal model of necrotic enteritis, supplementation of DoC in the diet (e.g., 1.5 g/kg) decreased intestinal inflammation and necrotic enteritis-associated intestinal cell death and apoptosis (32).
In vitro studies demonstrated synergistic antibiotic activity when Doc (2.5 mg/ml) was combined with vancomycin, or with vancomycin and furazolidone, to treat clinically important antibiotic-resistant pathogens (41). Synergism between DoC, or lithocholic acid, and tryptophan-derived antibiotics secreted by intestinal bacteria inhibited bacterial division of C. difficile strains (42). In this study, we assessed the antimicrobial activity of DoC against a collection of S. pneumoniae strains, including reference strains, S. pneumoniae isolated from cases of pneumococcal disease, and strains resistant to multiple antibiotics. The mechanistic basis for the observed sensitivity to DoC was investigated using knockout mutants in different autolytic mechanisms. We also adapted a model of nasopharyngeal colonization to investigate the efficacy of DoC to eradicate S. pneumoniae colonization and, finally, developed an in vivo prophylactic mouse model to demonstrate that administration of DoC inhibited S. pneumoniae colonization.
RESULTS
Deoxycholic acid (DoC) eradicates cultures of drug-resistant S. pneumoniae.
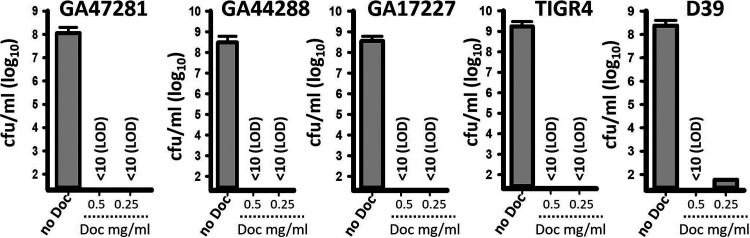
We challenged reference strains D39 and TIGR4 and drug-resistant S. pneumoniae 19A and 19F vaccine-serotype strains with increasing dosages of DoC and incubated them for 2 h. Whereas untreated bacteria grew at >108 CFU/ml, all five pneumococcal strains treated with DoC were killed within 2 h with 0.5 mg/ml (1.27 mM). Except for strain D39, all other vaccine serotype S. pneumoniae strains were also killed with 250 μg/ml of DoC (Fig. 1).
FIG 1.
Deoxycholic acid kills S. pneumoniae strains within 2 h of incubation. S. pneumoniae strains GA47281, GA44288, GA17227, TIGR4, and D39 were inoculated at a density of ∼5.15 × 108 CFU/ml in THY broth and left untreated (no Doc) or treated with 0.5 mg/ml or 0.25 mg/ml of deoxycholic acid (DoC). Bacteria were incubated for 2 h at 37°C in a 5% CO2 atmosphere, after which the cultures were serially diluted and plated onto blood agar plates to obtain the density (CFU/ml). Error bars represent the standard errors of the means calculated using data from at least three independent experiments. The limit of detection (LOD) was <10 CFU/ml.
We then assessed the antimicrobial efficacy of DoC (0.5 mg/ml) against 39 pneumococcal strains isolated from cases of pneumococcal disease. The strains represented all PCV13 vaccine serotypes, and at least two strains of each vaccine serotype were challenged. Table S1 in the supplemental material confirmed that 0.5 mg/ml (1.27 mM) of DoC incubated for 2 h killed all assessed S. pneumoniae strains. A MIC90 of 0.5 mg/ml DoC was established.
A similar MIC (0.5 mg/ml [1.27 mM]) was obtained with reference strain TIGR4, or EF3030, assessed in cation-activated Mueller-Hinton broth (CAMHB) with 3% of lysed horse blood (LHB) and according to CLSI guidelines to assess resistance (43), or nonsusceptibility, of S. pneumoniae strains (Fig. S1 and not shown). Whereas the density of TIGR4 inoculated in CAMHB and incubated overnight reached 6.7 × 1010 CFU/ml, those cultures treated with DoC (1.27 mM) had a median density of 1.2 × 104 CFU/ml, an ∼99.99% reduction in density after overnight incubation (Fig. S1).
DoC eradicates MDR S. pneumoniae colonization in a model with human pharyngeal cells.
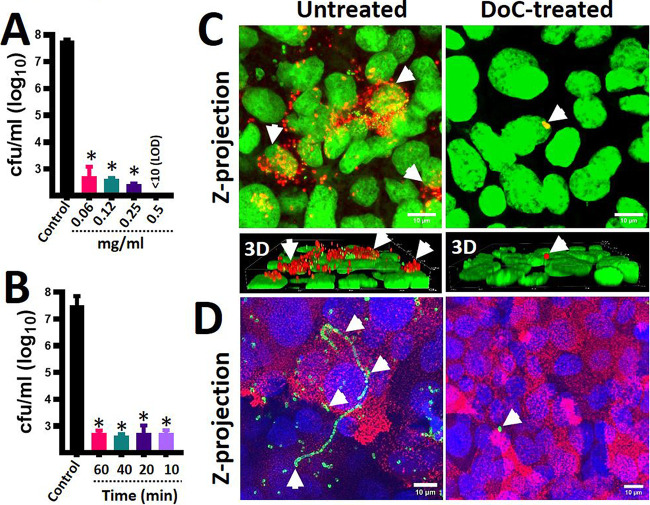
To assess the potential of DoC to eradicate pneumococcal colonization, we performed experiments using a model of colonization. To create a biotic substrate for pneumococci to colonize, we used polarized human pharyngeal cells that had been fixed with paraformaldehyde. These cell monolayers were infected with a multidrug-resistant (MDR) S. pneumoniae strain, GA47281, a vaccine strain serotype 19F bearing resistance to several antibiotics, including erythromycin, meropenem, and cefuroxime. Infected human cells were incubated for 4 h to allow attachment of S. pneumoniae, and then planktonic bacteria were removed. S. pneumoniae-colonized human cells were left untreated or challenged with different dosages of DoC and incubated for 2 h. All doses of DoC reduced the density of MDR pneumococci >90% after the 2 h of incubation compared to untreated human pharyngeal cells, which had a density of ∼1 × 107 CFU/ml of pneumococci (Fig. 2A). GA47281 bacteria were not recovered when pneumococci were treated with 0.5 mg/ml (1.27 mM). Confocal scanning laser microscopy, XY optical sections, and three-dimensional (3D) reconstructions of cells infected with S. pneumoniae confirmed that DoC had eradicated colonization by strain GA47281 (Fig. 2C) or colonization by strain TIGR4 (Fig. 2D) from human pharyngeal cells.
FIG 2.
Eradication of pneumococcal colonization with DoC using a model of human pharyngeal colonization. Polarized human pharyngeal Detroit 562 cells were immobilized with 2% paraformaldehyde and then infected with S. pneumoniae strain GA47281 (∼5.15 × 108 CFU/ml). Infected cells were incubated for 4 h, and planktonic cells were removed. (A and B) These S. pneumoniae-colonized human pharyngeal cells were left untreated (control) or (A) treated with different dosages of deoxycholic acid (DoC) and incubated for an additional 2 h period or (B) treated with DoC (0.5 mg/ml) and incubated for the indicated time. At the end of the incubation, the cultures were serially diluted and plated onto blood agar plates to obtain the density (CFU/ml). In panels A and B, error bars represent the standard errors of the means calculated using data from at least three independent experiments. *, P < 0.05 compared to the untreated control. LOD, limit of detection. (C and D) S. pneumoniae-colonized human pharyngeal cells (infected as described above) were left untreated or treated with DoC (0.5 mg/ml) and incubated for 2 h. (C and D) Preparations were fixed, and S. pneumoniae was stained with (C) an anti-S19-Alexa-555 labeled antibody (red) and the DNA was stained with TOPRO3 (green) or with (D) an anti-S4-Alexa-488 labeled antibody (green); cell membranes were labeled with WGA (red), and DNA was labeled with DAPI (blue). Preparations were analyzed with a confocal microscope. Panels show z-projections of z-stacks obtained from xy optical sections. Lower bottom of panel C shows a 3D digital reconstruction. The merge of the channels is shown in each panel. Arrows point out extracellular S. pneumoniae bacteria.
To determine the exposure time required to kill pneumococci, we performed a time course study treating MDR pneumococci colonizing human pharyngeal cells for up to 1 h with 0.5 mg/ml (1.27 mM). Interestingly, a 10-min exposure time was enough to kill most attached MDR pneumococci (Fig. 2B). The antimicrobial effect did not change with a longer exposure time of 60 min. These data indicate that 0.5 mg/ml (1.27 mM) of DoC rapidly eradicates MDR pneumococci that otherwise would have colonized human nasopharyngeal cells at a density of ∼107 CFU/ml (Fig. 2B).
Spontaneous resistance to DoC (500 μg/ml) was not developed by S. pneumoniae strains.
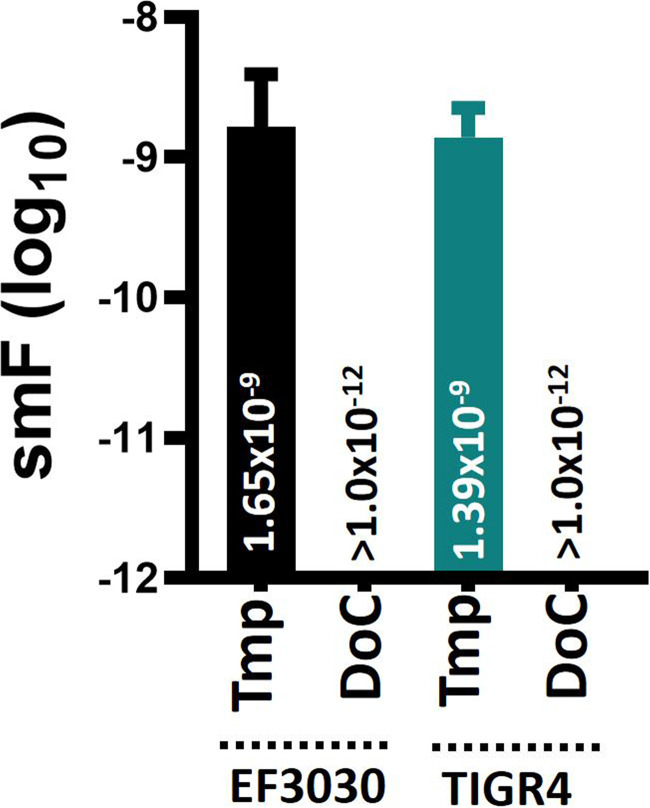
S. pneumoniae strains develop spontaneous resistance to some antibiotics at a spontaneous mutation frequency of >1 × 10−8 (44). We assessed spontaneous resistance to DoC using antibiotic-sensitive strains TIGR4 and EF3030. As expected, spontaneous resistance against trimethoprim developed at a frequency of ≥1.39 × 10−9 when either pneumococcal strain was assessed (Fig. 3). Spontaneous resistance to DoC, however, was not observed in any of the two strains tested, even at a population density of >1012 CFU/ml (Fig. 3).
FIG 3.
Spontaneous resistance mutation frequency of S. pneumoniae for trimethoprim and deoxycholic acid. Bacterial suspensions prepared with fresh cultures of S. pneumoniae strain EF3030 or TIGR4 were adjusted to a density of ∼108, ∼109, ∼1010, ∼1011, and ∼1012 CFU/ml and inoculated onto blood agar plates (BAP) containing trimethoprim (Tmp, 1 μg/ml) or deoxycholic acid (DoC, 0.5 mg/ml). Each inoculum was further diluted and plated onto plain BAP. All cultures were incubated for 20 h, after which bacteria were counted. The spontaneous mutation frequency (smF) was calculated by dividing the number of spontaneously resistant pneumococci, i.e., grown on BAP with trimethoprim or DoC, by the bacterial population. Error bars represent the standard errors of the means calculated using data from at least three independent experiments. The median smF is shown on each bar.
The mechanism of DoC-killing does not appear to trigger autolysis.
Since the above-described experiments revealed that a short exposure time was sufficient to kill pneumococci, we hypothesized that an irreversible autolytic mechanism may have been triggered by DoC. Autolysis is mainly driven by autolysins LytA and LytC (45, 46), but hydrogen peroxide produced by the pneumococcus, through enzymes SpxB and LctO, also contributes to lysis of pneumococci (47–49). To assess this hypothesis, we utilized strain R6 WT, its isogenic double mutants R6ΔlytAΔlytC, and R6ΔspxBΔlctO to assess the role of autolysis in the observed DoC-mediated bactericidal activity. Since autolysis can be measured by quantifying the release of extracellular DNA (eDNA), we first confirmed that the absence of the LytA and LytC autolysins, or SpxB and LctO, caused a decreased release of eDNA into the supernatant. After 4 h of incubation, the eDNA released by R6 WT strain reached a median of 2.24 × 106 pg/ml, whereas R6ΔspxBΔlctO released a statistically different 2-fold-decreased amount of eDNA (median, 1.09 × 106 pg/ml) in the supernatant (Fig. 4A). As expected, the autolysin double mutant R6ΔlytAΔlytC yielded an ∼14-fold-reduced amount of eDNA (median, 1.55 × 105 pg/ml) (Fig. 4A) compared to R6 WT.
FIG 4.
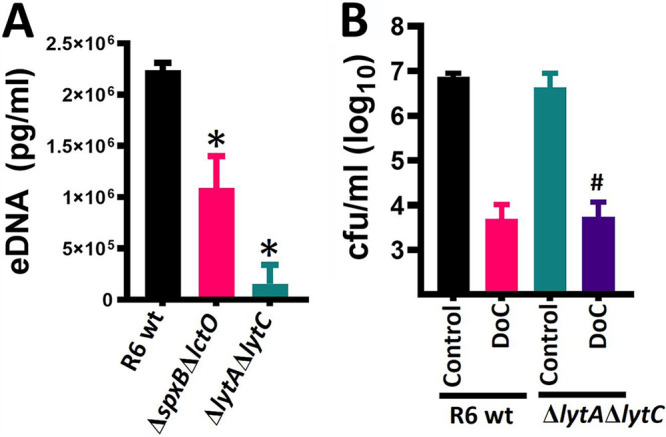
Deoxycholic acid kills pneumococci with a deficient autolytic mechanism. (A) S. pneumoniae strain R6 wt, or isogenic double mutant ΔlytAΔlytC or ΔspxBΔlctO were inoculated in six-well plates containing THY, and bacteria were incubated for 4 h. The supernatants were obtained and filter-sterilized, and the eDNA was purified. This eDNA was used as a template in species-specific quantitative PCRs (qPCRs) along with DNA standards for quantification purposes. *, P < 0.037 compared to R6 WT. (B) R6 WT or its isogenic ΔlytAΔlytC mutant was inoculated at a density of ∼5.15 × 108 CFU/ml in THY broth and left untreated (control) or treated with 0.5 mg/ml of DoC. Bacteria were incubated for 2 h at 37°C in a 5% CO2 atmosphere, after which the cultures were serially diluted and plated onto blood agar plates to obtain the density (CFU/ml). #, P = 0.33 compared to R6 WT strain treated with DoC. In panels A and B error bars represent the standard errors of the means calculated using data from at least three independent experiments.
These two mutant strains with an impaired autolysis phenotype were then challenged with DoC (0.5 mg/ml [1.27 mM]), and treated and untreated bacteria were incubated for 1 h. The results in Fig. 4B show a >90% significant reduction (i.e., killing) of the population of R6 pneumococci after 1 h of incubation. The density of the R6ΔspxBΔlctO isogenic mutant (not shown) or the R6ΔlytAΔlytC mutant (Fig. 4B) was similarly reduced after 1 h of incubation with DoC, indicating that the mechanism by which DoC kills pneumococci is not by triggering autolysis.
DoC does not affect viability of normal flora streptococci.
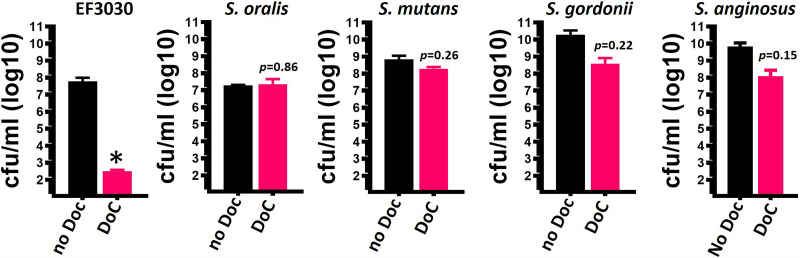
We next investigated whether the MIC90 of DoC that eradicates S. pneumoniae strains (0.5 mg/ml [1.27 mM]) within 2 h of incubation would have the same bactericidal effect against other streptococci that reside in the oral cavity. As a positive control, we utilized S. pneumoniae reference strain EF3030 (50, 51). The median density of EF3030 untreated cultures was 3.37 × 107 CFU/ml, whereas those treated with 0.5 mg/ml DoC for 2 h had a median density of 3.1 × 102 CFU/ml, and therefore DoC killed 99.99% of the bacterial population (Fig. 5). However, the same dose of DoC incubated for 2 h did not significantly affect the viability of S. oralis, S. mutans, of S. gordonii and induced a 2-log reduction of the density of S. anginosus (Fig. 5). An additional 16 different streptococcal species were treated with DoC (0.5 mg/ml [1.27 mM]) for 2 h, and except for S. pseudopneumoniae and S. salivarius, which were susceptible, cultures of all other species were not affected (Table 1).
FIG 5.
Deoxycholic acid (0.5 mg/ml) does not affect the viability of other streptococcal species. S. pneumoniae strain EF3030, S. oralis, S. mutans, S. gordonii, and S. anginosus were inoculated at a density of ∼5.15 × 108 CFU/ml in THY broth and left untreated (no DoC) or treated with 0.5 mg/ml deoxycholic acid (DoC). Bacteria were incubated for 2 h at 37°C in a 5% CO2 atmosphere, after which the cultures were serially diluted and plated onto blood agar plates to obtain the density (CFU/ml). Error bars represent the standard errors of the means calculated using data from at least three independent experiments. *, P < 0.04 compared to the untreated EF3030 control.
TABLE 1.
Antimicrobial activity of DoC against streptococcal species
| Strain | MIC90 (mg/ml)a |
|---|---|
| S. pneumoniae ATCC BAA-255 | 0.25 |
| S. australis ATCC 700641 | >1 |
| S. cristatus | >0.5 |
| S. infantis ATCC 700779 | >0.5 |
| S. intermedius ATCC 27335 | >0.5 |
| S. intestinalis ATCC 43492 | >1 |
| S. oligofermentus CDC SS-1725 | >1 |
| S. parasanguinis ATCC 15912 | >0.5 |
| S. peroris ATCC 700780 | >0.5 |
| S. pseudopneumoniae ATCC BAA-960 | 0.5 |
| S. salivarius ATCC 7073 | 0.5 |
| S. sanguinis ATCC 10556 | >1 |
| S. sinensis CDC SS-1726 | >0.5 |
| Dolosigranulum pigrum | >0.5 |
| S. sobrinus ATCC 33478 | >2 |
Strains were challenged with the MIC90 for S. pneumoniae strains (0.5 mg/ml). The limit of detection of this assay was 50 CFU/ml.
Standardizing the mouse model of pneumococcal carriage to assess the efficacy of DoC to eradicate nasopharyngeal colonization.
The bacterial inoculum utilized in a mouse model of pneumococcal colonization is usually ∼1 × 107 CFU, and nasal washes through the trachea were performed to assess colonization (52, 53). We first standardized the removal of a defined section of nasopharyngeal tissue consisting of the intact nasal septum (Fig. 6A) from colonized mice and confirmed by histological analysis the presence of typical nasopharyngeal tissue, including microvilli and pseudostratified, squamous epithelium overlaid by loose connective tissue including blood vessels with erythrocytes (Fig. 6B). Since our goal was to assess the efficacy of DoC to reduce and/or inhibit nasopharyngeal colonization, we investigated a low inoculum density of S. pneumoniae EF3030 that would sustain colonization of mice but avoided using a nonnatural (i.e., heavy) inoculum. Our experiments demonstrated that inoculating ∼1 × 105 CFU in the nostrils of mice allowed nasopharyngeal colonization for up to 4 days at a median density of 2.49 × 105 CFU/organ (Fig. 6D). We additionally removed the trachea and lungs and demonstrated consistent S. pneumoniae colonization of the trachea, at a low density of 9.0 × 102 CFU/organ, but colonization of the lungs was not observed (Fig. 6D). Encapsulated S. pneumoniae was detected using an anti-S19-Alexa-555 antibody and was identified both in nasopharyngeal homogenates (Fig. 6C) and colonizing the nasopharyngeal tissue (Fig. 6E). Microinvasion into the nasopharyngeal epithelium was also observed (Fig. 6E).
FIG 6.
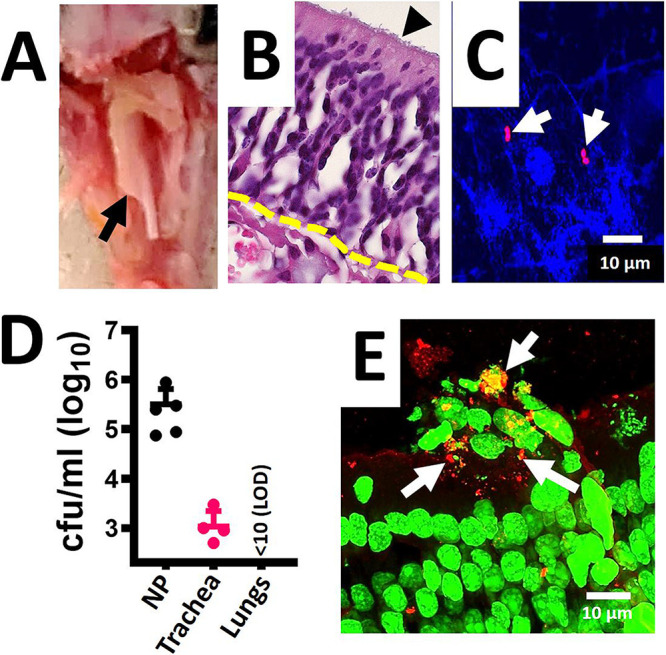
Standardizing the mouse model of pneumococcal nasopharyngeal carriage. C57BL/6 mice (n = 5) were intranasally inoculated with S. pneumoniae EF3030 (∼1 × 105 CFU). (A) After 48 h mice were euthanized, and the nasal bone was removed to expose the nasopharynx, arrow. The nasopharynx, trachea and lungs were removed. (B) The nasopharynges were sectioned (∼5 μm) and stained with hematoxylin and eosin. Arrowhead, microvilli; dotted line, connective tissue. (C and D) Nasopharyngeal (NP) tissue, trachea, and lungs were homogenized, and (C) NP homogenate was stained with DAPI and with an anti-S19-Alexa-555 antibody, or (D) homogenates were diluted and plated onto BAP with gentamicin (25 μg/ml) to obtain the bacterial density (CFU/ml). (E) NP tissue stained with TOTO-1 and with an anti-S19-Alexa-555 antibody; arrows, pneumococci (red). The micrographs in panels C and D are z-projections of z-stacks obtained from xy optical sections collected with a confocal microscope.
Deoxycholic acid (DoC) decreased in vivo colonization by S. pneumoniae strain EF3030 in a mouse model of nasopharyngeal carriage.
We then assessed the efficacy of DoC to eradicate colonization using three groups of mice (n = 8 each). Groups 1 and 2 drank regular water, while the drinking water of group 3 was supplemented with DoC at a concentration of 0.2 μg/ml (i.e., 0.02%) 6 days prior to infection, and it remained in their drinking water throughout the experiment (Fig. 7A). All three groups were then challenged intranasally (i.n.) with S. pneumoniae EF3030 (∼1 × 105 CFU), and 24 h postinfection, mice in groups 1 and 2 were treated twice a day i.n. with 10 μl of a phosphate-buffered saline (PBS) solution or DoC (2 mg/ml), respectively. Mice were euthanized 10 days after the prophylactic regimen was initiated (i.e., oral administration) in drinking water or 4 days after placebo or DoC topical nasopharyngeal treatment began (Fig. 7A). No apparent toxicity was observed (i.e., signs of inflammation, bleeding, and/or nasal discharge) in control mice or in those treated with DoC.
FIG 7.
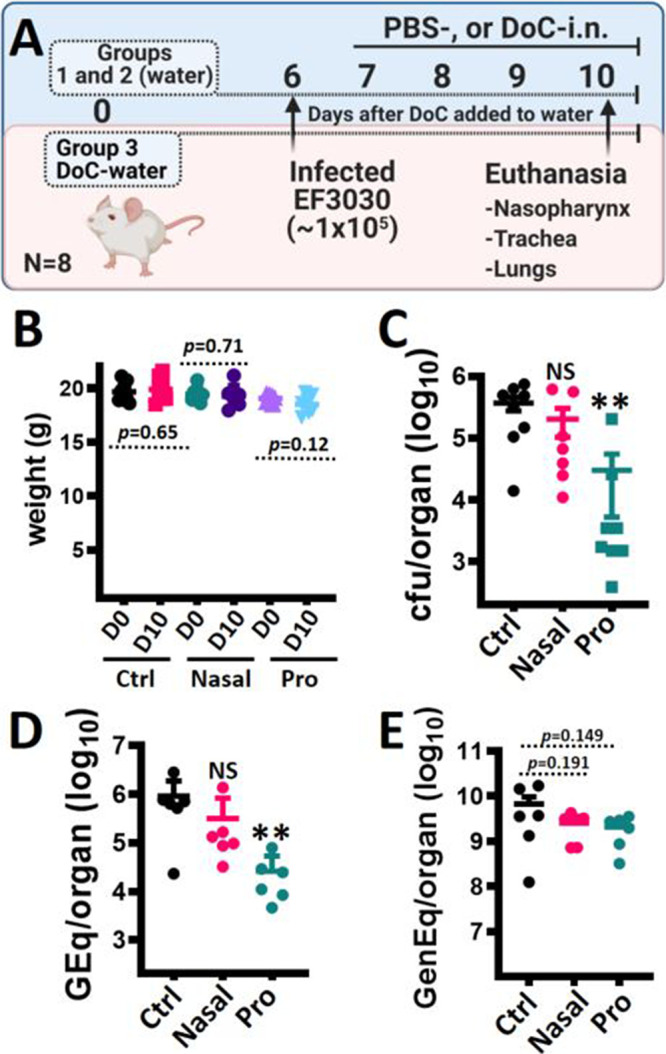
Prophylactic treatment with deoxycholic acid inhibits pneumococcal colonization in a mouse model of colonization. (A) Experimental design. Three groups of mice (n = 8 each) were utilized; groups 1 and 2 drank regular water throughout, whereas group 3 was prophylactically (pro) treated by adding DoC (0.2 μg/ml) to their drinking water at day 0. At day 6, all three groups were infected with S. pneumoniae EF3030. Then, 24 h postinoculation, mice in groups 1 and 2 were treated via intranasal (nasal) inoculation with PBS (Ctrl) or DoC (10 μg each nostril) two times a day for 4 days. All mice were sacrificed at day 10, and the nasopharynx, trachea, lungs, and blood were collected. (B) Mice in all three groups were weighed at days 0 and 10. (C) Nasopharyngeal specimens were homogenized, diluted, and plated onto BAP with gentamicin (25 μg/ml) to obtain bacterial density (CFU/ml). (D and E) DNA was extracted from nasopharyngeal homogenates and used in species-specific lytA-based qPCRs (D) or reactions targeting the mouse β-actin gene (E). In panels C and D, NS indicates P > 0.249, and ** indicates P < 0.003 compared with control mice.
Because of the oral and topical administration of DoC, we monitored the weight of mice daily, and no statistically significant difference in weight was observed between day zero and at the end of the experiment in groups 1, 2, and 3 (Fig. 7B) or in the weight of mice in the control group versus the weight of mice in the prophylactic group at the end of the experiment (P = 0.4463). The colonization density of S. pneumoniae was determined by dilution and plating of nasopharyngeal homogenates (Fig. 7C). The median density of S. pneumoniae in the control group was 4.05 × 105 CFU/ml (25th percentile, 2.50 × 105; 75th percentile, 5.60 × 105), whereas mice in the topical DoC-nostril group had a median density of 6.67 × 104 CFU/ml (25th percentile, 1.15 × 104; 75th percentile, 5.94 × 105). Although slightly reduced compared with the control group, this colonization density was not statistically different. However, mice in the prophylactic DoC administration group had a median S. pneumoniae density of 2.61 × 103 CFU/ml (25th percentile, 1.5 × 103; 75th percentile, 2.03 × 104), and therefore, a significant reduction of nasopharyngeal colonization density (e.g., 99.36% reduction) was achieved compared to the control group. We further extracted DNA from those nasopharyngeal homogenates, and the purified DNA was utilized as the template in S. pneumoniae-specific quantitative PCRs (qPCRs). qPCRs confirmed a statistically significant decreased density in the oral administration DoC-water group (median, 1.77 × 104 genome equivalents/ml) compared with the control group (median, 6.70 × 105 genome equivalents/ml) or with the topical DoC-nostril group (median, 1.15 × 105 genome equivalents/ml) (Fig. 7D). Additional qPCRs targeting the mouse β-actin gene confirmed that nasopharyngeal specimens from all three mouse groups contained similar loads of eukaryotic cells (Fig. 7E).
DISCUSSION
We demonstrated that the prophylactic treatment through the oral route with DoC protected mice from nasopharyngeal colonization with S. pneumoniae strain EF3030. We also described in the current study a rapid antimicrobial effect of DoC against S. pneumoniae strains, including reference strains, recent invasive isolates, and multidrug-resistant strains. DoC-susceptible strains included all PCV13 serotypes, strains bearing resistance to first-line antibiotics utilized to treat pneumococcal disease, such as beta-lactams and macrolides (54, 55), and last-resort antibiotics such as meropenem and linezolid. Killing of S. pneumoniae occurred with 1.27 mM, which is below the upper physiological limit (i.e., 2 mM) of free, unconjugated bile acids in the intestine, although the postprandial concentration of conjugated bile acids can be as high as 10 mM (56, 57). Remarkably, oral administration of DoC for 10 days by supplementing the drinking water of mice with 0.2 μg/ml (0.5 μM) inhibited nasopharyngeal colonization, reducing the pneumococcal density by ∼99%. Since adult mice (20 to 25 g) drink a minimum of 3 ml of water per day (58), this oral administration via drinking water reached a dosage of ∼0.6 mg/day (∼24 mg/kg/day).
DoC is a secondary bile acid synthesized by the intestinal microbiota from cholic acid and then rapidly absorbed in the intestine (33, 35); therefore, it is likely that the concentration of DoC in blood rapidly increased and remained at similar levels throughout the prophylactic treatment. Whether the level of DoC in circulation directly caused the reduction of the colonization density by means of the DoC-antimicrobial activity or by means of its immunomodulatory activities is under investigation in our laboratories. Bile produced by mammals has bacteriostatic activity keeping the sterility of the biliary tree, so an imbalance in the synthesis of bile acids, among other negative effects, is associated with the overgrowth of bacteria in the small intestine and with inflammation (59). For example, when the intestinal concentration of DoC increases, the synthesis and secretion of mucus increases as well, inducing the synthesis of immunoregulatory cytokines and the release of human β-defensins (59–61).
DoC is the most abundant secondary bile acid in serum of both mice and humans, with concentrations in healthy subjects ranging from 100 nM to 1 μM (62, 63). Individuals with deficiencies in bile salts have problems emulsifying fat, leading to intestinal disorders that have been treated by manipulating intestinal levels of bile acids (60). Cholic acid is used to treat patients with genetic deficiencies in the synthesis of bile acids due to single enzyme deficiencies; the typical dose is 10 to 15 mg/kg once daily, whereas DoC has been utilized in humans at a concentration of 15 mg/kg/day (36, 64). Thus, the prophylactic dosage of oral DoC that inhibited S. pneumoniae colonization in mice (24 mg/kg/day) was similar to that utilized to treat metabolic diseases in humans. More recent studies demonstrated that DoC (50 to 150 μM) and ursodeoxycholic acid (UDA) regulate colonic wound healing using a mouse model of colonic epithelial restitution in vivo by administering bile acids at a concentration of 30 mg/kg/day via rectal gavage (65). When administered to mice at a similar concentration, DoC and UDA prevented C. jejuni-induced colitis and C. difficile infection, respectively (31, 40). Whereas DoC did not affect viability of C. jejuni strains but enhanced an immune response against the pathogen, UDA (3.8 mM) directly decreased the viability of C. difficile and directly inhibited sporulation.
S. pneumoniae strains are “dissolved” in rabbit bile (66), and this is the basis of a phenotypic assay (i.e., bile solubility test) utilized to differentiate S. pneumoniae strains from other alpha-hemolytic streptococci (67, 68). At a more physiological concentration, such as that utilized in the current study (1.27 mM), DoC specifically killed S. pneumoniae strains but had little to no activity against other streptococci. Although the bile solubility test measures turbidity by a subjective visual method rather than by viability, bile solubility of streptococci using a semiquantitative assay correlated with our studies of bacterial viability after a challenge with DoC (69). Similar to our MIC studies, the semiquantitative assay identified S. pneumoniae strains as having the highest solubility in bile followed by strains with intermediate solubility, such as S. pseudopneumoniae, but all other streptococci were not soluble in bile (69).
Reports describing S. pneumoniae strains that were not soluble in DoC using the subjective visual readout are available; however, neither the semiquantitative assay nor our viability screening identified strains reduced in or lacking susceptibility to DoC (70). The possibility remains that some pneumococcal strains isolated from pneumococcal disease cases, or those colonizing healthy individuals, are naturally resistant to DoC. Spontaneous resistance to DoC when assessed using strains TIGR4 and EF3030 was not achieved even at bacterial populations of >1012 CFU/ml. The same strains, when challenged with trimethoprim, generated spontaneously resistant bacteria at a frequency of ≥1.39 × 10−9. A spontaneous resistance frequency to trimethoprim (44), similar to that found in the current study, or spontaneous mutation to optochin (71), has been reported for other S. pneumoniae strains.
Given the very short treatment with DoC (∼10 min) to reach a MIC in vitro, the current study assessed whether topical administration of DoC in the upper airways will result in eradication of colonization. However, mice infected with strain EF3030 and treated with DoC in the nares showed only a slight, but nonsignificant, reduction of the pneumococcal density. There are a number of reasons to explain the failure to eradicate colonization via the topical route. For example, the small nasal vestibule of mice may have resulted in failure of the DoC to reach all the nasopharyngeal tissue, and/or DoC may have been absorbed before reaching pneumococcal cells. Perhaps a lower pneumococcal carriage density, longer exposure to DoC in the upper airways, or a higher volume of DoC administered into the nostrils would have resulted in a further decrease in bacterial density. Microinvasion of pneumococcus into the nasopharyngeal epithelium observed in this study, and elsewhere (72), could have also been a factor for the failure of the topical route.
Regardless the route of administration, i.e., topical or in the drinking water, at the concentration utilized in the current study, animals treated with DoC did not show signs of toxicity. Certainly, the MIC90 (0.5 mg/ml; 1.27 mM) but not the dose used in the prophylactic treatment (0.2 μg/ml; 0.5 μM) induced release of lactate dehydrogenase (LDH), indicating toxicity of in vitro cultured human cells, including Detroit 562, A549, and Calu-3 cells (not shown). Other investigators have treated human T84 cells with 150 μM DoC with neither release of LDH nor disturbance of the transepithelial resistance (61). In our experiments, mice drinking DoC did not present a statistically significant body weight loss (Fig. 7), and that correlated with the lack of signs of toxicity. However, studies conducted with obese mice injected with DoC showed a significant reduction of body weight (73), and in normal C57BL/6J mice whose diet was supplemented with bile acids, this treatment protected mice from diet-induced increased body mass (74).
Earlier biochemical studies suggested that an autolysin(s) was responsible for the lysis of pneumococci in DoC (75). If this is true, such an autolysin should be other than the major LytA or LytC autolysins, since our experiments using two different autolysis-defective mutants demonstrated similar DoC susceptibility of the R6 WT strain, R6ΔlytAΔlytC, and R6ΔspxBΔlctO. This mechanism, however, appears to be exquisitely specific for S. pneumoniae strains, since when challenging other streptococci with the pneumococcus DoC MIC90, the majority of those strains were not susceptible. Studies are under way in our laboratories to identify such an enzyme and/or an additional mechanism(s).
In summary, we demonstrated in vitro antimicrobial activity of DoC against several pneumococcal strains, including multidrug-resistant strains, antimicrobial activity to reduce colonization of human nasopharyngeal cells, and in vivo activity that inhibited colonization in a mouse model of pneumococcal nasopharyngeal colonization. Because S. pneumoniae strains colonize billions of individuals, killing at least one million every year worldwide, the data within this study bear potential for future development of prophylactic interventions aimed to reduce pneumococcal colonization.
MATERIALS AND METHODS
Bacterial strains, culture media, and reagents.
The Streptococcus species are listed in Table 1, and the S. pneumoniae reference strains and isogenic mutant derivatives are listed in Table 2. All other S. pneumoniae strains are listed in Table S1. The strains were cultured on blood agar plates containing 5% sheep red blood cells (BAP) from frozen stocks made in medium containing skim milk, tryptone, glucose, and glycerin (STGG) (76). Animal experiments were cultured on BAP with gentamicin (25 μg/ml). The strains were inoculated in Todd-Hewitt broth containing 0.5% (wt/vol) yeast extract (THY) or in cation-adjusted Mueller-Hinton broth (CAMHB) with 3% lysed horse blood (LHB; Remel). Paraformaldehyde (PFA), gentamicin, tetracycline, trimethoprim, and sodium deoxycholate were sourced from Sigma.
TABLE 2.
Pneumococcal strains used in this study
| Strain | Descriptiona | Reference or source |
|---|---|---|
| TIGR4 | Invasive clinical isolate, capsular serotype 4, sensitive to antibiotics | 85 |
| D39 | Avery strain, clinical isolate, capsular serotype 2; sensitive to antibiotics | 87 |
| R6 | D39-derivative unencapsulated laboratory strain, sensitive to antibiotics | 87 |
| R6ΔspxBΔlctO | R6-derivative, hydrogen peroxide and autolysis deficient strain. | 88 |
| R6ΔlytAΔlytC | R6-derivative, autolysis deficient strain | 89 |
| EF3030 | Clinical isolate, capsular serotype 19F, sensitive to antibiotics | 50, 90 |
| S. pneumoniae ATCC 49619 | Invasive reference strain, recommended by the CLSIb for antimicrobial-sensitive test serotype 19F | American Type Culture Collection, 43 |
| GA44288 | Clinical isolate, capsular serotype 19A; ERY, TET, AMX, CXM, CLI, SXT, MEM, PEN, CRO, CTX | 91 |
| GA47281 | Clinical isolate, capsular serotype 19F; ERY, TET, AMX, CXM, CLI, SXT, MEM, PEN, CRO, CTX | 91 |
| GA16833 | Clinical isolate, capsular serotype 19F; ERY, TET, CXM | 91 |
| GA17227 | Clinical isolate, capsular serotype 23F; ERY, TET | 91 |
Resistance to amoxicillin (AMX), cefuroxime (CXM), ceftriaxone (CRO), cefotaxime (CTX), clindamycin (CLI), erythromycin (ERY), meropenem (MEM), penicillin (PEN), tetracycline (TET), trimethoprim-sulfamethoxazole (SXT).
CLSI, Clinical and Laboratory Standards Institute.
Preparation of inoculum for experiments.
Inoculum was prepared essentially as previously described (77, 78). Briefly, an overnight BAP culture of the strain was used to prepare a bacterial suspension in sterile phosphate-buffered saline (PBS; pH = 7.4), and the fresh bacterial suspension was inoculated to a final optical density at 600 nm (OD600) of ∼0.1. This suspension contained ∼5.15 × 108 CFU/ml. Aliquots of these suspensions were routinely diluted and plated to confirm bacterial counts (CFU/ml). To inoculate mice, S. pneumoniae strain EF3030 was inoculated in THY broth and grown until it reached an OD600 of ∼0.2 (i.e., early log phase), and then sterile glycerol was added to a final concentration of 10%, and aliquots were frozen at ∼80°C. An aliquot was removed from each batch to determine the density of the preparations.
Quantitative studies of the antimicrobial activity of DoC.
Studies of antimicrobial activity were performed using THY or CAMHB containing 3% LHB. Experiments using THY were performed as follows: a bacterial suspension was inoculated in 24-well polystyrene plates (Corning) at a final density of ∼ 5.15 × 108 CFU/ml and left untreated (control) or treated with DoC at various dosages and incubated for 2 h at 37°C in a 5% CO2 atmosphere. To remove bacteria that could have potentially attached to the substratum, the microplate was sonicated for 15 s in a Bransonic ultrasonic water bath (Branson, Danbury, CT) and then cultures were serially diluted and plated onto BAP.
To obtain the MIC recommended by the CLSI, we utilized the broth microdilution method (43). DoC was serially diluted in CAMHB containing 3% LHB in 96-well microtiter plates, and pneumococci, which had been adjusted to a turbidity corresponding to the 0.5 McFarland standard (∼1 × 108 CFU/ml), were inoculated and incubated for 20 h at 37°C. Untreated cultures and noninoculated medium were included as controls. In addition to reading the microplates as recommended by the CLSI, the untreated growth control and wells with the obtained MIC were serially diluted and plated as described before. As a control of the microdilution procedure, the MIC for tetracycline was assessed in parallel using reference strains S. pneumoniae ATCC 49619 and GA16833, which were sensitive (<1 μg/ml) and resistant (8 μg/ml), respectively.
Model of pneumococcal colonization on human pharyngeal cells.
This adhesion model on immobilized pharyngeal cells was developed by Marks et al. (79) and thereafter utilized in pathogenesis and biofilm research by different laboratories (79–81). Human pharyngeal Detroit 562 cells (ATCC CCL-198) were cultured in Dulbecco modified Eagle medium (DMEM; Gibco) supplemented with 10% non-heat-inactivated fetal bovine serum (FBS) (Atlanta Biologicals), 1% nonessential amino acids (Sigma), 1% glutamine (Sigma), penicillin (100 U/ml), and streptomycin (100 μg/ml), and the pH was buffered with HEPES (10 mM; Gibco). Cells were incubated at 37°C in a 5% CO2 humidified atmosphere until confluence for ∼7 to 10 days on an 8-well glass slide (Lab-Tek) or on CellBIND surface 24-well polystyrene plates (Corning) and then immobilized by fixation with 2% PFA for 15 min at room temperature. After extensive washes with sterile PBS, immobilized human pharyngeal cells were supplemented with cell culture medium without antibiotics and infected with an inoculum of the tested strain prepared as mentioned earlier. Infected human pharyngeal cells were incubated for 4 h at 37°C and with 5% CO2.
At the end of the incubation, planktonic pneumococci were removed, attached bacteria were gently washed two times with sterile PBS, and fresh cell culture medium with no antibiotics was added. Pneumococci attached to pharyngeal cells were challenged with DoC at different dosages and incubated for 2 h or treated with 0.5 mg/ml DoC and incubated for the indicated time at 37°C in a 5% CO2 atmosphere. To obtain the density of pneumococci in the 24-well plate model, pneumococci and cells were washed twice with PBS and then sonicated for 15 s in a Bransonic ultrasonic water bath (Branson, Danbury CT) followed by extensive pipetting to remove attached bacteria. The preparations were diluted and plated onto blood agar plates to obtain bacterial counts (CFU/ml).
To stain pneumococci adhered to cells on the 8-well glass slide, bacteria were fixed with 2% PFA as described before, and after three washes with PBS, the preparations were blocked with 2% bovine serum albumin (BSA) for 1 h at room temperature. These preparations were then incubated for 1 h with serotype-specific polyclonal antibodies (Statens Serum Institute, Denmark) (∼ 40 μg/ml) that had been previously labeled with Alexa-488 (anti-serotype 4-Alexa-488, to stain TIGR4) or Alexa-555 (anti-serogroup 19-Alexa-555, to stain GA47281) (Molecular Probes). Stained preparations were finally washed two times with PBS. TIGR4 experiments were additionally stained with wheat germ agglutinin conjugated to Alexa-555 (WGA; 5 μg/ml) and then mounted with ProLong Diamond antifade mounting medium containing DAPI (Molecular Probes), whereas GA47281 preparations were stained with TO-PRO-3 (1 μM), a carbocyanine monomer nucleic acid stain (Molecular Probes), for 15 min. Confocal images were obtained using a Nikon AX R confocal microscope and analyzed with ImageJ version 1.49k (National Institutes of Health, USA).
Investigating the spontaneous mutation frequency.
To determine the frequency of spontaneous mutation, BAP with 5% sheep red blood cells were prepared to contain either trimethoprim (1 μg/ml) or DoC (0.5 mg/ml). Fresh suspensions of S. pneumoniae strain EF3030, or TIGR4, made in PBS were prepared at a final density of ∼108, ∼109, ∼1010, ∼1011, and ∼1012 CFU/ml and inoculated on plain BAP or BAP containing trimethoprim or DoC. Bacterial suspensions were diluted and plated onto plain BAP to confirm the density of pneumococci. Inoculated plates were incubated at 37°C in a 5% CO2 atmosphere for ∼20 h. The spontaneous mutation frequency was then calculated by dividing the spontaneously resistant pneumococci, i.e., grown on BAP with trimethoprim or DoC, by the bacterial population.
Mouse model of pneumococcal nasopharyngeal carriage.
Three groups (n = 8 each) of inbred 6- to 7-week-old C57BL/6 mice (Charles River Laboratories) were utilized to assess in vivo antimicrobial activity of DoC. Two groups of mice drank regular water throughout. The drinking water of the third group of mice was supplemented with DoC to a final concentration of 0.2 μg/ml (i.e., 0.02%) starting at day 0 of the experiment, and DoC-containing water was provided ad libitum for the reminder of the experiment (10 days). Six days after DoC was added to the drinking water of mice in group 3, mice in all three groups were anesthetized with 2.5% isoflurane (vol/vol) over oxygen (2 liter/min) administered in an RC2 calibrated vaporizer (VetEquip Inc.) and then infected with 1 × 105 CFU of S. pneumoniae EF3030. Then, 24 h post-nasal inoculation of EF3030, mice in groups 1 and 2 were treated by nasal instillation with PBS or DoC (10 μg each nostril), respectively, two times a day for 4 days. Signs of inflammation, bleeding, and/or nasal discharge were monitored twice daily by visual inspection of the nose, body, cage, and bedding.
Mice were then sacrificed, and the nasopharynx, trachea, lungs, and blood were aseptically collected. Tissue homogenates were diluted in PBS and plated onto BAP with gentamicin. Aliquots of these homogenates were supplemented to a final concentration of 10% glycerol and kept at −80°C. The Institutional Animal Care and Use Committee (IACUC) at the University of Mississippi Medical Center approved the protocol used in this study (no. 1584); they oversaw the welfare, well-being, and proper care of all mice utilized in this study. All mouse experiments followed the guidelines summarized by the National Science Foundation Animal Welfare Act (AWA).
DNA extraction from nasopharyngeal homogenates and quantitative PCRs (qPCRs).
DNA was extracted from mouse nasopharyngeal homogenates using the Qiagen QIAmp minikit. Briefly, an aliquot (50 μl) of nasopharyngeal specimen was added to 100 μl of TE buffer containing 0.04 g/ml lysozyme and 75 U/ml of mutanolysin. Samples were then incubated for 1 h in a 37°C water bath. Following incubation, DNA was extracted from the samples following the recommended protocol from the manufacturer, eluted in 100 μl of buffer AE, and kept at −80°C until used. Following extraction, lytA-based qPCRs were performed with primers and probe sequences published by the CDC (82), the real-time PCR reagent QuantaBio PerfeCTa FastMix, and 2.5 μl of DNA template. Reactions were run in duplicate using a CFX96 real-time PCR detection system (Bio-Rad) at the following conditions: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 2 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Standard curves were generated under the same conditions as detailed in our previous studies (83, 84). Considering the genome size of reference strain TIGR4, 2.16 Mb (85), the approximate genome equivalent for each DNA standard was 4.29 × 105, 4.29 × 104, 4.29 × 103, 4.29 × 102, 4.29 × 101, 2.14 × 101, and 2.14 genome equivalents. qPCRs targeting the mouse β-actin gene were performed using primers JVS169 (tcctagcaccatgaagatcaagat) and JVS170 (ggttttgtcaaagaaagggtgta) and real-time reagent PerfeCTa SYBR green FastMix. Cycling conditions and the preparation of the standard curve were done essentially as described above, except that considering the genome size of the C57BL/6 mouse (86), the DNA standards were 1.26 × 108, 1.26 × 107, 1.26 × 106, and 1.26 × 105 genome equivalents. These DNA standards were prepared by amplifying the β-actin gene by conventional PCR with the primers listed above, and then the PCR product was purified using the QIAquick PCR purification kit, quantified, and serially diluted. The reaction efficiency of qPCRs (lytA and β-actin) was within the acceptable range of 90 to 110%.
Quantification of extracellular DNA (eDNA).
S. pneumoniae strains were inoculated into 24-well plates as detailed earlier and incubated for 4 h. The culture supernatants were then harvested by centrifugation for 15 min at 14,000 × g in a refrigerated centrifuge (Eppendorf, Hauppauge, NY) and filter-sterilized using a syringe filter (0.4 μm). DNA was purified from 200-μl aliquots of supernatant, as mentioned above, and used as the template in lytA-based qPCRs. For eDNA quantification purposes, standards containing 1 × 103, 1 × 102, 1 × 101, 1 × 10°, 1 × 10−1, 5 × 10−2, and 1 × 10−3 pg of chromosomal DNA purified from strain TIGR4 were run in parallel to generate a standard curve. The standard curve, and regression equation obtained, was then used to calculate final pg/ml using the CFX software (Bio-Rad, Hercules, CA).
Statistical analysis.
Statistical analysis was performed by the nonparametric two-tailed Student’s t test, the Mann-Whitney U test (comparing two groups), or one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison post hoc test, using the software GraphPad Prism version 9.0.0 (121).
ACKNOWLEDGMENTS
This study was in part supported by grants from the National Institutes of Health (NIH; 1R21AI144571-01 and 1R21AI151571-01A1 to J.E.V.). B.A. was supported by a Fulbright scholarship awarded by the U.S. Department of State. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH or the U.S. Department of State.
We thank Yih-Ling Tzeng and David Stephens from Emory University School of Medicine and Lesley McGee from the Centers for Disease Control and Prevention (CDC) for providing S. pneumoniae strains. We also thank Miriam Moscoso from “Centro de Investigaciones Biologicas” in Madrid, Spain, for providing the autolysin knockout mutant. Special thanks to Gene L. Bidwell (Cell and Molecular Biology) and Joshua R. Jefferson (Pharmacology and Toxicology) at UMMC for providing assistance with confocal microscopy and histology studies, respectively. We also thank Landon Murin from the Murrah-UMMC base pair program for his assistance with some laboratory procedures.
Footnotes
Supplemental material is available online only.
Contributor Information
Jorge E. Vidal, Email: jvidal@umc.edu.
Liise-anne Pirofski, Albert Einstein College of Medicine.
REFERENCES
- 1.Shak JR, Vidal JE, Klugman KP. 2013. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol 21:129–135. 10.1016/j.tim.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simell B, Auranen K, Kayhty H, Goldblatt D, Dagan R, O’Brien KL, Pneumococcal Carriage Group . 2012. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines 11:841–855. 10.1586/erv.12.53. [DOI] [PubMed] [Google Scholar]
- 3.Arguedas A, Trzciński K, O’Brien KL, Ferreira DM, Wyllie AL, Weinberger D, Danon L, Pelton SI, Azzari C, Hammitt LL, Sá-Leão R, Brandileone M-CC, Saha S, Suaya J, Isturiz R, Jodar L, Gessner BD. 2020. Upper respiratory tract colonization with Streptococcus pneumoniae in adults. Expert Rev Vaccines 19:353–366. 10.1080/14760584.2020.1750378. [DOI] [PubMed] [Google Scholar]
- 4.Dunne EM, Smith-Vaughan HC, Robins-Browne RM, Mulholland EK, Satzke C. 2013. Nasopharyngeal microbial interactions in the era of pneumococcal conjugate vaccination. Vaccine 31:2333–2342. 10.1016/j.vaccine.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 5.Krone CL, Wyllie AL, van Beek J, Rots NY, Oja AE, Chu MLJN, Bruin JP, Bogaert D, Sanders EAM, Trzciński K. 2015. Carriage of Streptococcus pneumoniae in aged adults with influenza-like-illness. PLoS One 10:e0119875. 10.1371/journal.pone.0119875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwambana BA, Barer MR, Bottomley C, Adegbola RA, Antonio M. 2011. Early acquisition and high nasopharyngeal co-colonisation by Streptococcus pneumoniae and three respiratory pathogens amongst Gambian new-borns and infants. BMC Infect Dis 11:175. 10.1186/1471-2334-11-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson KN, Grijalva CG, Chochua S, Hawkins PA, Gil AI, Lanata CF, Griffin MR, Edwards KM, Klugman KP, Vidal JE. 2018. Dynamics of colonization of Streptococcus pneumoniae strains in healthy Peruvian children. Open Forum Infect Dis 5:ofy039. 10.1093/ofid/ofy039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutcliffe CG, Grant LR, Cloessner E, Klugman KP, Vidal JE, Reid R, Colelay J, Weatherholtz RC, Chochua S, Jacobs MR, Santosham M, O’Brien KL, Hammitt LL. 2019. Association of laboratory methods, colonization density, and age with detection of Streptococcus pneumoniae in the nasopharynx. Am J Epidemiol 188:2110–2119. 10.1093/aje/kwz191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyllie AL, Rümke LW, Arp K, Bosch AATM, Bruin JP, Rots NY, Wijmenga-Monsuur AJ, Sanders EAM, Trzciński K. 2016. Molecular surveillance on Streptococcus pneumoniae carriage in non-elderly adults; little evidence for pneumococcal circulation independent from the reservoir in children. Sci Rep 6:34888. 10.1038/srep34888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Deursen AM, van den Bergh MR, Sanders EA, Carriage Pilot Study Group . 2016. Carriage of Streptococcus pneumoniae in asymptomatic, community-dwelling elderly in the Netherlands. Vaccine 34:4–6. 10.1016/j.vaccine.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Vu HTT, Yoshida LM, Suzuki M, Nguyen HAT, Nguyen CDL, Nguyen ATT, Oishi K, Yamamoto T, Watanabe K, Vu TD, Schmidt W-P, Phan HTL, Morimoto K, Le TH, Yanai H, Kilgore PE, Dang AD, Ariyoshi K. 2011. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J 30:11–18. 10.1097/INF.0b013e3181f111a2. [DOI] [PubMed] [Google Scholar]
- 12.Chochua S, D’Acremont V, Hanke C, Alfa D, Shak J, Kilowoko M, Kyungu E, Kaiser L, Genton B, Klugman KP, Vidal JE. 2016. Increased nasopharyngeal density and concurrent carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are associated with pneumonia in febrile children. PLoS One 11:e0167725. 10.1371/journal.pone.0167725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carr OJJ, Vilivong K, Bounvilay L, Dunne EM, Lai JYR, Chan J, Vongsakid M, Chanthongthip A, Siladeth C, Ortika B, Nguyen C, Mayxay M, Newton PN, Mulholland K, Do LAH, Dubot-Peres A, Satzke C, Dance DAB, Russell FM. 2021. Nasopharyngeal pneumococcal colonization density is associated with severe pneumonia in young children in the Lao PDR. J Infect Dis 10.1093/infdis/jiab239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Short KR, Reading PC, Wang N, Diavatopoulos DA, Wijburg OL. 2012. Increased nasopharyngeal bacterial titers and local inflammation facilitate transmission of Streptococcus pneumoniae. mBio 3:e00255-12. 10.1128/mBio.00255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyazika TK, Law A, Swarthout TD, Sibale L, Ter Braake D, French N, Heyderman RS, Everett D, Kadioglu A, Jambo KC, Neill DR. 2020. Influenza-like illness is associated with high pneumococcal carriage density in Malawian children. J Infect 81:549–556. 10.1016/j.jinf.2020.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Lastours V, Malosh R, Ramadugu K, Srinivasan U, Dawid S, Ohmit S, Foxman B. 2016. Co-colonization by Streptococcus pneumoniae and Staphylococcus aureus in the throat during acute respiratory illnesses. Epidemiol Infect 144:3507–3519. 10.1017/S0950268816001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grijalva CG, Griffin MR, Edwards KM, Williams JV, Gil AI, Verastegui H, Hartinger SM, Vidal JE, Klugman KP, Lanata CF. 2014. The role of influenza and parainfluenza infections in nasopharyngeal pneumococcal acquisition among young children. Clin Infect Dis 58:1369–1376. 10.1093/cid/ciu148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard LM, Zhu Y, Griffin MR, Edwards KM, Williams JV, Gil AI, Vidal JE, Klugman KP, Lanata CF, Grijalva CG. 2019. Nasopharyngeal pneumococcal density during asymptomatic respiratory virus infection and risk for subsequent acute respiratory illness. Emerg Infect Dis 25:2040–2047. 10.3201/eid2511.190157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai CC, Wang CY, Hsueh PR. 2020. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-CoV-2 agents? J Microbiol Immunol Infect 53:505–512. 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aykac K, Ozsurekci Y, Cura Yayla BC, Evren K, Lacinel Gurlevik S, Oygar PD, Yucel M, Karakoc AE, Alp A, Cengiz AB, Ceyhan M. 2021. Pneumococcal carriage in children with COVID-19. Hum Vaccin Immunother 17:1628–1634. 10.1080/21645515.2020.1849516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dananche C, Paranhos-Baccala G, Messaoudi M, Sylla M, Awasthi S, Bavdekar A, Sanghavi S, Diallo S, Pape JW, Rouzier V, Chou M, Eap T, Rakoto-Andrianarivelo M, Maeder M, Wang J, Ren L, Dash-Yandag B, Nymadawa P, Guillen R, Russomando G, Endtz H, Komurian-Pradel F, Vanhems P, Sanchez Picot V. 2020. Serotypes of Streptococcus pneumoniae in children aged <5 years hospitalized with or without pneumonia in developing and emerging countries: a descriptive, multicenter study. Clin Infect Dis 70:875–883. 10.1093/cid/ciz277. [DOI] [PubMed] [Google Scholar]
- 22.Weiser JN, Ferreira DM, Paton JC. 2018. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol 16:355–367. 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis SM, Deloria-Knoll M, Kassa HT, O’Brien KL. 2013. Impact of pneumococcal conjugate vaccines on nasopharyngeal carriage and invasive disease among unvaccinated people: review of evidence on indirect effects. Vaccine 32:133–145. 10.1016/j.vaccine.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Klugman KP. 2001. Efficacy of pneumococcal conjugate vaccines and their effect on carriage and antimicrobial resistance. Lancet Infect Dis 1:85–91. 10.1016/S1473-3099(01)00063-9. [DOI] [PubMed] [Google Scholar]
- 25.O’Brien KL, Dagan R. 2003. The potential indirect effect of conjugate pneumococcal vaccines. Vaccine 21:1815–1825. 10.1016/S0264-410X(02)00807-1. [DOI] [PubMed] [Google Scholar]
- 26.Simonsen L, Taylor RJ, Schuck-Paim C, Lustig R, Haber M, Klugman KP. 2014. Effect of 13-valent pneumococcal conjugate vaccine on admissions to hospital 2 years after its introduction in the USA: a time series analysis. Lancet Respir Med 2:387–394. 10.1016/S2213-2600(14)70032-3. [DOI] [PubMed] [Google Scholar]
- 27.Feikin DR, Kagucia EW, Loo JD, Link-Gelles R, Puhan MA, Cherian T, Levine OS, Whitney CG, O’Brien KL, Moore MR, the Serotype Replacement Study Group . 2013. Serotype-specific changes in invasive pneumococcal disease after pneumococcal conjugate vaccine introduction: a pooled analysis of multiple surveillance sites. PLoS Med 10:e1001517. 10.1371/journal.pmed.1001517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singleton RJ, Hennessy TW, Bulkow LR, Hammitt LL, Zulz T, Hurlburt DA, Butler JC, Rudolph K, Parkinson A. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 297:1784–1792. 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- 29.Weinberger DM, Malley R, Lipsitch M. 2011. Serotype replacement in disease after pneumococcal vaccination. Lancet 378:1962–1973. 10.1016/S0140-6736(10)62225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler ES, Shrihari S, Hykes BL Jr, Handley SA, Andhey PS, Huang YS, Swain A, Droit L, Chebrolu KK, Mack M, Vanlandingham DL, Thackray LB, Cella M, Colonna M, Artyomov MN, Stappenbeck TS, Diamond MS. 2020. The intestinal microbiome restricts alphavirus infection and dissemination through a bile acid-type I IFN signaling axis. Cell 182:901–918.e18. 10.1016/j.cell.2020.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun X, Winglee K, Gharaibeh RZ, Gauthier J, He Z, Tripathi P, Avram D, Bruner S, Fodor A, Jobin C. 2018. Microbiota-derived metabolic factors reduce campylobacteriosis in mice. Gastroenterology 154:1751–1763.e2. 10.1053/j.gastro.2018.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Latorre JD, Bansal M, Abraha M, Al-Rubaye B, Tellez-Isaias G, Hargis B, Sun X. 2019. Microbial metabolite deoxycholic acid controls Clostridium perfringens-induced chicken necrotic enteritis through attenuating inflammatory cyclooxygenase signaling. Sci Rep 9:14541. 10.1038/s41598-019-51104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyer JL. 2013. Bile formation and secretion. Compr Physiol 3:1035–1078. 10.1002/cphy.c120027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahlstrom A, Sayin SI, Marschall HU, Backhed F. 2016. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24:41–50. 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Foley MH, O'Flaherty S, Barrangou R, Theriot CM. 2019. Bile salt hydrolases: gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog 15:e1007581. 10.1371/journal.ppat.1007581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woollett LA, Buckley DD, Yao L, Jones PJ, Granholm NA, Tolley EA, Tso P, Heubi JE. 2004. Cholic acid supplementation enhances cholesterol absorption in humans. Gastroenterology 126:724–731. 10.1053/j.gastro.2003.11.058. [DOI] [PubMed] [Google Scholar]
- 37.Kamalpour S, Leblanc K Jr.. 2016. Injection adipolysis: mechanisms, agents, and future directions. J Clin Aesthet Dermatol 9:44–50. [PMC free article] [PubMed] [Google Scholar]
- 38.Amore R, Amuso D, Leonardi V, Leva F, Sibaud AC, Guida A, Costa E, Terranova F, Amodeo V, Gkritzalas K. 2018. Evaluation of safe and effectiveness of an injectable solution acid deoxycholic based for reduction of localized adiposities. Plast Reconstr Surg Glob Open 6:e1794. 10.1097/GOX.0000000000001794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winston JA, Rivera AJ, Cai J, Thanissery R, Montgomery SA, Patterson AD, Theriot CM. 2020. Ursodeoxycholic acid (UDCA) mitigates the host inflammatory response during Clostridioides difficile Infection by altering gut bile acids. Infect Immun 88:e00045-20. 10.1128/IAI.00045-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonald JAK, Mullish BH, Pechlivanis A, Liu Z, Brignardello J, Kao D, Holmes E, Li JV, Clarke TB, Thursz MR, Marchesi JR. 2018. Inhibiting growth of Clostridioides difficile by restoring valerate, produced by the intestinal microbiota. Gastroenterology 155:1495–1507.e15. 10.1053/j.gastro.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olivera C, Le VVH, Davenport C, Rakonjac J. 2021. In vitro synergy of 5-nitrofurans, vancomycin and sodium deoxycholate against Gram-negative pathogens. J Med Microbiol 70. 10.1099/jmm.0.001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang JD, Myers CJ, Harris SC, Kakiyama G, Lee IK, Yun BS, Matsuzaki K, Furukawa M, Min HK, Bajaj JS, Zhou H, Hylemon PB. 2019. Bile acid 7alpha-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: role of secondary bile acids. Cell Chem Biol 26:27–34.e4. 10.1016/j.chembiol.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing. 30th Informational Supplement. M100-S30. CLSI, Wayne, PA. [Google Scholar]
- 44.Henderson-Begg SK, Livermore DM, Hall LM. 2006. Effect of subinhibitory concentrations of antibiotics on mutation frequency in Streptococcus pneumoniae. J Antimicrob Chemother 57:849–854. 10.1093/jac/dkl064. [DOI] [PubMed] [Google Scholar]
- 45.Eldholm V, Johnsborg O, Haugen K, Ohnstad HS, Havarstein LS. 2009. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology (Reading) 155:2223–2234. 10.1099/mic.0.026328-0. [DOI] [PubMed] [Google Scholar]
- 46.Havarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol Microbiol 59:1297–1307. 10.1111/j.1365-2958.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- 47.Wu X, Gordon O, Jiang W, Antezana BS, Angulo-Zamudio UA, Del Rio C, Moller A, Brissac T, Tierney ARP, Warncke K, Orihuela CJ, Read TD, Vidal JE. 2019. Interaction between Streptococcus pneumoniae and Staphylococcus aureus generates (.)OH radicals that rapidly kill Staphylococcus aureus strains. J Bacteriol 201:e00474-19. 10.1128/JB.00474-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bryant JC, Dabbs RC, Oswalt KL, Brown LR, Rosch JW, Seo KS, Donaldson JR, McDaniel LS, Thornton JA. 2016. Pyruvate oxidase of Streptococcus pneumoniae contributes to pneumolysin release. BMC Microbiol 16:271. 10.1186/s12866-016-0881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Echlin H, Frank M, Rock C, Rosch JW. 2020. Role of the pyruvate metabolic network on carbohydrate metabolism and virulence in Streptococcus pneumoniae. Mol Microbiol 114:536–552. 10.1111/mmi.14557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Junges R, Maienschein-Cline M, Morrison DA, Petersen FC. 2019. Complete genome sequence of Streptococcus pneumoniae serotype 19F strain EF3030. Microbiol Resour Announc 8:e00198-19. 10.1128/MRA.00198-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott EJ II, Luke-Marshall NR, Campagnari AA, Dyer DW. 2019. Draft genome sequence of pediatric otitis media isolate Streptococcus pneumoniae strain EF3030, which forms in vitro biofilms that closely mimic in vivo biofilms. Microbiol Resour Announc 8:e01114-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puchta A, Verschoor CP, Thurn T, Bowdish DM. 2014. Characterization of inflammatory responses during intranasal colonization with Streptococcus pneumoniae. J Vis Exp 2014:e50490. 10.3791/50490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keller LE, Luo X, Thornton JA, Seo KS, Moon BY, Robinson DA, McDaniel LS. 2015. Immunization with pneumococcal surface protein K of nonencapsulated Streptococcus pneumoniae provides protection in a mouse model of colonization. Clin Vaccine Immunol 22:1146–1153. 10.1128/CVI.00456-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, Cooley LA, Dean NC, Fine MJ, Flanders SA, Griffin MR, Metersky ML, Musher DM, Restrepo MI, Whitney CG. 2019. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 200:e45–e67. 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim WS, Baudouin SV, George RC, Hill AT, Jamieson C, Le Jeune I, Macfarlane JT, Read RC, Roberts HJ, Levy ML, Wani M, Woodhead MA, Pneumonia Guidelines Committee of the BTS Standards of Care Committee . 2009. BTS guidelines for the management of community acquired pneumonia in adults: update 2009. Thorax 64(Suppl 3):1–55. 10.1136/thx.2009.121434. [DOI] [PubMed] [Google Scholar]
- 56.Northfield TC, McColl I. 1973. Postprandial concentrations of free and conjugated bile acids down the length of the normal human small intestine. Gut 14:513–518. 10.1136/gut.14.7.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vertzoni M, Archontaki H, Reppas C. 2008. Determination of intralumenal individual bile acids by HPLC with charged aerosol detection. J Lipid Res 49:2690–2695. 10.1194/jlr.D800039-JLR200. [DOI] [PubMed] [Google Scholar]
- 58.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, Mastroeni P, Scott P, Raisen C, Mottram L, Fairweather NF, Wren BW, Parkhill J, Dougan G. 2009. Antibiotic treatment of clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun 77:3661–3669. 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sipka S, Bruckner G. 2014. The immunomodulatory role of bile acids. Int Arch Allergy Immunol 165:1–8. 10.1159/000366100. [DOI] [PubMed] [Google Scholar]
- 60.Keating N, Keely SJ. 2009. Bile acids in regulation of intestinal physiology. Curr Gastroenterol Rep 11:375–382. 10.1007/s11894-009-0057-8. [DOI] [PubMed] [Google Scholar]
- 61.Lajczak NK, Saint-Criq V, O'Dwyer AM, Perino A, Adorini L, Schoonjans K, Keely SJ. 2017. Bile acids deoxycholic acid and ursodeoxycholic acid differentially regulate human beta-defensin-1 and −2 secretion by colonic epithelial cells. FASEB J 31:3848–3857. 10.1096/fj.201601365R. [DOI] [PubMed] [Google Scholar]
- 62.Choucair I, Nemet I, Li L, Cole MA, Skye SM, Kirsop JD, Fischbach MA, Gogonea V, Brown JM, Tang WHW, Hazen SL. 2020. Quantification of bile acids: a mass spectrometry platform for studying gut microbe connection to metabolic diseases. J Lipid Res 61:159–177. 10.1194/jlr.RA119000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danese E, Negrini D, Pucci M, De Nitto S, Ambrogi D, Donzelli S, Lievens PM, Salvagno GL, Lippi G. 2020. Bile acids quantification by liquid chromatography-tandem mass spectrometry: method validation, reference range, and interference study. Diagnostics (Basel) 10:462. 10.3390/diagnostics10070462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Jones PJ, Woollett LA, Buckley DD, Yao L, Granholm NA, Tolley EA, Heubi JE. 2006. Effects of chenodeoxycholic acid and deoxycholic acid on cholesterol absorption and metabolism in humans. Transl Res 148:37–45. 10.1016/j.lab.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 65.Mroz MS, Lajczak NK, Goggins BJ, Keely S, Keely SJ. 2018. The bile acids, deoxycholic acid and ursodeoxycholic acid, regulate colonic epithelial wound healing. Am J Physiol Gastrointest Liver Physiol 314:G378–G387. 10.1152/ajpgi.00435.2016. [DOI] [PubMed] [Google Scholar]
- 66.Wadsworth A. 1903. The agglutination of the Pneumococcus with certain normal and immune sera. J Med Res 10:228–242. [PMC free article] [PubMed] [Google Scholar]
- 67.Sadowy E, Hryniewicz W. 2020. Identification of Streptococcus pneumoniae and other Mitis streptococci: importance of molecular methods. Eur J Clin Microbiol Infect Dis 39:2247–2256. 10.1007/s10096-020-03991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Satzke C, Turner P, Virolainen-Julkunen A, Adrian PV, Antonio M, Hare KM, Henao-Restrepo AM, Leach AJ, Klugman KP, Porter BD, Sa-Leao R, Scott JA, Nohynek H, O’Brien KL, WHO Pneumococcal Carriage Working Group . 2013. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 32:165–179. 10.1016/j.vaccine.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 69.Slotved HC, Facklam RR, Fuursted K. 2017. Assessment of a novel bile solubility test and MALDI-TOF for the differentiation of Streptococcus pneumoniae from other mitis group streptococci. Sci Rep 7:7167. 10.1038/s41598-017-07772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mundy LS, Janoff EN, Schwebke KE, Shanholtzer CJ, Willard KE. 1998. Ambiguity in the identification of Streptococcus pneumoniae. Optochin, bile solubility, quellung, and the AccuProbe DNA probe tests. Am J Clin Pathol 109:55–61. 10.1093/ajcp/109.1.55. [DOI] [PubMed] [Google Scholar]
- 71.Cortes PR, Orio AG, Regueira M, Pinas GE, Echenique J. 2008. Characterization of in vitro-generated and clinical optochin-resistant strains of Streptococcus pneumoniae isolated from Argentina. J Clin Microbiol 46:1930–1934. 10.1128/JCM.02318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weight CM, Venturini C, Pojar S, Jochems SP, Reine J, Nikolaou E, Solorzano C, Noursadeghi M, Brown JS, Ferreira DM, Heyderman RS. 2019. Microinvasion by Streptococcus pneumoniae induces epithelial innate immunity during colonisation at the human mucosal surface. Nat Commun 10:3060. 10.1038/s41467-019-11005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park SH, Hyun MR, Kim SW. 2019. Effect of a formulation containing low-dose sodium deoxycholate on local fat reduction. Aesthetic Plast Surg 43:1657–1662. 10.1007/s00266-019-01514-2. [DOI] [PubMed] [Google Scholar]
- 74.Fromme T, Huttinger K, Maurer S, Li Y, Gantert T, Fiamoncini J, Daniel H, Westphal S, Klingenspor M. 2019. Bile acid supplementation decreases body mass gain in C57BL/6J but not 129S6/SvEvTac mice without increasing energy expenditure. Sci Rep 9:131. 10.1038/s41598-018-37464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mosser JL, Tomasz A. 1970. Choline-containing teichoic acid as a structural component of pneumococcal cell wall and its role in sensitivity to lysis by an autolytic enzyme. J Biol Chem 245:287–298. 10.1016/S0021-9258(18)63393-9. [DOI] [PubMed] [Google Scholar]
- 76.O’Brien KL, Bronsdon MA, Dagan R, Yagupsky P, Janco J, Elliott J, Whitney CG, Yang YH, Robinson LG, Schwartz B, Carlone GM. 2001. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J Clin Microbiol 39:1021–1024. 10.1128/JCM.39.3.1021-1024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vidal JE, Ludewick HP, Kunkel RM, Zahner D, Klugman KP. 2011. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun 79:4050–4060. 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu X, Jacobs NT, Bozio C, Palm P, Lattar SM, Hanke CR, Watson DM, Sakai F, Levin BR, Klugman KP, Vidal JE. 2017. Competitive dominance within biofilm consortia regulates the relative distribution of pneumococcal nasopharyngeal density. Appl Environ Microbiol 83:e00953-17. 10.1128/AEM.00953-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marks LR, Parameswaran GI, Hakansson AP. 2012. Pneumococcal interactions with epithelial cells are crucial for optimal biofilm formation and colonization in vitro and in vivo. Infect Immun 80:2744–2760. 10.1128/IAI.00488-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vidal JE, Howery KE, Ludewick HP, Nava P, Klugman KP. 2013. Quorum-sensing systems LuxS/autoinducer 2 and Com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect Immun 81:1341–1353. 10.1128/IAI.01096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wholey WY, Kochan TJ, Storck DN, Dawid S. 2016. Coordinated bacteriocin expression and competence in Streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog 12:e1005413. 10.1371/journal.ppat.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carvalho M. d G S, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R, Sampson JS. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45:2460–2466. 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakai F, Sonaty G, Watson D, Klugman KP, Vidal JE. 2017. Development and characterization of a synthetic DNA, NUversa, to be used as a standard in quantitative polymerase chain reactions for molecular pneumococcal serotyping. FEMS Microbiol Lett 364:fnx173. 10.1093/femsle/fnx173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sakai F, Chochua S, Satzke C, Dunne EM, Mulholland K, Klugman KP, Vidal JE. 2015. Single-plex quantitative assays for the detection and quantification of most pneumococcal serotypes. PLoS One 10:e0121064. 10.1371/journal.pone.0121064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 86.Sarsani VK, Raghupathy N, Fiddes IT, Armstrong J, Thibaud-Nissen F, Zinder O, Bolisetty M, Howe K, Hinerfeld D, Ruan X, Rowe L, Barter M, Ananda G, Paten B, Weinstock GM, Churchill GA, Wiles MV, Schneider VA, Srivastava A, Reinholdt LG. 2019. The genome of C57BL/6J “Eve”, the mother of the laboratory mouse genome reference strain. G3 (Bethesda) 9:1795–1805. 10.1534/g3.119.400071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McDevitt E, Khan F, Scasny A, Thompson CD, Eichenbaum Z, McDaniel LS, Vidal JE. 2020. Hydrogen peroxide production by Streptococcus pneumoniae results in alpha-hemolysis by oxidation of oxy-hemoglobin to met-hemoglobin. mSphere 5:e01117-20. 10.1128/mSphere.01117-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moscoso M, Garcia E, Lopez R. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol 188:7785–7795. 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andersson B, Dahmen J, Frejd T, Leffler H, Magnusson G, Noori G, Eden CS. 1983. Identification of an active disaccharide unit of a glycoconjugate receptor for pneumococci attaching to human pharyngeal epithelial cells. J Exp Med 158:559–570. 10.1084/jem.158.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schroeder MR, Lohsen S, Chancey ST, Stephens DS. 2019. High-level macrolide resistance due to the mega element [mef(E)/mel] in Streptococcus pneumoniae. Front Microbiol 10:868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hanke CR, Grijalva CG, Chochua S, Pletz MW, Hornberg C, Edwards KM, Griffin MR, Verastegui H, Gil AI, Lanata CF, Klugman KP, Vidal JE. 2016. Bacterial density, serotype distribution and antibiotic resistance of pneumococcal strains from the nasopharynx of Peruvian children before and after pneumococcal conjugate vaccine 7. Pediatr Infect Dis J 35:432–439. 10.1097/INF.0000000000001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download iai.00463-21-s0001.pdf, PDF file, 0.2 MB (164KB, pdf)