ABSTRACT
Bordetella pertussis is a highly contagious bacterium that is the causative agent of whooping cough (pertussis). Currently, acellular pertussis vaccines (aP, DTaP, and Tdap) are used to prevent pertussis disease. However, it is clear that the aP vaccine efficacy quickly wanes, resulting in the reemergence of pertussis. Furthermore, recent work performed by the CDC suggest that current circulating strains are genetically distinct from strains of the past. The emergence of genetically diverging strains, combined with waning aP vaccine efficacy, calls for reevaluation of current animal models of pertussis. In this study, we used the rat model of pertussis to compare two genetically divergent strains Tohama 1 and D420. We intranasally challenged 7-week-old Sprague-Dawley rats with 108 viable Tohama 1 and D420 and measured the hallmark signs/symptoms of B. pertussis infection such as neutrophilia, pulmonary inflammation, and paroxysmal cough using whole-body plethysmography. Onset of cough occurred between 2 and 4 days after B. pertussis challenge, averaging five coughs per 15 min, with peak coughing occurring at day 8 postinfection, averaging upward of 13 coughs per 15 min. However, we observed an increase of coughs in rats infected with clinical isolate D420 through 12 days postchallenge. The rats exhibited increased bronchial restriction following B. pertussis infection. Histology of the lung and flow cytometry confirm both cellular infiltration and pulmonary inflammation. D420 infection induced higher production of anti-B. pertussis IgM antibodies compared to Tohama 1 infection. The coughing rat model provides a way of characterizing disease manifestation differences between B. pertussis strains.
KEYWORDS: Bordetella pertussis, cough, DTaP, plethysmography, rat
INTRODUCTION
Whooping cough (pertussis) is a respiratory disease that is caused by the bacterium Bordetella pertussis. Pertussis is characterized by severe leukocytosis, bronchopneumonia, hypoglycemia, and paroxysmal cough (1, 2). During the catarrhal stage, B. pertussis colonizes the upper respiratory epithelium. Once colonization occurs, B. pertussis releases toxins such as pertussis toxin (PT) and adenylate cyclase toxin (ACT), as well as others. The catarrhal stage is followed by the paroxysmal stage, which is characterized by severe coughing episodes. Innate and adaptive immune responses result in clearance of the bacterium, allowing for the patient or host to convalesce. Despite extensive research, a full understanding of B. pertussis pathogenesis remains elusive for reasons, such as vaccine pressure, a lack of predictive models, and strain evolution (3). One of the major reasons for this gap in knowledge is the lack of a suitable animal model that shares common disease symptoms as seen in humans. Besides nonhuman primates, rats are the only other known model of pertussis that share similar coughing manifestation as seen in humans (4). As of now, the origin or the trigger of the cough has yet to be determined (5). The rat model of pertussis offers a potential animal model that can be used to evaluate B. pertussis pathogenesis.
In 1938, Hornibrook and Ashburn were the first to report that rats infected with B. pertussis induced cough-like paroxysms and bronchopneumonia, as seen in humans (6). Hornibrook and Ashburn found that young rats could be infected and the bacterium could be cultured from the lungs (6). Infected rats produced coughs that could be heard from a distance of 20 feet (6). Of the 31 infected rats, 26 that did not succumb to the infection had pathology indicative of inflammation in the lungs, most notably early neutrophil infiltration, followed by recruitment of mononuclear cells (6). The subsequent studies utilized intrabronchial inoculation of B. pertussis encased in agar beads and confirmed leukocytosis and paroxysmal cough by sound activated tape recorders from 5 to 21 days postchallenge (7, 8). Further development of the rat model led to the evaluation of PT-negative strains and the evaluation of coughs (9). Strain BP357, which is deficient in PT resulted in low cough induction (9). Further evaluation of the rat model demonstrated leukocytosis, weight loss, and paroxysmal cough in Sprague-Dawley rats during the course of B. pertussis infection (10). During the development phase of the acellular pertussis vaccine (aP and DTaP), the rat model was used to test vaccine efficacy of various acellular pertussis formulations and was used to validate protection against the onset of leukocytosis and cough (11). Recently, an intranasal rat model has been used to evaluate bacterial factors responsible for cough in Bordetella bronchiseptica (12). BspR, which is an anti-sigma factor of B. bronchiseptica, plays some role in cough induction upon infection (12). Collectively, these studies show that the pertussis rat model can be used to critically evaluate B. pertussis pathogenesis and disease progression.
In the 1940s, whole-cell pertussis (wP and DTP) vaccines were introduced to protect against B. pertussis infection (13). Widespread use of DTP in the United States led to a 90% decrease in the number of reported B. pertussis infections (13). Despite the efficacy of this vaccine, serious adverse side effects ensued, leading to the development of acellular pertussis vaccines. The antigens that are included in the current aP vaccines contain: filamentous hemagglutinin (FHA), fimbriae (FIM), PT, and pertactin (PRN). After the switch from the wP to the aP vaccines, there has been a significant increase in the number of pertussis cases in the United States and Europe (14). While nationwide vaccine coverage is 95% in the United States, the incidence of B. pertussis infections rose in the past 10 years as the number of aP-vaccinated-only population has increased in size. (15). While vaccine coverage remains high, the population dynamics are changing, and more people are aP immunized as the new generations are born and as the wP-only generations age. Numerous studies have demonstrated waning efficacy of aP vaccines in parallel with the emergence of genetically divergent strains of B. pertussis (3, 16–21). Increased surveillance of B. pertussis has led to the identification of clinical isolates that do not express PRN, FIM, and even PT, the hallmark toxin of the organism (22). One plausible hypothesis to explain these observations is that circulating B. pertussis strains have evolved due to acellular vaccine pressure. While we know the current strains are genetically different, we do not know whether this genetic variability affects the virulence, disease burden, toxicity, or fitness of the pathogen (3).
Early rat studies were performed with intranasal administration of B. pertussis for infection (6). Subsequent rat challenge studies used B. pertussis encased in agar beads for intrabronchial instillation (7–11, 23). While the agar bead infection method was successful at establishing infection, we aimed to utilize the simplicity of intranasal administration. We sought to reinvestigate the coughing rat model of pertussis to compare the pathogenesis of reference strain Tohama 1 to the recent clinical isolate D420, which has been extensively studied in the baboon model of pertussis (24–30). Tohama 1 was first isolated from a case of whooping cough in 1954, while D420 was isolated in 2002 from a critically ill infant in Texas (25, 31). Although Tohama 1 is now a reference strain that has been widely used since Sato and Sato developed the aP, but recent data have shown that this strain is an considered an outlier (32, 33). Tohama 1 does express PRN but has lower expression of PT and ACT (34). When Tohama 1 was used as a challenge strain in baboons, the baboons did not exhibit symptoms of pertussis despite being infected (26). This led to the selection of recent clinical isolate D420 as the baboon challenge strain, which readily infected and caused disease (26). With 48 years of potential genetic divergence, we sought to understand the differences in pathogenesis between these two commonly studied strains. D420 is known to infect mice and baboons, and it belongs to the clade (CDC013) of strains that represented 50% of isolates recovered in the United States in 2000 (3). However, it is important to note that D420 has an intact pertactin gene, and it does express the PRN protein, as confirmed by shotgun proteomic analysis (data not shown).
In the present study, we aimed to reestablish an upper respiratory tract infection model in rats following intranasal challenge with Tohama I and D420. We hypothesized that the recent isolate D420 would induce a more severe disease profile compared to Tohama 1, since previous research investigating pathogenesis in rhesus macaques noted that Tohama 1-infected animals did not exhibit overt disease symptoms (26). Sprague-Dawley rats were intranasally challenged with B. pertussis, and we characterized their disease progression profile over a 12-day infection. Cough was critically assessed utilizing whole-body plethysmography (WBP). Bacterial colonization, leukocytosis, and serological responses were measured as a result of infection. Rats challenged with D420 had increased coughing, greater bacterial burden in the respiratory tract, and a more robust IgM antibody response compared to rats challenged with Tohama 1. The coughing rat model of pertussis can shed light on the pathogenesis of B. pertussis and will likely be a useful tool for pertussis vaccine evaluation.
RESULTS
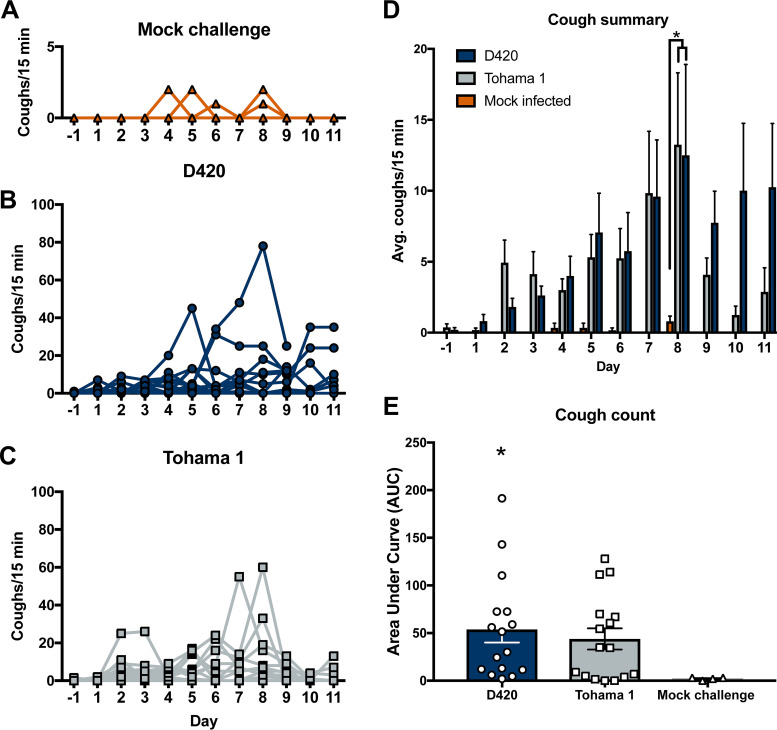
D420-infected rats have increased number of coughs compared to strain Tohama 1 over the entire course of infection.
Over the past decade it has become clear that improvements to the acellular pertussis vaccine strategy are needed due to the increases in PRN mutants, genomic divergence, and epidemiological data. We believe that in order to improve pertussis vaccines we need to better understand the pathogenesis of B. pertussis in an animal model that shares similar clinical manifestations of pertussis seen in humans. Here, we intranasally infected 7-week-old Sprague-Dawley rats with 108 CFU of B. pertussis strain D420 or Tohama 1 (Fig. 1) in an effort establish infection and observe B. pertussis induced cough. For negative controls (no bacterial challenge), rats were intranasally administered sterile phosphate-buffered saline (PBS). Paroxysmal cough is the hallmark symptom of pertussis and is thought to play a major role in transmission of the organism to a new host. To quantify respiratory function during infection, we utilized whole-body plethysmography (WBP). WBP instrumentation consists of specialized containment chambers which monitor box flow, temperature, and airflow changes to measure respiratory function. Counting coughs via WBP provides an extremely accurate and unbiased way of counting coughs based on cough waveforms. Rat containment chambers were placed inside a laminar flow hood and connected to a computer that would monitor the rats breathing (see Fig. S1). Early studies reported that B. pertussis infection in young rats induced coughs that were audible by ear (6), and in our preliminary studies this was apparent to us as well (data not shown). Studies in the 1990s of paroxysmal cough in rats were quantified using an analog sound recording device (7). We hypothesized that rats infected with the recent isolate D420 would induce more coughs than animals infected with the reference strain Tohama 1 because rhesus macaques infected with Tohama 1 did not exhibit overt disease symptoms but D420 induced robust coughing in baboons (26). B. pertussis-infected rats developed cough at days 2 to 11 (Fig. 2B and C), while mock-infected rats (Fig. 2A) only had a few isolated coughs, unrelated to infection. At days 1 to 3 postinfection, the average cough count for rats infected with Tohama 1 or D420 was fewer than 5 coughs per 15 min. The average cough count doubled by day 7 postinfection, with peak coughing occurring at day 8 postinfection and with an average of 13 coughs per 15 min of monitoring. After day 8, the average cough count for rats infected with D420 remained above 10 coughs per 15 min, while the rats infected with Tohama 1 averaged fewer than 5 coughs. To summarize the cough data, the average cough count over the course of infection is shown (Fig. 2D). We observed a significant increase in the number of coughs at day 8 postinfection of B. pertussis-infected rats compared to the mock-infected challenge control. Rats infected with D420 coughed a total of 949 times, whereas rats infected with Tohama 1 coughed a total of 724 times over the entire 12-day infection. To analyze differences in cough count between strains, we utilized area under the curve (AUC) analysis (Fig. 2E). AUC analysis allows us to quantify the number of coughs over the entire course of infection per rat. Using this analysis approach, we note a significant increase in the number of coughs for rats infected with D420 compared to the mock-challenged rats over the 12-day infection, while there was no significant difference observed in Tohama 1-infected rats compared to mock-challenged rats (noninfected).
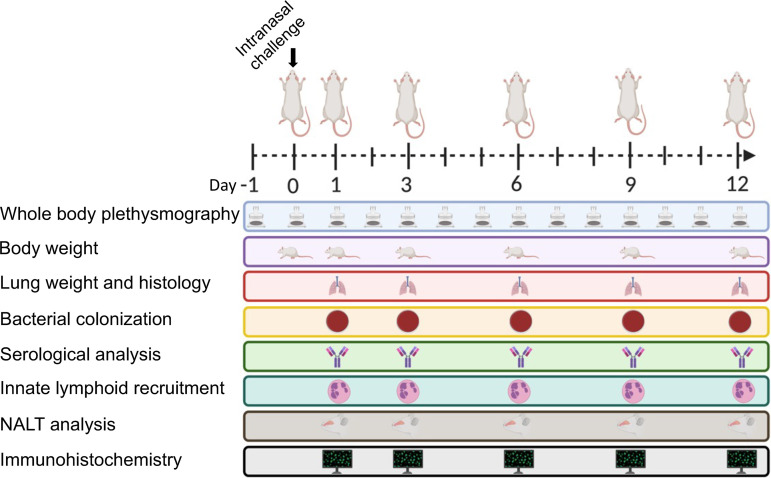
FIG 1.
Experimental design of intranasal infection of B. pertussis. Schematic representation of Sprague-Dawley rats intranasally infected with B. pertussis. Whole-body plethysmography was used to measure cough and analyze lung function over the course of a 12-day infection. Body weight, bacterial burden of the respiratory tract, innate lymphoid recruitment, and antibody titers were measured at the days 1, 3, 6, 9, and 12 postchallenge. The left lobe of the lung and the NALT were sectioned and stained with H&E for histological analysis, and immunohistochemistry was performed to visualize the bacteria in the left lobe of the lung and nasal cavity. (Figure 1 was created with BioRender.com.)
FIG 2.
Infection of B. pertussis induces cough in Sprague-Dawley rats. Coughs were measured each day of the 12-day infection using whole-body plethysmography. Coughs were counted for mock challenge rats (A), rats infected with D420 (B), and rats infected with Tohama 1 (C). (D) Summary of results (see panels A to C) shown as means ± the standard errors of the mean (SEM; n = 4 to 8). (E) AUC, represented as averages of coughs per 15 min for each rat. P values were determined by two-way ANOVA, followed by a Bonferroni comparison test and an unpaired Student t test for AUC (*, P < 0.05 [D420 compared to the mock-challenged control group], #, P < 0.05 [Tohama 1 compared to the mock challenged control group]).
B. pertussis infection causes pulmonary distress.
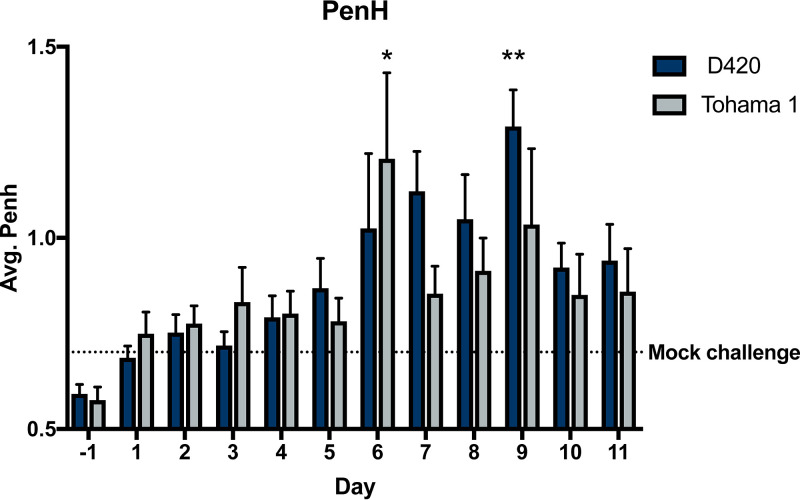
Infection with B. pertussis leads to mucus production, lung damage, and invasion of cellular infiltrates into the bronchioles of the lung (35). The lungs of infants with B. pertussis exhibit edema, necrotizing bronchiolitis, and inflammation in the lung leading to respiratory distress (35). WBP was used to quantify the pulmonary distress over the course of infection. Pulmonary distress was measured by calculating the enhanced pause (PenH) of the animal (Fig. 3). The higher the PenH value, the increased respiratory distress of the animal. PenH corresponds to the resistance associated with peak inspiratory height and peak expiratory height and taking into account time between early and late expiration per breath (see Fig. S2). With increased constriction, the expiratory peak becomes prominent and the area of the peak expiratory height becomes a larger percentage of the complete expiratory area. Other factors that contribute to the increased PenH is a decrease in relaxation time (Tr) in relation to expiratory time (Te). Previous research has shown that severe inflammation in the lungs of mice directly correlated with an increase in PenH (36). More recently, SARS-CoV-2 infected mice were observed to have subtle but significant increase in PenH postchallenge (37, 38). Compared to the mock-challenged rats, both D420- and Tohama 1-infected rats displayed an increase in PenH (Fig. 3). We observed a significant increase at day 9 with rats infected with D420 and at day 6 with rats infected with Tohama 1 compared to mock-challenged rats.
FIG 3.
B. pertussis infection impairs respiratory capacity. Over the course of infection, Sprague-Dawley rat respiratory function was analyzed using whole-body plethysmography. Each day at 5 p.m., rat bronchial restriction was determined utilizing WBP to calculate the PenH. The results are shown as means ± the SEM (n = 4 to 8). P values were determined by two-way ANOVA, followed by Dunnett’s comparison test (*, P < 0.05; **, P < 0.01 [compared to the mock challenge group]).
Histological assessment of the lung and nasal cavity.
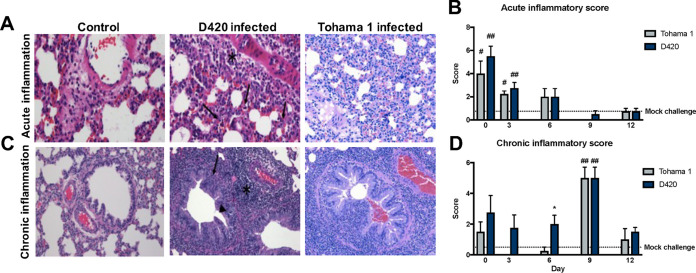
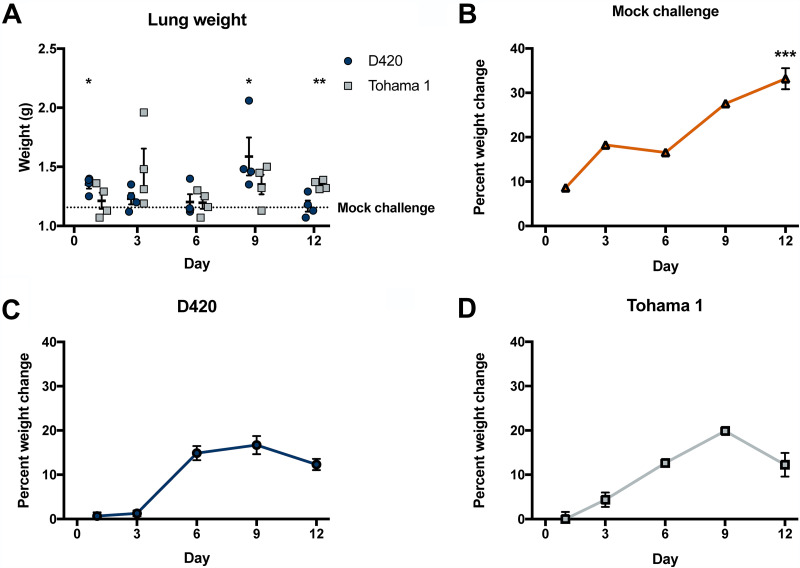
To further evaluate the inflammation in the lung, we utilized histology to confirm both acute and chronic inflammation in response to B. pertussis infection. At days 1, 3, 6, 9, and 12 postchallenge, the left lobes were collected, sectioned, and stained with hematoxylin and eosin (H&E) (Fig. 4A and C). Whole-lung images were taken to assess cellular infiltration (see Fig. S3). Once stained, the blinded slides were then scored by a board-certified pathologist. Mock-challenged animals exhibited minimal inflammatory infiltrates consisting of focal accumulations of mononuclear cells in the parenchyma and occasional infiltrates of neutrophils surrounding blood vessels (Fig. 4A). Rats infected with Tohama 1 and D420 showed significant increases in their acute inflammation scores compared to mock-challenged animals at days 1 and 3 postinfection (Fig. 4B). The highest acute inflammation score occurred at day 1 postinfection (Fig. 4B). Rats exhibited mild to moderate neutrophil infiltration of the parenchyma, blood vessels, and bronchioles (Fig. 4A). Markers of acute inflammation resolved after day 6 postchallenge; however, infected rats at day 9 exhibited obvious differences in the lungs associated with mild to moderate infiltration of mononuclear cells and higher chronic inflammation compared to mock-challenged animals (Fig. 4C). At day 12 postinfection, we notice moderate resolution of inflammation in both D420- and Tohama 1-infected rats (Fig. 4B and D). We also observed a significant increase in lung weight, which can be associated with elevated inflammation, at days 1 and 9 postchallenge in D420-infected rats compared to the mock-challenged controls (Fig. 5A). At day 12 postchallenge, rats infected with Tohama 1 showed a significant increase in lung weight compared to the mock-challenged rats (Fig. 5A). These results confirm that B. pertussis infection induces inflammation in the lungs of coughing Sprague-Dawley rats.
FIG 4.
B. pertussis infection induces acute and chronic inflammation in the lungs of Sprague-Dawley rats. The left lobe of the lung was sectioned and stained with H&E from rats infected with D420 or Tohama 1 or from the PBS control. (A) Representative images of acute inflammation demonstrating neutrophil recruitment around surrounding blood vessel (asterisk) and parenchyma (arrows) at ×400 magnification. (B) Average acute inflammatory score of the lung based on the predominance of neutrophils in the parenchyma, blood vessels, and airways. (C) Representative images of chronic inflammation showing mononuclear cells surrounding the blood vessel (asterisk), lamina propria (arrow), and bronchioles (arrowhead) at ×200 magnification. (D) Average chronic inflammatory score of the lungs characterized by mononuclear infiltrates in the parenchyma, blood vessels, and airway. Histological assessment was determined blinded with no knowledge of the treatment groups. The results are shown as means ± the SEM (n = 4). P values were determined by one-way ANOVA, followed by Dunnett’s comparison test (*, P < 0.05 [compared between challenge groups]; #, P < 0.05, ##, P < 0.01 [compared to mock challenge]).
FIG 5.
B. pertussis infection induces inflammation of the lung and a decrease in weight gain. (A) After euthanasia, the lung weight was measured and recorded. To observe any weight changes over the course of infection, we calculated the percent weight change for mock-challenged (B), D420-infected (C), and Tohama 1-infected (D) rats. Rat body weights were recorded before infection, as well as immediately posteuthanasia. The percent weight change was calculated by taking the differences between starting weight and end weight and dividing by the initial weight and then multiplying by 100. Results are shown as means ± the SEM (n = 1 to 4). Only one mock-challenge rat weight was measured at days 1, 3, 6, and 9. Weights of 4 mock-challenge rats were recorded at day 12 postinfection. P values were determined by one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [(A) compared to mock-challenge group; (B to D) compared to both infection groups]).
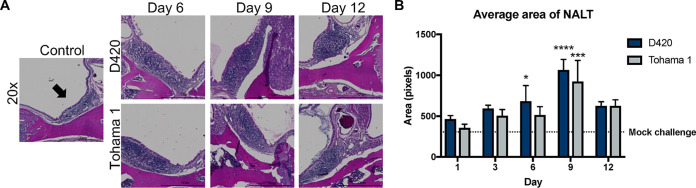
Due to the fact that B. pertussis is a mucosal pathogen, we sought out to investigate any phenotypic changes or inflammation in the nasal-associated lymphoid tissues (NALT) of B. pertussis-infected rats via histology. NALT helps elicit immunity against airborne and mucosal pathogens (39). The NALT is located above the hard palate, and studies have also shown that with the introduction of antigen via vaccination or bacterial components, cellular expansion of the NALT ensues (40–44). The NALT was stained with H&E, and we observed an increase in cellular infiltrate and a size enlargement in the NALT of B. pertussis-infected rats compared to the mock-challenged rats (Fig. 6A). ImageJ analysis was used to measure the area of both the left and right NALT of each animal (Fig. 6B). These data could mark the potential cellular expansion of the NALT upon infection with B. pertussis. Further analysis of the expanding cell populations in the NALT will warrant more analysis especially in the context of vaccine immunity.
FIG 6.
Analysis of the nasal associated lymphoid tissue (NALT). The skull was sectioned, allowing staining of the NALT with H&E. (A) Representative 20× images of the NALT. Arrows indicate the NALT. (B) ImageJ analysis of the area of both left and right NALT. The dotted line represents the average area of the NALT in the mock-challenged group. Results are shown as means ± the SEM (n = 4). P values were determined by two-way ANOVA (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001 [compared to the mock-challenge group]).
D420-infected rats demonstrate an increased bacterial burden in the respiratory tract.
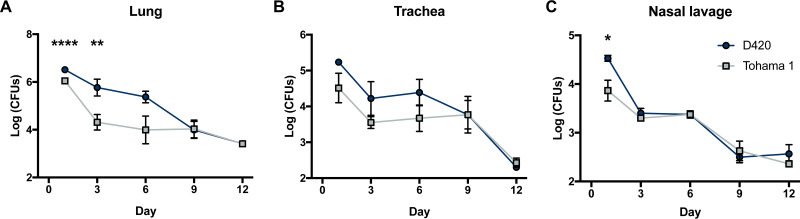
We determined the bacterial burden in the respiratory tract for Tohama I- and D420-challenged rats at days 1, 3, 6, 9, and 12. Over the coughing timeline, we observed an overall decrease in viable bacteria in the lung, trachea, and nasal cavity. In the lung, we observed a significant increase in the recovered bacteria in D420-infected rats compared to Tohama 1-infected rats at both days 1 and 3 (Fig. 7A). The same trend is also seen in the nasal cavity at day 1 postinfection (Fig. 7C). We observed no difference in the viable bacteria recovered from the trachea (Fig. 7B). Previous pertussis studies utilizing the rat model of pertussis noted weight loss from intrabronchial infection of B. pertussis strain 18-323 (10). B. pertussis infection is sufficient to cause weight loss in rats (10). To confirm this using intranasal infection, body weights were measured before challenge and upon euthanasia to calculate the percent weight change. We observed a significant decrease in weight gain in rats infected with B. pertussis strain D420 and Tohama 1 compared to the mock-challenged animals at day 12 postinfection (Fig. 5B to D).
FIG 7.
Analysis of bacterial burden over the course of B. pertussis infection. Bacteria were quantified by serially diluted CFU following intranasal challenge. CFU counts were determined from lung homogenate (A), trachea (B), and nasal lavage (C). The results are shown as means ± the SEM (n = 4). P values were determined by two-way ANOVA corrected with the Bonferroni comparison test (*, P < 0.05; **, P < 0.01; ****, P < 0.0001 [compared between D420 and Tohama 1]).
Next, we aimed to compare the bacterial colonization between outbred mice and rats in an effort to relatively compare the bacterial burden between models. We utilized the bacterial burden data in CD1 mice and compared them to our data in rats (45). To address the body weight differences between the mice and rats, we used the CFU/organ and divided these values by their respective body weights for a crude comparison. Our data suggest that despite having a higher initial challenge dose in the rat, the bacterial burden remaining in the respiratory tract is higher in a mouse compared to a rat later during infection (see Fig. S4). In general, the rat model is likely a lower challenge dose per body weight model of pertussis compared to mouse models.
Immunofluorescence confirms colonization of B. pertussis in the lungs and nasal cavity.
We next wanted to assess the location of bacterial colonization in the nasal cavity and visualize any bacteria left behind following flushing of the nares. We sectioned the skulls after flushing the nasal cavity and utilized immunofluorescence (IF). After visually scanning the nasal cavity using confocal microscopy, we found bacteria in the nasal turbinates and the NALT (Fig. 8A and C). We also confirmed that the bacteria were found between the epithelial cells (Fig. 8B). The relative locations where the bacteria were found in the nasal cavity can be seen in Fig. S5A in the supplemental material. Our results show that B. pertussis remained in the nasal cavity over the entire course of infection even after flushing the nares (see Fig. S5B). The flushing of the nasal cavity underrepresents the total number of B. pertussis in the nasal cavity. In the lung, B. pertussis colonizes the bronchioles, while the alveolar spaces remain noncolonized (Fig. 8D and F). IF was performed on the nasal cavities and lungs of mock-challenged animals to access nonspecific binding (see Fig. S6). To assess any potential differences between the strains we blindly manually counted the labeled microcolonies in the lung and nasal cavity. It is also worthy to note, we found no differences in microcolony counts in the lung or nasal cavity between the strains (see Fig. S5C and D) despite our observed differences in bacterial burden by CFU enumeration. CFU is likely a better indicator of overall bacterial burden, but IF revealed that some B. pertussis remains in the airway following flushing of the nares.
FIG 8.
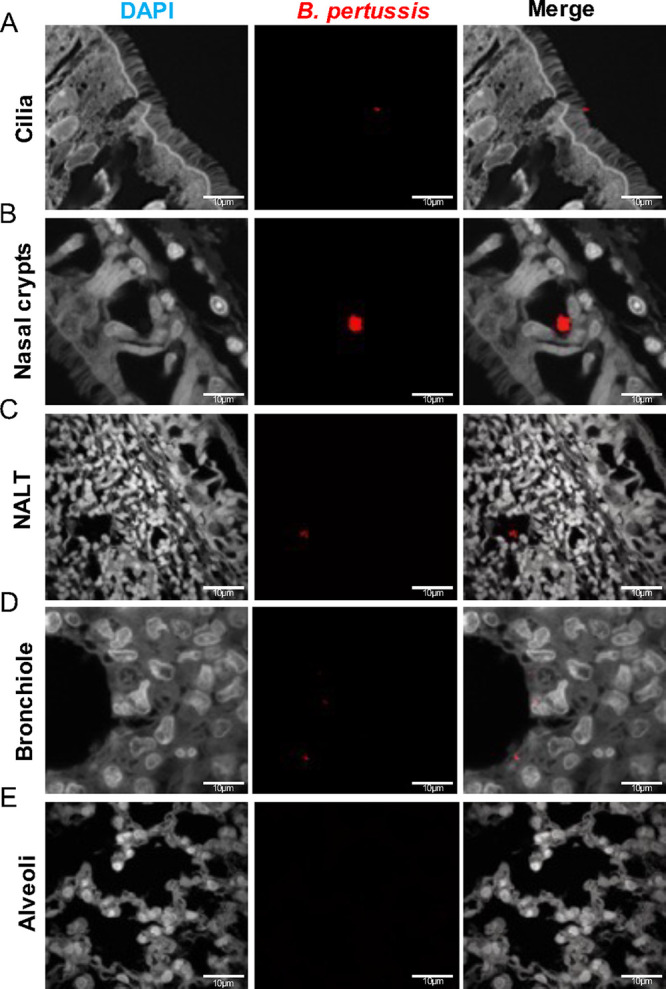
Immunofluorescence (IF) staining of B. pertussis localization in the respiratory tract. B. pertussis was labeled using a polyclonal antibody to FHA and counter-tagged with a fluorescently conjugated antibody (Texas Red). Sections were counterstained with DAPI. (A to C) Representative images of B. pertussis in the nasal cavity over the course of infection. B. pertussis was found captured in the cilia of the nasal cavity, as well as of the NALT. (D and E) Representative images of the bronchioles and alveoli of infected rats. B. pertussis was found localized in the bronchioles over the course of infection and absent in the alveoli.
Measurement of recruitment of neutrophils in response to B. pertussis infection.
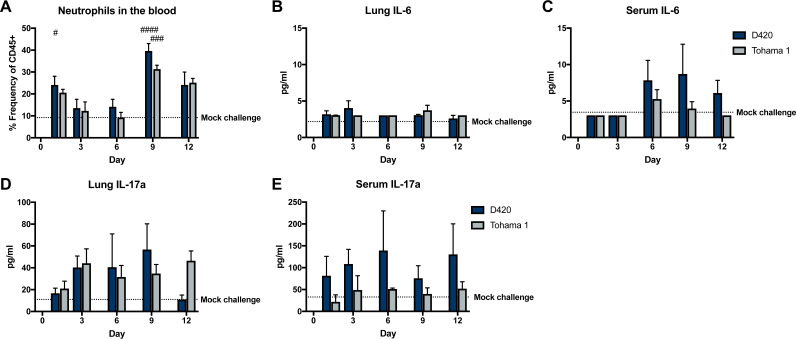
Previous studies utilizing the rat model of pertussis noted leukocytosis and an increase in the number of total white blood cells when agar-encased B. pertussis was used for infection (8). Neutrophils circulating through the blood were measured during intranasal infection by antibody staining and flow cytometry, since B. pertussis infection results in various amounts of neutrophilia (35, 46). One caveat in analyzing cell populations via flow cytometry in the rats is the limited availability of rat specific antibodies compared to mice. Recent research by Barnett-Vanes et al. has developed a flow cytometry panel to analyze cell populations in a model of lipopolysaccharide-induced pulmonary inflammation (47). The percentages of CD45+ (CD161− B220− CD43+ His48Hi) neutrophils in the blood were assessed in B. pertussis-infected rats compared to mock-challenged animals (Fig. 9A). Specifically, a significant increase was observed in the number of circulating neutrophils in the blood of rats infected with D420 at days 1 and 9, with Tohama 1-infected rats only having a significant increase in response to mock challenge at day 9 (Fig. 9A). Next, cytokine concentrations in the lung and serum were measured to identify factors that could contribute to the recruitment of neutrophils, specifically interleukin-6 (IL-6) and IL-17. IL-6 is a potent inducer of Th17 polarization and has been found elevated during murine infection with B. pertussis (45, 48–50). Surprisingly, we did not observe any significant changes in IL-6 due to B. pertussis infection (Fig. 9B and C). It has been well documented in the mouse model that the production of IL-17 in pertussis plays a role in the increase in circulating neutrophils (51). There were no significant changes in IL-17a in the sera or lungs of B. pertussis-infected rats compared to mock-challenged rats (Fig. 9D and E). Th1 and Th2 cytokines and chemokines in the lung and serum were also measured (see Fig. S7 to S10). We did not observe an increase in the induction of cytokines by infection overall; however, in the serum, rats infected with D420 had a significant increase in IL-5 compared to Tohama 1-infected rats at day 12 postinfection (see Fig. S7). There was a significant increase in IL-5 and TNF-α at day 6 postinfection in the lung in rats infected with Tohama 1 compared to D420 (see Fig. S8). We also observed significant increases in chemokines MIP-1a, RANTES, MCP-3, MIP-2, and IP10 in rats infected with D420 at day 12 postchallenge in the serum compared to rats infected with Tohama 1 (see Fig. S19). In the lung, MIP-2 and IP10 were increased in rats infected with D420 compared to the mock-challenged controls (see Fig. S10). Despite slight differences highlighted above we did not observe a massive difference in the cytokine and chemokine responses during infection.
FIG 9.
Neutrophil recruitment and proinflammatory cytokine production after challenge determined by flow cytometry and multiplex immunoassay analysis. (A) Blood neutrophil count of Sprague-Dawley rats infected with Bordetella pertussis. At days 1, 3, 6, 9, and 12 postinfection, rats were euthanized, and blood was collected via cardiac puncture. Flow cytometry was used to quantify the amount of neutrophils (CD45+ CD161− B220– CD43+ His48hi) in the blood. Quantification of the percentage of single, CD45+ cells was performed. The dotted line represents the average frequency of CD45+ neutrophils in the mock challenge animals over the course of the study. (B and C) Analysis of cytokine IL-6 from the supernatant of lung homogenate and serum. (D and E) Analysis of cytokine IL-17 in serum and lung homogenate supernatant. Cytokines were analyzed using a ProcartaPlex multiplex immunoassay kit. Results are shown as means ± the SEM (n = 4). The dotted line represents the average mock-challenged cytokine levels measured. P values were determined by one-way ANOVA, followed by a Tukey comparison test (#, P < 0.05; ##, P < 0.01; ####, P < 0.0001 [compared to mock challenge]).
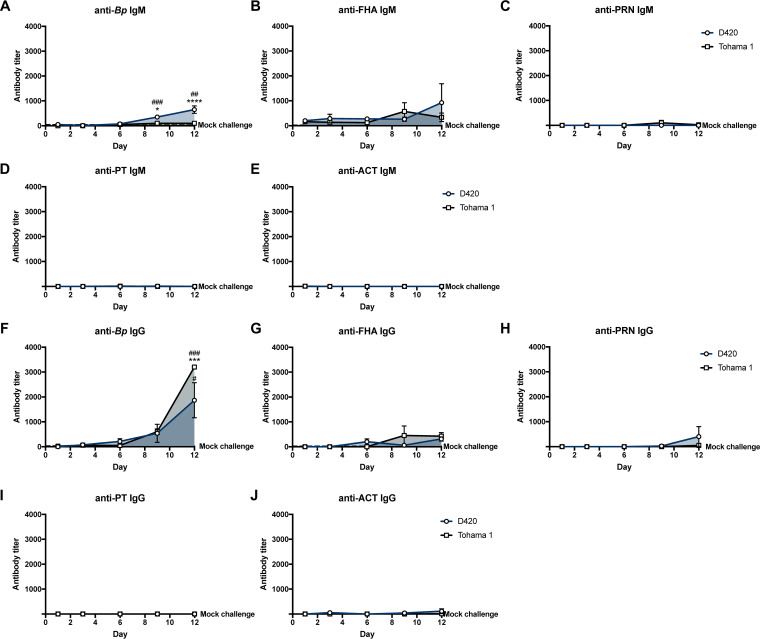
Serological analysis of B. pertussis-specific antibodies.
Enzyme-linked immunosorbent assays (ELISAs) were performed to determine the serological response generated from B. pertussis infection. We measured IgM and IgG antibody titers in the serum to whole bacterium and B. pertussis-associated virulence factors at days 1, 3, 6, 9, and 12 postinfection. In previous studies utilizing the rat model, intrabronchial infection induced the generation of IgG antibody titers to sonicated B. pertussis at 28 days postinfection (8). Intranasal infection of strain D420 has generated significant anti-PT IgG antibody titers in the baboon model at days 17 to 19 postinfection (26). However, analysis of antibody titers generated against an intranasal pertussis infection in the rat has yet to be determined, and we wanted to characterize the primary antibody response to B. pertussis (see Fig. S11). We observed a significant 10-fold increase in the anti-B. pertussis IgM antibody response at day 9 and an almost 20-fold increase 12 days postchallenge in rats infected with D420 compared to the mock challenge, while we also saw a significant 6-fold increase in anti-B. pertussis IgM at day 12 postinfection compared to the amount of anti-B. pertussis IgM from Tohama 1 infection (Fig. 10A). At day 12 postinfection, we observed that the B. pertussis infection induced significant production of anti-B. pertussis IgG in the serum compared to mock-challenged animals (Fig. 10F). At day 12 postinfection, Tohama 1-infected rats showed a significant increase, nearly double anti-B. pertussis IgG antibody titers compared to rats infected with D420. AUC analysis revealed a significant increase in anti-B. pertussis IgM antibodies from rats infected with D420 compared to Tohama 1-infected rats (see Fig. S12A). However, no difference was observed in the AUC values of the anti-B. pertussis IgG antibody response over the total course of infection between the bacterial challenge groups (see Fig. S12B). It is also important to note that we also did not see a significant increase in the generation of antibodies titers to PT, ACT, PRN, and FHA in B. pertussis-infected rats compared to mock-challenged animals (Fig. 10B, E, and G to J). We also note that IgA antibodies were not detected to either whole bacteria or PT in the lung and nasal cavity at days 1 and 9 postchallenge (data not shown).
FIG 10.
Measurement of serum antibody titers over the course of B. pertussis infection. ELISA was used to compare serological responses from rats intranasal challenge with B. pertussis. Total IgM and IgG serum antibody titers were measured from challenged rats against whole B. pertussis bacterium (A and F), FHA (B and G), PRN (C and H), PT (D and I), and ACT (E and J). The dotted line represents the average serum titer for mock-challenged rats. P values were determined by two-way ANOVA corrected with Bonferroni comparison test (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001 [compared between infection groups]). For comparison to the mock-challenged control group, one-way ANOVA was used, followed by a Tukey comparison test (##, P < 0.01; ###, P < 0.001).
DISCUSSION
To date, humans are the only known reservoir for B. pertussis, making the development of a suitable animal model challenging. Multiple models have been studied to recapitulate similar disease symptoms seen in humans, as well as provide insight into the pathogenesis of B. pertussis. We set out to reinvestigate the rat model of pertussis that was originally introduced in 1938 (6). In our study, we have confirmed that intranasal administration of B. pertussis led to bacterial colonization of the upper and lower respiratory tracts (Fig. 7). Through intranasal instillation, bacteria were able to infect the nasal cavity, trachea, and lungs postchallenge. This is important since infants with pertussis experience pneumonia more frequently than do adolescents and adults (35). Mouse models of pertussis also rely on depositing large numbers of bacteria into the lung to establish infection (2). Here, our data show that through intranasal infection, we can establish an infection of both the upper and lower respiratory tracts, resembling the infection seen in adolescents and adults with pertussis (52, 53).
Previous reports have shown B. pertussis infection of rats induced paroxysmal coughs, and these coughs were counted by analog audio recordings (7). Utilizing WBP, we are able to precisely and digitally count coughs and measure the respiratory capacity for the first time with rats infected with B. pertussis (Fig. 2 and 3). Here, coughs induced from infection appeared at days 1 to 2 postchallenge, with the number of coughs gradually increasing with peak cough occurring at day 8 postchallenge. Rats challenged with intrabronchial instillation of B. pertussis noted similar peak cough counts approximately 10 days postchallenge (54). This is also one of the first studies utilizing WBP to count coughs in rats with bacterial infection. We did observe some infected rats that did not overtly cough. This reflects the frequency of coughs in adolescents and adults (55). Previous studies using baboons also show that B. pertussis infection elicits severe coughs lasting over 2 weeks postchallenge, with peak coughs per hour occurring at day 4 postchallenge (26). It is also important to note that at peak coughing, the bacterial burden was low, which was also observed in previous research on B. pertussis-infected rats (7). The data suggest that other factors could be playing a role in cough production despite the bacterial infection being resolved. Given that B. pertussis-infected rats coughed, we also investigated transmission of B. pertussis from infected 7-week-old rats (actively coughing) cohoused with more susceptible 3-week-old rats, and we have observed no evidence of transmission via negative qPCR (IS481) analysis of swabs from the nasal cavity and no active coughing in the naive 3-week-old rats. We also performed a challenge study with B. bronchiseptica strain KM22 in 7-week-old Sprague-Dawley rats, and challenged rats coughed more violently and earlier after bacterial challenge compared to B. pertussis-infected rats. B. bronchiseptica-infected rats coughed the most when the bacterial loads in the respiratory tract were at their highest, which could potentially lead to transmission of the bacterium. TLR4-deficient rats could be investigated to evaluate transmission of B. pertussis (56). We also observed that the increased bronchial restriction coincides with the increased coughs during days 6 to 11 postchallenge (Fig. 3). The decreased lung capacity and increase in cough could be further associated with an increase of chronic inflammation in the lung (Fig. 4). Our data suggest that the coughing rat model of pertussis can be used to potentially evaluate the mechanism of cough, which still requires further investigation.
Initial studies characterizing the serum antibody response to rats infected with B. pertussis noted an increase antibody response to endotoxin correlated with the small number of cultivable organisms in the lung following intrabronchial challenge (7). Hall et al. demonstrated that rats infected with B. pertussis become seropositive in response to whole-cell B. pertussis lysate, FHA, and PT by 28 days after intrabronchial challenge (8). Anti-FHA antibody titers were not detected until day 16 postchallenge, whereas anti-PT antibody titers were not detected until day 28 postchallenge (23). In our study, we characterized the antibody response following intranasal challenge of B. pertussis over the whole infection timeline. Using B. pertussis-coated ELISA plates, we detected anti-B. pertussis IgG antibody titers in rats infected with B. pertussis at day 9 postchallenge, with a significant increase compared to mock challenge observed at day 12 (Fig. 10). We also measured a significant increase in anti-B. pertussis IgM antibody titers in rats infected with D420 compared to the mock-challenged and Tohama 1-infected animals (Fig. 10). One potential hypothesis that could explain this observation is the genetic differences between the two strains as D420 harbors the ptxP3 allele, leading to an increase in PT production (25). PT has been shown to suppress B. pertussis antibody responses in serum and limit the expression of antigen-presenting receptors (57–60).
With the increased interest in developing new pertussis vaccines, it is imperative for the development of animal models to evaluate the pathogenic potential of current emerging strains, as well as to identify any potential differences in disease burden between genetically divergent strains. Sato and Sato first used B. pertussis strain Tohama 1 in the development of the acellular pertussis vaccine, and Tohama 1 is still being used in pertussis research today, notably due to its genetic malleability (61). D420 has been instrumental in the development of the baboon model and has also been used to study vaccine efficacy in regard to transmission in the baboon model (29). We hypothesize that the observed differences could be due to the increased amount of PT through the ptxP3 allele of D420 (25). Strains, such as D420, harboring the ptxP3 allele lead to an increase in virulence, PT production, and prevalence (62, 63). A previous study utilizing the intranasal mouse model has shown an increase in bacterial colonization with ptxP3 strains (64). Parton et al. illustrated that infection with a PT-deficient strain was unable to induce cough (9). It is possible that there are other factors beyond PT that can account for the observed virulence differences between Tohama I and D420. In the rat model, unlike the mouse, we also did not measure any significant increases in IL-17 and IL-6 in the lung and serum after challenge. IL-17 and IL-6 are increased following B. pertussis infection in mice and baboons (30, 48, 65). Further investigation is needed to explain the observed differences. One potential approach to evaluate differences between strains is the use of a neonatal rat model of pertussis. Studying B. pertussis in neonates may be useful in distinguishing subtle differences between strains. Hornibrook et al. demonstrated that B. pertussis-infected young 3-week-old rats demonstrated 65% mortality, while 7-week-old rats do not succumb to infection with a similar dose (6).
In summary, we have demonstrated that the rat model of pertussis can be used as a tool to further study B. pertussis pathogenesis and recapitulate some of the similar symptoms of pertussis as seen in humans. Pertussis is often described as a “100-day cough” because adolescents and adults experience an extended duration of coughing (66). Coughing episodes in infants with pertussis can lead to vomiting, choking, gagging, and apneic episodes that can lead to seizures (67). Mice have not been used to evaluate coughing manifestation in pertussis until recently, and baboons are expensive and limited to specialized facilities (68). Use of the coughing rat model of pertussis provides feasibility through low husbandry costs, the ready availability of animals, and its ease of use as a model. The immunological tools available in murine research is vast, but as basic research continue to evolve, so, too, will the resources and immunological tools available for rats. Recently, flow cytometry markers have been developed to study innate immune cells and adaptive immune cells, such as B and T cells (47). In other studies, we have used ELISpot assays to investigate antigen-specific immune populations as well. The increasing availability of antibodies has also allowed investigation of systemic and mucosal serological responses. Future studies utilizing the coughing rat model of pertussis will capitalize on the growing immunological toolbox in regard to vaccination against pertussis.
In this study, we carefully compared two established strains that have been used in the pertussis field, Tohama 1 and D420. In future studies, we aim to evaluate whole-cell and acellular vaccine-mediated immunity against D420. Armed with this model, we can further our understanding of pathogenesis, host response during pertussis, genetic divergence between strains, and vaccine-mediated immunity.
MATERIALS AND METHODS
Bordetella pertussis strains and growth conditions.
B. pertussis strain Tohama 1 was graciously provided by Peter Sebo (Czech Academy of Sciences), and strain D420 was acquired from the CDC Pertussis Lab provided by Maria L. Tondella and Michael Weigand. B. pertussis strain Tohama 1 and D420 were cultured on Bordet-Gengou (BG) agar (Remel, catalog no. R45232) that was supplemented with 15% defibrinated sheep blood (Hemostat Laboratories, catalog no. DSB500) (69). Bacteria were cultured on BG plates for 48 h at 36°C. B. pertussis was transferred from BG plates with polyester swabs (Puritan, catalog no. 22-029-574) and transferred into 20 ml of Stainer-Scholte liquid media (SSM) in new, 125-ml flasks (Thermo Fisher Scientific, catalog no. FB50000125) (70). The liquid cultures were grown for 24 h at 36°C inside a shaking incubator at 180 rpm.
Intranasal challenge with B. pertussis.
Seven-week-old ∼170-g female Sprague-Dawley rats (Charles River, catalog no. 001CD) were used for challenge. B. pertussis was grown as described above. The rats were then anesthetized with ketamine and xylazine (50 to 100 and 5 to 10 mg/kg, respectively) and challenged with 108 CFU (100 μl intranasally), administering two 50-μl doses, one in each nostril (Fig. 1). The body weight of each rat was recorded both before challenge and immediately after euthanasia. Rats were euthanized at days 1, 3, 6, 9, and 12 after bacterial challenge. Mock-challenge animals (no bacteria) were administered 100 μl of sterile endotoxin-free PBS (Thermo Fisher Scientific, catalog no. TMS012A) intranasally. Upon euthanasia, blood was collected via cardiac puncture and transferred into EDTA (BD, catalog no. 365974) and serum separation (BD, catalog no. 026897) tubes. Blood collected in the EDTA tube was used for flow cytometric analysis, while blood collected in the serum separation tubes were used to isolate the serum via centrifugation (15,000 × g for 3 min) and used for serological and cytokine analysis. Lungs and tracheas were excised and homogenized to determine the bacterial burden. Upon removal, the wet weight of the lungs was recorded. Lungs were then collected in gentleMACS C tubes (Miltenyi Biotec) and homogenized using a gentleMACS dissociator (catalog no. 130-095-927). The trachea was homogenized using a Polytron homogenizer. To determine the bacterial burden in the nares, 2 ml of sterile 1× PBS was flushed through the nares, and then samples were collected for plating. Serial dilutions of the homogenates and nasal collections were plated on BG plates supplemented with ceftibuten (Sigma-Aldrich, catalog no. SML0037) at 10 μg/ml to decrease the growth of normal rat respiratory tract flora.
Analysis of coughing and bronchiole restriction using whole-body plethysmography.
To quantify respiratory function during infection, we utilized a Buxco FinePointe Whole Body Plethysmograph instrument (WBP; DSI). FinePointe software was used to collect, analyze, and report the breathing data. Every day postchallenge for 12 consecutive days at 5:00 p.m., the rats were placed inside designated chambers to acclimate for 5 min. We chose this time not only due to rats being nocturnal but also because at this time the animals were most awake and active, as confirmed by video camera (data not shown). After acclimation, each rat’s respiratory profile was recorded for 15 min. Each chamber is fitted with a transducer that measured the changes in box flow and airflow of the subject. The chambers were also fitted with a screen pneumotach that allowed airflow in and out of the chamber that can be recorded. Coughs were counted during the designated 15 min, and the enhanced pause (PenH) was calculated to signify bronchiole restriction. Coughs were counted based on large box flow changes of the subject and changes in both humidity and temperature of the air flowing in and out of the subject with classical cough-like waveforms. Cough detection algorithm was applied using a patented fuzzy logic criteria to determine whether the event was a cough (71). When analyzing the number of coughs, each cough in a multicough event was counted individually.
Histological assessment of the lung and NALT.
Upon euthanasia, the rat skull was excised, and the mandible was removed. The skulls were fixed in 10% formalin (Fisher Scientific, catalog no. SF98-4) for 48 h at 26°C. After fixation, the formalin was removed, and the skulls were frozen at −80°C until decalcification. Skulls were decalcified using Richard-Allan Scientific decalcifying solution (Thermo Scientific, catalog no. 8340-1) at room temperature for 24 to 48 h and then embedded in paraffin. Samples were sectioned and stained with H&E. A BioTek Lionheart Fx was used to scan and image the NALT. ImageJ was used to trace and measure the area of both the left and the right NALT using the images from the Lionheart Fx. The left lobe of the lung was excised and fixed in 10% formalin 48 h at 26°C. The left lobe was then embedded in paraffin and stained with H&E by the WVU Pathology Department. H&E-stained sections were used to characterize and score acute and chronic inflammation of the lung. All scorings were done by a board-certified pathologist (iHisto). Individual scores were based on a standard qualitative scoring criterion: 0, none; 1, minimal (rare); 2, mild (slight); 3, moderate; 4, marked; and 5, severe. Chronic inflammation was characterized by mononuclear infiltrates of the parenchyma, blood vessels, and airway. Acute inflammation scores were assigned due to the presence of neutrophils in the parenchyma, blood vessels, and airway. All examination and scoring were performed blindly since no knowledge of the treatment groups was known.
Imaging of B. pertussis in the lung and nasal cavity.
Detection of B. pertussis in the nasal cavity and lung was quantified via IF and confocal imaging. The left lobe of the lung and nasal cavity were preserved and sectioned as described above. Sectioned samples underwent deparaffination and rehydration using xylene and ethanol (70 to 100%). Antigen retrieval was performed by incubating samples in citrate buffer at 98°C for 20 min. Samples were blocked using 5% bovine serum albumin (Fisher Scientific, catalog no. 159008) for 1 h and primarily labeled utilizing a polyclonal rabbit FHA antibody (a gift from Erik Hewlett) diluted in 1× PBS. Secondary labeling occurred utilizing an anti-rabbit IgG conjugated with Texas Red (Fisher Scientific, catalog no. T2767) diluted in 1× PBS. Samples were then covered in mounting media (Prolong Gold Antifade reagent with [4′,6′-diamidino-2-phenylindole], catalog no. 8961). Samples were imaged using a Nikon A1R confocal microscope. Images were analyzed on DAPI channel and at wavelength 650 nm for Texas Red acquisition. Images were acquired using a 100× oil immersion lens (100×/1.40 Nikon Plan APO). To identify any potential differences in IF between the two strains, all IF images were deidentified, and the microcolonies were manually counted blindly by four volunteers with two to three fields of view used per sample.
Flow cytometry analysis of phagocytes.
Neutrophil recruitment in the blood was evaluated by flow cytometry. Blood samples upon collection were then lysed with PharmLyse buffer (BD Biosciences, catalog no. 555899) for 20 min at room temperature, with slight vortexing throughout. The remaining cells were resuspended in RPMI–10% fetal bovine serum (FBS) to neutralize the lysis buffer, centrifuged five times at 1,000 × g, and then washed with RPMI–10% FBS again. Blood was then resuspended in 1% FBS–PBS–5 mM EDTA. Samples were next blocked with anti-CD32 (BD Pharmingen, catalog no. 550270) antibody for 30 min at 4°C. After incubation, the cells were stained with the appropriate antibody markers: CD45-Alexa Fluor 700 (BioLegend, catalog no. 202218), CD161-APC (BioLegend, catalog no. 205606), CD45R-PE-Cy7 (eBioscience, catalog no. 25-0460-82), His48-FITC (eBioscience, catalog no. 11-0570-82), CD43-PE (BioLegend, catalog no. 202812), and CD3-VioGreen (Miltenyi Biotec, catalog no. 130-119-125) (47). After the addition of antibodies, the cells were incubated 1 h at 4°C in the dark. To prepare the lung samples for flow cytometry, the lung homogenate was pushed through a 70-μm-pore-size cell strainer (BioDesign Cell MicroSives, catalog no. N70R), creating a single-cell suspension. The suspension was then centrifuged at 1,000 × g for 5 min. After removal of the supernatant, the pellet was resuspended in PharmLyse buffer, and the cells were incubated at 37°C for 2 min. After incubation, the cells were centrifuged at 1,000 × g for 5 min, and the supernatant was removed, blocked, and labeled with antibody, as described above. The lung and blood samples were centrifuged at 1,000 × g for 5 min, and the pellets were resuspended in 0.4% paraformaldehyde and stored overnight at 4°C. Samples were washed with 1× PBS and resuspended in 1× PBS for analysis. Cell samples were analyzed on a LSR Fortessa, and samples were gated and analyzed using FlowJo v10.
Lung and serum cytokine and chemokine analysis.
Lung homogenate samples were centrifuged at 19,000 × g for 4 min, and the supernatant was removed and stored at −80°C. Quantitative analysis of cytokines in the serum and lung homogenate was performed using a ProcartaPlex multiplex immunoassay kit (Cytokine & Chemokine 22-Plex Rat ProcartaPlex panel, catalog no. EPX220-30122-901) according to the manufacturer’s instructions.
Serological analysis.
Antibody titers of infected rats were measured by ELISA. B. pertussis-specific ELISA plates were coated with 50 μl of 108 B. pertussis grown, as mentioned above for infection. To measure antibody titers to FHA (Enzo ALX-630-123-0100), PRN (GSK), PT (List Biological Laboratories, catalog no. 180), and ACT (a gift from Erik Hewlett), we coated plates with 50 μl of each antigen at 1 μg/ml. Once coated, the plates were incubated overnight at 4°C. The plates were washed with 1× PBS-Tween 20 and then blocked with 5% skim milk for 2 h at 37°C. The plates were washed again, and the sera from the challenge studies were serially diluted down the ELISA plate, followed by incubation for 2 h at 37°C. After incubation, the plates were washed and coated with 100 μl of secondary goat anti-rat IgG (Southern Biotech, catalog no. 3030-04) and IgM (Southern Biotech, catalog no. 3020-04) antibody at a dilution of 1:2,000 in PBS–5% milk. Once coated, the plate was incubated for 1 h at 37°C. The plates were washed again with PBS-Tween 20, and 100 μl of p-nitrophenyl phosphate substate (Thermo Scientific, catalog no. 37620), prepared according to the manufacturer’s instructions, was added. The plate was then developed for 30 min at room temperature to determine the titers of IgG and IgM. A BioTek Synergy H1 microplate reader was used to measure the colorimetric signal of the ELISA plate at A450. Positive antibody titers were determined as any values that were higher than the baseline. The baseline was set as double the average value of the blank; no serum added to these wells.
Generating Nightingale Rose plots in Python.
Data from individual mice were averaged in Microsoft Excel and log transformed. Values were formatted in Excel to be compatible with Python. Data were imported from .csv files using the pandas package and plotted using the “Barpolar” representation feature in the plotly.graph_objects module. Samples with a titer of <50 were assigned a value of 0.
Statistical analysis.
All data were analyzed using GraphPad Prism 7. The minimum number of biological replicates for the challenge studies was 4. For statistical comparisons between multiple groups over the entire course of the infection, a two-way analysis of variance (ANOVA) was used. One-way ANOVA was used for comparisons between groups for each individual day with Tukey’s post hoc test. Unpaired Student t tests were used for AUC analysis. Specific follow-up statistical tests are annotated in the figure legends.
Ethics statement.
All studies were performed in accordance with West Virginia University Institutional Animal Care and Use Committee approved protocol 1811019148.6.
Data availability.
Data requests for figures provided can be addressed to the corresponding author.
ACKNOWLEDGMENTS
The preliminary work on this project was in part supported by the Vaccine Development Center at WVU-HSC through research challenge grant HEPC.dsr.18.6 from the Division of Science and Research, WV Higher Education Policy Commission. The project was also supported by NIH R01AI137155 (F.H.D.) and CDC Contract (Broad Agency Announcement) 75D301-19-R-67835 (F.H.D.). Flow cytometry experiments were performed in the West Virginia University Flow Cytometry Core Facility, which is supported the National Institutes of Health equipment grant S10OD016165 and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grants P30GM103488 (CoBRE) and P20GM103434 (INBRE).
J.M.H., D.T.B., and J.K. performed bacterial challenge. J.M.H., S.M.K., and J.K. monitored rat coughing and respiratory function by whole-body plethysmography. All authors participated in the animal experiments. J.M.H. and E.S.-K. contributed to flow cytometry panel design and analysis. J.M.H. and T.Y.W. prepared and ran flow cytometry samples. M.A.W. and M.A.D. performed cytokine analysis. J.M.H. analyzed H&E histology slides. J.M.H. and G.J.B. performed and analyzed immunohistochemistry. J.M.H. performed ELISAs. J.M.H., M.B., and F.H.D. contributed to experimental design. J.M.H. wrote manuscript with critical revisions from all authors.
We thank Kathleen Brundage (WVU Flow Cytometry and Single Cell Core Facility) for assisting in flow cytometry and equipment instruction. We thank Amanda Ammer (Microscope Imaging Facility) for support in imaging the IF slides. We also thank James Coad, Jacqueline Karakiozis, and Brice Hickey (Pathology/Histology Core Facility) for the preparation of lung and nasal cavity for IF and performing H&E on the lung and nasal cavity.
Footnotes
Supplemental material is available online only.
Contributor Information
F. Heath Damron, Email: fdamron@hsc.wvu.edu.
Guy H. Palmer, Washington State University
REFERENCES
- 1.Mattoo S, Cherry JD. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18:326–382. 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melvin JA, Scheller EV, Miller JF, Cotter PA. 2014. Bordetella pertussis pathogenesis: current and future challenges. Nat Rev Microbiol 12:274–288. 10.1038/nrmicro3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weigand MR, Peng Y, Loparev V, Batra D, Bowden KE, Burroughs M, Cassiday PK, Davis JK, Johnson T, Juieng P, Knipe K, Mathis MH, Pruitt AM, Rowe L, Sheth M, Tondella ML, Williams MM. 2017. The history of Bordetella pertussis genome evolution includes structural rearrangement. J Bacteriol 199:e00806-16. 10.1128/JB.00806-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elahi S, Holmstrom J, Gerdts V. 2007. The benefits of using diverse animal models for studying pertussis. Trends Microbiol 15:462–468. 10.1016/j.tim.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Cherry JD. 2015. The history of pertussis (whooping cough), 1906–2015: facts, myths, and misconceptions. Curr Epidemiol Rep 2:120–130. 10.1007/s40471-015-0041-9. [DOI] [Google Scholar]
- 6.Hornibrook JW, Ashburn LL. 1939. A study of experimental pertussis in the young rat. Public Heal Rep 54:439. 10.2307/4582826. [DOI] [Google Scholar]
- 7.Woods DE, Franklin R, Cryz SJ, Ganss M, Peppler M, Ewanowich C. 1989. Development of a rat model for respiratory infection with Bordetella pertussis. Infect Immun 57:1018–1024. 10.1128/iai.57.4.1018-1024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall E, Parton R, Wardlaw AC. 1994. Cough production, leukocytosis, and serology of rats infected intrabronchially with Bordetella pertussis. J Med Microbiol 40:205–213. 10.1099/00222615-40-3-205. [DOI] [PubMed] [Google Scholar]
- 9.Parton R, Hall E, Wardlaw AC. 1994. Responses to Bordetella pertussis mutant strains and to vaccination in the coughing rat model of pertussis. J Med Microbiol 40:307–312. 10.1099/00222615-40-5-307. [DOI] [PubMed] [Google Scholar]
- 10.Hall E, Parton R, Wardlaw AC. 1997. Differences in coughing and other responses to intrabronchial infection with Bordetella pertussis among strains of rats. Infect Immun 65:4711–4717. 10.1128/iai.65.11.4711-4717.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall E, Parton R, Wardlaw AC. 1998. Responses to acellular pertussis vaccines and component antigens in a coughing rat model of pertussis. Vaccine 16:1595–1603. 10.1016/S0264-410X(98)80001-7. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura K, Shinoda N, Hiramatsu Y, Ohnishi S, Kamitani S, Ogura Y, Hayashi T, Horiguchi Y. 2019. BspR/BtrA, an anti-σ factor, regulates the ability of Bordetella bronchiseptica to cause cough in rats. mSphere 4:e00093-19. 10.1128/mSphere.00093-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Libster R, Edwards KM. 2012. Re-emergence of pertussis: what are the solutions? Expert Rev Vaccines 11:1331–1346. 10.1586/erv.12.118. [DOI] [PubMed] [Google Scholar]
- 14.Hill HA, Elam-Evans LD, Yankey D, Singleton JA, Dietz V. 2016. Vaccination coverage among children aged 19–35 months—United States, 2015. MMWR Morb Mortal Wkly Rep 65:1065–1071. 10.15585/mmwr.mm6539a4. [DOI] [PubMed] [Google Scholar]
- 15.CDC. 2011. National, state, and local area vaccination coverage among children aged 19–35 months—United States. MMWR 61(35):689–696. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm6135a1.htm. [PubMed] [Google Scholar]
- 16.Klein NP, Bartlett J, Fireman B, Baxter R. 2016. Waning Tdap effectiveness in adolescents. Pediatrics 137:e20153326. 10.1542/peds.2015-3326. [DOI] [PubMed] [Google Scholar]
- 17.Klein NP, Bartlett J, Rowhani-Rahbar A, Fireman B, Baxter R. 2012. Waning protection after fifth dose of acellular pertussis vaccine in children. N Engl J Med 367:1012–1019. 10.1056/NEJMoa1200850. [DOI] [PubMed] [Google Scholar]
- 18.Klein NP, Bartlett J, Fireman B, Rowhani-Rahbar A, Baxter R. 2013. Comparative effectiveness of acellular versus whole-cell pertussis vaccines in teenagers. Pediatrics 131:e1716–e1722. 10.1542/peds.2012-3836. [DOI] [PubMed] [Google Scholar]
- 19.Sheridan SL, Frith K, Snelling TL, Grimwood K, McIntyre PB, Lambert SB. 2014. Waning vaccine immunity in teenagers primed with whole cell and acellular pertussis vaccine: recent epidemiology. Expert Rev Vaccines 13:1081–1106. 10.1586/14760584.2014.944167. [DOI] [PubMed] [Google Scholar]
- 20.Sheridan SL, Ware RS, Grimwood K, Lambert SB. 2012. Number and order of whole cell pertussis vaccines in infancy and disease protection. JAMA 308:454. 10.1001/jama.2012.6364. [DOI] [PubMed] [Google Scholar]
- 21.Tartof SY, Lewis M, Kenyon C, White K, Osborn A, Liko J, Zell E, Martin S, Messonnier NE, Clark TA, Skoff TH. 2013. Waning immunity to pertussis following 5 doses of DTaP. Pediatrics 131:e1047–e1052. 10.1542/peds.2012-1928. [DOI] [PubMed] [Google Scholar]
- 22.Williams MM, Sen KA, Weigand MR, Skoff TH, Cunningham VA, Halse TA, Tondella ML. 2016. Bordetella pertussis strain lacking pertactin and pertussis toxin. Emerg Infect Dis 22:319–322. 10.3201/eid2202.151332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall E, Parton R, Wardlaw AC. 1999. Time-course of infection and responses in a coughing rat model of pertussis. J Med Microbiol 48:95–98. 10.1099/00222615-48-1-95. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MTG, Churcher CM, Bentley SD, Mungall KL, Cerdeño-Tárraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, et al. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica. Nat Genet 35:32–40. 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 25.Boinett CJ, Harris SR, Langridge GC, Trainor EA, Merkel TJ, Parkhill J. 2015. Complete genome sequence of Bordetella pertussis D420. Genome Announc 3. 10.1128/genomeA.00842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warfel JM, Beren J, Kelly VK, Lee G, Merkel TJ. 2012. Nonhuman primate model of pertussis. Infect Immun 80:1530–1536. 10.1128/IAI.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warfel JM, Merkel TJ. 2014. The baboon model of pertussis: effective use and lessons for pertussis vaccines. Expert Rev Vaccines 13:1241–1252. 10.1586/14760584.2014.946016. [DOI] [PubMed] [Google Scholar]
- 28.Pinto MV, Merkel TJ. 2017. Pertussis disease and transmission and host responses: insights from the baboon model of pertussis. J Infect 74:S114–S119. 10.1016/S0163-4453(17)30201-3. [DOI] [PubMed] [Google Scholar]
- 29.Warfel JM, Zimmerman LI, Merkel TJ. 2014. Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proc Natl Acad Sci USA 111:787–792. 10.1073/pnas.1314688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warfel JM, Merkel TJ. 2013. Bordetella pertussis infection induces a mucosal IL-17 response and long-lived Th17 and Th1 immune memory cells in nonhuman primates. Mucosal Immunol 6:787–796. 10.1038/mi.2012.117. [DOI] [PubMed] [Google Scholar]
- 31.Kasuga T, Nakase Y, Ukishima K, Takatsu K. 1954. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Kitasato Arch Exp Med 27:57–62. [PubMed] [Google Scholar]
- 32.Caro V, Bouchez V, Guiso N. 2008. Is the sequenced Bordetella pertussis strain Tohama I representative of the species? J Clin Microbiol 46:2125–2128. 10.1128/JCM.02484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato H, Sato Y. 1985. Protective antigens of Bordetella pertussis mouse-protection test against intracerebral and aerosol challenge of B. pertussis. Dev Biol Stand 61:461–467. [PubMed] [Google Scholar]
- 34.Barbier M, Boehm DT, Sen-Kilic E, Bonnin C, Pinheiro T, Hoffman C, Gray M, Hewlett E, Damron FH. 2017. Modulation of pertussis and adenylate cyclase toxins by sigma factor RpoE in Bordetella pertussis. Infect Immun 85. 10.1128/IAI.00565-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paddock CD, Sanden GN, Cherry JD, Gal AA, Langston C, Tatti KM, Wu K, Goldsmith CS, Greer PW, Montague JL, Eliason MT, Holman RC, Guarner J, Shieh W, Zaki SR. 2008. Pathology and pathogenesis of fatal Bordetella pertussis infection in infants. Clin Infect Dis 47:328–338. 10.1086/589753. [DOI] [PubMed] [Google Scholar]
- 36.Verheijden KAT, Henricks PAJ, Redegeld FA, Garssen J, Folkerts G. 2014. Measurement of airway function using invasive and noninvasive methods in mild and severe models for allergic airway inflammation in mice. Front Pharmacol 5:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leist SR, Dinnon KH, Schäfer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ, Gully KL, Scobey T, Brown AJ, Sheahan TP, Moorman NJ, Boucher RC, Gralinski LE, Montgomery SA, Baric RS. 2020. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183:1070–1085. 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinnon KH, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586:560–566. 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takaki H, Ichimiya S, Matsumoto M, Seya T. 2018. Mucosal immune response in nasal-associated lymphoid tissue upon intranasal administration by adjuvants. J Innate Immun 10:515–521. 10.1159/000489405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asanuma H, Thompson AH, Iwasaki T, Sato Y, Inaba Y, Aizawa C, Kurata T, Tamura S. 1997. Isolation and characterization of mouse nasal-associated lymphoid tissue. J Immunol Methods 202:123–131. 10.1016/S0022-1759(96)00243-8. [DOI] [PubMed] [Google Scholar]
- 41.Debertin AS, Tschernig T, Tönjes H, Kleemann WJ, Tröger HD, Pabst R. 2003. Nasal-associated lymphoid tissue (NALT): frequency and localization in young children. Clin Exp Immunol 134:503–507. 10.1111/j.1365-2249.2003.02311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirano T, Kodama S, Moriyama M, Kawano T, Suzuki M. 2009. The role of Toll-like receptor 4 in eliciting acquired immune responses against nontypeable Haemophilus influenzae following intranasal immunization with outer membrane protein. Int J Pediatr Otorhinolaryngol 73:1657–1665. 10.1016/j.ijporl.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 43.Petukhova G, Naikhin A, Chirkova T, Donina S, Korenkov D, Rudenko L. 2009. Comparative studies of local antibody and cellular immune responses to influenza infection and vaccination with live attenuated reassortant influenza vaccine (LAIV) utilizing a mouse nasal-associated lymphoid tissue (NALT) separation method. Vaccine 27:2580–2587. 10.1016/j.vaccine.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 44.Zuercher AW, Coffin SE, Thurnheer MC, Fundova P, Cebra JJ. 2002. Nasal-associated lymphoid tissue is a mucosal inductive site for virus-specific humoral and cellular immune responses. J Immunol 168:1796–1803. 10.4049/jimmunol.168.4.1796. [DOI] [PubMed] [Google Scholar]
- 45.Boehm DT, Hall JM, Wong TY, DiVenere A, Sen-Kilic E, Bevere JR, Bradford SD, Blackwood CB, Elkins C, DeRoos KA, Gray MC, Cooper CG, Varney ME, Maynard JA, Hewlett EL, Barbier M, Damron FH. 2018. Evaluation of adenylate cyclase toxoid antigen in acellular pertussis vaccines using a Bordetella pertussis challenge model in mice. Infect Immun 86. 10.1128/IAI.00857-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawal M, Cohen M, Irazuzta JE, Kumar R, Kirton C, Brundler M-A, Evans CA, Wilson JA, Raffeeq P, Azaz A, Rotta AT, Vora A, Vohra A, Abboud P, Mirkin LD, Cooper M, Dishop MK, Graf JM, Petros A, Klonin H. 2009. Fulminant pertussis: a multi-center study with new insights into the clinico-pathological mechanisms. Pediatr Pulmonol 44:970–980. 10.1002/ppul.21082. [DOI] [PubMed] [Google Scholar]
- 47.Barnett-Vanes A, Sharrock A, Birrell MA, Rankin S. 2016. A single 9-colour flow cytometric method to characterize major leukocyte populations in the rat: validation in a model of LPS-Induced pulmonary inflammation. PLoS One 11:e0142520. 10.1371/journal.pone.0142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boehm DT, Wolf MA, Hall JM, Wong TY, Sen-Kilic E, Basinger HD, Dziadowicz SA, Gutierrez M, de la P, Blackwood CB, Bradford SD, Begley KA, Witt WT, Varney ME, Barbier M, Damron FH. 2019. Intranasal acellular pertussis vaccine provides mucosal immunity and protects mice from Bordetella pertussis. NPJ Vaccines 4:1–12. 10.1038/s41541-019-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Goel T, Goodfield LL, Muse SJ, Harvill ET. 2011. Decreased leukocyte accumulation and delayed Bordetella pertussis clearance in IL-6–/– mice. J Immunol 186:4895–4904. 10.4049/jimmunol.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf MA, Boehm DT, DeJong MA, Wong TY, Sen-Kilic E, Hall JM, Blackwood CB, Weaver KL, Kelly CO, Kisamore CA, Bitzer GJ, Bevere JR, Barbier M, Damron FH. 2021. Intranasal immunization with acellular pertussis vaccines results in long-term immunity to Bordetella pertussis in mice. Infect Immun 89. 10.1128/IAI.00607-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreasen C, Carbonetti NH. 2009. Role of neutrophils in response to Bordetella pertussis infection in mice. Infect Immun 10.1128/IAI.01150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solans L, Locht C. 2018. The role of mucosal immunity in pertussis. Front Immunol 9:3068. 10.3389/fimmu.2018.03068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Graaf H, Ibrahim M, Hill AR, Gbesemete D, Vaughan AT, Gorringe A, Preston A, Buisman AM, Faust SN, Kester KE, Berbers GAM, Diavatopoulos DA, Read RC. 2020. Controlled human infection with Bordetella pertussis induces asymptomatic, immunizing colonization. Clin Infect Dis 71:403–411. 10.1093/cid/ciz840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wardlaw AC, Hall E, Parton R. 1993. Coughing rat model of pertussis. Biologicals 21:27–29. 10.1006/biol.1993.1024. [DOI] [Google Scholar]
- 55.Kilgore PE, Salim AM, Zervos MJ, Schmitt H-J. 2016. Pertussis: microbiology, disease, treatment, and prevention. Clin Microbiol Rev 29:449–486. 10.1128/CMR.00083-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rolin O, Smallridge W, Henry M, Goodfield L, Place D, Harvill ET. 2014. Toll-like receptor 4 limits transmission of Bordetella bronchiseptica. PLoS One 9:e85229. 10.1371/journal.pone.0085229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connelly CE, Sun Y, Carbonetti NH. 2012. Pertussis toxin exacerbates and prolongs airway inflammatory responses during Bordetella pertussis infection. Infect Immun 80:4317–4332. 10.1128/IAI.00808-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carbonetti NH, Artamonova GV, Andreasen C, Dudley E, Mays RM, Worthington ZV. 2004. Suppression of serum antibody responses by pertussis toxin after respiratory tract colonization by Bordetella pertussis and identification of an immunodominant lipoprotein. Infect Immun 72:3350–3358. 10.1128/IAI.72.6.3350-3358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shumilla JA, Lacaille V, Hornell TMC, Huang J, Narasimhan S, Relman DA, Mellins ED. 2004. Bordetella pertussis infection of primary human monocytes alters HLA-DR expression. Infect Immun 72:1450–1462. 10.1128/IAI.72.3.1450-1462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martino A, Volpe E, Auricchio G, Colizzi V, Baldini PM. 2006. Influence of pertussis toxin on CD1a isoform expression in human dendritic cells. J Clin Immunol 26:153–159. 10.1007/s10875-006-9009-3. [DOI] [PubMed] [Google Scholar]
- 61.Weiss AA, Falkow S. 1982. Plasmid transfer to Bordetella pertussis: conjugation and transformation. J Bacteriol 152:549–552. 10.1128/jb.152.1.549-552.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mooi FR, Van Loo IHM, Van Gent M, He Q, Bart MJ, Heuvelman KJ, De Greeff SC, Diavatopoulos D, Teunis P, Nagelkerke N, Mertsola J. 2009. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis 15:1206–1213. 10.3201/eid1508.081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Advani A, Gustafsson L, Carlsson RM, Donnelly D, Hallander HO. Clinical outcome of pertussis in Sweden: association with pulsed-field gel electrophoresis profiles and serotype. APMIS 115:736–742. 10.1111/j.1600-0463.2007.apm_628.x. [DOI] [PubMed] [Google Scholar]
- 64.King AJ, Van Der Lee S, Mohangoo A, Van Gent M, Van Der Ark A, Van De Waterbeemd B. 2013. Genome-wide gene expression analysis of Bordetella pertussis isolates associated with a resurgence in pertussis: elucidation of factors involved in the increased fitness of epidemic strains. PLoS One 8:e66150. 10.1371/journal.pone.0066150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andreasen C, Powell DA, Carbonetti NH. 2009. Pertussis toxin stimulates IL-17 production in response to Bordetella pertussis infection in mice. PLoS One 4:e7079. 10.1371/journal.pone.0007079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee GM, Lett S, Schauer S, Lebaron C, Murphy TV, Rusinak D, Lieu TA. 2004. Societal costs and morbidity of pertussis in adolescents and adults. Clin Infect Dis 39:1572–1580. [DOI] [PubMed] [Google Scholar]
- 67.Cherry JD. 2016. Pertussis in young infants throughout the world. Clin Infect Dis 63:S119–S122. 10.1093/cid/ciw550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hiramatsu Y, Suzuki K, Nishida T, Onoda N, Satoh T, Akira S, Ikawa M, Ikeda H, Kamei J, Derouiche S, Tominaga M, Horiguchi Y. 2020. Lipooligosaccharide, Vag8, and pertussis toxin of Bordetella pertussis cooperatively cause coughing in mice. bioRxiv https://www.biorxiv.org/content/10.1101/2020.12.25.424096v1.
- 69.Bordet J, Gengou O. 1906. Le microbe de la coqueluche. Les Ann L’Institut Pasteur 20:731–741. [Google Scholar]
- 70.Stainer DW, Scholte MJ. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol 63:211–220. 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- 71.Lomask J, Larson R. Cough/sneeze analyzer and method. 2006. US patent USOO7104962B2. US patent 7,104,962 B2. Application granted, 12 September 2006.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download iai.00304-21-s0001.pdf, PDF file, 1.0 MB (976.5KB, pdf)
Supplemental material. Download iai.00304-21-s0002.xlsx, XLSX file, 0.06 MB (58.5KB, xlsx)
Data Availability Statement
Data requests for figures provided can be addressed to the corresponding author.