Abstract
A CREB-CREB binding protein (CBP) complex was used as bait to screen a mouse embryo cDNA library in yeast. One of the strongest interactions identified the histone binding protein RbAp48. RbAp48 also interacted weakly with CBP alone but did not interact with phosphorylated or nonphosphorylated CREB. CBP (or its homologue p300) from HeLa cell nuclear extracts coimmunoprecipitated with RbAp48 and its homologue RbAp46 and bound to a glutathione S-transferase–RbAp48 fusion protein. This interaction was stimulated by the addition of phosphorylated CREB and allowed the association of core histones and mononucleosomes in an acetylation-dependent manner. RbAp48 lowered the Km of CBP histone acetylase activity and facilitated p300-mediated in vitro transcription of a chromatinized template in the presence of acetylcoenzyme A. These data indicate that the association of phosphorylated CREB with CBP promotes the binding of RbAp48 and its homologue RbAp46, allowing the formation of a complex that facilitates histone acetylation during transcriptional activation.
The signaling mechanism responsible for activating genes through the cyclic-AMP-regulated enhancer (CRE) represents one of the most intensively studied transcriptional pathways (27). Following the activation of certain G protein-coupled receptors, the catalytic subunit of protein kinase A (PKA) is released from the regulatory subunit and is transported to the cell nucleus, where it phosphorylates a unique site in the CRE binding transcription factor CREB. CREB is phosphorylated at this same site by many additional protein kinases, including those activated by calcium-calmodulin and growth factors. Thus, the transcription factor CREB has been proposed to serve as a fairly general signal-activated transcriptional mediator, linking a variety of signal transduction pathways to genes containing CRE sequences (40).
Phosphorylation allows CREB to interact with the coactivator CREB binding protein (CBP) or its homologue p300 (6, 7). CBP associates with a wide variety of additional transcriptional activators as well, suggesting that it serves as a transcriptional “integrator” (for a review, see reference 42). Thus, there appears to be a hierarchy of posttranscriptional modifications and protein-protein interactions that permit transcriptional signal integration—extracellular signals of various types converge on the CREB transcription factor, and distinct transcription factors converge by simultaneously interacting with CBP.
How CBP transmits the activation signal to gene promoters remains unresolved. Evidence from several laboratories has suggested that CBP interacts with the basal transcription factors TFIIB and TFIID (8, 21, 44). In addition, Nakajima et al. have shown that CBP contacts the RNA polymerase II holoenzyme through interactions with RNA helicase A (29). Thus, one model for CBP function is to bridge DNA binding transcription factors to components of the basal transcriptional machinery. Alternatively, CBP might alter some of these proteins through posttranslational modifications (15).
Other evidence suggests that transcriptional activation mediated through CBP occurs only in the context of chromatin (19, 20). The involvement of chromatin in CBP function is consistent with the findings that this coactivator and several associated proteins, including PCAF, SRC-1, and pCIP, have the ability to acetylate the amino-terminal tails of histone proteins in a manner that may lead to some, as-yet-uncharacterized, change in nucleosome structure (2, 5, 30, 43, 47, 53). A multistep model initially proposed by Roeder and coworkers suggests that CBP contributes to the first step of transcriptional initiation while other coactivator complexes, such as TRAP, DRIP, and ARC, mediate subsequent steps (10, 28, 36). More recent evidence suggests that p300 functions at a stage subsequent to chromatin disruption (23).
Of all the transcription factor-CBP associations, only the interaction with phosphorylated CREB has been studied in detail. Nuclear magnetic resonance analysis has revealed that the interaction of the two proteins introduces structure into both components of the complex (37). After binding to CBP, phosphorylated CREB adopts a bihelical configuration with the helical axes approximately perpendicular to one another. One prediction from this finding is that new protein interaction surfaces might be generated upon CREB-CBP binding. We have taken advantage of this possibility by developing a yeast “three-hybrid” assay that uses a CREB-CBP complex to screen cDNA expression libraries. Such a screen is possible because the PKA site in CREB is phosphorylated in yeast and allows the CREB-CBP interaction (41). In this report, we describe an interaction of the phosphorylated CREB-CBP complex with histone binding protein RbAp48.
RbAp48 and its homologue RbAp46 were initially identified as retinoblastoma binding proteins (35). Subsequently, these proteins were characterized as components of at least four distinct nucleosome-modifying complexes, the nuclear histone deacetylases (HDACs), the Drosophila nucleosome-remodeling factor NURF, chromatin assembly factor 1 (CAF-1), and Hat1, a type B (cytoplasmic) histone acetylase involved in chromatin assembly (13, 18, 25, 33, 46, 49, 51, 55). In general, the functions of the RbAp48-like proteins in these complexes remain undetermined. Two exceptions to this generalization are in the context of the human cytoplasmic histone acetyltransferase Hat1 or its yeast homologue Hat1p, where RbAp46 and Hat2p appear to link the enzymes to their target, histone H4 (33, 50). Thus, although RbAp48 association with nuclear transcriptional coactivators has not been described, there is abundant evidence that these histone binding factors interact with related classes of proteins. Moreover, this function is consistent with the model in which a critical function of transcriptional coactivators is to direct the targeting of histone acetyltransferases to specific promoters.
Our studies demonstrate that the association of RbAp48 with the coactivator CBP is stimulated by phosphorylated CREB. The binding of RbAp48 to the CREB-CBP complex then allows an interaction with core histones and mononucleosomes. The binding of histone particles to the CBP-RbAp48 complex depends upon their acetylation state—acetylation by CBP blocks the ability of histones or mononucleosomes to associate with the coactivator. In addition, RbAp48 increases the histone acetyltransferase activity of CBP, probably by enhancing the affinity of the coactivator for its substrate. This ability of RbAp48 to increase CBP histone acetyltransferase activity is reflected by its capacity to stimulate p300-mediated transcription of a reconstituted chromatin template in vitro. Furthermore, this stimulation is enhanced by the addition of acetyl coenzyme A (AcCoA), supporting the functional significance of RbAp48-targeted histone acetylation.
MATERIALS AND METHODS
Plasmids.
The bait plasmid for the three-hybrid screen was constructed from the pBTMYeA backbone. The parental plasmid pBTM116 was a gift from Stan Hollenberg (Oregon Health Sciences University), and pAD4 was obtained from Michael Wigler (Cold Spring Harbor Laboratory). The plasmid pYeA was created by digesting the alcohol dehydrogenase promoter, polylinker, and terminator from pAD4 with BamHI and cloning them into pYEP24 (New England Biolabs). To create pBTMYeA, the alcohol dehydrogenase promoter, terminator, and polylinker of pYeA were digested with BamHI, blunt ended, and cloned into the PvuII site of pBTM116. To generate the three-hybrid bait plasmid, VP16-CREB1–283 (41) was digested with BamHI and ligated into the BamHI site of pBTMYeA. LexA-CBP461–682 (41) was digested with EcoRI and BamHI, blunt ended, and ligated into the LexA-CREBYeA plasmid using a NotI linker, creating LexA-CREB-YeACBP. LexA-CREBM1 encodes amino acids (aa) 1 to 283 and contains a Ser133Ala mutation. The remaining bait plasmids used in secondary screens were generated similarly. An E9.5 mouse embryo VP16 fusion cDNA library was a gift from Stan Hollenberg. The carboxyl-terminal portion of RbAp48 recovered from the library screen, aa 273 to 425, was subcloned into pGEXKG (Pharmacia) and pET28a (Novagen) by PCR. Full-length RbAp48 and RbAp46 in pGEX2T3 were obtained from Bruce Stillman (Cold Spring Harbor Laboratory) and subcloned into pET28a by PCR. pGEXKG-CBP551–682 was kindly provided by Roland Kwok (University of Michigan).
Proteins.
Glutathione S-transferase (GST)-RbAp48273–425, GST-RbAp48, and GST-CBP551–682 were expressed in bacteria and purified by glutathione-Sepharose affinity chromatography (Sigma). His-tagged RbAp48, RbAp46, or RbAp48273–425 was expressed in strain BL21(DE3) and purified by Ni-nitrilotriacetic acid affinity chromatography (Qiagen). HeLa cell nuclear extracts were prepared from HeLa-S3 cells provided by the National Cell Culture Center, Minneapolis, Minn. (1). CREB in which all of the cysteines were mutated to serines was purified as described previously (38) and phosphorylated with PKA for 1 h at 30°C as described by Laurance et al. (22). PKA was a gift from Richard Maurer (Oregon Health Sciences University). Flag-tagged CBP and p300 were expressed in baculovirus-infected SF9 cells and purified using an M2 Flag affinity matrix (Sigma). Core histone octamers were isolated from chicken blood (9). Mononucleosomes were generated by digestion of purified chicken chromatin with micrococcal nuclease (Sigma), 2 U/ml, in 0.3 mM CaCl2 at 37°C for 40 min. Precautions were taken to avoid overdigestion of the chicken chromatin to minimize the production of free core histones. The chicken core particles were confirmed to be hypoacetylated by TAU gel analysis as described by Tse et al. (48). Gal4-VP16 was expressed in Escherichia coli and purified as previously described (31).
Yeast three-hybrid screen.
The LexA-CREB-YeACBP bait plasmid was transformed into the L40 yeast strain using standard small-scale transformation protocols (14, 39). This bait strain was then subsequently transformed with approximately 50 μg of the library plasmid. The yeast cells were allowed to grow at 30°C on Leu-, Trp-, His-, Ura-, and Lys-deficient plates, and colonies were picked daily for β-galactosidase assays. Plasmids from yeast that were positive for production of both histidine and β-galactosidase were isolated and sequenced. Secondary screens were performed using LexA-CREB, LexA-CBP, LexA-CREBM1, and LexA-CREBM1-YeACBP as bait.
GST pull-down assays.
GST, GST-RbAp48273–425, GST-RbAp48, and GST-CBP551–682 were coupled to glutathione-Sepharose beads (Pharmacia) and blocked with bovine serum albumin (BSA). Equimolar amounts of GST or GST fusion proteins were used in pull-down assays. HeLa cell nuclear extracts, recombinant RbAp48 or RbAp46, purified CREB, phosphorylated CREB, chicken core histones, and mononucleosomes were added to binding buffer HEG100 (20 mM HEPES [pH 7.6], 10% glycerol, 100 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 10 μM NaF, 10 μM Na3VO4) plus protease inhibitors (Complete; Boehringer Mannheim) for 1 h at 4°C. The beads were washed three times with HEG100 buffer, boiled in 15 μl of 5× sodium dodecyl sulfate (SDS) loading buffer, and electrophoresed on an SDS–6 to 15% polyacrylamide gel. After transfer to a polyvinylidene difluoride membrane, the bound fraction was detected by Western blotting using anti-CBP451–682 (which recognizes both CBP and p300), anti-CREB (New England Biolabs), anti-RbAp48/46 15G12 (GeneTex), or anti-histone H4 BWA3 and anti-histone H3 LG2-1 (generous gifts from Marc Monestier, Temple University) antibodies.
Coimmunoprecipitations.
Anti-RbAp48 or anti-RbAp46 antibody 15G12 was coupled to protein G-Sepharose (Pharmacia) and blocked with BSA. The beads were used to precipitate RbAp48 from HeLa cell nuclear extracts in HEG100 buffer for 1 h at 4°C. Normal mouse immunoglobulin G (Sigma) served as a control for background binding. The beads were washed three times with HEG100 buffer, boiled in 15 μl of 5× SDS loading buffer, and electrophoresed on an SDS–6% polyacrylamide gel. After transfer to a polyvinylidene difluoride membrane, the bound fraction was assayed for CBP by Western blotting using anti-CBP451–682 antibody.
Histone acetylation assays.
A 0.2-pmol sample of full-length CBP was preincubated with 1 pmol of His-tagged RbAp48 protein or BSA (used as a control) in HEG100 buffer on ice for 30 min and then mixed with purified chicken core histones or mononucleosomes in 30 μl of a reaction buffer containing 10 mM Tris-HCl (pH 8.0), 10% glycerol, 0.1 mM EDTA, 10 mM sodium butyrate, and [3H]AcCoA (Amersham) and incubated for 30 min at 30°C. The entire reaction mixture was spotted onto a phosphocellulose filter (Gibco BRL). The filter was then washed three times with sodium bicarbonate buffer, and the 3H signal from the transferred acetyl group was quantified by scintillation. CBP autoacetylation was subtracted from the total counts.
Chromatin assembly and in vitro transcription assays.
S190 extracts and Drosophila core histones were generous gifts from W. Lee Kraus (Cornell University). The DNA template containing five Gal4 DNA-binding sites upstream from the adenovirus major late promoter (MLP) encoding a 390-nucleotide (nt) G-free transcription cassette (26) and the Gal4-VP16 construct were gifts from Danny Reinberg (University of Medicine and Dentistry of New Jersey). The control DNA template encoding a 170-nt G-free transcription cassette placed downstream from the adenovirus MLP was a gift from Richard Maurer (Oregon Health Sciences University).
Chromatin was assembled as previously described (17, 31). Gal4-VP16 (200 nM) was added to the reaction mixture subsequent to chromatin assembly, and the mixture was incubated for 30 min. Where indicated, full-length p300 (25 nM) was added after Gal4-VP16 and this remodeled assembly mixture was incubated for a further 30 min at 30°C. The chromatin was then purified over a Sepharose CL-4B column (Pharmacia).
In vitro transcription reactions were performed with HeLa cell nuclear extracts as previously described (31). Briefly, naked or chromatinized DNA was incubated with nuclear extracts in the presence or absence of recombinant RbAp46 (272 nM) and AcCoA (1 μM) for 30 min at 30°C. The templates were then transcribed at 30°C for 45 min upon the addition of ribonucleoside triphosphates and RNase T1. The adenovirus MLP-driven 170-nt template was added to the transcription mixture as an RNA recovery control. The purified RNA products were resolved on a 5% acrylamide–6 M urea gel and analyzed by autoradiography.
RESULTS
Identification of RbAp48 by a yeast three-hybrid screen.
Although the yeast two-hybrid assay has identified many important protein-protein interactions, the high false-positive rate often limits its usefulness as a screening approach. This has particularly been the case for the transcriptional coactivator CBP (unpublished observations). One mechanism for generating spurious interactions could result if the bait component is capable of adopting a variety of three-dimensional configurations. As discussed above, the association of phosphorylated CREB with CBP introduces secondary structure into both components of the complex. Thus, this interaction might stabilize structures that are important for specific protein-protein interactions and decrease the occurrence of nonphysiological protein configurations. Alternatively, the association of phosphorylated CREB and CBP may generate new protein interaction surfaces that are not found in either component in isolation. It is possible that critical transcriptional effectors interact with CBP only, or preferentially, in the context of these novel induced structures. We have taken advantage of these possibilities by developing a yeast three-hybrid assay that uses a CREB-CBP complex as bait.
We had previously shown that LexA-CREB (containing the activation domain of CREB1–283 fused to the LexA DNA binding domain) becomes phosphorylated at Ser133 in yeast, thereby allowing an interaction with VP16-CBP (41). Ser133 is the residue that is recognized by PKA and other kinases that have the ability to mediate CREB activation. Mutation of Ser133 to Ala (designated LexA-CREBM1) prevented the interaction, supporting the idea that CBP binding to the CREB activation domain in yeast depends upon phosphorylation. The kinase responsible for this phosphorylation event is unknown, although it could be PKA, which is constitutively active in yeast (41). To develop the yeast three-hybrid assay, we cloned LexA-CREB and a nucleus-localized fragment of CBP (representing the CREB binding domain, aa 461 to 682, fused to the simian virus 40 Tag nuclear localization signal) into plasmid BTMYeA, a derivative of pYeA (Fig. 1a). The two fusion proteins were cloned into the same expression vector to reduce the use of selectable markers and to ensure that the expression ratio of both components of the bait remained constant. The two-component bait plasmid (LexA-CREB-YeACBP) was introduced into yeast strain L40 and used to screen a VP16-cDNA library representing embryonic day 9.5 mouse mRNAs. In marked contrast to our results obtained by using the same fragment of CBP in standard two-hybrid assays, which have shown very little selectivity, virtually all of the cDNAs obtained by using the yeast three-hybrid screen encoded proteins involved in transcriptional regulation (unpublished observation).
FIG. 1.
Yeast three-hybrid screen. (a) Illustration of two-component bait plasmid LexA-CREB-YeACBP. LexA-CREB (containing the activation domain of CREB, aa 1 to 283, fused to the LexA DNA binding domain) and a nucleus-localized fragment of CBP (representing the CREB binding domain, aa 461 to 682) were cloned into plasmid pBTMYeA, a derivative of pYeA. The bait plasmid is ampicillin resistant and enables Trp production. P, promoter; T, terminator; NLS, nuclear localization signal. (b) Interaction of VP16-RbAp48 with the CREB-CBP complex and CBP alone but not with CREB. LexA-CBP, LexA-CREB, and LexA-CREBM1 are LexA fusion constructs encoding aa 461 to 682 of CBP, aa 1 to 283 of CREB, or aa 1 to 283 of CREB with a Ser133Ala mutation, respectively. LexA-CREBM1-YeACBP is a two-component bait plasmid identical to LexA-CREB-YeACBP with the exception of the described Ser133Ala mutation. The levels of interaction with VP16-RbAp48, indicated by the plus and minus signs, were determined by assaying the growth of the transformants on a His-negative background.
One of the strongest positives identified in the screen was the histone recognition factor RbAp48 (Fig. 1b). When subjected to a secondary screen, VP16-RbAp48 was found to bind weakly to LexA-CBP alone but did not interact with LexA-CREB, LexA-CREBM1, or LexA-CREBM1-YeACBP (Fig. 1b). These studies indicate that the interaction requires the CBP component. Because the CBP fragment does not bind to DNA, association with the LexA-CREB component is required to generate an efficient target for VP16-RbAp48. It is not possible in these assays to assess the absolute affinities of the interactions of VP16-RbAp48 with LexA-CBP and LexA-CREB-YeACBP, however, because the two bait plasmids confer different levels of background activity.
Interaction between RbAp48 and CBP in mammalian cells.
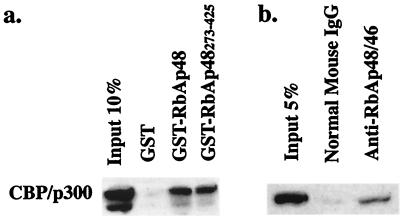
The portion of RbAp48 isolated in the yeast three-hybrid screen extended from residue 273 to residue 425, which includes carboxyl-terminal WD repeats 4 through 7. A GST fusion protein containing this portion of RbAp48, GST-RbAp48273–425, and a GST protein containing full-length RbAp48 were expressed in bacteria, coupled to glutathione-agarose beads, and incubated with HeLa cell nuclear extracts. Both GST-RbAp48 fusion proteins bound to full-length CBP, as determined by Western blotting using an antibody directed against the CREB binding domains of CBP and its homologue p300 (Fig. 2a), confirming the interaction identified in yeast. No binding of CBP was detected in the presence of GST alone. The amino-terminal portion of RbAp48 was incapable of binding to CBP (data not shown). To determine whether the interaction of CBP and RbAp48 exists in vivo, we immunoprecipitated HeLa cell nuclear extracts with a monoclonal antibody directed against RbAp48 and performed a Western blot assay using an antibody directed against CBP (Fig. 2b). These studies showed that RbAp48 complexes contain CBP. Although these experiments suggest that the association of CBP and RbAp48 occurs in vivo, they do not show that the interaction is direct. Moreover, it is possible that some unknown component of the extracts, such as a transcription factor, could promote the appropriate CBP interaction interface to allow RbAp48 binding.
FIG. 2.
Interaction between RbAp48 and CBP in HeLa cells. (a) GST pull-down assay. A GST fusion protein encoding RbAp48273–425, which contains carboxyl-terminal WD repeats 4 through 7 (GST-RbAp48273–425), and a GST fusion protein containing full-length RbAp48 (GST-RbAp48) were coupled to glutathione-agarose beads and incubated with HeLa cell nuclear extracts. CBP bound to the GST-RbAp48 fusion proteins was visualized by Western blotting. (b) Association of CBP and RbAp48 in vivo. Endogenous RbAp48 was immunoprecipitated from HeLa cell nuclear extracts using a monoclonal antibody against RbAp48. CBP coimmunoprecipitated with RbAp48 and was visualized by Western blotting. Normal mouse immunoglobulin G (IgG) was used as a negative control.
The association of RbAp48 and CBP is augmented by phosphorylated CREB.
We next performed GST pull-down assays using GST-CBP551–682 and GST alone to determine whether recombinant RbAp48 interacts with CBP directly. His-tagged RbAp48 and RbAp46 were expressed in bacteria, purified on a nickel affinity column, and incubated with the two GST fusion proteins. Both RbAp48 and RbAp46 bound directly to GST-CBP551–682 (Fig. 3a). The carboxyl-terminal portion of the RbAp48 protein that was identified in the yeast three-hybrid screen was also able to bind directly to GST-CBP551–682 (data not shown).
FIG. 3.
Augmentation of RbAp48-CBP interaction by phosphorylated CREB. (a) Direct binding of RbAp48 and RbAp46 to CBP. GST pull-down assays were performed with GST or GST-CBP551–682 and recombinant His-tagged RbAp48 or -46. RbAp48 or -46 bound to GST-CBP551–682 was visualized by Western blotting. (b) Binding of RbAp48 to the phosphorylated CREB-CBP complex. GST- or GST-CBP551–682-coupled beads were incubated with His-tagged RbAp48 in the presence of nonphosphorylated (+CREB) or phosphorylated (+pCREB) CREB. The binding of RbAp48 and CREB to GST-CBP551–682 was visualized by Western blotting.
Our initial yeast assays demonstrated that RbAp48 associates more strongly with the phosphorylated CREB-CBP complex than with CBP alone. The interaction of RbAp48 with LexA-CBP was easily detectable in yeast, however, and as indicated above, purified recombinant RbAp48 bound to GST-CBP551–682 in vitro. One explanation for our failure to identify RbAp48 in a standard two-hybrid screen using LexA-CBP as bait may relate to the large number of false positives. The addition of phosphorylated CREB could potentially decrease the abundance of aberrant CBP structures that generate these false interactions. Alternatively, phosphorylated CREB might contribute directly to the RbAp48 interaction interface, either by inducing a necessary structure in CBP or by providing protein interaction sites itself.
Because we are unable to compare the affinities of RbAp48 for the LexA-CBP and LexA-CREB-YeACBP constructs in yeast due to their different background activities, we cannot distinguish between these models. To resolve this issue, we assayed the binding of purified recombinant CBP and RbAp48 in vitro in the presence of nonphosphorylated or phosphorylated CREB. As previously reported, CREB binds to GST-CBP551–682 only after phosphorylation (Fig. 3b). Consistent with our studies with yeast, we did not detect binding of nonphosphorylated or phosphorylated CREB to GST-RbAp48 (data not shown). Surprisingly, we found that the addition of phosphorylated CREB greatly increased the binding of RbAp48 to GST-CBP551–682 (Fig. 3b). Binding of RbAp48 to GST alone was not detected, even in the presence of phosphorylated CREB. This is consistent with the hypothesis that phosphorylated CREB induces or stabilizes a particular structure in CBP that favors interaction with RbAp48. Alternatively, it is possible that RbAp48 also interacts with CREB structures that have been induced by association with CBP.
RbAp48 bridges CBP to histone proteins.
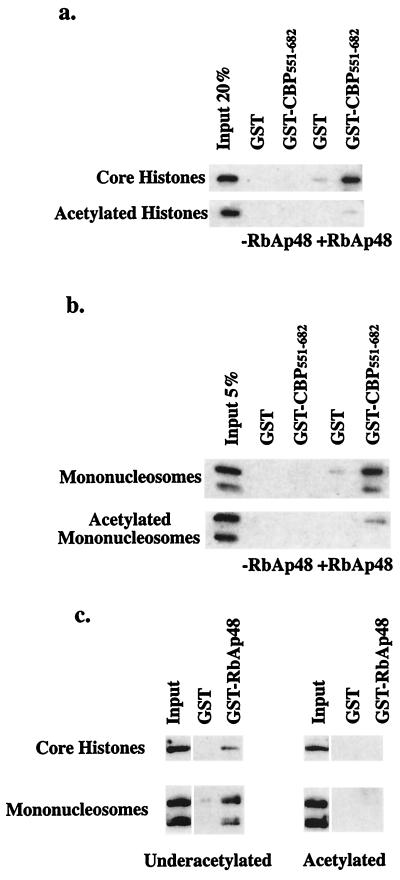
The physical association of CBP and RbAp48 and the functional characterization of these proteins as histone acetyltransferases and histone binding factors suggest that RbAp48 allows CBP to associate with its histone substrates. As described above, a similar function has been proposed for RbAp46 in the context of cytoplasmic Hat1 (50). To test whether RbAp48 bridges CBP to histones, we isolated hypoacetylated core histones and mononucleosomes from chicken blood and performed pull-down assays using GST-CBP551–682 and GST alone. These GST proteins were incubated with chicken core histones or mononucleosomes in the presence or absence of recombinant RbAp48. Bound proteins were detected by Western blotting using antibodies specific for RbAp48, H3, or H4. RbAp48 associated with GST-CBP551–682 but not GST alone (data not shown). As shown in Fig. 4a, the binding of histone octamers to GST-CBP551–682 was dramatically enhanced by the addition of RbAp48. Histone octamer binding was also stimulated by RbAp46 (data not shown). These results suggest that CBP, RbAp48, and core histones are capable of forming a stable ternary complex. Relatively little binding of histone proteins to CBP was detected in the absence of RbAp48.
FIG. 4.
Bridging of CBP to histone proteins by RbAp48. GST- or GST-CBP551–682-coupled beads were incubated with chicken core histones (a) or mononucleosomes (b) in the presence or absence of full-length RbAp48. Bound core histone octamers were assayed by Western blotting using antibody specific for histone H4 (core histones). Bound mononucleosomes were detected by two antibodies against either H3 or H4 (mononucleosomes).
Similar results were obtained when experiments were performed using intact mononucleosomes rather than core histones (Fig. 4b). We interpret these data to indicate that histone binding factor RbAp48 targets nucleosomal components to the CBP histone acetyltransferase. Although the conditions utilized in these studies should maintain mononucleosome integrity, we cannot completely rule out the possibility that some of the histone binding resulted from the release of core histones from the mononucleosome particles, however.
RbAp48 increases CBP histone acetylation activity.
Given the association of CBP, RbAp48, and core histones, one obvious question is whether RbAp48 affects CBP histone acetyltransferase activity. Acetylation assays were performed using purified, baculovirus-expressed full-length CBP, excess [3H]AcCoA, a fivefold molar excess of RbAp48 or BSA over CBP, and various concentrations of chicken core histones as the substrate. Neither RbAp48 nor BSA had intrinsic histone acetylase activity, and neither was a substrate for the CBP acetyltransferase (data not shown). Because CBP acetylates itself, data were corrected by subtracting the self-acetylation activity. RbAp48 treatment did not affect CBP self-acetylation (data not shown).
Results from three independent experiments demonstrated that RbAp48 augmented the acetyltransferase activity of CBP at low histone concentrations but not at higher histone concentrations. When the data were analyzed by the Michaelis-Menten equation, RbAp48 was found to lower the Km by three- to fourfold but had relatively little effect on the Vmax (Table 1). This mode of activation is consistent with a model wherein RbAp48 bridges the histone substrates to the CBP enzyme. RbAp48 also increased the ability of CBP to acetylate intact nucleosomes (data not shown). The ability of RbAp48 to augment CBP histone acetyltransferase activity is reminiscent of its effect on cytoplasmic Hat1.
TABLE 1.
Increase in CBP histone acetylation activity caused by RbAp48a
| Treatment | Avg CBP HATb activity
|
|
|---|---|---|
| Vmax (1/s) | Km (μM) | |
| CBP + BSA | 0.38 | 16.8 |
| 0.29 | 13.9 | |
| 0.31 | 15.0 | |
| CBP + RbAp48 | 0.25 | 5.1 |
| 0.26 | 9.4 | |
| 0.23 | 4.0 | |
Various concentrations of chicken core histones were acetylated by baculovirus-expressed full-length CBP that had been pretreated with a fivefold molar excess of RbAp48 (CBP + RbAp48) or an equivalent amount of BSA (CBP + BSA) in the presence of excess [3H]AcCoA. Data were corrected by subtracting the self-acetylation of CBP and analyzed by the Michaelis-Menten equation. The Km and Vmax from three independent experiments are shown.
HAT, histone acetyltransferase.
Histone binding to the CBP-RbAp48 complex is blocked by acetylation.
To assess the specificity of the RbAp48-histone interaction, we examined the effect of acetylation on histone binding. Chicken core histones are largely underacetylated (3) but can be acetylated by incubation with baculovirus-expressed CBP. GST pull-down assays were performed using GST-CBP551–682 or GST in the presence or absence of RbAp48 and acetylated versus underacetylated histones. Compared to the underacetylated core histones, the acetylated histones bound to the GST-CBP551–682-RbAp48 complex relatively poorly (Fig. 5a). Mock-acetylated histones (treated with CBP in the absence of AcCoA) showed the same binding pattern as the underacetylated histone proteins (data not shown). Similar experiments were performed using acetylated versus underacetylated mononucleosomes (Fig. 5b). These experiments showed that only underacetylated mononucleosomes interacted with the GST-CBP551–682-RbAp48 complex. Although the CREB binding motif of CBP did not interact with histones, the possibility exists that full-length CBP contains an additional domain that contacts histone particles directly. However, because GST-RbAp48 was able to directly bind to either core histones or mononucleosomes in an acetylation-sensitive manner (Fig. 5c), we think it more likely that the binding of histone particles to the CBP-RbAp48 complex occurs through RbAp48. Thus, the association of histones with the CBP-RbAp48 complex is dependent on their acetylation status, such that the acetylated histone particles fail to associate or readily dissociate from the CBP-RbAp48 complexes.
FIG. 5.
Blockage of histone binding to the CBP-RbAp48 complex by acetylation. Core histone octamers (a) and mononucleosomes (b) were acetylated by CBP. GST pull-down assays were performed using GST or GST-CBP551–682 in the presence or absence of RbAp48 with acetylated versus underacetylated histone particles. (c) Both hypoacetylated (underacetylated) and CBP-acetylated (acetylated) core histones or mononucleosomes were incubated with GST- or GST-RbAp48-coupled beads directly. Bound core histone octamers were assayed by Western blotting using an antibody specific for histone H4 (histones). Bound mononucleosomes were detected by two antibodies against either H3 or H4 (mononucleosomes).
RbAp46 facilitates p300-mediated transcription of a chromatinized template.
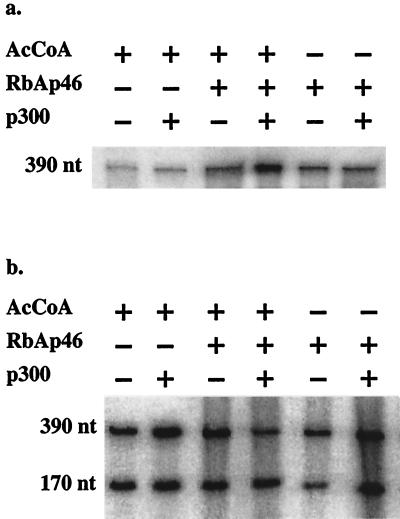
We used a reconstituted chromatin template to examine the functional contributions of RbAp46 to acetylation-dependent, p300-mediated transcription. Productive transcription assays (31) were performed on a chromatinized template that contained five Gal4 DNA-binding sites upstream from the adenovirus MLP and a 390-nt G-free cassette (26). A Gal4-VP16 fusion protein was used to remodel the chromatin templates. We assayed the effects of adding p300 and RbAp46 in the presence and absence of AcCoA. The addition of p300 in the presence or absence of AcCoA did not significantly stimulate transcription above the levels obtained with Gal-VP16 alone (Fig. 6a and data not shown). Supplementation of the transcription reaction mixtures with RbAp46 modestly increased the synthesis of the 390-nt product in the presence or absence of exogenous p300 (Fig. 6a). It should be noted, however, that the HeLa cell nuclear extract contains significant levels of both CBP and p300, so it is not possible to assay the contribution of RbAp46 in the complete absence of a coactivator. Unfortunately, stripping the extracts of CBP/p300 removes other factors required for transcription (54). A similar increase was detected in the presence of RbAp46 and AcCoA. The highest level of expression was seen in the presence of AcCoA, RbAp46, and p300 (Fig. 6a), which suggests that the activity of RbAp46 depends upon its ability to augment the CBP histone acetyltransferase function. This stimulation of transcription was also observed when RbAp48 was substituted for RbAp46 (data not shown).
FIG. 6.
Facilitation of p300-mediated transcription of a chromatinized template by RbAp46. (a) In vitro transcription assays performed using Gal-VP16-remodeled chromatin templates. The DNA template contains five Gal4 DNA binding sites upstream from the adenovirus MLP and a 390-nt G-free transcription cassette. Addition of AcCoA alone or p300 and AcCoA did not significantly enhance transcription. Addition of p300 and RbAp46 modestly activated transcription, while addition of p300, RbAp46, and AcCoA significantly increased the transcription levels of the 390-nt transcript. Similar results were obtained in four independent assays. (b) In vitro transcription assays using a naked DNA template. The DNA template contains five Gal4 DNA binding sites upstream from the adenovirus MLP and a 390-nt G-free transcription cassette. As an RNA recovery control, a DNA template encoding a 170-nt G-free transcription cassette placed downstream from the adenovirus MLP was used in the productive-transcription assays. When results were normalized with the MLP control, no significant increase in transcription was detected among the different manipulations. These results were obtained in three independent assays.
As a control, in vitro transcription assays were also performed using naked DNA templates. Neither 5× Gal4-MLP- nor MLP-driven transcription was significantly stimulated by the addition of AcCoA, RbAp46, and p300 (Fig. 6b). This contrast in the effects of RbAp46 in the presence of naked versus chromatinized templates supports the hypothesis that RbAp46 mediates the selective targeting of CBP acetyltransferase activity toward histones.
DISCUSSION
As a prototypical transcriptional coactivator, CBP and its homologue p300 have important roles in gene regulation, potentially integrating signals from diverse transcriptional pathways. Consequently, CBP and p300 have been implicated in a variety of cellular processes, including regulation of the cell cycle, differentiation, DNA repair, and apoptosis. Although there is evidence that CBP and p300 interact with components of the basal transcriptional machinery (8, 21, 29, 44), recent studies suggest that the major activities of these coactivators occur only in the presence of chromatin (19, 20).
Two models for this chromatin-dependent activity can be envisioned. First, the intrinsic histone acetylation activity of CBP (or the activities of associated histone acetyltransferases) may modify the amino-terminal tails of histone proteins in a manner that may lead to changes in nucleosome structure. These nucleosomal changes could result in chromatin decondensation, which may be required for the initial steps in transcriptional initiation. In support of this model, studies have correlated the histone acetylation activity of CBP with its ability to activate transcription (24). It should be noted, however, that no direct link between histone acetylation and any structural change in chromatin has been demonstrated. In a second model, CBP may interact with proteins that have the potential to mediate chromatin remodeling. While such interactions have been proposed (16), there is no evidence that these associations contribute to the chromatin-remodeling process. Although neither of these models has been proven, they have a theme in common, i.e., that chromatin-modifying proteins (histone acetyltransferases, chromatin remodeling factors) may be localized to specific promoters through the CBP and p300 coactivators.
The current report proposes that the nucleosome-modifying activities of a coactivator can, in fact, be augmented by transcription factor binding. We envisage that this occurs through the formation of new protein interaction surfaces. This idea is supported by nuclear magnetic resonance analysis of the phosphorylated CREB-CBP complex, which shows that structure is induced into both components of the complex upon their interaction (37). We had previously determined that an uncharacterized kinase in yeast is capable of recognizing Ser133 of CREB, the same site that is critical for the interaction with CBP. This allowed us to generate CREB-CBP complexes for use as bait to screen cDNA expression libraries. One of the strongest interactors identified in this screen was histone-binding factor RbAp48. Subsequent control studies were performed to determine whether both components of the bait were required for the RbAp48 interaction. These experiments indicated that while RbAp48 can bind to CBP alone, the association is much stronger in the presence of phosphorylated CREB. Thus, we conclude that RbAp48 binds preferentially to the complex of the activated transcription factor and its coactivator. This model is somewhat reminiscent of the transcription factor-mediated activation of the peroxisome proliferator-activated receptor γ coactivator PGC-1 recently reported by Puigserver et al. (34).
RbAp48 and its homologue RbAp46 have been characterized as components of four distinct complexes involved in nucleosome assembly or modification, the type B histone acetyltransferases, CAF-1, the HDACs, and NURF (13, 18, 25, 33, 46, 49, 51, 55). For example, the type B histone acetyltransferases have been shown to contain two subunits, the catalytic subunit Hat1p and the RbAp48-like factor Hat2p (33). Hat2p enhances Hat1p activity by increasing the affinity for histone H4 (33). Hat2p is structurally related to Cac3p, the small subunit of CAF-1 (18). In humans, this activity is provided by RbAp48 (51). In contrast, human Hat1 is associated with RbAp46 (50). Both RbAp46 and RbAp48 copurify with HDAC1 and HDAC2 (13, 46, 55). Finally, the ATP-dependent nucleosomal remodeling factor NURF contains a 55-kDa subunit (p55) that is highly related to both RbAp proteins (25). Although the NURF complex does not contain histone acetyltransferase activity, recombinant p55 associates with factors in Drosophila nuclear extracts that can acetylate histones H3 and H4 (25). Our studies indicate that the CREB-CBP complex associates with both RbAp48 and RbAp46.
Despite their association with various histone-modifying complexes, the functional roles of the RbAp48 and RbAp46 proteins remain unclear. For example, CAF-1 complexes lacking RbAp48 still can associate with histones while human Hat1 lacking RbAp46 cannot (50). It is perhaps more puzzling that the RbAp48/46 proteins appear to interact with a region of histone H4 (aa 31 to 40) predicted from the crystallographic structure of the nucleosome to be inaccessible for binding (50). The type B acetyltransferases and CAF-1 are involved in de novo chromatin assembly; thus, the participation of RbAp48 in histone recognition in these instances is easy to rationalize. The involvement of RbAp48 in transcriptional regulation is obviously more problematic because the target histone sites are embedded within chromatin. Nonetheless, our data indicate that RbAp48 bound to CBP can associate with histone proteins in the context of nucleosomes and the same is presumably true of the RbAp48 and RbAp46 proteins associated with the HDACs. Although we cannot conclusively exclude the possibility of a slow partial dissociation of the mononucleosomes in our sample preparation (12), the demonstration that RbAp48 stimulates both CBP-mediated acetylation of nucleosomes and transcription from chromatin templates supports our hypothesis that the nucleosome may be capable of adopting an alternative configuration that is permissive for RbAp48 binding.
The Vmax and Km values for CBP enzymatic activity were not markedly different from those measured for other histone acetyltransferases. The Vmax for CBP was slightly higher than that for GCN5, for example (s−1 = 0.3 versus 0.08), possibly due to the fact that the GCN5 fragment examined represented only the catalytic domain (45). In the absence of RbAp48, the Km values of GCN5 and CBP were very similar (28 versus 16 μM). Addition of RbAp48 lowered the Km of CBP for the histone substrate to about 6 μM. Somewhat unexpected was the finding that RbAp48 bound to CBP recognized underacetylated but not acetylated histones. Although it has not been tested explicitly, the RbAp48 and RbAp46 proteins in the HDACs would be expected to recognize predominantly acetylated histones. It is possible that the binding specificities of RbAp48 and RbAp46 depend upon whether they are associated with histone acetylases or deacetylases.
The ability of RbAp48 to augment CBP histone acetyltransferase function appears to be reflected by an increase in transcriptional activity. Using in vitro transcription assays, we were able to demonstrate that transcription from a reconstituted chromatin template was stimulated upon addition of p300, RbAp46, and AcCoA. This enhancement is specific for chromatin templates, suggesting that RbAp48 and CBP act in concert to alter the acetylation state of nucleosomal histones. These assays utilized a Gal-VP16 activator to remodel the chromatin template. VP16 binds to CBP and p300 (52), and p300 activation of Gal-VP16 has been observed by other investigators (20). Addition of p300 and AcCoA alone did not activate transcription above the basal levels achieved by Gal-VP16 in our assays, and addition of p300 and RbAp46 only modestly stimulated transcription. The requirement for all three components implicates CBP's histone acetylation function in transcriptional activation. The acetyltransferase activity of CBP is not restricted to histones. It is possible that RbAp48 targets the acetylation function of CBP to histones, as opposed to other targets (11, 15).
Finally, it remains to be determined whether the ability of phosphorylated CREB to augment RbAp48 binding to CBP is shared by other transcriptional activators. Many additional transcription factors interact with the CREB binding domain of CBP, some dependent upon phosphorylation and some in a phosphorylation-independent manner. Three such phosphorylation-independent factors, c-myb, SREBP, and Drosophila cubitus interruptus, have been proposed to interact with CBP through an alpha-helical interface related to that of phosphorylated CREB in the CREB-CBP complex (4, 32), suggesting that similar protein interaction surfaces can be generated. Whether complexes containing these other transcription factors also bind RbAp48 is unknown.
ACKNOWLEDGMENTS
Q. Zhang and N. Vo contributed equally to this study.
We thank Phyllis Goldman for construction of yeast bait constructs and help with the three-hybrid assay; Stan Hollenberg for the VP16 cDNA library; Danny Reinberg, Alejandra Loyola, and Gary LeRoy for instructions on chromatin assembly and in vitro transcription; and John Denu, W. Lee Kraus, Roland Kwok, Richard Mauer, Marc Monestier, Bruce Stillman, and Michael Wigler for reagents.
This work was supported by grants from the NIH.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 12.1.1–12.1.9. [Google Scholar]
- 2.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Brotherton T W, Covault J, Shires A, Chalkley R. Only a small fraction of avian erythrocyte histone is involved in ongoing acetylation. Nucleic Acids Res. 1981;9:5061–5073. doi: 10.1093/nar/9.19.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardinaux J-R, Notis J C, Zhang Q, Vo N, Craig J C, Fass D M, Brennan R G, Goodman R H. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol Cell Biol. 2000;20:1546–1552. doi: 10.1128/mcb.20.5.1546-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 6.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 7.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 8.Felzien L K, Farrell S, Betts J C, Mosavin R, Nabel G J. Specificity of cyclin E-Cdk2, TFIIB, and E1A interactions with a common domain of the p300 coactivator. Mol Cell Biol. 1999;19:4241–4246. doi: 10.1128/mcb.19.6.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng H P, Scherl D S, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–7831. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 10.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 12.Hansen J C, Ausio J, Stanik V H, van Holde K E. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 13.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 14.Hill J, Donald K A, Griffiths D E, Donald G. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 16.Johnston H, Kneer J, Chackalaparampil I, Yaciuk P, Chrivia J. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J Biol Chem. 1999;274:16370–16376. doi: 10.1074/jbc.274.23.16370. [DOI] [PubMed] [Google Scholar]
- 17.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 19.Kraus W L, Kadonaga J T. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraus W L, Manning E T, Kadonaga J T. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 22.Laurance M E, Kwok R P, Huang M S, Richards J P, Lundblad J R, Goodman R H. Differential activation of viral and cellular promoters by human T-cell lymphotropic virus-1 tax and cAMP-responsive element modulator isoforms. J Biol Chem. 1997;272:2646–2651. doi: 10.1074/jbc.272.5.2646. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, Imhof A, Collingwood T N, Urnov F D, Wolffe A P. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 1999;18:5634–5652. doi: 10.1093/emboj/18.20.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Balbas M A, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci USA. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 27.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 28.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 30.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 31.Orphanides G, LeRoy G, Chang C H, Luse D S, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 32.Parker D, Rivera M, Zor T, Henrion-Caude A, Radhakrishnan I, Kumar A, Shapiro L H, Wright P E, Montminy M, Brindle P K. Role of secondary structure in discrimination between constitutive and inducible activators. Mol Cell Biol. 1999;19:5601–5607. doi: 10.1128/mcb.19.8.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parthun M R, Widom J, Gottschling D E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 34.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman B M. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 35.Qian Y W, Wang Y C, Hollingsworth R E, Jr, Jones D, Ling N, Lee E Y. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 36.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 37.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 38.Richards J P, Bachinger H P, Goodman R H, Brennan R G. Analysis of the structural properties of cAMP-responsive element-binding protein (CREB) and phosphorylated CREB. J Biol Chem. 1996;271:13716–13723. doi: 10.1074/jbc.271.23.13716. [DOI] [PubMed] [Google Scholar]
- 39.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 40.Shaywitz A J, Greenberg M E. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 41.Shih H M, Goldman P S, DeMaggio A J, Hollenberg S M, Goodman R H, Hoekstra M F. A positive genetic selection for disrupting protein-protein interactions: identification of CREB mutations that prevent association with the coactivator CBP. Proc Natl Acad Sci USA. 1996;93:13896–13901. doi: 10.1073/pnas.93.24.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shikama M, Lyon J, Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 43.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 44.Swope D L, Mueller C L, Chrivia J C. CREB-binding protein activates transcription through multiple domains. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 45.Tanner K G, Trievel R C, Kuo M H, Howard R M, Berger S L, Allis C D, Marmorstein R, Denu J M. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 46.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 47.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 48.Tse C, Georgieva E I, Ruiz-Garcia A B, Sendra R, Hansen J C. Gcn5p, a transcription-related histone acetyltransferase, acetylates nucleosomes and folded nucleosomal arrays in the absence of other protein subunits. J Biol Chem. 1998;273:32388–32392. doi: 10.1074/jbc.273.49.32388. [DOI] [PubMed] [Google Scholar]
- 49.Tyler J K, Bulger M, Kamakaka R T, Kobayashi R, Kadonaga J T. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 51.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Grossman S R, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 54.Yie J, Senger K, Thanos D. Mechanism by which the IFN-beta enhanceosome activates transcription. Proc Natl Acad Sci USA. 1999;96:13108–13113. doi: 10.1073/pnas.96.23.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]