I. Introduction
ANNOTATION-EFFICIENT deep learning refers to methods and practices that yield high-performance deep learning models without the use of massive labeled training datasets. This paradigm has recently attracted attention from the medical imaging research community because (1) it is difficult to collect large, representative medical imaging datasets given the diversity of imaging protocols, imaging devices, and patient populations, (2) it is expensive to acquire accurate annotations from medical experts even for moderately-sized medical imaging datasets, and (3) it is infeasible to adapt data-hungry deep learning models to detect and diagnose rare diseases whose low prevalence hinders data collection.
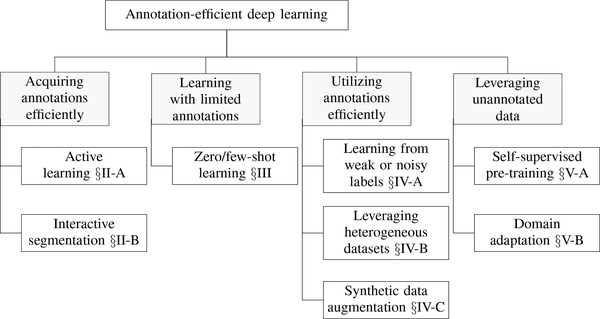
The challenge of annotation-efficient deep learning has been approached from various angles in the medical imaging literature, but the relevant publications are scattered across numerous sources and there exist many gaps that require further research. This special issue addresses these deficiencies by presenting a collection of state-of-the-art research articles spanning the major topics of annotation-efficient deep learning, which are diagrammed in Figure 1.
Fig. 1.
Overview of the topics covered in this special issue on annotation-efficient deep learning.
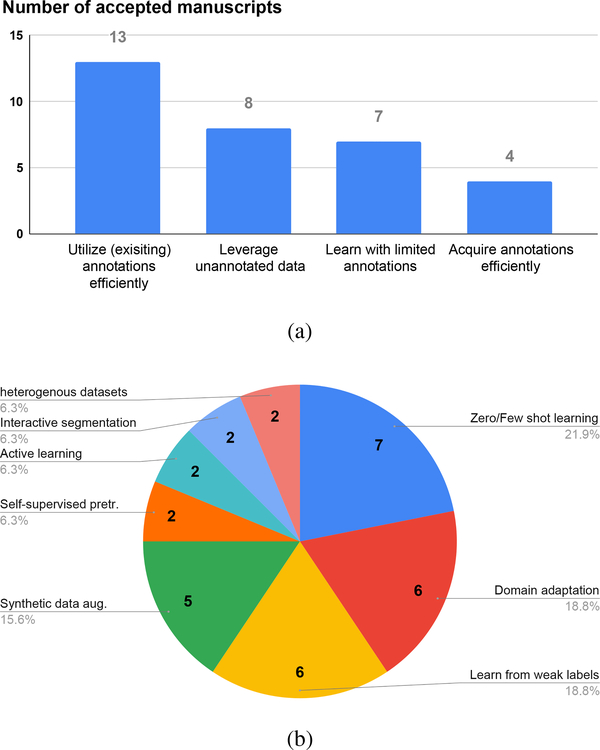
The call for papers attracted significant interest from the medical imaging community. A total of 101 manuscripts were submitted and 32 were finally accepted for publication. The submissions were subjected to a rigorous review process, in which each manuscript was refereed by 3 to 6 experts in the field and underwent typically two rounds of revision. Figure 2 graphs the number of accepted manuscripts related to each topic and its sub-topics. Most of the articles fall in the categories of leveraging unannotated data and utilizing annotations efficiently. The most popular sub-topics include zero/few-shot learning, domain adaptation, learning from weak and noisy labels, and synthetic data augmentation. In the remainder of this editorial, we will first categorize and summarize the articles and then highlight potential opportunities for future research.
Fig. 2.
Number of articles related to each of the four major topics (a) and all sub-topics (b).
II. Acquiring Annotations Efficiently
Active learning and interactive segmentation are two common practices for acquiring annotations in a budget-efficient manner. The former determines which data samples should be annotated, while the latter shortens the annotation session. The two approaches are complementary, and they enable substantial reductions in annotation time and cost.
A. Active Learning
Active learning aims to select the most informative and representative samples for experts to annotate, thereby minimizing the total number of samples that must be annotated to train performant models. The effectiveness of an active learning method largely depends on its criteria for determining the informativeness and representativeness of an unlabeled sample. The more uncertain the sample’s prediction is, the more information its label offers. Minimizing the number of annotated samples requires that the labeled samples be distinct from one another. Therefore, uncertainty and diversity are two natural metrics for informativeness and representativeness [1], [2], upon which the two articles featured in this special issue present two methods. Nath et al. [3] argue that duplicating uncertain samples in the labeled training dataset helps decrease the overall model uncertainty, and they regulate the amount of duplication based on mutual information between data pairs of the training and unlabeled pool. Mahapatra et al. [4] propose a self-supervised method for training a classifier to select informative samples based on saliency maps that embody both uncertainty and diversity.
B. Interactive Segmentation
Smart interactive segmentation tools play an essential role in reducing the manual burden of producing high-quality annotated data. In this special issue, two teams have made advances in this topic. Ma et al. [5] explore the use of boundary-aware multiagent reinforcement learning to interactively and iteratively refine 3D image segmentations. The approach is able to combine different types of user interactions, such as point clicks and scribbles, via an efficient “super-voxel-clicking” design. Feng et al. [6] investigate few-shot learning to arrive at better medical image segmentation using only limited supervision. The approach has the potential to reduce the annotation burden and fix some common issues in few-shot learning methods by enabling the user to make minor corrections interactively.
III. Learning With Limited Annotations
The ability to analyze medical images with little or no training data is highly desirable. For example, Huang et al. [7] present an unsupervised deep learning method for multicontrast MR image registration, using a coarse-to-fine network architecture consisting of affine coarse and deformable fine transformations to achieve end-to-end registration. For the task of semantic segmentation, compiling large-scale (non-synthetic) medical image datasets with pixel-level annotations is time-consuming and often prohibitively expensive, and it may be impossible to balance all the relevant classes in the training set. Although semi-supervised approaches aim to relax the level of supervision to bounding boxes and image-level tags, these models still require copious training samples and are prone to sub-optimal performance on unseen classes.
A. Zero/Few-Shot Learning
By contrast, the few-shot learning paradigm attempts to utilize a few annotated “support” samples to learn representations of unseen classes, denoted “query” samples. The few-shot learning paradigm was initially focused on classification and later applied to segmentation. In the context of volumetric ultrasound segmentation, Chanti et al. [8] introduce a decremental update of the objective function to guide model convergence in the absence of copious annotated data. Their update strategy switches from weakly supervised training to a few-shot setting. Also in the context of volumetric segmentation, in this case of the heart, Wang et al. [9] propose a few-shot learning framework that combines ideas from semi-supervised learning and self-training. The key to their success lies in cascaded learning; specifically, in the selection and evolution of high-quality pseudo labels produced by an auto-encoder network that learns shape priors. Feng et al. [6] introduce interactive learning into the few shot learning strategy for image segmentation. In their approach, an expert indicates error regions as an error mask that corrects the predicted segmentation mask from which the network learns to produce better segmentations. Paul et al. [10] propose a zero-shot learning strategy for the diagnosis of chest radiographs, leveraging multi-view semantic embedding and incorporating self-training to deal with noisy labels. Cui et al. [11] propose a unified framework for generalized low-shot medical image segmentation that deals both with data scarcity and lack of annotations, which is the case for rare diseases. Using distance metric learning, their model learns a multimodal mixture representation for each category and effectively avoids overfitting the extremely limited data.
Finally, a recent trend in image segmentation is combining traditional model-based contour segmentation methods with deep learning methods to accrue the benefits of both approaches [12]. Accordingly, Lu et al. [13] formulate anatomy segmentation as a contour evolution process, using Active Contour Models whose evolution is governed by graph convolutional networks. The model requires only one labeled image exemplar and supports human-in-the loop editing. Their one-shot learning scheme leverages additional unlabeled data through loss functions that measure the global shape and appearance consistency of the contours.
IV. Utilizing Annotations Efficiently
Acquiring additional strongly-annotated data is arguably the best approach to improving deep learning models, but this practice may not always be feasible due to limited budgets or shortages of medical expertise. Consequently, one may resort to using annotations that are cheaper or faster to obtain. The resulting weak and noisy annotations require proper learning methodologies (Section IV-A). Another approach to this challenge is to leverage related annotated datasets, which can increase the effective size of training sets (Section IV-B). Synthetic data augmentation (Section IV-C) is yet another popular approach by which the training sets are amplified by creating artificial examples and corresponding annotations.
A. Learning From Weak or Noisy Labels
Enabling learning from weak or noisy labels can be especially useful for medical imaging as the collection of quality labels is time-consuming, cumbersome, and often requires expert knowledge. This was one of the most popular topics of this special issue, probably due to the availability of weak or noisy labels that are routinely recorded during clinical workflows, such as measurements taken by clinicians (e.g., RECIST) or image-level labels provided in clinical reporting. Authors have addressed the topic by exploiting both “weak” labels (e.g., image-level labels [14] instead of pixel/voxel-wise labels) or “noisy” labels transferred from other modalities, and demonstrated promise by effectively combining supervision from such weak or noisy signals to train medical imaging models.
The research direction most investigated in this special issue is the utilization of weak image-level labels. For example, Liu et al. [15] employ a self-supervised attention mechanism to learn from image-level labels for pediatric bone age assessment. Quyang et al. [16] exploit weak supervision from image-level labels to train abnormality localization models for chest X-ray diagnosis. Tardy and Mateus [17] use image-level labels from mammography to detect abnormalities in a weakly self-supervised fashion. Wang et. al [18] combine weakly-supervised and semi-supervised learning to train models from both image-level and densely-labeled images. Both attention guidance and multiple-instance learning are utilized to effectively learn from both types of annotated data for the purposes of adenocarcinoma prediction in abdominal CT. Another interesting research direction is presented by Zhao and Yin [19] who explore the use of point annotations for weakly supervised cell segmentation in microscopy images. Using one point per cell, they train segmentation models that perform comparably to models trained on dense annotation maps. The utilization of noisy labels is explored by Ding et al. [20], who use weak-supervision for automated vessel detection in ultra wide-field fundus images. They exploit a deep neural network pre-trained on a different imaging modality, fluorescein angiography, together with multi-model registration to iteratively train their model and simultaneously refine the registration.
B. Leveraging Heterogeneous Datasets
Another promising approach to mitigating annotation costs in medical imaging is to leverage multiple heterogeneous datasets. These datasets might have been acquired for other purposes, but they can be combined to build more robust models. Two articles explore this approach. Yan et al. [21] train a lesion detection ensemble from multiple heterogeneous lesion datasets in a multi-task approach (their test dataset, including manually annotated 3D lesions in CT, has been publicly released). The ensemble is utilized for proposal fusion and missing annotations are mined from partially labeled datasets to improve the overall detection sensitivity. Li et al. [22] exploit intra-domain and inter-domain knowledge to improve cardiac segmentation models. They evaluate their method on a multi-modality dataset, demonstrating improvements over previous semi-supervised and domain adaptation methods. Both articles are promising with regard to the future development of AI models because it is natural to combine information about human anatomy acquired for different purposes. These approaches may be able to leverage features across datasets in order to build more powerful and robust models, as well as to reduce drastically the cost of curating task-specific datasets.
C. Synthetic Data Augmentation
Synthetic data augmentation aims to generate artificial yet realistic-looking images, thereby enabling the construction of large and diverse datasets. This practice is particularly desirable for organ segmentation where acquiring large labeled datasets is expensive and time-consuming and, even more importantly, for lesion segmentation where data collection is hindered by the low prevalences of underlying diseases and conditions. This special issue features five articles demonstrating that the addition of synthetic data to real data either improves segmentation performance or enables a comparable level of performance while reducing the need for real annotated data. These articles cover various medical imaging modalities, including MR scans, CT scans, ultrasound images, microscopic images, and retinal images. The proposed methods differ in the training data required to synthesize images and their corresponding segmentation masks. Relying on variants of the CycleGAN, three articles exploit image synthesis for the task of image segmentation, but take different measures to circumvent the use of segmentation masks from the target domain. Specifically, Gilbert et al. [23] use a pre-existing 3D shape model of the heart, Wang et al. [24] require segmentation masks from a domain only related to the target domain, and Yao et al. [25] rely on a handcrafted model to generate synthetic COVID lesions. The other two articles do require segmentation masks from the desired domain in order to generate synthetic data. In particular, Liu et al. [26] generate synthetic segmentation masks through an active appearance model, which is trained using segmentation masks from the target domain, and Guo et al. [27] rely on brain atlases, which makes their method suitable for applications where high-quality atlases are available for training. Despite the promising performances of the aforementioned methods, the requirement for atlases or shape and appearance models of the target diseases or organs may limit applicability.
V. Leveraging Unannotated DATA
Unannotated images contain a wealth of knowledge that can be leveraged in various settings, such as self-supervised learning (Section V-A) and unsupervised domain adaptation (Section V-B). The former utilizes unannotated data to pre-train the model weights, whereas the latter leverages unannotated data from a target domain to mitigate the distribution shifts between the training and test datasets.
A. Self-Supervised Learning
Self-supervised learning has recently gained prominence for its capacity to learn generalizable and transferable representations without the use of any expert annotation. The idea is to pretrain models on pretext tasks (e.g., rotation [28], inpainting [29], and contrasting [30]), where supervisory signals are automatically derived directly from the unlabeled data themselves (avoiding expert annotation costs), and then fine-tune (transfer) the pretrained models in a supervised manner so as to achieve annotation efficiency for the target tasks (e.g., segmenting organs and localizing diseases). Self-supervised learning often leverages the underlying structures and intrinsic properties of the unlabeled data, a feature that is particularly attractive in medical imaging. To take advantage of the special property of pathology, Koohbanani et al. [31] design pathology-specific pretext tasks and demonstrate annotation efficiency for the target domain in the small data regime. To exploit the semantics of anatomical patterns embedded in medical images, Haghighi et al. [32] pretrain their model using chest CT and demonstrate its across-domain capability to contrast-enhanced CT and MRI. They further show that, as an add-on strategy, their method complements existing self-supervised methods (e.g., inpainting [29], context restoration [33], rotation [28], and models genesis [34], [35]), thus boosting performance.
B. Domain Adaptation
Deep learning models struggle to generalize when the target domain exhibits a data distribution shift with respect to the source dataset used for training. This challenge is even more pronounced in the field of medical imaging where variations in ethnicities, scanner devices, and imaging protocols lead to large data distribution shifts. Unsupervised domain adaption has emerged as an effective approach to improving the tolerance of deep learning models to the distribution shifts in medical imaging datasets. The special issue features six articles on this topic. Mobiny et al. [36] tackle the distribution shift through an episodic training strategy where the training data of each episode are generated so as to mimic a distribution shift with respect to the original training dataset. The other five articles improve the CycleGAN in domain adaptation. Tomczak et al. [37] include auxiliary segmentation and reconstruction tasks. Ju et al. [38] exercise consistency regularization during training. Tomar et al. [39] introduce a self-attentive spatial normalization block in the generator networks. Koehler et al. [40] use task-specific probabilities to focus the attention of the transformation module on more relevant regions. In these articles, domain adaptation is leveraged for various applications, including reorienting MR images, staining unstained white blood cell images, cross-modality heart chamber segmentation between unpaired MR and CT scans, nucleus detection in cross-modality microscopy images, lung nodule detection in cross-protocol CT datasets, and various medical vision applications in cross-domain retinal imaging.
VI. Research Opportunities
It was inspiring to see the numerous submissions to this special issue and the quality of the accepted manuscripts. However, there remain unanswered questions and unaddressed issues that offer many enticing research opportunities.
a). Quantifying annotation efficiency:
Annotations generally refer to the ground truth information that is used in training, validating, and testing models. In medical imaging, this information is mostly provided (subjectively) by experts, but it may also derive from objective conditions obtained through tests (e.g., the malignancy of a tumor) and from medical concepts (diseases and conditions) automatically extracted from clinical notes and diagnostic reports. In a broader sense in self-supervised learning, it could also be a part of the data that are to be predicted based on other parts of the data or the original data that are to be restored from their transformed versions [35]. Annotations can be acquired at the patient, image, and pixel levels. Therefore, they require different levels of effort/cost and offer different levels of semantics/power as supervisory signals in training models. With regard to annotation, Method A is said to be more efficient than Method B if, compared with Method B, Method A (1) achieves better performance with equal annotation cost, (2) offers equal performance with lower annotation cost, or (3) provides equal performance with equal annotation cost, but reduces the training time. Currently, the prevailing literature generally assumes the same annotation cost for each and every sample (e.g., patient, image, or lesion), but costs could vary dramatically from one sample to another. It is also important to understand the trade-off between annotation cost and supervisory power in different settings. For example, for proton therapy, a few lesions with carefully-delineated masks may have more supervisory power than many lesions coarsely bounded by boxes. There is a need for new concepts, algorithms, and tools to analyse annotation efficiency across different contexts.
b). Annotating patients efficiently:
This matter is essential to medical image analysis, and resolving it can greatly accelerate the development of deep learning models. The special issue offers only four articles on this topic, leaving it under-investigated. In particular, the two articles on active learning primarily focus on the tasks of image segmentation and classification, leaving other important tasks such as lesion detection largely unexplored. This application bias is not peculiar; in fact, it applies to the entire medical imaging literature as well. It could be due to the fact that finding an optimal set of samples that are informative and representative is inherently difficult [41], [42]. With regard to interactive segmentation, the issue features two articles with promising task-specific solutions. However, generic interactive segmentation tools remain difficult to build for medical imaging applications. The type of user interactions employed (points, scribbles, bounding boxes, polygons, etc.) are often based on the targeted anatomy, which tends to hinder generalization to new anatomical structures. Building more comprehensive datasets with which to train general-purpose interactive models on several target anatomies and/or imaging modalities may be a good way forward. It would be desirable to develop tools that integrate active learning with interactive segmentation, as active learning can select the most important samples for annotation while interactive segmentation can shorten the annotation session for each sample. Active learning also may be embedded within interactive segmentation to suggest which parts of the image should be segmented next, thereby further accelerating the annotation process. Such tools will prove to be not only valuable for clinical purposes but also indispensable for building massive, strongly-annotated datasets—essential infrastructure for research, without which the annotation efficiency of a method can hardly be quantitatively determined or benchmarked.
c). Learning by zero/few shots:
A majority of the articles contained in this special issue focus on zero- or few-shot learning for medical image classification and segmentation, reflecting the eagerness in the community to progress beyond data-intensive machine learning methods. The promise of this technology is especially appealing in the healthcare domain where the collection of large and varied annotated datasets is difficult and sometimes hindered by regulations, a problem that is aggravated when studying rare diseases for which the acquisition of training examples becomes even more difficult. In future research, it may be beneficial to consider the use of multi-modality information in designing few shot learning methods. One could force the embeddings learned from multiple modalities (e.g., X-rays, CT, MRI, and reports) to be matched in a shared space, thereby encouraging collaborative learning via joint- and cross-supervision. Furthermore, analogous to the saying that training to identify counterfeit currency begins with studying genuine money, an effective approach to zero- or few-shot learning for diagnosing diseases and abnormal conditions in medical imaging could begin with learning dense (normal) anatomical embeddings. Successful zero- and few-shot learning will bring us closer to human abilities, i.e., a clinician learning to identify/diagnose certain diseases by studying just a few textbook examples.
d). Synthesizing annotations:
Artificially generating realistic-looking images with associated ground truth information helps relieve laborious human annotation and facilitates the creation of large datasets. This is particularly attractive for image segmentation where acquiring carefully-delineated masks is tedious and time-consuming as well as for localization where collecting rare diseases and conditions is challenging. Hand-crafted or trained generative models have proven to be powerful in creating “realistic” images and videos. Nevertheless, for the purposes of medical imaging, special attention and care must be given to potential artifacts. Embedding physiological and anatomical knowledge as well as the physical principles of imaging modalities into the synthesis process may prove to be critical.
e). Innovating architectures:
Recent advances in network architecture design and search have proven successful in natural language processing and computer vision. However, it is not clear whether these new architectures are more annotation-efficient than their predecessors. For instance, Tan and Le [43] show that transformer architectures [44] as well as a new family of efficient models, called EfficientNet v2.0, benefit from a significantly larger version of ImageNet, which raises concerns about the annotation efficiency of new architectures. Further research is required to study the annotation efficiency of such models, particularly in the context of medical imaging. It would be intriguing to investigate if, by reducing the need for annotated data, annotation-efficient methods can engender new architectural advancements. For instance, Caron et al. [45] demonstrate that self-supervised training is effective in reducing the amount of annotated visual data required to train transformers. It is worth studying how such recent architectural advancements benefit from other annotation-efficient paradigms in the context of medical imaging. Although the medical imaging community tends to adopt deep architectures developed for computer vision, given the differences between medical and natural images, to maximize annotation utilization [46], [47], [48], it would be worth designing or automatically searching for architectures that exploit the particular opportunities of medical imaging for annotation efficiency [49].
f). Exploiting clinical information:
Medical concepts (diseases and conditions) extractable from clinical notes and reports may be capitalized as (weak) patient-level annotation [14]. Indeed, natural language processing (NLP) has already been utilized to harvest image-level annotations from diagnostic reports in order to build medical imaging datasets and used to develop machine learning models [50], but the vast resources of clinical notes and diagnostic reports that are available in electronic health records of hospital systems have yet to be fully mined and well utilized. Learning representations jointly from both images and texts (clinical notes and diagnostic reports) will likely be a very active area of research in the near future, although patient privacy and other regulatory constraints must be carefully considered [51].
g). Integrating data and annotation from multiple sources:
Datasets created at different institutions tend to be annotated differently even when addressing the same clinical issue. There remains a need for learning methods that can seamlessly integrate data and annotation from different sources. Federated learning (FL) has recently emerged as an effective solution for addressing the growing concerns about data privacy when integrating data and annotation from different providers. FL trains a model using data from various sites without breaching patient privacy and other regulations [52], thereby making more data available for AI model development. FL has yet to seamlessly handle the unavoidable heterogeneity of data across different sites. Semi-supervised [53] and unsupervised/self-supervised [54] approaches have already been successfully used in the context of FL. Nevertheless, it will be interesting to see how other annotation-efficient methods can be combined with FL as well as how FL and other methods that integrate data and annotation from multiple sources can relieve the annotation burden.
h). Mining common knowledge:
Currently, the dominant approach in deep learning—supervised learning—offers expert-level and sometimes even super-expert-level performance. Models trained via supervised learning have also demonstrated remarkable capacity in knowledge transfer across domains [55], [56], but at their core, they are trained to be “specialists” [57] on (target) tasks that can be annotated by experts. There are many diseases and conditions in medical imaging as well as common medical knowledge that can hardly be annotated even by willing experts. Self-supervised learning has proven to be promising in training models to be “generalists” on various pretext tasks so as to reduce expert annotation effort on target tasks. Typically, the semantics of expert-provided annotation is strong but narrow (task-specific), while that of machine-generated annotation in self-supervised learning is weak but general. Fundamental to annotation efficiency is learning common generalizable knowledge. Therefore, self-supervised learning will inevitably overtake supervised learning in extracting generalizable and transferable knowledge; i.e., self-supervised representation learning followed by (supervised) transfer learning is poised to become the most practical paradigm towards annotation efficiency. This is particularly true for medical imaging, because medical images harbor rich semantics about human anatomy, thus offering a unique opportunity for deep semantic representation learning. Yet harnessing the powerful semantics associated with medical images remains largely unexplored [58], [59]. Furthermore, medical images are often augmented by clinical notes and reports, making it even more attractive to learn generic semantic representations jointly from both images and reports via self-supervision.
i). Reusing knowledge in trained models:
Research and development in deep learning across academia and industry have resulted in numerous models trained on various datasets in supervised, self-supervised, unsupervised, or federated manners for diverse clinical objectives. These trained models retain a large body of knowledge, and properly reusing this knowledge could reduce annotation efforts and accelerate training cycles, thereby increasing annotation efficiency. However, current practice in reusing knowledge from (existing) pretrained models for new tasks is very limited. Therefore, advanced methods are needed for transferring, reusing, and distilling the knowledge [60] from pretrained models as well as integrating the knowledge from multiple pretrained models with the same or distinct architectures.
j). Demonstrating annotation efficiency in practice:
Methods and techniques are being developed from various perspectives to circumvent the annotation dearth in medical imaging; their value and effectiveness needs to be quantitatively evaluated and benchmarked. This calls for an infrastructure of massive, strongly-annotated datasets in well-defined domains (e.g., pulmonary embolism [61], [62], colon cancer [63], [64], and cardiovascular disease [65], [66]), without which the annotation efficiency of a method cannot be adequately understood. As a result, we expect large-scale competitions and challenges to be organized. It would be encouraging to see more reports that annotation-efficient methods and practices have been employed in the design and development of commercial products.
There is no doubt that deep learning has dramatically transformed medical imaging; annotation-efficient deep learning remains the Holy Grail of medical imaging.
Acknowledgments
We would like to thank all the authors who submitted manuscripts to this special issue. We greatly appreciate the valuable efforts of the many expert reviewers who contributed to the selection of articles that have shaped the issue. We would also like to thank Fatemeh Haghighi, Jeffrey Liang, Julia Liang, and Zongwei Zhou for their valuable comments on earlier drafts. We are grateful to the Editor-in-Chief, Professor Leslie Ying, for approving our proposal to edit this special issue and for her guidance along the way. Our special thanks go to the Managing Editor, Professor Rutao Yao, for his timely and helpful support.
This work was funded in part by the National Institutes of Health (NIH) under Award Number R01HL128785.
Contributor Information
Nima Tajbakhsh, VoxelCloud, Inc., Los Angeles, USA.
Holger Roth, NVIDIA, Inc., Bethesda, MD, USA.
Demetri Terzopoulos, University of California, Los Angeles, and VoxelCloud, Inc., Los Angeles, CA, USA.
Jianming Liang, Arizona State University, Scottsdale, AZ, USA.
References
- [1].Zhou Z, Shin JY, Gurudu SR, Gotway MB, and Liang J, “Active, continual fine tuning of convolutional neural networks for reducing annotation efforts,” Medical Image Analysis, vol. 71, p. 101997, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhou Z, Shin J, Feng R, Hurst RT, Kendall CB, and Liang J, “Integrating active learning and transfer learning for carotid intima-media thickness video interpretation,” Journal of Digital Imaging, vol. 32, no. 2, pp. 290–299, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nath V, Yang D, Landman BA, Xu D, and Roth HR, “Diminishing uncertainty within the training pool: Active learning for medical image segmentation,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [4].Mahapatra D, Poellinger A, Shao L, and Reyes M, “Interpretability-driven sample selection using self supervised learning for disease classification and segmentation,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [5].Ma C, Xu Q, Wang X, Jin B, Zhang X, Wang Y, and Zhang Y, “Boundary-aware supervoxel-level iteratively refined interactive 3D image segmentation with multi-agent reinforcement learning,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [6].Feng R, Zheng X, Gao T, Chen J, Wang W, Chen DZ, and Wu J, “Interactive few-shot learning: Limited supervision, better medical image segmentation,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [7].Huang W, Yang H, Liu X, Li C, Zhang I, Wang R, Zheng H, and Wang S, “A coarse-to-fine deformable transformation framework for unsupervised multi-contrast MR image registration with dual consistency constraint,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [8].Al Chanti D, Duque VG, Crouzier M, Nordez A, Lacourpaille L, and Mateus D, “IFSS-Net: Interactive few-shot siamese network for faster muscle segmentation and propagation in volumetric ultrasound,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [9].Wang W, Xia Q, Hu Z, Yan Z, Li Z, Wu Y, Huang N, Gao Y, Metaxas D, and Zhang S, “Few-shot learning by a cascaded framework with shape-constrained pseudo label assessment for whole heart segmentation,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [10].Paul A, Shen TC, Lee S, Balachandar N, Peng Y, Lu Z, and Summers RM, “Generalized zero-shot chest X-ray diagnosis through trait-guided multi-view semantic embedding with self-training,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cui H, Wei D, Ma K, Gu S, and Zheng Y, “A unified framework for generalized low-shot medical image segmentation with scarce data,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [12].Minaee S, Boykov YY, Porikli F, Plaza AJ, Kehtarnavaz N, and Terzopoulos D, “Image segmentation using deep learning: A survey,” IEEE Transactions on Pattern Analysis and Machine Intelligence, pp. 1–20, 2021. [DOI] [PubMed] [Google Scholar]
- [13].Lu Y, Zheng K, Li W, Wang Y, Harrison AP, Lin C, Wang S, Xiao J, Lu L, Kuo C-F et al. , “Contour transformer network for one-shot segmentation of anatomical structures,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [14].Rahman Siddiquee MM, Zhou Z, Tajbakhsh N, Feng R, Gotway MB, Bengio Y, and Liang J, “Learning fixed points in generative adversarial networks: From image-to-image translation to disease detection and localization,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 2019, pp. 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu C, Xie H, and Zhang Y, “Self-supervised attention mechanism for pediatric bone age assessment with efficient weak annotation,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [16].Ouyang X, Karanam S, Wu Z, Chen T, Huo J, Zhou XS, Wang Q, and Cheng J-Z, “Learning hierarchical attention for weakly-supervised chest X-ray abnormality localization and diagnosis,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [17].Tardy M and Mateus D, “Looking for abnormalities in mammograms with self-and weakly supervised reconstruction,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [18].Wang Y, Tang P, Zhou Y, Shen W, Fishman EK, and Yuille AL, “Learning inductive attention guidance for partially supervised pancreatic ductal adenocarcinoma prediction,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [19].Zhao T and Yin Z, “Weakly supervised cell segmentation by point annotation,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [20].Ding L, Kuriyan AE, Ramchandran RS, Wykoff CC, and Sharma G, “Weakly-supervised vessel detection in ultra-widefield fundus photography via iterative multi-modal registration and learning,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yan K, Cai J, Zheng Y, Harrison AP, Jin D, Tang Y.-b., Tang Y-X, Huang L, Xiao J, and Lu L, “Learning from multiple datasets with heterogeneous and partial labels for universal lesion detection in CT,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [22].Li K, Wang S, Yu L, and Heng P-A, “Dual-Teacher++: Exploiting intra-domain and inter-domain knowledge with reliable transfer for cardiac segmentation,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [23].Gilbert A, Marciniak M, Rodero C, Lamata P, Samset E, and McLeod K, “Generating synthetic labeled data from existing anatomical models: An example with echocardiography segmentation,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang L, Guo D, Wang G, and Zhang S, “Annotation-efficient learning for medical image segmentation based on noisy pseudo labels and adversarial learning,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [25].Yao Q, Xiao L, Liu P, and Zhou SK, “Label-free segmentation of COVID-19 lesions in lung CT,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu J, Shen C, Aguilera N, Cukras C, Hufnagel RB, Zein WM, Liu T, and Tam J, “Active cell appearance model induced generative adversarial networks for annotation-efficient cell segmentation and identification on adaptive optics retinal images,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Guo P, Wang P, Yasarla R, Zhou J, Patel VM, and Jiang S, “Anatomic and molecular MR image synthesis using confidence guided cnns,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gidaris S, Singh P, and Komodakis N, “Unsupervised representation learning by predicting image rotations,” arXiv preprint arXiv:1803.07728, 2018. [Google Scholar]
- [29].Pathak D, Krahenbuhl P, Donahue J, Darrell T, and Efros AA, “Context encoders: Feature learning by inpainting,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 2536–2544. [Google Scholar]
- [30].Chen T, Kornblith S, Norouzi M, and Hinton G, “A simple framework for contrastive learning of visual representations,” arXiv preprint arXiv:2002.05709, 2020. [Google Scholar]
- [31].Koohbanani NA, Unnikrishnan B, Khurram SA, Krishnaswamy P, and Rajpoot N, “Self-Path: Self-supervision for classification of pathology images with limited annotations,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [32].Haghighi F, Taher MRH, Zhou Z, Gotway MB, and Liang J, “Transferable visual words: Exploiting the semantics of anatomical patterns for self-supervised learning,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen L, Bentley P, Mori K, Misawa K, Fujiwara M, and Rueckert D, “Self-supervised learning for medical image analysis using image context restoration,” Medical Image Analysis, vol. 58, p. 101539, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhou Z, Sodha V, Siddiquee MMR, Feng R, Tajbakhsh N, Gotway MB, and Liang J, “Models genesis: Generic autodidactic models for 3D medical image analysis,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2019, pp. 384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhou Z, Sodha V, Pang J, Gotway MB, and Liang J, “Models genesis,” Medical Image Analysis, vol. 67, p. 101840, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mobiny A, Yuan P, Cicalese PA, Moulik SK, Garg N, Wu CC, Wong K, Wong ST, He TC, and Nguyen HV, “Memory-augmented capsule network for adaptable lung nodule classification,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [37].Tomczak A, Ilic S, Marquardt G, Engel T, Forster F, Navab N, and Albarqouni S, “Multi-task multi-domain learning for digital staining and classification of leukocytes,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [38].Ju L, Wang X, Zhao X, Bonnington P, Drummond T, and Ge Z, “Leveraging regular fundus images for training UWF fundus diagnosis models via adversarial learning and pseudo-labeling,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [39].Tomar D, Lortkipanidze M, Vray G, Bozorgtabar B, and Thiran J-P, “Self-attentive spatial adaptive normalization for cross-modality domain adaptation,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PubMed] [Google Scholar]
- [40].Koehler S, Hussain T, Blair Z, Huffaker T, Ritzmann F, Tandon A, Pickardt T, Sarikouch S, Latus H, Greil G et al. , “Unsupervised domain adaptation from axial to short-axis multi-slice cardiac MR images by incorporating pretrained task networks,” IEEE Transactions on Medical Imaging, vol. 40, no. 7, pp. ??–??, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chakraborty S, Balasubramanian V, Sun Q, Panchanathan S, and Ye J, “Active batch selection via convex relaxations with guaranteed solution bounds,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 37, no. 10, pp. 1945–1958, 2015. [DOI] [PubMed] [Google Scholar]
- [42].Zhou Z, Shin J, Zhang L, Gurudu S, Gotway M, and Liang J, “Fine-tuning convolutional neural networks for biomedical image analysis: Actively and incrementally,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 7340–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tan M and Le QV, “EfficientNetV2: Smaller models and faster training,” arXiv preprint arXiv:2104.00298, 2021. [Google Scholar]
- [44].Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S et al. , “An image is worth 16×16 words: Transformers for image recognition at scale,” arXiv preprint arXiv:2010.11929, 2020. [Google Scholar]
- [45].Caron M, Touvron H, Misra I, Jégou H, Mairal J, Bojanowski P, and Joulin A, “Emerging properties in self-supervised vision transformers,” arXiv preprint arXiv:2104.14294, 2021. [Google Scholar]
- [46].Zhou Z, “Towards annotation-efficient deep learning for computer-aided diagnosis,” Ph.D. dissertation, Arizona State University, 2021. [Google Scholar]
- [47].Zhou Z, Siddiquee MMR, Tajbakhsh N, and Liang J, “UNet++: A nested U-net architecture for medical image segmentation,” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer, 2018, pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].——, “UNet++: Redesigning skip connections to exploit multiscale features in image segmentation,” IEEE Transactions on Medical Imaging, vol. 39, no. 6, pp. 1856–1867, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Imran A-A-Z, “From fully-supervised, single-task to scarcely-supervised, multi-task deep learning for medical image analysis,” Ph.D. dissertation, University of California, Los Angeles, 2021. [Google Scholar]
- [50].Wang X, Peng Y, Lu L, Lu Z, Bagheri M, and Summers RM, “ChestX-Ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 2097–2106. [Google Scholar]
- [51].Topol EJ, “High-performance medicine: The convergence of human and artificial intelligence,” Nature Medicine, vol. 25, no. 1, pp. 44–56, 2019. [DOI] [PubMed] [Google Scholar]
- [52].Rieke N, Hancox J, Li W, Milletari F, Roth HR, Albarqouni S, Bakas S, Galtier MN, Landman BA, Maier-Hein K et al. , “The future of digital health with federated learning,” NPJ Digital Medicine, vol. 3, no. 1, pp. 1–7, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yang D, Xu Z, Li W, Myronenko A, Roth HR, Harmon S, Xu S, Turkbey B, Turkbey E, Wang X et al. , “Federated semi-supervised learning for COVID region segmentation in chest CT using multi-national data from china, italy, japan,” Medical Image Analysis, vol. 70, p. 101992, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].van Berlo B, Saeed A, and Ozcelebi T, “Towards federated unsupervised representation learning,” in Proceedings of the Third ACM International Workshop on Edge Systems, Analytics and Networking, 2020, pp. 31–36. [Google Scholar]
- [55].Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, and Liang J, “Convolutional neural networks for medical image analysis: Full training or fine tuning?” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1299–1312, 2016. [DOI] [PubMed] [Google Scholar]
- [56].Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, and Summers RM, “Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1285–1298, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].LeCun Y and Misra I, “Self-Supervised Learning: The Dark Matter of Intelligence.” [Online]. Available: https://ai.facebook.com/blog/self-supervised-learning-the-dark-matter-of-intelligence/
- [58].Haghighi F, Taher MRH, Zhou Z, Gotway MB, and Liang J, “Learning semantics-enriched representation via self-discovery, self-classification, and self-restoration,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2020, pp. 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Feng R, Zhou Z, Gotway MB, and Liang J, “Parts2Whole: Self-supervised contrastive learning via reconstruction,” in Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning. Springer, 2020, pp. 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hinton G, Vinyals O, and Dean J, “Distilling the knowledge in a neural network,” arXiv preprint arXiv:1503.02531, 2015. [Google Scholar]
- [61].Tajbakhsh N, Gotway MB, and Liang J, “Computer-aided pulmonary embolism detection using a novel vessel-aligned multi-planar image representation and convolutional neural networks,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2015, pp. 62–69. [Google Scholar]
- [62].Tajbakhsh N, Shin JY, Gotway MB, and Liang J, “Computer-aided detection and visualization of pulmonary embolism using a novel, compact, and discriminative image representation,” Medical image analysis, vol. 58, p. 101541, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tajbakhsh N, Gurudu SR, and Liang J, “A comprehensive computer-aided polyp detection system for colonoscopy videos,” in International Conference on Information Processing in Medical Imaging. Springer, 2015, pp. 327–338. [DOI] [PubMed] [Google Scholar]
- [64].——, “Automated polyp detection in colonoscopy videos using shape and context information,” IEEE transactions on medical imaging, vol. 35, no. 2, pp. 630–644, 2015. [DOI] [PubMed] [Google Scholar]
- [65].Shin J, Tajbakhsh N, Hurst RT, Kendall CB, and Liang J, “Automating carotid intima-media thickness video interpretation with convolutional neural networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 2526–2535. [Google Scholar]
- [66].Tajbakhsh N, Shin JY, Hurst RT, Kendall CB, and Liang J, “Automatic interpretation of carotid intima–media thickness videos using convolutional neural networks,” in Deep Learning for Medical Image Analysis. Elsevier, 2017, pp. 105–131. [Google Scholar]