Abstract
Cricket paralysis virus is a member of a group of insect picorna-like viruses. Cloning and sequencing of the single plus-strand RNA genome revealed the presence of two nonoverlapping open reading frames, ORF1 and ORF2, that encode the nonstructural and structural proteins, respectively. We show that each ORF is preceded by one internal ribosome entry site (IRES). The intergenic IRES is located 6,024 nucleotides from the 5′ end of the viral RNA and is more active than the IRES located at the 5′ end of the RNA, providing a mechanistic explanation for the increased abundance of structural proteins relative to nonstructural proteins in infected cells. Mutational analysis of this intergenic-region IRES revealed that ORF2 begins with a noncognate CCU triplet. Complementarity of this CCU triplet with sequences in the IRES is important for IRES function, pointing to an involvement of RNA-RNA interactions in translation initiation. Thus, the cricket paralysis virus genome is an example of a naturally occurring, functionally dicistronic eukaryotic mRNA whose translation is controlled by two IRES elements located at the 5′ end and in the middle of the mRNA. This finding argues that eukaryotic mRNAs can express multiple proteins not only by polyprotein processing, reinitiation and frameshifting but also by using multiple IRES elements.
Cricket paralysis virus (CrPV) was isolated in 1970 by Carl Reinganum (47), who observed that laboratory colonies of Australian field crickets (Teleogryllus oceanicus and T. commodus) contained some early-instar nymphs which developed a paralysis of the hind legs, became uncoordinated, and died. Electron microscopic sections of paralyzed insects revealed many virus-like particles in crystalline arrays reminiscent of those observed in picornavirus-infected cells. Although originally isolated from crickets, CrPV has a wide host range, infecting insects which belong to Diptera, Lepidoptera, Orthoptera, and Heteroptera species (8). Importantly, it also replicates in cultured cells from various insect species including Drosophila SL2 cells (37, 55).
More recently, CrPV has been classified as a member of a group of insect picorna-like viruses which also includes Drosophila C virus (DCV) (24), Plautia stali intestine virus (PSIV) (53), himetobi P virus (HiPV) (40), and Rhopalosiphum padi virus (RhPV) (36). Insect picorna-like viruses share with mammalian picornaviruses many physical and morphological properties of the viral structural proteins (37, 56, 59), and the presence of a single plus-strand RNA genome with a genome-linked protein at the 5′ end (25) and polyadenosine residues at the 3′ end (9). In contrast to earlier reports (26), recent cloning and sequencing of several insect picorna-like viruses has revealed that the organization of these viral genomes differs from that of human picornaviruses (8, 28). Specifically, picornavirus RNA genomes consist of a single open reading frame (ORF) encoding a polyprotein which is posttranslationally processed to give rise to both structural (encoded in the N-terminal part of the polyprotein) and nonstructural viral proteins (49). In contrast, certain insect picorna-like virus genomes encode two distinct polyproteins. In these genomes, the polyprotein precursors to the nonstructural viral proteins are encoded by the upstream ORF (ORF1), while the structural protein precursors are encoded by the downstream ORF (ORF2) (24, 36, 40, 53). Furthermore, it has been noted that structural proteins accumulate in vast excess over nonstructural proteins in cells infected with such insect picorna-like viruses (37, 38). In contrast, cells infected with human picornaviruses produce equimolar amounts of structural and nonstructural proteins (49). The differential expression of the two insect virus polyproteins from a single mRNA has led to the suggestion that the two ORFs might be under independent translational control. By analogy to picornavirus polyproteins, which are known to be translated by an internal initiation mechanism, we speculated that both ORFs in picorna-like virus mRNAs might be translated by internal initiation. Consistent with this idea, ORF2 in the PSIV RNA genome is translated cap independently in the rabbit reticulocyte lysate (RRL) (52).
Here we show that the CrPV RNA genome encodes two large, nonoverlapping ORFs with viral nonstructural and structural polyprotein precursors encoded by the upstream and downstream ORFs, respectively. Each ORF is preceded by an internal ribosome entry site (IRES), demonstrating that the CrPV RNA genome is a naturally occurring eukaryotic RNA that is functionally dicistronic. Mutational analysis of the downstream IRES has revealed that the first three nucleotides in the CrPV downstream ORF are CCU, which differs from the canonical AUG start codon at all three positions. This finding, together with the fact that complementarity of the CCU codon with an upstream sequence in the IRES must be maintained to preserve IRES function, suggests an unusual mechanism of translation initiation mediated by the intragenic region (IGR) of CrPV.
MATERIALS AND METHODS
Viruses and cells.
A CrPV stock was originally obtained from Chris Hellen and Eckard Wimmer (State University of New York at Stony Brook, Stony Brook, N.Y.). By comparison to genomic sequences of several independent CrPV isolates kindly provided by Peter Christian (Commonwealth Scientific and Industrial Research Organisation, Canberra, Australia), the CrPV isolate used in this study was similar to one isolated from Telegryllus commodus, Victoria, Australia (23). Viruses were propagated in cultured Drosophila Schneider's line 2 (SL2) cells. For infection, the medium was removed and the cells were overlaid with 150 to 250 μl of virus suspension per flask of 6 × 106 cells. After 1 h, the cells were overlaid with 4.5 ml of Schneider's Drosophila medium containing 2% serum. The cells were harvested 48 h after infection by sedimentation, and virus was released by repeated cycles of freezing and thawing.
Cloning and sequencing of the CrPV genome.
Total RNA was prepared from mock- and CrPV-infected SL2 cells by using the guanidinium method (2). RNA was resuspended in TES solution (10 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1% sodium dodecyl sulfate [SDS]), precipitated twice with 0.3 M sodium acetate (pH 5.2)–ethanol, resuspended in water, and stored at −20°C.
First-strand cDNA, representing the 3′ end of the viral genome, was produced using an anchored oligo(dT) primer whose 3′ sequences were complementary to the published 3′-end sequence of CrPV (26). Reverse transcription was carried out in a reaction mixture containing 20 mM Tris-HCl (pH 8.3), 20 mM KCl, 6 mM MgCl2, 10 mM dithiothreitol, 0.5 mM each deoxynucleoside triphosphate (dNTP), 80 μg of actinomycin D per ml, 2 μg of RNA, 3 pmol of DNA primer, and either 0.1 U of avian myeloblastosis virus reverse transcriptase (Life Sciences Inc.) per μl or 9.5 U of Superscript reverse transcriptase (Gibco/BRL) per μl. One-eighth of each reverse transcription product was used for PCR amplification with Vent polymerase (New England Biolabs). The reaction mixtures contained 1× ThermoPol buffer (New England Biolabs), 0.2 mM each dNTP, and 20 μmol of primers selected based on published 3′-end sequences (26). The remainder of the viral genome was cloned and sequenced by sequential application of the 5′ rapid amplification of cDNA ends method (11) with reagents purchased from Gibco/BRL. Reverse transcription was carried out in reaction mixtures containing 20 mM Tris-Cl (pH 8.4), 50 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.4 mM each dNTP, 0.5 μg of RNA, 2.5 pmol of cDNA primer, and 8 U of Superscript II reverse transcriptase per μl. First-strand cDNA was purified on Glassmax spin columns (Gibco/BRL), and one-fifth of the total product was used for dC tailing. Tailing reaction mixtures contained 10 mM Tris-HCl (pH 8.4), 25 mM KCl, 1 mM MgCl2, 0.2 mM dCTP, and 0.4 U of terminal deoxynucleotidyltransferase per μl. One-fifth of the tailed product was used for PCR amplification in reaction mixtures containing 20 mM Tris-Cl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dNTP, 100 to 400 nM each primer, and 0.04 U of Taq DNA polymerase (Promega) per μl. Amplification reactions were performed under the following conditions: 94°C for 1 min, 50°C for 0.5 to 1 min, and 72°C for 2 to 3 min for 35 cycles, followed by 10 min at 72°C. Amplified DNA products were purified and inserted into pGEM-3 (Promega). Automated sequencing was performed by Macromolecular Resources (Fort Collins, Colo.) and by the PAN facility at Stanford University (Stanford, Calif.).
Cloning of the 5′NCR and IGR of CrPV.
The 5′ noncoding region (5′NCR) of CrPV was obtained by PCR using primers 5′AGAATTCTTTAATAAGTGTTGTGCAGATTAATCTGCACAGCACTAGCC3′, which includes nucleotides 1 to 41 of the viral genome, and 5′GCCATGGTCACATTGTAAGAATCGGTTACTCC3′, which is complementary to nucleotides 682 to 706 of the viral genome. For amplification of the IGR, primers 5′CGGAATTCAAAGCAAAAATGTGATCTTGCTT3′ (nucleotides 6025 to 6047 of the CrPV genome) and 5′CATGCCATGGTATCTTGAAATGTAGCAGGTAAA3′ (complementary to nucleotides 6210 to 6232) were used; these 5′ and 3′ primers contained EcoRI and NcoI restriction sites, respectively. PCR was performed using the Advantage cDNA PCR kit (Clontech), and PCR products were cloned directly into the pCR2.1 cloning vector (Invitrogen). Stop codons and nucleotide insertions were introduced into the IGR by PCR using altered 3′ primers carrying the desired mutations, listed in Table 1. The complete nucleotide sequences of all PCR-amplified fragments were confirmed by DNA sequencing.
TABLE 1.
Sequences of oligonucleotides used to introduce mutations into the CrPV IGR
| IGR mutant | Oligodeoxynucleotide sequencea |
|---|---|
| 1 | CATGCCATGGTATCTTGAAATGTAGCAGGTAATTATCTTAGGTTTTTCGA |
| 2 | CATGCCATGGTATCTTGAAATGTAGCAGGTTAATTTCTTAGGTT |
| 3 | CATGCCATGGTATCTTGAAATGTAGCTTATAAATTTCTTAGGT |
| 4 | CATGCCATGGTATCTTGAAATGTTTAAGGTAAATTTCTTAGGT |
| 5 | CATGCCATGGTATCTTGAAATTAAGCAGGTAAATTTCTTAGGT |
| 6 | CATGCCATGGTATCTTGTTATGTAGCAGGTAAATTTC |
| 7 | CATGCCATGGTATCTTAAAATGTAGCAGGTAAATTTCTTAG |
| 8 | CATGCCATGGTTTATTGAAATGTAGCAGGTAAATTTC |
| 9 | CATGCCATGGTATCTTGAAATGTTAGCAGGTAAATTTCTTAGGTT |
| 10 | CATGCCATGGTATCTTGAAATGTGTAGCAGGTAAATTTCTTAGGTT |
| 11 | CATGCCATGGTATCTTGAAATGTTGTAGCAGGTAAATTTCTTAGGT0T |
| 12 | CATGCCATGGTATCTTGAAATGTAGCATAGGTAAATTTCTTAGGTT |
| 13 | CATGCCATGGTATCTTGAAATGTTAGCATAGGTAAATTTCTTAGGTT |
| 14 | CATGCCATGGTATCTTGAAATGTAGCACCTAAATTTCTTAGGT |
| 15 | CATGCCATGGTATCTTGAAATGTAGCACCTAAATTTCTTAGGTTTTTCGACTAGGTAATCTGA |
Introduced mutations are highlighted in bold.
Construction of dicistronic luciferase reporter constructs.
The pCR2.1 vectors containing the desired CrPV fragments (see above) were digested with EcoRI and NcoI; the resulting EcoRI-NcoI fragments were subcloned into the intercistronic region of a dicistronic luciferase reporter construct, described previously (22). For transcription in vitro, dicistronic luciferase constructs were digested with BamHI and linear DNA was transcribed with T7 RNA polymerase using the RiboMAX protocol (Promega) as described previously (17).
Dicistronic vectors containing the IGR linked to its natural coding region were constructed as follows. First, a BglII-PstI restriction fragment corresponding to nucleotides 5919 to 8571 of the CrPV genome (i.e., IGR-ORF2) was cloned into pGEM3 (Promega), which was digested with BamHI and PstI. Subsequently, an EcoRI-XbaI restriction fragment derived from a dicistronic luciferase construct containing the wild-type encephalomyocarditis virus (EMCV) IRES fused in frame to Fluc, or an XhoI-XbaI restriction fragment containing the ΔEMCV sequences upstream of Fluc, was cloned into the SmaI site of pGEM3 upstream of the inserted IGR-CrPV sequences to yield plasmids T7 EMCV/Fluc-IGR/CrPVORF2 and T7 ΔEMCV/Fluc-IGR/CrPVORF2, respectively. For in vitro transcription, these dicistronic constructs were digested with HindIII.
For expression in insect cells, dual luciferase constructs containing no inserted sequences, the 5′NCR, or the IGR were digested with HindIII and BamHI and the dicistronic fragments were inserted into the polylinker of pRmHa3, a vector containing the copper-inducible promoter of the Drosophila metallothionein gene (7).
In vitro translation.
Uncapped dicistronic RNAs were translated in the RRL or the wheat germ extract (Promega), as recommended, in the presence of 154 mM potassium acetate. Products of translation reactions were measured enzymatically using the dual luciferase reporter assay system (Promega) or by incorporation of [35S]methionine-[35S]cysteine (1175 Ci/mmol; New England Nuclear) followed by SDS-polyacrylamide gel electrophoresis and fluorography.
DNA and RNA transfections.
Transfections of simian virus 40 promoter-containing plasmids were performed using the lipofectin (GIBCO/BRL) reagent. Briefly, SL2 cells were plated at 4 × 106 cells per 60-mm plate and transfected using 4 μg of DNA mixed with 20 μl of Lipofectin. Serum-containing medium was added after 24 h, and extracts were prepared after 72 h. Cells were pelleted, resuspended in 50 μl of passive lysis buffer (Promega), and lysed by applying three freezing-thawing cycles. Cellular debris were sedimented, and 20 μl of the soluble extract was assayed using dual luciferase assay reagents.
Transfections of metallothionein promoter-containing plasmids were performed as described above, except that the promoter was induced 12 h after transfection by addition of 0.5 mM cupric sulfate to the medium. The cells were harvested 48 h later.
For RNA transfections, 3 × 106 SL2 cells were plated per 35-mm dish and incubated overnight. The cells were washed once using serum-free medium. Then 400 or 800 ng of capped RNA was mixed with 40 μl of lipofectin and transfected into cells as specified by the manufacturer, and the cells were harvested after 3 to 4 h. The cells were washed in phosphate-buffered saline, sedimented, resuspended in 50 μl of passive lysis buffer, and processed for dual luciferase measurements as described above.
Northern analysis.
Total RNA was extracted from cells using the Trizol reagent as recommended (GIBCO/BRL). Poly(A)-containing RNA was selected using the Oligotex mRNA kit (Qiagen). RNA was quantitated by spectrophotometry, and 10 μg of poly(A) RNA was separated in formaldehyde-containing agarose gels. Radiolabeled probes were generated using the RadRrime kit (GIBCO/BRL). Hybridization was carried out in ExpressHyb solution (Clontech) as recommended.
Nucleotide sequence accession number.
The sequence data have been submitted to GenBank under accession no. AF218039.
RESULTS
CrPV contains a dicistronic RNA genome whose organization is similar to that of other insect picorna-like viruses.
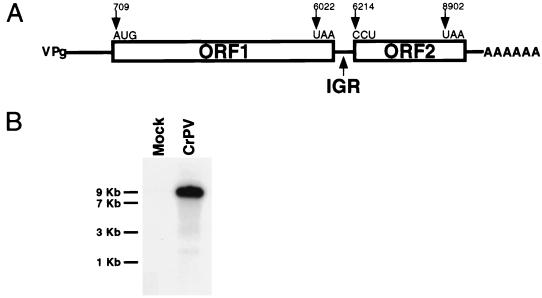
We have constructed a full-length cDNA copy of the CrPV RNA genome and have determined its nucleotide sequence. Analysis of data predicted a genome organization (Fig. 1A) which is conserved among the insect picorna-like viruses (24, 36, 40, 53). In contrast to picornavirus genomes, which encode a single polyprotein, the CrPV genome contains two large ORFs separated by a 189-nucleotide IGR (Fig. 1A). As in picornavirus genomes, the 5′NCR preceding the upstream ORF (ORF1) is burdened with many AUG codons followed by numerous stop codons in all three reading frames. Comparative analysis of amino acid sequence motifs suggests that ORF1 of CrPV encodes proteins with RNA helicase (32, 54), 3C-like proteinase (57), and RNA-dependent RNA polymerase (27, 42) activities with amino acid identities of 19, 14, and 20%, respectively, to the hepatitis A virus proteins and with identities of 65, 53, and 68%, respectively, to the Drosophila C virus proteins (data not shown). Recently, partial sequence analysis and crystal structure determination of CrPV has mapped the structural proteins to the downstream ORF (ORF2) (59). Therefore, unlike in picornavirus genomes, the structural proteins are encoded from a second ORF located at the 3′ end of the viral genome.
FIG. 1.
Properties of the CrPV RNA genome. (A) Schematic of the general organization of the CrPV genome. Lines and boxes designate untranslated and translated portions of the viral RNA, respectively. IGR denotes the region which separates ORF1 and ORF2. The positions, in nucleotides, of the start and stop codons of each ORF are indicated. The initiation and termination codons of each ORF, as predicted by conceptual translation of the viral RNA using DNA Strider or determined experimentally (see Results), are shown. (B) CrPV contains a single plus-strand RNA genome. Northern blot analysis of poly(A)-containing RNA isolated from mock- or CrPV-infected SL2 cells, using a hybridization probe that was specific to viral RNA, is shown. The migration of RNA molecular size standards (in kilobases) is shown at left.
The predicted dicistronic genome organization argues that ORF2 was translated (i) from a subgenomic mRNA species, (ii) from ribosomes that failed to terminate at the stop codon of ORF1 (readthrough mechanism), (iii) from ribosomes that initially entered the genomic mRNA at the 5′ end but were subsequently transferred to ORF2 (shunting mechanism), or (iv) by a second downstream IRES. Because only a single polyadenylated viral mRNA species could be detected in CrPV-infected cells (Fig. 1B) (39), the structural proteins are likely to be translated from the genomic RNA, excluding only the first possibility.
The CrPV genome contains two IRES elements.
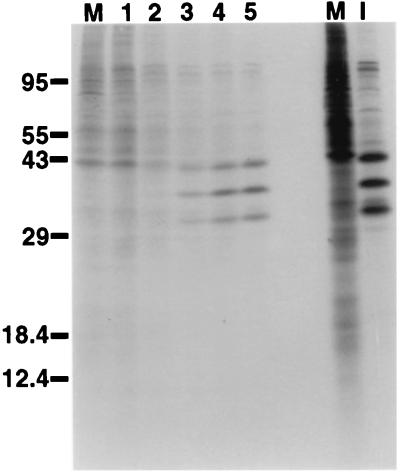
To examine the translation of the CrPV mRNA, cultured Drosophila SL2 cells were infected with CrPV and pulse-labeled with [35S]methionine-[35S]cysteine at different times after infection. As can be seen in Fig. 2, translation of host cell mRNAs was inhibited by approximately 3 h after infection. However, viral mRNA was still efficiently translated between 3 and 5 h after infection, suggesting that viral mRNA can compete with cellular mRNAs for the translation apparatus or that viral mRNA is translated by a mechanism that escapes the translational inhibition experienced by the cellular mRNAs. These scenarios are reminiscent of events that take place in picornavirus-infected cells, in which the eukaryotic translation initiation factors eIF-4G or the eIF-4E-binding proteins are degraded or modified, respectively (10, 15), resulting in the inhibition of cap-dependent translation of cellular mRNAs but allowing translation of IRES-containing viral mRNAs. In contrast to picornavirus-infected cells, CrPV-infected cells produced viral structural proteins, migrating between 29,000 and 43,000 Da (Fig. 2), in supramolecular excess over nonstructural proteins (37, 38, 49).
FIG. 2.
Expression of CrPV proteins in infected cells. SL2 cells were infected with CrPV at a multiplicity of infection of 100 PFU per cell and, at the indicated times (in hours) after infection, labeled with [35S]methionine for 10 min. Labeled extracts from mock-infected cells (M) and cells infected for 6 h (I) are shown on the right. Soluble extracts were prepared (50) and analyzed in SDS-polyacrylamide gels. An autoradiograph of the gel is shown. The migration of proteins with known molecular masses (in kilodaltons) is shown on the left.
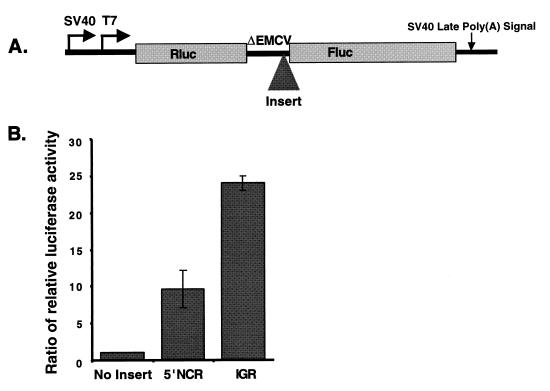
We wished to examine whether ORF1 and ORF2 in CrPV RNA were each preceded by IRES elements that could independently regulate the translation of ORF1 and ORF2. The cDNA sequences encoding the 5′NCR (nucleotides 1 to 709) or the IGR (nucleotides 6022 to 6232) of CrPV were inserted between the Renilla luciferase (Rluc) and firefly luciferase (Fluc) coding regions in expression plasmids (Fig. 3A) that can direct the synthesis of dicistronic mRNAs either in tissue culture cells, using the simian virus 40 promoter, or in vitro, using the promoter for T7 RNA polymerase (22). A landscape of RNA structures (ΔEMCV) was inserted between the two cistrons to prevent translation of the downstream ORF from reinitiation or readthrough of ribosomes (44) which had traversed the first Rluc cistron (22). Expression plasmids that contained the CrPV IGR sequences, the CrPV 5′NCR sequences, or no inserted sequences were transfected into SL2 cells, and the accumulation of the translation products from both cistrons was monitored by an enzymatic luciferase assay. Figure 3B shows that dicistronic mRNAs containing the viral 5′NCR and the IGR in the intercistronic spacer region allowed the accumulation of the translation product of the second cistron, Fluc, to approximately 10- and 25-fold-higher levels, respectively, than did control dicistronic mRNAs. Northern analysis showed that the size and integrity of all intracellularly expressed dicistronic mRNAs remained intact (data not shown).
FIG. 3.
Accumulation of translation products encoded by plasmids expressing dicistronic mRNAs that contain the CrPV 5′NCR or the CrPV IGR in the intercistronic spacer region in cultured cells. (A) Diagram of the dual luciferase dicistronic construct. The locations of promoters for RNA polymerase II (simian virus 40 [SV40]) and for T7 RNA polymerase (T7) and sequences mediating polyadenylation [SV40 Late Poly(A) Signal] are indicated. Rluc and Fluc encode Renilla luciferase and firefly luciferase, respectively. ΔEMCV designates the cDNA that encodes highly structured RNA sequences to prevent translational readthrough or reinitiation (see Materials and Methods). The shaded triangle, labeled Insert, indicates the position at which the CrPV 5′NCR or CrPV IGR sequences were inserted. (B) The viral 5′NCR and IGR mediate the translation of second cistrons in dicistronic mRNAs. Luciferase activities in extracts from SL2 cells, transfected with expression vectors that contain no added insertion or insertions of the viral 5′NCR or the viral IGR in the intergenic spacer region (see panel A), are displayed. The ratio of relative luciferase activity represents the Fluc/Rluc ratios in reference to the construct with no insert, whose Fluc/Rluc ratio was set to 1. The mean values from three independent experiments are shown. Error bars indicate standard deviations.
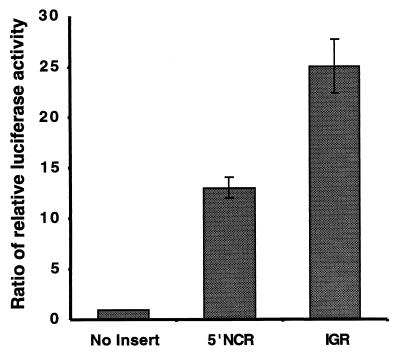
To address the concern that splicing or cryptic promoter activities in the inserted viral cDNAs generated minor mRNA species that lacked the upstream Rluc cistron and could therefore mediate translation of the second cistron, Fluc, by a 5′-end-dependent mechanism, capped dicistronic mRNAs were synthesized in vitro by T7 RNA polymerase and transfected into SL2 cells. The synthesis of Rluc and Fluc was examined using enzymatic assays. The results in Fig. 4 show that both the viral 5′NCR and the viral IGR mediated translation of the second cistron of dicistronic RNAs, transfected into cultured cells, with an efficiency similar to that observed in the DNA transfection experiments in Fig. 3. Interestingly, in embryo and ovary extracts from Drosophila melanogaster, the IGR IRES functions with an efficiency similar to that of the Drosophila Antennapedia IRES (Y. S. Lie and P. M. Macdonald, personal communication).
FIG. 4.
Accumulation of translation products of dicistronic mRNAs that contain the CrPV 5′NCR or the CrPV IGR in the intercistronic spacer region in cultured cells. Shown are the ratios of relative luciferase activities in SL2 cells which were transfected with in vitro-synthesized dicistronic mRNAs (see the legend to Fig. 3B).
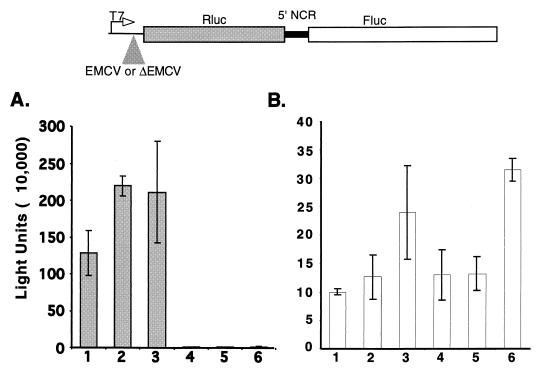
To test the possibility that ribosomal subunits could have been tethered from the 5′ end of the dicistronic mRNA to the 5′NCR IRES sequences (13, 61), entry of ribosomal subunits to the 5′ end of the dicistronic mRNAs was blocked by the insertion of a landscape of RNA structures. Translation of dicistronic mRNAs that contained either an active EMCV IRES (EMCV) or RNA structures that do not function as an IRES (ΔEMCV) preceding the first cistron was monitored in the RRL. Figure 5 shows that the presence of RNA structures (ΔEMCV) upstream of the Rluc cistron abolished the synthesis of Rluc protein (compare columns 1 to 3 with columns 4 to 6). In contrast, translation of the second cistron mediated by the 5′NCR of CrPV was not inhibited in the latter RNAs (columns 4 to 6).
FIG. 5.
Translation of dicistronic mRNAs containing the viral 5′NCR in the intercistronic spacer region in the RRL. RNAs containing an active (EMCV) or inactive (ΔEMCV) EMCV IRES preceding the first cistron in the dicistronic mRNAs (top) were translated at 5 ng/μl (columns 1 and 4), 10 ng/μl (columns 2 and 5), or 20 ng/μl (columns 3 and 6) in the RRL. The average Renilla light units (A) and firefly luciferase light units (B) of three independent experiments are shown.
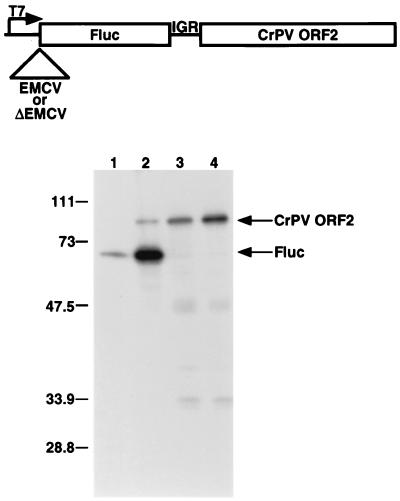
Similar experiments were performed to determine whether the CrPV IGR could confer translational initiation of a second cistron independently of the translation of the first cistron in a dicistronic mRNA. In this case, the second cistron of the dicistronic mRNAs contained a truncated ORF2 of CrPV linked to its naturally preceding IGR element (Fig. 6). In RNAs that contained the EMCV IRES upstream of the first cistron Fluc, both translation products were synthesized, although the EMCV IRES mediated more efficient translation than did the IGR (lanes 1 and 2). Replacing the EMCV IRES with the ΔEMCV element as leader of Fluc completely abolished the synthesis of Fluc (lanes 3 and 4). However, the synthesis of ORF2 was slightly increased under these conditions (lanes 3 and 4), supporting the hypothesis that the IGR element functions as an IRES and not as a sequence element which allows reinitiation or promotes the shunting of ribosomal subunits. These data indicate that both the 5′NCR and the IGR sequences in the CrPV genome contain IRES activities that function both in cultured cells and in the RRL. Moreover, under all experimental conditions examined so far, the IGR IRES was more active than the 5′NCR IRES; however, the IGR IRES was approximately threefold less active than the well-characterized EMCV IRES in the RRL (data not shown).
FIG. 6.
Translation of dicistronic mRNAs containing the viral IGR in the intercistronic spacer region in the RRL. RNAs containing an active (EMCV) or inactive (ΔEMCV) encephalomyocarditis IRES preceding the first cistron in dicistronic Fluc-IGR-CrPV ORF2 mRNAs (top) were translated in the RRL in the presence of [35S]methionine-[35S]cysteine. Translation products were visualized after SDS-polyacrylamide gel electrophoresis. An autoradiograph of the gel is shown. Lanes 1 and 2 show the reaction products initiated with 10 and 20 ng of EMCV-containing dicistronic RNAs per μl, respectively. Lanes 3 and 4 show the translation products produced in reactions containing 10 and 20 ng of ΔEMCV-containing dicistronic RNAs per μl, respectively. The migration of protein molecular mass standards is indicated at left in kilodaltons. The positions of the CrPV ORF2 and Fluc proteins are indicated by arrows.
The IGR IRES functions in wheat germ extracts.
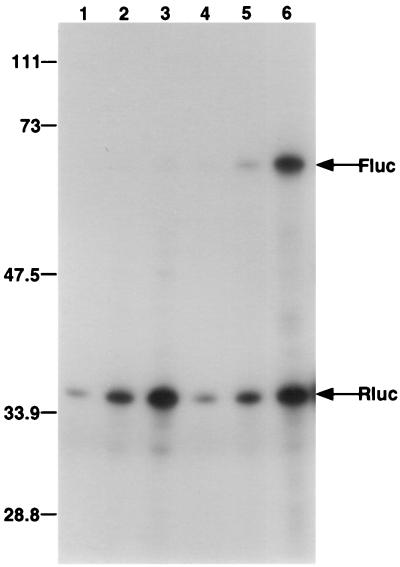
Although translation of capped mRNAs ensues efficiently in wheat germ translation extracts, no IRES has been shown to be functional in this translation system (21). Because the CrPV IRES elements were evolutionarily divergent from IRES elements present in mammalian RNA viruses or vertebrate mRNAs, we tested whether the CrPV IRES elements were active in wheat germ extracts. Different amounts of in vitro-transcribed, dicistronic mRNAs lacking or containing the two CrPV IRES elements in the intercistronic spacer region were translated in the wheat germ extracts, and luciferase synthesis was monitored by incorporation of radiolabeled amino acids. While the CrPV 5′NCR IRES was not active in this translation system (data not shown), the CrPV IGR IRES stimulated Fluc translation 40-fold at the highest RNA concentration tested relative to the control dicistronic RNA containing no inserted sequences (Fig. 7). This finding shows that the wheat germ extract is capable of mediating internal initiation on at least the CrPV IGR.
FIG. 7.
Translation of dicistronic mRNAs containing the viral IGR in the intercistronic spacer region in the wheat germ lysate. The RNAs described in Fig. 3 were translated in the presence of [35S]methionine-[35S]cysteine in wheat germ extracts at a final concentration of 5 ng/μl (lanes 1 and 4), 10 ng/μl (lanes 2 and 5), or 20 ng/μl (lanes 3 and 6). Dicistronic mRNAs that encoded Rluc in the first cistron and Fluc in the second cistron, either lacking an IRES (lanes 1 to 3) or containing the IGR IRES in the intercistronic spacer, were used in the translation assay. Translation products were separated by SDS-polyacrylamide gel electrophoresis, and an autoradiograph of the gel is shown. The migration of protein molecular mass standards (in kilodaltons) and the positions of the Fluc and Rluc proteins are indicated.
ORF2 begins with noncognate CCU triplet.
Inspection of the predicted ORF2 failed to reveal any cognate AUG or weakly cognate GUG or CUG codons that could be used as the initiation codon for ORF2; this feature has been observed in the other picorna-like insect viruses DCV, RhPV, PSIV and HiPV (24, 36, 40, 53). To determine the first triplet of the CrPV ORF2, mRNAs which contained the IGR IRES linked to the 5′-proximal codons in ORF2 were translated in the RRL, translation products were purified, and N-terminal amino acids were determined by microcapillary reverse-phase high-pressure liquid chromatography nanoelectrospray tandem mass spectroscopy (Harvard Microchemistry Facility). This analysis revealed that the N-terminal amino acid of ORF2 was alanine, presumably encoded by GCU at nucleotides 6217 to 6219, suggesting that either the preceding codon, CCU, or the GCU6217–6219 codon itself functions as start codon.
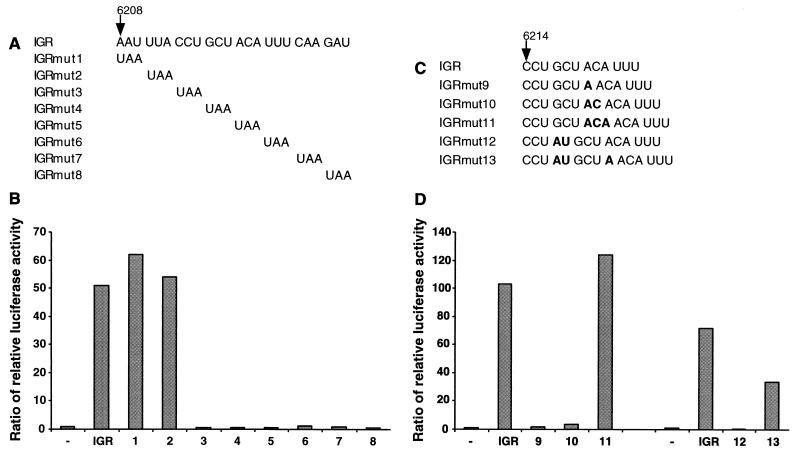
To address this question, a series of stop codon mutations was systematically introduced into the CrPV IGR in the region spanning nucleotides 6208 to 6231. All mutated CrPV IGR elements were tested for IRES activity in the RRL in the context of dual luciferase dicistronic mRNAs. Changing either the AAU codon at positions 6208 to 6210 or the UUA codon at positions 6211 to 6213 to a nonsense codon (Fig. 8A, IGRmut1 and IGRmut2) had no effect on IGR activity (Fig. 8B). In contrast, mutating the CCU6214–6216 or GCU6217–6219 or any of the next four downstream codons to a UAA nonsense codon (Fig. 8A, IGRmut3 through IGRmut8) abolished IRES activity (Fig. 8B). The mutated IRES elements could be inactive either because the IRES structure was disrupted or because a nonsense codon was introduced into the coding region, consistent with the notion that either the CCU6214–6216 or the GCU6217–6219 codon is the start site for translation.
FIG. 8.
Translational activities of mutagenized CrPV IGR elements. (A) Description of IGR stop codon insertions (IGRmut1 to IGRmut8). (B) Dicistronic dual luciferase RNAs bearing IGR stop codon mutations (IGRmut1 to IGRmut8) were translated in the RRL at a final concentration of 20 ng/μl. The ratio of relative luciferase activity was calculated as described in the legend to Fig. 3B. (C) Description of IGR nucleotide insertions (IGRmut9 to IGRmut13). (D) The translation of dicistronic RNAs bearing nucleotide insertions (IGRmut9 to IGRmut13) was determined and is displayed as in panel B.
To distinguish whether the CCU6214–6216 or the GCU6217–6219 codon was the initiation codon for the CrPV ORF2, 1 to 3 nucleotides were inserted into the CrPV IGR at various positions to alter the translational reading frame (Fig. 8C). The mutated IGR elements were tested for IRES activity in the context of dual luciferase RNAs as described above. Insertion of 1 or 2 nucleotides immediately downstream of the GCU codon (Fig. 8C, IGRmut9 and IGRmut10) eliminated the accumulation of Fluc (Fig. 8D). However, insertion of 3 nucleotides at the same position (Fig. 8C, IGRmut11), predicted to maintain the original reading frame, restored Fluc accumulation (Fig. 8D). Therefore, the GCU codon is part of the CrPV ORF2. Insertion of 2 nucleotides immediately upstream of the GCU codon (Fig. 8C, IGRmut12) eliminated the accumulation of Fluc (Fig. 8D); however, the insertion of a single additional nucleotide downstream of the GCU codon (Fig. 8C, IGRmut13) partially restored Fluc synthesis (Fig. 8D). Therefore, the 2-nucleotide insertion in IGRmut12 was unlikely to abolish translation via structural alteration of the IGR IRES but, instead, did so by disrupting the reading frame. These results argue that the CCU6214–6216 is the first triplet of the CrPV ORF2.
The CCU triplet is part of an inverted-repeat sequence element whose integrity is important for IRES activity.
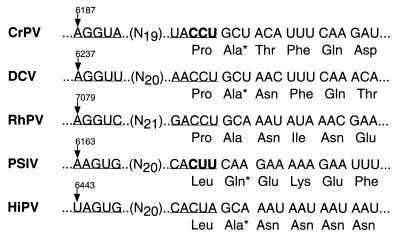
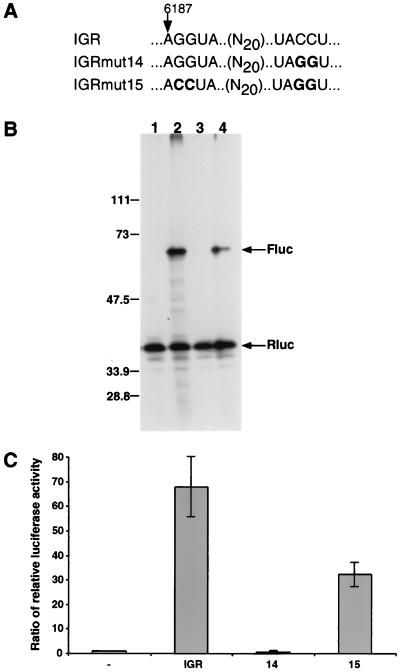
A 5-nucleotide inverted-repeat sequence previously noted in other insect picorna-like viruses (Fig. 9) was also present in the CrPV IGR; the length and the spacing between the repeated sequences elements were conserved among the viral genomes (Fig. 9). Specifically, nucleotides 6187 to 6191 of the CrPV IGR are complementary to nucleotides 6212 to 6216. Conspicuously, CCU6214–6216 is contained within one of the inverted-repeat elements (Fig. 9). Furthermore, the two elements of the inverted repeat are located in predicted loop regions of two adjacent hairpin structures, implying possible tertiary interactions between the inverted-repeat elements. To test a role of the conserved inverted repeat in IGR IRES activity, two mutated IGR elements were constructed in which the inverted-repeat sequences were altered. In IGRmut14 (Fig. 10A), the downstream half of the inverted repeat was altered from UACCU6212–6216 to UAGGU6212–6216, destroying the predicted complementarity of the inverted repeat. IGRmut15 (Fig. 10A) contains the UACCU-to-UAGGU change of IGRmut14 along with mutations that change the upstream repeat from AGGUA6187–6191 to ACCUA6187–6191; as a result, the complementarity of the inverted repeat elements is restored. These mutated IGRs were inserted into dual luciferase dicistronic RNAs, and their translational efficiencies were compared to those of wild-type IGR elements. All dicistronic mRNAs expressed the first Rluc cistron with similar efficiency (Fig. 10B, lanes 1 to 4). Dicistronic mRNAs lacking an IRES between the luciferase cistrons failed to mediate translation of the second Fluc cistron (lane 1). The wild-type IGR mediated the translation of Fluc (lane 2). Destruction of the complementarity of the inverted repeat in IGRmut14 prevented translation of the second Fluc cistron (lane 3); in contrast, the compensatory mutations of IGRmut15 restored activation of Fluc translation (lane 4). Quantitation by enzymatic assays (Fig. 10C) showed that IGRmut15 restored translation to within 50% of that seen with the wild-type IGR. Similar results were seen with IGR elements in which UUACCU6211–6216 was changed to GACUGA6211–6216, resulting in an inactive IGR; the activity of the mutated IGR could be restored by a compensatory UCAGU6187–6191 mutation in the GACUGA6211–6216-containing IGR (data not shown). These results demonstrate that interaction between the inverted-repeat sequences is essential for IGR IRES activity and, furthermore, that certain variations of the sequence of the inverted repeats may be tolerated if complementarity is maintained.
FIG. 9.
Nucleotide alignments between sequences surrounding the beginning of ORF2 in CrPV (accession no. AF218039), DCV (accession no. AF0014388), RhPV (accession no. AF0022937), PSIV (accession no. AB006531), and HiPV (accession no. AB017037). A predicted inverted-repeat element, conserved among those sequences, is underlined. Amino acids labeled with an asterisk were identified as N-terminal amino acids by sequence analysis. Codons highlighted in bold were identified as start codons using mutational analyses.
FIG. 10.
Translational activities in the RRL of IGR IRES elements with mutations in the inverted-repeat element. (A) Sequences of wild-type IGR and mutated IGRmut14 and IGRmut15. (B) Translation of dicistronic luciferase mRNAs containing no insert (lane 1), wild-type IGR (lane 2), mutated IGRmut14 (lane 3), or IGRmut15 (lane 4) between the first (Rluc) and second (Fluc) cistrons. RNAs were present at final concentrations of 20 ng/μl. Radiolabeled translation products were visualized after separation on SDS-polyacrylamide gels. An autoradiograph of the gel is shown. The migration of protein molecular mass standards (in kilodaltons) is indicated at left. The positions of the Fluc and Rluc proteins are indicated by arrows. (C) Enzymatic quantitation of the IRES translational efficiencies. Fold activation represents the Fluc/Rluc ratios in reference to the construct with no insert (lane −), whose Fluc/Rluc ratio was set to 1. The mean values from three independent experiments are shown. Error bars show standard deviations.
DISCUSSION
Polycistronic transcripts in eukaryotic cells.
Polycistronic transcripts are commonly found in bacteria, in the protist trypanosome and in the nematode Caenorhabditis elegans (5). However, unlike bacterial transcripts, polycistronic transcripts in trypanosomes (45) and C. elegans (30, 58) are processed by trans-splicing events (41) that generate functionally monocistronic mRNAs which contain a capped 5′ end. It is thought that the cap structure provides stability to the RNA and ensures efficient translation in both invertebrate (33) and vertebrate (29) mRNAs.
In higher eukaryotic cells, most, but not all, mRNAs encode one protein product. For example, expression of two ORFs in certain retroviral RNA genomes (20) and in polyamine-regulated synthesis of mammalian ornithine decarboxylase antizyme mRNA (34) occurs by ribosomal frameshifting, a mechanism in which ribosomes pause at defined structural elements in the coding region and then continue translation in an altered reading frame (14). Instances of truly dicistronic mRNAs in which the two cistrons are separated by intergenic nucleotide spacer regions have been described in the Drosophila stoned locus (1), the Drosophila Adh/Adhr locus (6), the vertebrate growth/differentiation factor 1 gene (31), and the mammalian SNURF-SNRPN gene (16); however, in none of these cases is the mechanism of translation of the downstream cistron known. The mammalian SNRPN gene locus, for example, encodes a dicistronic mRNA whose two protein products, SNURF and SmN, have been implicated in a developmental disease, the Prader-Willi syndrome, which results from the loss of function of the paternally inherited gene (3). The two ORFs in the 1.6-kb SNURF-SNRPN mRNA are separated by a 150-nucleotide intercistronic spacer region. While the synthesis of both SNURF and SmN proteins from the dicistronic mRNA has been documented, it is unclear whether the second cistron is translated by leaky scanning, reinitiation, or internal initiation.
With the discovery that picornavirus genomes contain IRES elements in their 5′NCRs, it was possible to test the idea that chimeric RNAs which contain IRES elements between multiple cistrons are translated by the eukaryotic translation apparatus. The finding that several IRES elements in an mRNA molecule can be used as start sites for translation (12, 35, 62) raised the possibility that naturally occurring mRNAs with multiple IRES elements may exist. Our study of the CrPV genome provides an example of such a functionally dicistronic RNA.
Regulation of the CrPV genome by two IRES elements.
The CrPV genome encodes two nonoverlapping ORFs, both of which are independently controlled by individual IRES elements. In all systems in which these two IRESs were studied, the downstream IGR IRES was consistently more active than the upstream 5′NCR IRES, providing a mechanistic explanation for the previously noted increased expression of viral structural proteins relative to the nonstructural proteins in infected cells (37, 38). The utility of a dicistronic genome and the use of dual IRES elements of different strengths for differential gene expression for an RNA virus could be economical, facilitating the production of the relatively large amounts of structural proteins required for the formation of the viral capsid while allowing nonstructural proteins, which function enzymatically in the replication of the viral genome, to be produced sparingly. The molecular basis of the differential activities of the 5′NCR and IGR IRES elements is not yet clear but could involve one or more possible mechanisms. For example, the IGR IRES might compete more effectively than the 5′NCR IRES for components of the translational machinery; competition among IRES elements in cis has been previously noted (43). Alternatively, the activity of the 5′NCR IRES may be modulated by a requirement for some limiting factor or may be directly repressed by a viral or cellular protein. Further characterization of these IRES elements is required to distinguish among these possibilities.
An unusual mode of translation initiation at the CrPV IGR IRES.
Mutational analyses have shown that a noncognate CCU, rather than an cognate AUG or weakly cognate GUG- or CUG-like codon, is the first triplet of the CrPV ORF2. Translational start codons which differ from AUG at one position (18), usually at the first nucleotide, such as GUG (19) and CUG (4, 60), or the second nucleotide, such as ACG (48), have been described. In contrast, the CrPV downstream ORF2 is an example of a cistron whose start codon differs from the canonical AUG start codon at all three positions. Our study has shown that the N-terminal amino acid of CrPV ORF2 is an alanine, encoded by a GCU codon. Sequencing of DCV (24) and PSIV (52) VP2 proteins, which represent the N terminus of ORF2, also revealed that the N-terminal amino acid of DCV or PSIV VP2 was encoded by GCU (in DCV) and CAA (in PSIV), which encode alanine and glutamine, respectively. More recently, the N terminus of ORF2 in HiPV was shown to be a GCA-encoded alanine (40).
Extensive mutagenesis has shown that the CCU codon, immediately preceding the GCU-encoding alanine codon, is required to set the translational reading frame in CrPV (Fig. 8 and 10). How might the IGR IRES promote translation initiation at a CCU codon? One possibility is that some sequence or structural element of the CrPV IGR IRES facilitates the CCU initiator tRNAMet interactions in the absence of sequence complementarity at any position; in this case, methionine would be the N-terminal amino acid in ORF2, which is then posttranslationally removed by aminopeptidases. Alternatively, initiator tRNAMet might not be needed for translational initiation. This notion is supported by preliminary toeprinting experiments indicating that the CCU triplet is correctly positioned in the ribosomal P site in the absence of initiator tRNAMet and eukaryotic initiation factor eIF2 and by in vitro translation assays showing that 80S ribosomes can be assembled in the presence of nonhydrolyzable GTP analog and the compound edeine, which normally inhibits AUG start codon recognition by the scanning 43S ternary complex (Wilson et al., submitted). That initiator tRNAMet is also not required for the translational initiation of ORF2 in PSIV has been very recently reported by Sasaki and Nakashima (51).
A variety of translation strategies in RNA viruses.
RNA viruses have evolved a remarkable variety of ways to translate their RNA genomes into structural and nonstructural proteins. Production of subgenomic mRNAs, synthesis of polyprotein precursors from single mRNAs, ribosomal readthrough, and frameshifting are all ways to produce more than one protein product from a single RNA genome. The identification of CrPV RNA as a dicistronic mRNA regulated by two independent IRES elements expands this repertoire.
ACKNOWLEDGMENTS
Joan E. Wilson, Marguerite J. Powell, and Susan E. Hoover contributed equally to this work.
We thank Karla Kirkegaard for critical reading of the manuscript.
This work was supported by NIH grants R01 GM55979 and R01 AI 25105 (to P.S.). J.E.W. is a recipient of a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
REFERENCES
- 1.Andrews J, Smith M, Merakovsky J, Coulson M, Hannan F, Kelly L E. The stoned locus of Drosophila melanogaster produces a dicistronic transcript and encodes two distinct polypeptides. Genetics. 1996;143:1699–1711. doi: 10.1093/genetics/143.4.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates, Inc.; 1994. [Google Scholar]
- 3.Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood E M, Lugo T G, Kretzner L, King M W, Street A J, Witte O N, Eisenman R N. Functional analysis of the AUG- and CUG-initiated forms of the c-Myc protein. Mol Biol Cell. 1994;5:597–609. doi: 10.1091/mbc.5.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal T. Gene clusters and polycistronic transcription in eukaryotes. Bioessays. 1998;20:480–487. doi: 10.1002/(SICI)1521-1878(199806)20:6<480::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Brogna S, Ashburner M. The Adh-related gene of Drosophila melanogaster is expressed as a functional dicistronic messenger RNA: multigenic transcription in higher organisms. EMBO J. 1997;16:2023–2031. doi: 10.1093/emboj/16.8.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunch T A, Grinblat Y, Goldstein L S. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christian P D, Scotti P D. Picornalike viruses of insects. In: Miller L K, Ball L A, editors. The insect viruses. New York, N.Y: Plenum Publishing Corp.; 1998. pp. 301–336. [Google Scholar]
- 9.Eaton B T, Steacie A D. Cricket paralysis virus RNA has a terminal poly(A) J Gen Virol. 1980;50:167–171. [Google Scholar]
- 10.Ehrenfeld E. Picornavirus inhibition of host cell protein synthesis. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. Vol. 19. New York, N.Y: Plenum Press; 1984. pp. 177–221. [Google Scholar]
- 11.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fussenegger M, Mazur X, Bailey J E. pTRIDENT, a novel vector family for tricistronic gene expression in mammalian cells. Biotechnol Bioeng. 1998;57:1–10. doi: 10.1002/(sici)1097-0290(19980105)57:1<1::aid-bit1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 13.Futterer J, Kiss-Laszlo Z, Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 14.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 15.Gingras A C, Svitkin Y, Belsham G J, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray T A, Saitoh S, Nicholls R D. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA. 1999;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller A A, Sarnow P. In vitro selection of a 7-methyl-guanosine binding RNA that inhibits translation of capped mRNA molecules. Proc Natl Acad Sci USA. 1997;94:8521–8526. doi: 10.1073/pnas.94.16.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hann S R. Regulation and function of non-AUG-initiated proto-oncogenes. Biochimie. 1994;76:880–886. doi: 10.1016/0300-9084(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 19.Imataka H, Olsen H S, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- 21.Jackson R J. A comparative view of initiation site selection mechanisms. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 71–112. [Google Scholar]
- 22.Johannes G, Carter M S, Eisen M B, Brown P O, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson K N, Christian P D. A molecular taxonomy for cricket paralysis virus including two new isolates from Australian populations of Drosophila (Diptera: Drosophilidae) Arch Virol. 1996;141:1509–1522. doi: 10.1007/BF01718251. [DOI] [PubMed] [Google Scholar]
- 24.Johnson K N, Christian P D. The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J Gen Virol. 1998;79:191–203. doi: 10.1099/0022-1317-79-1-191. [DOI] [PubMed] [Google Scholar]
- 25.King L A, Moore N F. Evidence for the presence of a genome-linked protein in two insect picornaviruses, cricket paralysis and Drosophila C viruses. FEMS Microbiol Lett. 1988;50:41–44. [Google Scholar]
- 26.King L A, Pullin J S K, Stanway G, Almond J W, Moore N F. Cloning of the genome of cricket paralysis virus: sequence of the 3′ end. Virus Res. 1987;6:331–344. [Google Scholar]
- 27.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. . (Erratum, 28:546.) [DOI] [PubMed] [Google Scholar]
- 28.Koonin E V, Gorbalenya A E. An insect picornavirus may have genome organization similar to that of caliciviruses. FEBS Lett. 1992;297:81–86. doi: 10.1016/0014-5793(92)80332-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S J. Expression of growth/differentiation factor 1 in the nervous system: conservation of a bicistronic structure. Proc Natl Acad Sci USA. 1991;88:4250–4254. doi: 10.1073/pnas.88.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luking A, Stahl U, Schmidt U. The protein family of RNA helicases. Crit Rev Biochem Mol Biol. 1998;33:259–296. doi: 10.1080/10409239891204233. [DOI] [PubMed] [Google Scholar]
- 33.Maroney P A, Denker J A, Darzynkiewicz E, Laneve R, Nilsen T W. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA. 1995;1:714–723. [PMC free article] [PubMed] [Google Scholar]
- 34.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins J F, Gesteland R F, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molla A, Jang S K, Paul A V, Reuer Q, Wimmer E. Cardioviral internal ribosomal entry site is functional in a genetically engineered dicistronic poliovirus. Nature. 1992;356:255–257. doi: 10.1038/356255a0. [DOI] [PubMed] [Google Scholar]
- 36.Moon J S, Domier L L, McCoppin N K, D'Arcy C J, Jin H. Nucleotide sequence analysis shows that Rhopalosiphum padi virus is a member of a novel group of insect-infecting RNA viruses. Virology. 1998;243:54–65. doi: 10.1006/viro.1998.9043. [DOI] [PubMed] [Google Scholar]
- 37.Moore N F, Kearns A, Pullin J S K. Characterization of cricket paralysis virus-induced polypeptides in Drosophila cells. J Virol. 1980;33:1–9. doi: 10.1128/jvi.33.1.1-9.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore N F, Reavy B, Pullin J S K. Processing of cricket paralysis virus induced polypeptides in Drosophila cells: production of high molecular weight polypeptides by treatment with iodoacetamide. Arch Virol. 1981;68:1–8. doi: 10.1007/BF01315161. [DOI] [PubMed] [Google Scholar]
- 39.Moore N F, Tinsley T W. The small RNA-viruses of insects. Arch Virol. 1982;72:229–245. doi: 10.1007/BF01315220. [DOI] [PubMed] [Google Scholar]
- 40.Nakashima N, Sasaki J, Toriyama S. Determining the nucleotide sequence and capsid-coding region of himetobi P virus: a member of a novel group of RNA viruses that infect insects. Arch Virol. 1999;144:2051–2058. doi: 10.1007/s007050050726. [DOI] [PubMed] [Google Scholar]
- 41.Nilsen T W. trans-splicing: an update. Mol Biochem Parasitol. 1995;73:1–6. doi: 10.1016/0166-6851(94)00107-x. [DOI] [PubMed] [Google Scholar]
- 42.O'Reilly E K, Kao C C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 43.Ohlmann T, Jackson R J. The properties of chimeric picornavirus IRESes show that discrimination between internal translation initiation sites is influenced by the identity of the IRES and not just the context of the AUG codon. RNA. 1999;5:764–778. doi: 10.1017/s1355838299982158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peabody D S, Berg P. Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol Cell Biol. 1986;6:2695–2703. doi: 10.1128/mcb.6.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry K, Agabian N. mRNA processing in the Trypanosomatidae. Experientia. 1991;47:118–128. doi: 10.1007/BF01945412. [DOI] [PubMed] [Google Scholar]
- 46.RajBhandary U L. More surprises in translation: initiation without the initiator tRNA. Proc Natl Acad Sci USA. 2000;97:1325–1327. doi: 10.1073/pnas.040579197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinganum C, O'Loughlin G T, Hogan T W. A non-occluded virus of the field crickets Teleogryllus oceanicus and T. commodus (Orthoptera: Gryllidae) J Invertebr Pathol. 1970;16:214–220. [Google Scholar]
- 48.Riechmann J L, Ito T, Meyerowitz E M. Non-AUG initiation of AGAMOUS mRNA translation in Arabidopsis thaliana. Mol Cell Biol. 1999;19:8505–8512. doi: 10.1128/mcb.19.12.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rueckert R R. Piconaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 609–654. [Google Scholar]
- 50.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 51.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci USA. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasaki J, Nakashima N, Saito H, Noda H. An insect picorna-like virus, Plautia stali intestine virus, has genes of capsid proteins in the 3′ part of the genome. Virology. 1998;244:50–58. doi: 10.1006/viro.1998.9094. [DOI] [PubMed] [Google Scholar]
- 54.Schmid S R, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 55.Scotti P D. Cricket paralysis virus replicates in cultured Drosophila cells. Intervirology. 1976;6:333–342. doi: 10.1159/000149489. [DOI] [PubMed] [Google Scholar]
- 56.Scotti P D, Longworth J F, Plus N, Crozier G, Reinganum C. The biology and ecology of strains of an insect small RNA virus complex. Adv Virus Res. 1981;26:117–143. doi: 10.1016/s0065-3527(08)60422-4. [DOI] [PubMed] [Google Scholar]
- 57.Seipelt J, Guarne A, Bergmann E, James M, Sommergruber W, Fita I, Skern T. The structures of picornaviral proteinases. Virus Res. 1999;62:159–168. doi: 10.1016/s0168-1702(99)00043-x. [DOI] [PubMed] [Google Scholar]
- 58.Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Operons in C. elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 59.Tate J, Liljas L, Scotti P, Christian P, Lin T, Johnson J E. The crystal structure of cricket paralysis virus: the first view of a new virus family. Nat Struct Biol. 1999;6:765–774. doi: 10.1038/11543. [DOI] [PubMed] [Google Scholar]
- 60.Vagner S, Touriol C, Galy B, Audigier S, Gensac M C, Amalric F, Bayard F, Prats H, Prats A C. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 62.Zhu J, Musco M L, Grace M J. Three-color flow cytometry analysis of tricistronic expression of eBFP, eGFP, and eYFP using EMCV-IRES linkages. Cytometry. 1999;37:51–59. [PubMed] [Google Scholar]