Abstract
With the COVID-19 pandemic still wreaking havoc worldwide, new variants being discovered every month in some parts of the globe due to the mutating nature of the virus. There is no specific solution for this highly transmissible disease. In search of a lead molecule for the discovery and development of drug, extensive research is being conducted throughout the world. Many synthetic drugs are already in clinical trials and some are utilized for the treatment of this viral infection. Apart from synthetic drugs, phytocompounds from plants act as a potential drug candidate which can inhibit the growth of virus and thus able to prevent the viral infection. In this study, 26 ligands (bioactive compounds) from Boswellia serrata (an important medicinal plant) were tested against SARS-CoV-2 by using computational method. Selected ligands were shortlisted using Lipinski’s rule and then subjected to molecular docking against one of the main proteins of SARS-CoV-2, i.e., Mpro. Out of these compounds, Euphane, Ursane, α-Amyrin, Phytosterols, and 2,3-Dihydroxyurs-12-en-28-oic acid were potential to inhibit the Mpro activity with binding energies of − 10.47 kcal/mol, − 10.41 kcal/mol, − 9.99 kcal/mol, − 9.94 kcal/mol and − 9.72 kcal/mol respectively. A comparative study was performed using the best five ligands against four possible drug targets of SARS-CoV-2. It was found that Euphane showed highest negative binding energy against all the four crucial targets of SARS-CoV-2. Further, in-vitro experimentation is required to validate the use of Euphane as a potent drug against SARS-CoV-2.
Keywords: SARS-CoV-2, Boswellia serrata, Molecular docking, Phytocompounds, Autodock
Introduction
COVID-19 is one of the most transmissible and deadly diseases in recent years, resulting in the millions of deaths worldwide (Dong et al. 2020). Still, there is no targeted drug or effective treatment developed for this virus because of its pathogenesis and mutation efficiency. Therefore, it is of utmost priority to identify and utilize effective antiviral agents that can be approved as drug candidates against SARS-CoV-2 in order to curb the spread of this pandemic (Jin et al. 2020; Garg et al. 2020; Garg and Roy 2020).
SARS-CoV-2 is an enclosed ss-RNA virus with a non-segmented genome of almost 30 kb in size (Andersen et al. 2020). The structure of SARS-CoV-2 is spherical and belongs to genus betacoronavirus (Lu et al. 2020). SARS-CoV and SARS-CoV-2 share the same spike protein’s structure attached to the viral envelope. It recognizes and bind to Angiotensin-Converting Enzyme-2 (ACE2) and host cell's protease enzyme (Zhou et al. 2020). SARS-CoV-2 has fourteen Open Reading Frames (ORFs) which codes for four structural proteins: spike (S), nucleocapsid (N), envelope (E), and membrane (M) proteins (Li et al. 2020). It also has 5′ and 3′ untranslated regions (UTR), a poly (A) tail and many unknown non-structural ORFs. The S protein, around 150 kDa in size, is responsible for binding of virus to the receptors present on the host cells, which then allows the virus to enter into the host (Kirchdoerfer et al. 2016). N protein is the sole protein that fuses with the RNA genome. It aids in the complete virion transformation by assembling the virus via the process of budding. The confirmation of virus’ envelope is due to the most profuse structural protein (M protein) which is about 25–30 kDa in size and consists of three transmembrane domains (Garg et al. 2020; Garg and Roy 2020). The E protein (8–12 kDa) is also involving in the assembling and budding of virus (Venkatagopalan et al. 2015). In addition, ORF codes for two polyproteins that build the complex involved in viral replication. These then proliferate into new viral particles after being processed further proteolytically by coronavirus main protease (Mpro) enzyme and papain-like protease (PLpro) enzyme (Hilgenfeld 2014).
Mpro is one of the main enzymes of coronaviruses essential to the viral replication and transcription process. SARS-CoV-2 Mpro is similar to SARS-CoV Mpro with about 96% structural similarity (Ullrich and Nitsche 2020). Thus, drug development studies aim to find therapeutic candidates that target this enzyme in order to inhibit viral growth. (Jin et al. 2020). In-silico drug design technologies were used to develop an inhibitor against Mpro of SARS-CoV-2. The active site of SARS-CoV-2 Mpro is determined by the catalytic dyad which is responsible for the maturation of virus (Mirza and Froeyen 2020).
Plants are the most abundant source of bioactive compounds which are used for the potential drug development (Roy and Bharadvaja 2017; Roy 2018; Roy et al. 2018; Roy and Bhatia 2021). Ayurveda has proved to be a fast-growing branch of medicine for treatment of various diseases due to its advantages, such as inexpensiveness, efficiency with no adverse effects, and ability to cater exponentially growing population (Mirza and Froeyen 2020). Boswellia serrata is an Ayurvedic herb belonging to Burseraceae family and commonly known as 'Indian Frankincense' or 'Indian Olibanum.' This plant is native to the dry states of Peninsular India, such as Gujarat, Andhra Pradesh, Jharkhand, Madhya Pradesh and, Chhattisgarh. This traditional plant found in the Eastern part of the globe is proven to have anti-inflammatory effects, helps in relieving congestion due to colds and flu, act as a disinfectant, and even reduce stress and anxiety (Al-Yasiry and Kiczorowska 2016). Biological activities of this plant are due to the presence of various bioactive compounds which includes monoterpenes, diterpenes, sterols, triterpenes, tetracyclic triterpenes like euphane, and pentacyclic triterpenes (Sultana et al. 2013).
In-silico analysis using molecular docking contributes immensely contributes to the drug development process by first screening the vast database of compounds and then reducing the burden of in vitro experiments (Garg and Roy 2020; Roy and Bhatia 2021; Bhatia et al. 2021). These tools apply inbuilt scientific calculation algorithms to the interactions of docked complex between receptor and ligand/drug molecule. The present study aims to find the potential of bioactive compounds of Boswellia serrata against Mpro enzyme of SARS-CoV-2 using computational approach. To determine the interactions of selected molecules against Mpro, computational techniques such as ADME analysis and molecular docking analysis were performed.
Materials and methods
Protein selection and preparation
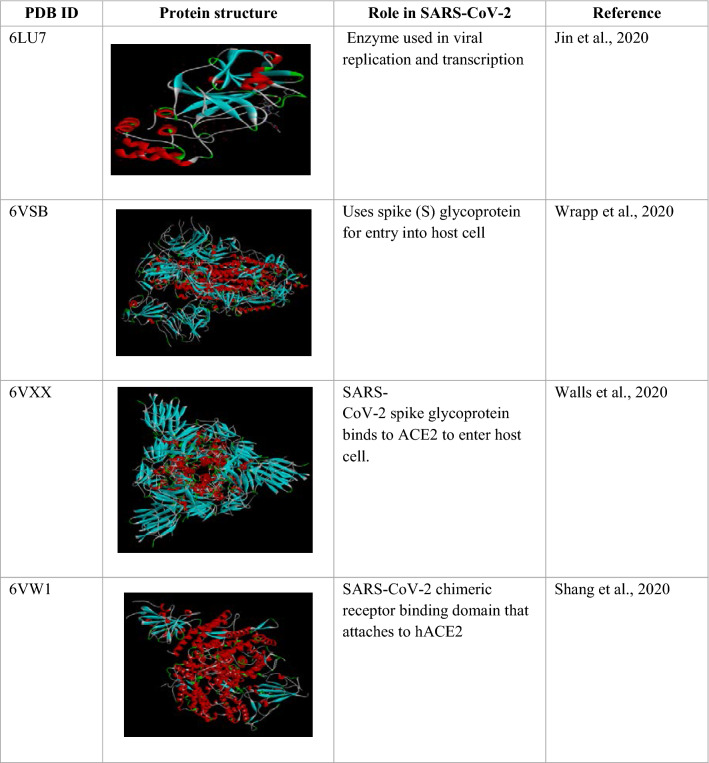
The 3D structure of Mpro enzyme of SARS-CoV-2 from RCSB Protein Data Bank (PDB id: 6LU7) was extracted. Structure consists of a homodimer composed of two unique protein chains-A consisting of 306 amino acids complexed with an inhibitor N3 molecule. Other targets including prefusion SARS-CoV-2 spike protein (PDB id: 6VSB), SARS-CoV-2 spike glycoprotein (PDB id: 6VXX) and SARS-CoV-2 chimeric receptor-binding domain (PDB id: 6VW1) were also taken into consideration for a comparative study (Table 1).
Table 1.
Protein structure and role in SARS-CoV-2
Selection of phytocompounds from Boswellia serrata
A total of 26 bioactive compounds from Boswellia serrata were selected. 3D structures for docking purposes were obtained in '.pdb' format. All the information on phytocompounds was collected from the IMPPAT: Indian Medicinal Plants, Phytochemistry and Therapeutics Database (Mohanraj et al. 2018).
ADME analysis
For evaluating compounds as potential ligands, their Absorption, Distribution, Metabolism, and Excretion properties (ADME) were determined. Lipinski’s rule was applied using a web-based tool named SwissADME (https://www.swissadme.ch/). Any ligands showing violations of Lipinski’s rule were eliminated from further studies.
Active site prediction
To predict the active binding site for most stable ligand–protein interaction, Computed Atlas for Surface Topography of proteins (CASTp) was used (Tian et al. 2018). 10 residues were found in the active binding pocket of Mpro which were identified as follows: THR24, THR26, PHE140, ASN142, GLY143, CYS145, HIS163, HIS164, GLU166, HIS172.
Molecular docking
Molecular docking is a vital computational tool that is utilized for prescreening of compounds in the drug discovery process. It helps in the selection and filtration of potential inhibitors for targeted drug discovery by assessing the binding affinity of protein–ligand complex. Before the process of docking, preparation of protein and ligands was done. Autodock 4.2 was used for docking analysis. Preparing the optimized protein structure involved removing of inhibitor N3 and H2O molecules, adding polar hydrogens, and calculating Kollman charges and Gasteiger charges. Energy minimization of protein was done by using Swiss PDB Viewer. A grid box of 54 × 53 × 38 was calibrated as per the active site prediction of protein with 0.375 Å spacing with the center X: − 11.164, Y: 16.901, Z: 65.46. The Docking Log Files (DLG) were produced for checking the binding energies further search parameter was assigned as “Genetic algorithm” and Lamarckian GA to run the output. The docked complex with maximum negative binding energy was then chosen to visualized the 2D structure to evaluate the protein–ligand interactions. Then for comparative study, three other receptors (6VSB, 6VXX, and 6VW1) of SARS-CoV-2 were docked against selected ligands. Grid box dimensions of 60 × 60 × 60 and coordinates X: 204.457, Y: 199.799, Z: 246.898, 80 × 80 × 80, coordinates X: 223.221, Y: 190.083, Z: 263.907 and 75 × 75 × 75, coordinates X: 85, Y: − 10, Z: 180 respectively were selected with each having grid point spacing of 0.375 Å.
Results and discussion
ADME assessment
All ligands were evaluated based on their physicochemical properties, i.e., ADME analysis before docking analysis. Compounds which did not violate two or more parameters of Lipinski's rule were then selected for further analysis. Compound must possess a molecular weight < 500 Da, Log P < 5, H-bond donors < 5, and hydrogen bond acceptors < 10 in order to follow Lipinski's rule (Table 2).
Table 2.
ADME analysis
| S. no | Compound name | PubChem ID | Analysis | |
|---|---|---|---|---|
| 1 | 11-Keto-β-boswellic acid methyl ester | CID: 101,695,788 | MW | 468.71 g/mol |
| L | 7.39 | |||
| HD | 0 | |||
| HA | 3 | |||
| V | 1 | |||
| 2 | AC1L4F91 | CID: 155,934 | MW | 472.71 |
| L | 6.06 | |||
| HD | 3 | |||
| HA | 3 | |||
| V | 1 | |||
| 3 | α-Campholenic-Acid | CID: 117,235 | MW | 168.24 |
| L | 2.45 | |||
| HD | 1 | |||
| HA | 1 | |||
| V | 0 | |||
| 4 | Euphane | CID: 12,312,921 | MW | 414.76 |
| L | 9.52 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 1 | |||
| 5 | Phytosterols | CID: 12,303,662 | MW | 414.72 |
| L | 8.02 | |||
| HD | 1 | |||
| HA | 1 | |||
| V | 1 | |||
| 6 | α-Amyrin | CID: 225,688 | MW | 426.73 |
| L | 8.02 | |||
| HD | 1 | |||
| HA | 1 | |||
| V | 1 | |||
| 7 | (−)-Camphene | CID: 440,966 | MW | 136.24 |
| L | 3.00 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 0 | |||
| 8 | (+)-α-Phellandrene | CID: 443,160 | MW | 136.24 |
| L | 3.16 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 0 | |||
| 9 | (1S,2R,4S)-(−)-Bornyl acetate | CID: 442,460 | MW | 196.29 |
| L | 2.76 | |||
| HD | 0 | |||
| HA | 2 | |||
| V | 0 | |||
| 10 | 11-Keto-β-boswellic acid | CID: 9,847,548 | MW | 470.69 |
| L | 6.27 | |||
| HD | 2 | |||
| HA | 3 | |||
| V | 1 | |||
| 11 | 3-Acetyl-β-boswellic acid | CID: 11,386,458 | MW | 498.75 |
| L | 7.66 | |||
| HD | 1 | |||
| HA | 3 | |||
| V | 1 | |||
| 12 | 3-Carene | CID: 26,049 | MW | 136.24 |
| L | 3.00 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 0 | |||
| 13 | 3-O-Acetyl-11-Keto-β-Boswellic-Acid | CID: 11,168,203 | MW | 512.73 |
| L | 6.84 | |||
| HD | 1 | |||
| HA | 4 | |||
| V | 2 | |||
| 14 | α-Boswellic acid | CID: 637,234 | MW | 456.71 |
| L | 7.23 | |||
| HD | 2 | |||
| HA | 2 | |||
| V | 1 | |||
| 15 | α-Campholytic-Aci | CID: 11,091 | MW | 154.21 |
| L | 2.06 | |||
| HD | 1 | |||
| HA | 1 | |||
| V | 0 | |||
| 16 | α-Terpinene | CID: 7462 | MW | 136.24 |
| L | 3.31 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 0 | |||
| 17 | β-Boswellic acid | CID: 168,928 | MW | 456.71 |
| L | 7.09 | |||
| HD | 2 | |||
| HA | 2 | |||
| V | 1 | |||
| 18 | β-Pinene | CID: 14,896 | MW | 136.24 |
| L | 3.00 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 0 | |||
| 19 | Dipentene | CID: 22,311 | MW | 136.24 |
| L | 3.31 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 0 | |||
| 20 | l-Arabinose | CID: 439,195 | MW | 150.13 |
| L | − 2.58 | |||
| HD | 4 | |||
| HA | 5 | |||
| V | 0 | |||
| 21 | l-Idose | CID: 11,030,410 | MW | 180.16 |
| L | − 3.22 | |||
| HD | 5 | |||
| HA | 6 | |||
| V | 0 | |||
| 22 | Myrcene | CID: 31,253 | MW | 136.24 |
| L | 3.48 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 0 | |||
| 23 | P-Cymene | CID: 7463 | MW | 134.22 |
| L | 3.12 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 0 | |||
| 24 | Rhamnose | 19,233 | MW | 164.16 |
| L | − 2.35 | |||
| HD | 4 | |||
| HA | 5 | |||
| V | 0 | |||
| 25 | Serratol | 101,618,281 | MW | 290.49 |
| L | 5.96 | |||
| HD | 1 | |||
| HA | 1 | |||
| V | 1 | |||
| 26 | Ursane | CID: 9,548,870 | MW | 412.75 |
| L | 9.13 | |||
| HD | 0 | |||
| HA | 0 | |||
| V | 1 | |||
MW molecular weight, L lipophilicity, HD H bond donor, HA H bond acceptor, V violations
Molecular docking analysis
A molecular docking process was then performed with the selected ligands after ADME analysis. Ligands were ranked according to their binding energies, i.e., one with the least binding energy was ranked at the top, which reflected the maximum stability of the docked complex (Table 3). It was then compared with the control N3 (native inhibitor) which revealed that ten compounds had more binding affinity to the protein because of its higher binding energy as compared to N3 (− 8.15 kcal/mol).
Table 3.
Molecular docking results of 26 ligands with 6LU7
| S no | Ligands | CID | Binding energy (ΔG) (kcal/mol) | Ligand efficiency | Inhibition constant (uM) | Intermolecular energy | Vdw H-bond desolvation |
|---|---|---|---|---|---|---|---|
| 1 | Euphane | 12,312,921 | − 10.47 | − 0.35 | 0.021 | − 11.96 | − 11.95 |
| 2 | Ursane | 9,548,870 | − 10.41 | − 0.35 | 0.023 | − 10.41 | − 10.4 |
| 3 | α-Amyrin | 225,688 | − 9.99 | − 0.32 | 0.047 | − 10.29 | − 10.3 |
| 4 | Phytosterols | 12,303,662 | − 9.94 | − 0.33 | 0.052 | − 12.02 | − 11.97 |
| 5 | 2,3-Dihydroxyurs-12-en-28-oic acid | 155,934 | − 9.72 | − 0.29 | 0.074 | − 10.91 | − 10.99 |
| 6 | 11-Keto-β-boswellic acid methyl ester | 101,695,788 | − 9.48 | − 0.28 | 0.112 | − 10.07 | − 10.09 |
| 7 | 11-Keto-β-boswellic acid | 9,847,548 | − 9.2 | − 0.27 | 0.181 | − 10.09 | − 10.12 |
| 8 | α-Boswellic acid | 637,234 | − 8.94 | − 0.27 | 0.28 | − 9.83 | − 9.88 |
| 9 | 3-Acetyl-β boswellic acid | 11,386,458 | − 8.77 | − 0.24 | 0.37 | − 9.97 | − 10.04 |
| 10 | β-Boswellic acid | 168,928 | − 8.55 | − 0.26 | 0.537 | − 9.45 | − 9.750 |
| 11 | 3-O-Acetyl-11-keto-β-Boswellic Acid | 11,168,203 | − 8.08 | − 0.22 | 1.2 | − 9.27 | − 9.3 |
| 12 | serratol | 101,618,281 | − 7.34 | − 0.35 | 4.19 | − 7.93 | − 7.87 |
| 13 | (1S,2R,4S)-(−)-Bornyl acetate | 442,460 | − 6.24 | − 0.45 | 26.83 | − 6.83 | − 6.68 |
| 14 | ( +)-α-Phellandrene | 443,160 | − 5.41 | − 0.54 | 108.79 | − 5.71 | − 5.7 |
| 15 | β-Pinene | 14,896 | − 5.18 | − 0.52 | 159.39 | − 5.18 | − 5.18 |
| 16 | α-Terpinene | 7462 | − 4.93 | − 0.49 | 243.5 | − 5.23 | − 5.22 |
| 17 | (−)-Camphene | 440,966 | − 4.91 | − 0.49 | 250.93 | − 4.91 | − 4.91 |
| 18 | α-Campholytic acid | 110,918 | − 4.9 | − 0.45 | 0.256 | − 5.5 | − 5.3 |
| 19 | α-Campholenic acid | 117,235 | − 4.81 | − 0.40 | 296.55 | − 5.71 | − 5.40 |
| 20 | Dipentene | 22,311 | − 4.71 | − 0.47 | 355.29 | − 5 | − 5 |
| 21 | 3-Carene | 26,049 | − 4.6 | − 0.46 | 427.54 | − 4.6 | − 4.58 |
| 22 | P-Cymene | 7463 | − 4.57 | − 0.46 | 446.76 | − 4.8 | − 4.87 |
| 23 | l-Idose | 11,030,410 | − 4.49 | − 0.37 | 508.69 | − 6.28 | − 5.86 |
| 24 | Myrcene | 31,253 | − 4.36 | − 0.44 | 638.37 | − 5.55 | − 5.56 |
| 25 | l-Arabinose | 439,195 | − 3.83 | − 0.38 | 1560 | − 5.02 | − 4.72 |
| 26 | Rhamnose | 19,233 | − 3.63 | − 0.33 | 2170 | − 6.02 | − 5.72 |
| 27 | N3 (Control) | − 8.13 | − 0.17 | 1.07 | − 13.54 | − 13.34 |
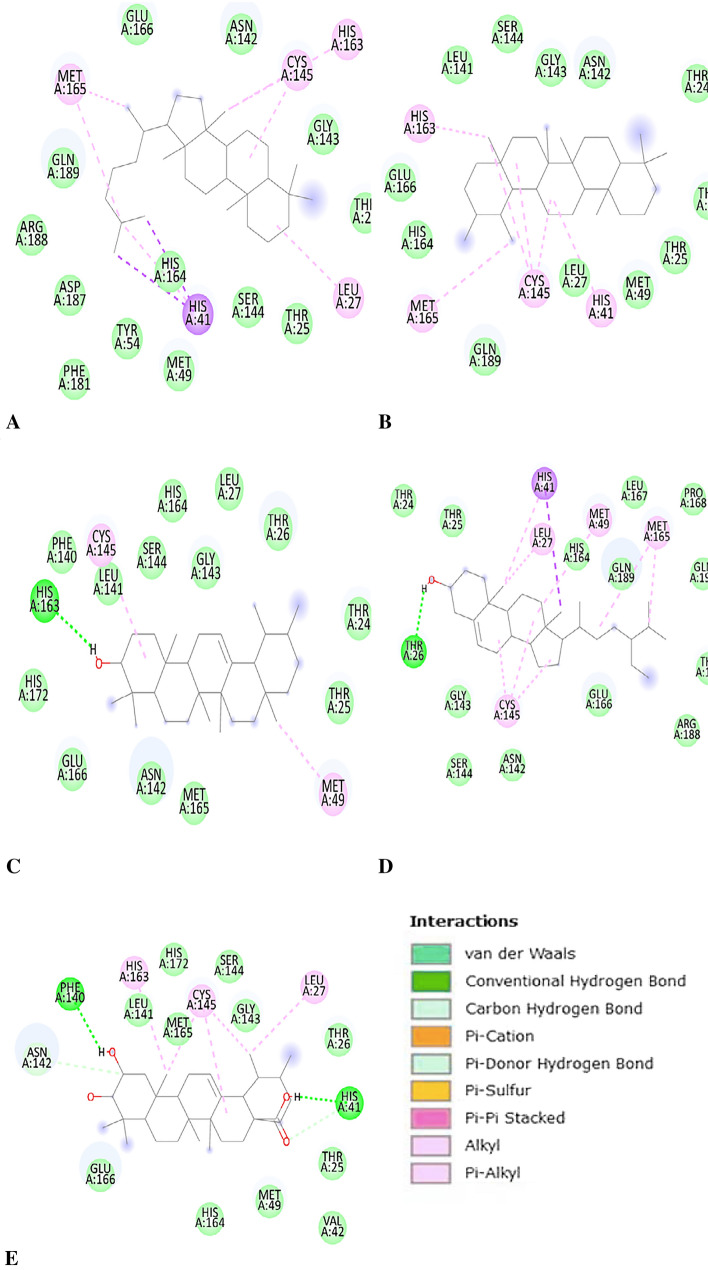
Euphane showed binding energy of − 10.47 kcal/mol, and four different types of bonding interactions were observed, including van der Waals, pi-sigma, alkyl, and pi-alkyl. HIS163, MET165, CYS155, and LEU27 were bonded through pi-alkyl and alkyl bonds, and the remaining residues were weakly bound by van der Waals interactions (Fig. 1).
Fig. 1.
A Interaction of Euphane with Mpro. B Interaction of Ursane with Mpro. C Interaction of α-Amyrin with Mpro. D Interaction of Phytosterol with Mpro. E Interaction of 2,3-dihydroxyurs-12-en-28-oic acid with Mpro
Ursane showed binding energy of − 10.41 kcal/mol. Only three types of hydrophobic interactions were identified. HIS163, CYS145, HIS41, and MET165 are involved in Pi-alkyl and alkyl bonding, while the remaining twelve residues interact with the ligand via van der Waal interactions (Fig. 1).
α-Amyrin-Mpro docked complex showed binding energy of − 9.99 kcal/mol. HIS163 was involved in the H-bonding and CYS145, and MET49 were interacting via the Alkyl bond. Thirteen other residues were interested in maintaining the stability of complex via van der Waals forces (Fig. 1).
Phytosterol indicated the lowest binding energy of − 9.94 kcal/mol. Five different types of bonding between amino acid residues and ligand atoms were observed. THR26 was involved in H-bonding. HIS41 was bonded with the ligand via Pi-sigma and Pi-alkyl bonds. MET49, LEU27, CYS145, and MET165 were also engaged in Pi-alkyl and alkyl interactions. The other thirteen residues in the vicinity stabilize the complex via van der Waal forces (Fig. 1).
2,3-Dihydroxyurs-12-en-28-oic acid showed binding energy of − 9.72 kcal/mol. Five kinds of bonding interactions were seen. PHE140 and HIS41 were associated with hydrogen-bond formations. HIS163, CYS145, and LUE27 formed alkyl and pi-alkyl bonds with the ligand and helped in stabilizing the complex. ASN142 and HIS41 form carbon-hydrogen bonds with the ligand. Twelve other residues could be visualized interacting via van der Waals forces around the ligand (Fig. 1).
The antiviral drug ritonavir, which has already surpassed in-vitro screening and undergoes clinical trials reported to have amino acid residue interactions with Mpro where (thiazoly-5-yl) methylcarbamate oxygen was involved in hydrogen bonding with GLY143 and CYS145 and interacted with THR25, THR26, and LEU27 residues. The other atoms were stabilized at the active site by Pi alkyl and Pi sigma bonds with HIS41 (Kanhed et al. 2021).
One study reported molecular docking analysis of phytocompounds present in Caesalpinia minax. Docking analysis revealed that 5,7-dimethoxyflavanone-4′-O-d-glucopyranoside has binding energy of − 9.23 kcal/mol. It was stabilized by hydrophobic interactions with other residues such as PHE140, LEU141, ASN142. The inhibitor α-ketoamide is a standard inhibitor with an IC50 value of 0.67 ± 0.18 μM against Mpro enzyme of SARS-CoV-2 and binding energy of − 8.24 kcal/mol (Zhang et al. 2020). In another study, similar amino acids were involved in the interaction where residues ASN142, CYS145, and GLU166 were bonded via hydrogen bonding and hydrophobic interactions via THR26, HIS41, PHE140, LEU141, GLY143, SER144, HIS163, HIS164, MET165, PRO168, ASP187, GLN189, THR190 and GLN192 (Gurung et al. 2020). The top five compounds which showed maximum binding affinities i.e. Euphane, Ursane, α-Amyrin, Phytosterol, and 2,3-Dihydroxyurs-12-en-28-oic were further selected for comparative analysis.
Comparative analysis of SARS-CoV-2 spike proteins with selected ligands
After obtaining binding energy of top five compounds docked with Mpro, the compounds with maximum binding energies were then docked against three other protein receptors of SARS-CoV-2, namely 6VSB, 6VXX, and 6VW1, respectively, using Autodock (Table 4). The results were then verified using PatchDock server, which is an open-access tool with a geometry complementarity of protein–ligand molecule-based algorithm online. (http://bioinfo3d.cs.tau.ac.il/PatchDock/). Its principle is built around the geometry of complementarity criteria for docking (Table 5). Patchdock evaluates the features of docked complexes which suggests the optimum molecular interaction between protein and ligand that reports the maximum interaction area covered by the ligand with protein having least steric hindrance. Atomic Contact Energy (ACE) is also evaluated which is defined as the amount of desolvation-free energy required to transfer the ligand from exterior (water) to the protein's interior. PatchDock analysis also validated better efficacy of Euphane amongst the other compounds of Boswellia serrata. Euphane has demonstrated the most potential ligand with highest average patch dock score compared to other phytocompounds.
Table 4.
Comparative analysis of molecular docking analysis of bioactive compounds against four different target protein receptors of SARS-CoV-2
| Compound | Binding energy (kcal/mol) 6LU7 |
Binding energy (kcal/ mol) 6VSB |
Binding energy (kcal/mol) 6VXX |
Binding energy (kcal/mol) 6VW1 |
|---|---|---|---|---|
| α-Amyrin | − 9.99 | − 7.26 | − 9.97 | − 7.9 |
| Euphane | − 10.47 | − 7.62 | − 10.26 | − 7.97 |
| Ursane | − 10.41 | − 6.15 | − 10.77 | − 8.37 |
| Phytosterol | − 9.94 | − 5.44 | − 9.44 | − 8.53 |
| 2,3-Dihydroxyurs-12-en-28-oic acid | − 9.72 | − 5.7 | − 10.33 | − 6.19 |
Table 5.
Validation of manual docking results using online docking software PatchDock
| Compound | Target receptors of SARS-CoV-2 | |||||||
|---|---|---|---|---|---|---|---|---|
| 6LU7 | 6VSB | 6VXX | 6VW1 | |||||
| Score | ACE Value | Score | ACE Value | Score | ACE Value | Score | ACE Value | |
| α-Amyrin | 4812 | − 262.95 | 6026 | − 241.79 | 6234 | − 156.20 | 6126 | − 231.86 |
| Euphane | 5034 | − 253.99 | 6168 | − 197.48 | 6426 | − 166.23 | 6362 | − 226.26 |
| Ursane | 4702 | − 225.18 | 6010 | − 257.70 | 6098 | − 208.65 | 5978 | − 231.58 |
| Phytosterol | 5006 | − 293.50 | 6418 | − 292.27 | 6246 | − 85.60 | 6208 | − 170.06 |
| 2,3-Dihydroxyurs-12-en-28-oic acid | 4804 | − 245.21 | 6184 | − 245.83 | 6082 | − 163.73 | 5772 | − 218.39 |
The Patchdock software was used in various docking analysis. In a recent study on in-silico docking analysis of Nigellidine against spike glycoprotein, Patchdock software was used, and results were analyzed and compared to other phytocompounds of Nigella sativa (Maiti et al. 2020). The ACE value reported for nigellidine with spike glycoprotein was − 340.50. Different values included − 181.08 and − 269.93 for thymoquinone and dithymoquinone, respectively. Nigellicine was also compared but showed lower affinity than nigellidine. Maiti et al. (2020) concluded that nigellidine was a more potent drug against SARS-CoV-2 than thymoquinone, dithymoquinone, and nigellicine. Another study reported inhibition of SARS-CoV-2 spike glycoprotein by tea flavonoids. The top 20 docked results obtained by Patchdock for each complex were selected based on their geometric complementarity analysis and those with the most negligible ACE value. The ligands chosen for the study showed high binding affinities against spike protein (Maiti and Banerjee 2021). The highest ACE value was reported for theaflavin digallate (− 465.17) and theaflavin monogallate (− 434.42), followed by Epigallocatechin 3-gallate (− 407.58). On the other hand, a drug already in use to treat COVID-19, hydroxychloroquine, showed an ACE value of only − 293.32 (Maiti and Banerjee 2021). In a study, Tucatinib, a drug developed for HER2-positive breast cancer, was repurposed against SARS-CoV-2. The docking results with the Mpro enzyme using PatchDock reported docking score of 5640 and ACE value of − 348.62 (Alsalme et al. 2020).
Conclusion
Studies have been conducted in order to find potential drug candidate against SARS-CoV-2. Most of the studies are targeted on finding a cure by using chemically synthesized or previously used drugs (drug repurposing). Since time immemorial, bioactive compounds from plants have also shown great efficacy in controlling diseases, thus utilizing phytocompounds for drug development can help combat COVID-19. From the present study, it can be concluded that the top five ligands, except α-Amyrin, bind to the catalytic dyad amino acid residues of Mpro by different bonding interactions. A comparative study using AutoDock and PATCHDOCK revealed the affinity of top five ligands. It was found that Euphane possesses most significant inhibitory potential against all of four receptors. Further in-vitro experiments are required to validate potential of Euphane as a clinical drug for the treatment of COVID-19.
Funding
Dr. Arpita Roy is thankful to Sharda University for providing seed fund (Seed fund-4 2001 (SUSF2001/12)).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alsalme A, Pooventhiran T, Al-Zaqri N, Rao DJ, Rao SS, Thomas R. Modelling the structural and reactivity landscapes of tucatinib with special reference to its wavefunction-dependent properties and screening for potential antiviral activity. J Mol Model. 2020;26:341. doi: 10.1007/s00894-020-04603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Yasiry A, Kiczorowska B. Frankincense—therapeutic properties. Postępy Higieny i Medycyny Doświadczalnej. 2016;70:380–391. doi: 10.5604/17322693.1200553. [DOI] [PubMed] [Google Scholar]
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–462. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia KS, Garg S, Anand A, Roy A. Evaluation of different phytochemicals against BRCA2 receptor. Biointerface Res Appl Chem. 2021;22:1670–1681. doi: 10.33263/BRIAC122.16701681. [DOI] [Google Scholar]
- Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S, Roy A. In-silico analysis of selected alkaloids against main protease (Mpro) of SARS-CoV-2. Chem Biol Interact. 2020;332:109309. doi: 10.1016/j.cbi.2020.109309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S, Anand A, Lamba Y, Roy A. Molecular docking analysis of selected phytochemicals against SARS-CoV-2 M pro receptor. Vegetos. 2020;33:766–781. doi: 10.1007/s42535-020-00162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung AB, Ali MA, Lee J, Farah MA, Al-Anazi KM. Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in-silico approach. Life Sci. 2020;255:117831. doi: 10.1016/j.lfs.2020.117831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kanhed AM, Patel DV, Teli DM, Patel NR, Chhabria MT, Yadav MR. Identification of potential Mpro inhibitors for the treatment of COVID-19 by using systematic virtual screening approach. Mol Divers. 2021;25:383–401. doi: 10.1007/s11030-020-10130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S, Banerjee A. Epigallocatechin-gallate and theaflavin-gallate interaction in SARS CoV-2 spike-protein central-channel with reference to the hydroxychloroquine interaction: bioinformatics and molecular docking study. Drug Dev Res. 2021;82:86–96. doi: 10.1002/ddr.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S, Banerjee A, Nazmeen A, Kanwar M, Das S. Active-site molecular docking of Nigellidine with nucleocapsid-NSP2-MPro of COVID-19 and to human IL1R-IL6R and strong antioxidant role of Nigella-sativa in experimental rats. J Drug Target. 2020 doi: 10.1080/1061186X.2020.1817040. [DOI] [PubMed] [Google Scholar]
- Mirza M, Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J Pharm Anal. 2020;10:320–328. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraj K, Karthikeyan BS, Vivek-Ananth RP, Chand R, Aparna SR, Mangalapandi P, Samal A. IMPPAT: A curated database of Indian medicinal plants, phytochemistry and therapeutics. Sci Rep. 2018;8:4329. doi: 10.1038/s41598-018-22631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A. Role of medicinal plants against Alzheimer’s disease. Int J Complement Altern Med. 2018;11:205–208. doi: 10.15406/ijcam.2018.11.00398. [DOI] [Google Scholar]
- Roy A, Bharadvaja N. Establishment of the shoot and callus culture of an important medicinal plant Plumbago zeylanica. Adv Plants Agric Res. 2017;7:00274. doi: 10.15406/apar.2017.07.00274. [DOI] [Google Scholar]
- Roy A, Bhatia KS. In silico analysis of plumbagin against cyclin-dependent kinases receptor. Vegetos. 2021;34:50–56. doi: 10.1007/s42535-020-00169-8. [DOI] [Google Scholar]
- Roy A, Krishnan L, Bharadvaja N. Qualitative and quantitative phytochemical analysis of Centella asiatica. Nat Prod Chem Res Article. 2018;6:1–4. doi: 10.4172/2329-6836.1000323. [DOI] [Google Scholar]
- Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana A, Padmaja AR. Boswellia serrata Roxb. a traditional herb with versatile pharmacological activity: a review. Int J Pharm Sci Res. 2013;4:2106–2117. doi: 10.13040/IJPSR.0975-8232.4(6).2106-17. [DOI] [Google Scholar]
- Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S, Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett. 2020;30:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatagopalan P, Daskalova SM, Lopez LA, Dolezal KA, Hogue BG. Coronavirus envelope protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A, Park Y, Tortorici M, Wall A, McGuire A, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett K, Goldsmith J, Hsieh C, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]