Abstract
Centrosomes are multifunctional organelles tasked with organizing the microtubule cytoskeleton required for genome stability, intracellular trafficking, and ciliogenesis. Contributing to the diversity of centrosome functions are cell cycle-dependent oscillations in protein localization and post-translational modifications. Less understood is the role of centrosome-localized messenger RNA (mRNA). Since its discovery, the concept of nucleic acids at the centrosome was controversial, and physiological roles for centrosomal mRNAs remained muddled and underexplored. Over the past decades, however, transcripts, RNA-binding proteins, and ribosomes were detected at the centrosome in various organisms and cell types, hinting at a conservation of function. Indeed, recent work defines centrosomes as sites of local protein synthesis, and defined mRNAs were recently implicated in regulating centrosome functions. In this review, we summarize the evidence for the presence of mRNA at the centrosome and the current work that aims to unravel the biological functions of mRNA localized to centrosomes.
Keywords: RNA localization, centrosome, MTOC, co-translational tranport, post-transcriptional regulation, local translation, mRNA
Introduction
To generate spatial enrichments of specific proteins, cells deploy a variety of strategies, including protein trafficking or local protein synthesis. RNA localization is the process by which mRNAs are enriched at subcellular locales. Often, RNA localization is coupled to translational control, whereby localizing RNAs are translationally repressed until they reach their final destinations. Thus, RNA localization offers an efficient means to generate spatially defined gene enrichments (Martin and Ephrussi, 2009).
Although perhaps best known for its role in developing embryos and oocytes, or highly polarized cells, such as neurons, RNA localization is a fairly ubiquitous post-transcriptional mechanism of gene regulation capable of altering acute cellular responses, like cell migration or division (Kislauskis et al., 1997; Groisman et al., 2000; Holt and Bullock, 2009; Katz et al., 2012). For example, the first observation of a localized mRNA was of β-actin mRNA in an ascidian embryo, later also found at the leading edge of migratory chicken fibroblasts (Jeffery et al., 1983; Lawrence and Singer, 1986). Many excellent reviews about RNA localization are available (Jansen, 2001; Martin and Ephrussi, 2009; Buxbaum et al., 2015; Das et al., 2021). Here, we wish to specifically address the topic of RNA localization to centrosomes. While the localization of RNA to centrosomes is now irrefutable, investigation into its biological significance is finally gaining momentum.
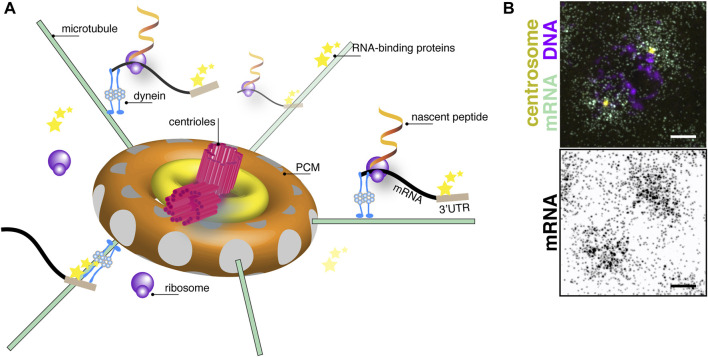
The centrosome is a structured organelle comprising the centrioles, a pair of microtubule-based barrel-shaped structures at the center of the centrosome, and an encompassing protein matrix known as the pericentriolar material (PCM; Figure 1A) (Nigg and Raff, 2009). Although not restricted or compartmentalized by a membrane, the PCM is an organized yet dynamic structure that regulates centrosomal function as the primary microtubule-organizing center of most cells (Mennella et al., 2014; Woodruff et al., 2014). Importantly, cell cycle-dependent oscillations in centrosome structure and composition render the centrosome responsive to cellular demands (e.g., ciliogenesis) or developmental contexts (Khodjakov and Rieder, 1999; Palazzo et al., 2000; Nigg and Raff, 2009). Thus, centrosomes must modulate their activities rapidly, and RNA localization coupled to translational control represents one efficient means to do so.
FIGURE 1.
The centrosome as a center for translational control. (A) Cartoon schematic of centrosome organization showing a central pair of centrioles (magenta) with 9-fold radial symmetry of microtubule triplets surrounded by subconcentric rings of PCM (yellow and orange toroids). Microtubule filaments (green lines) are anchored with their minus-ends docked within γ-tubulin ring complexes (grey circles) embedded within the PCM. A model mRNA (black) recognized by RNA-binding proteins (stars) binding to its 3′-untranslated region (UTR; brown box) is shown undergoing active translation by ribosomes (purple circles). Nascent peptides (fiery ribbons) emerge near the centrosome. Note, objects are not drawn to scale. (B) Image shows single molecule fluorescence in situ hybridization for plp mRNA (green) in a prophase (DNA, magenta) syncytial Drosophila embryo. plp mRNA coalesces around centrosomes (yellow). Image below shows an inverted display of the mRNA channel to maximize contrast. Bar: 5 μm. Image courtesy of Dr. Junnan Fang, Ph.D.
The presence of nucleic acids at the centrosome held dubious importance despite several studies prior to the 2000s hinting at mRNA association with the centrosomes (for historical overview, we recommend (Marshall and Rosenbaum, 2000; Alliegro, 2011)). Progress in the field was hindered by conflicting results arguing about the incidence and relevance of mRNA at centrosomes. Early efforts to interrogate RNA at centrosomes typically involved 1) co-purifying RNA from isolated centrosomes, 2) detection of bulk RNA through non-specific labeling, or 3) treating cells or isolated centrosomes with RNase digestion and monitoring changes to centrosome size and/or microtubule nucleation. The “hammer” approach of RNase treatment led to contradictory findings (reviewed in Marshall and Rosenbaum, 2000), and progress in the field stalled. More refined and physiological approaches less prone to artifacts are now being deployed to systematically investigate defined mRNAs using a full suite of modern technologies. Recent work offers the titillating first glance at RNA function at centrosomes.
Why mRNAs reside at centrosomes is a question under active investigation. Increasing evidence, as we will discuss, supports the idea that some mRNAs are locally translated at the centrosome where their products are needed to tune centrosome activity. Still, other models are possible. For example, local RNA might contribute to centrosome structure or simply “hitch a ride” as a cargo to be passaged on to specific cellular lineages (Lambert and Nagy, 2002; Alliegro et al., 2006; Lerit and Gavis, 2011; Ryder and Lerit, 2018; Woodruff et al., 2018). While these remain intriguing possibilities, this review will focus on the centrosome as a center for translational control, as supported by recent advances.
Delivering the Message: How RNAs Localize
RNA localization is an efficient means to generate local enrichments of protein activities, as a single mRNA may serve as a template for the synthesis of multiple translation products. Thus, localizing mRNA is more cost-effective in terms of cellular resources than localizing individual proteins. Further, because RNA localization is typically coupled to translational control, RNAs packaged within transport granules, or ribonucleoprotein (RNP) complexes, may be considered more inert as compared to their protein products, such as cell fate determinants, proteolytic enzymes, or other factors requiring precise, restricted activity (Das et al., 2021).
RNA localization occurs primarily through three distinct mechanisms: diffusion and entrapment, best exemplified by nanos mRNA in the Drosophila oocyte (Forrest and Gavis, 2003); protection from degradation, as typified by Hsp83 mRNA in the Drosophila embryo (Ding et al., 1993); or active transport, as observed for many RNAs, including ASH1 mRNA in budding yeast (Long et al., 1997; Takizawa et al., 1997; Bertrand et al., 1998). Active transport involves the trafficking of a cargo, such as an RNP, connected via adaptor proteins to molecular motors, which translocate on actin or microtubule cytoskeletal tracks (Bullock, 2011). Because centrosomes serve as sites of microtubule nucleation with microtubule minus-ends embedded within the PCM (Wu and Akhmanova, 2017), centrosomes are structurally suited as a hub for mRNA transport. RNA localization to the centrosome is, however, remarkably specific. As evidenced by genome-wide screens, relatively few transcripts reside at the centrosome (Lecuyer et al., 2007; Wilk et al., 2016; Chouaib et al., 2020; Kwon et al., 2021; Safieddine et al., 2021). The unique distributions of specific mRNAs rely upon cis-sequences, nascent peptides, and/or structural motifs within the RNA being recognized by trans-acting RNA-binding proteins. Multiple events through the RNA lifetime, including splicing, influence RNA localization patterns (Hachet and Ephrussi, 2004; Palacios et al., 2004).
You’ve Got Mail: mRNA at the Centrosome
While non-specific labeling approaches and biochemical purification allowed early researchers to discover RNA at the centrosome, technological advances, including those in mRNA detection and transcriptomics, facilitated the identification of transcripts at the centrosome. Traditional in situ hybridization, which permits subcellular resolution of RNA distributions, remains a mainstay approach to localize mRNAs to centrosomes (Raff et al., 1990; Groisman et al., 2000; Lambert and Nagy, 2002; Kingsley et al., 2007; Lecuyer et al., 2007; Sepulveda et al., 2018; Bergalet et al., 2020). Single molecule fluorescent in situ hybridization approaches offer superior resolution and quantitative advantages (Figure 1B; Femino et al., 1998; Raj et al., 2008; Chouaib et al., 2020; Ryder et al., 2020; Ryder and Lerit, 2020; Kwon et al., 2021; Safieddine et al., 2021). Together, such approaches resolved the localization of specific mRNAs to centrosomes in diverse model systems, including Ilyanassa, Spisula, Drosophila, Xenopus, zebrafish, and mammalian cell lines (Raff et al., 1990; Han et al., 1997; Lambert and Nagy, 2002; Alliegro et al., 2006; Lecuyer et al., 2007; Sepulveda et al., 2018). Genetically encoded RNA aptamer tags, such as the MS2/MCP system, permit visualization of endogenous RNA dynamics in live cells (Bertrand et al., 1998). Recently, CRISPR/CAS-9 genome engineering was utilized to integrate MS2 stem loops into endogenous loci of the centrosome-localized ASPM and NUMA1 mRNAs to monitor physiological and dynamic live trafficking. This study revealed ASPM and NUMA1 mRNAs undergo directed transport toward centrosomes at velocities consistent with an active transport mechanism. Once localized, the RNAs remain anchored near centrosomes (Safieddine et al., 2021). As a whole, these localization-based approaches revealed several specific mRNAs are enriched at centrosomes in Drosophila and mammalian cells, showcasing the localization of mRNA to centrosomes as an evolutionarily conserved phenomenon.
While seeing is believing, transcriptomics approaches offer added insight for unbiased discovery of centrosome enriched mRNAs and subsequent bioinformatics analysis of shared features, including consensus motifs (Blower et al., 2007; Sharp et al., 2011). Presently, transcriptomics approaches remain relatively underutilized in identifying centrosome-specific mRNAs. Expanding transcriptomics approaches may uncover a consensus motif sufficient for mRNA targeting to centrosomes, or conserved motifs overrepresented in centrosome-localized RNAs. Nevertheless, recent advances in proximity RNA profiling, spatial transcriptomics, and related approaches will surely expand the parts list of mRNAs enriched at centrosomes (Jan et al., 2014; Li et al., 2018; Fazal et al., 2019; Alon et al., 2021; Engel et al., 2021; Rao et al., 2021). Once centrosome-associated mRNAs are identified, they must be validated by localization and, ideally, subjected to functional analysis.
Prioritizing Messages in the Inbox
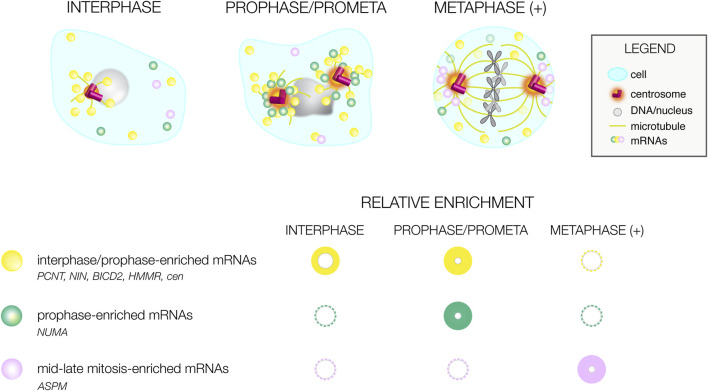
The use of genome-wide RNA localization screening strategies identified conserved mRNAs residing at centrosomes of divergent species (Table 1). Such transcripts should be prioritized for functional analysis, as the conservation of their localization argues for functional relevance. Many of these mRNAs show unique distributions that correlate with cell cycle stage, arguing RNA localization to the centrosome is a dynamic process (Figure 2). Most RNAs appear to preferentially enrich at prophase centrosomes, while relatively few RNAs reside at centrosomes during mid-to-late mitosis (metaphase (+), Figure 2). Moreover, these marked preferences for RNA localization to interphase/early mitotic centrosomes are conserved in human and Drosophila cells, perhaps to support the local synthesis of centrosomal proteins required before mitosis (Sepulveda et al., 2018; Ryder et al., 2020; Ryder and Lerit, 2020; Safieddine et al., 2021). Consistent with this idea, localization of RNAs to centrosomes generally precedes or correlates with the time at which they are translated (Tanenbaum et al., 2015; Safieddine et al., 2021).
TABLE 1.
Conserved mRNAs localizing to centrosomes.
Alphabetical list of mRNAs showing centrosome localization in two or more organisms from two or more independent studies, where “human” refers to human cell culture experiments. *, Centrosomal localization of cen mRNA is observed in multiple species of Drosophila: melanogaster, simulans, and mojavensis and to a lesser extent in virilis (Bergalet et al., 2020).
FIGURE 2.
Cell cycle-dependent variances in mRNA distributions. Illustration of differential mRNA distributions of conserved centrosome-enriched mRNAs during interphase, prophase, and metaphase-to-late mitosis (metaphase (+)). Below, a graphical summary of mRNA distributions at the same cell cycle stages as reported by (Sepulveda et al., 2018; Ryder et al., 2020; Safieddine et al., 2021). The size and intensity of the circle correlates with the prevalence of mRNA localization; representative mRNAs are listed.
Additional conserved targets are informed by transcriptomic analysis of mRNAs associated with taxol-stabilized microtubules from Xenopus and cultured mammalian cells, which identified >100 common transcripts (Blower et al., 2007). Further, several of the transcripts identified as associating with taxol-stabilized microtubules are shared among those RNAs localizing to bona fide centrosomes, including cyclin B1 (CCNB1/cyc B), ninein (NIN), hyaluronan-mediated motility receptor (RHMMR/HMMR), and nuclear mitotic apparatus protein 1 (NUMA1) mRNAs, indicative of partial overlap among these datasets (Blower et al., 2007; Sharp et al., 2011; Pascual et al., 2020). Such congruency is consistent with dynamic, microtubule-dependent localization.
Among these conserved transcripts, the first mRNA identified at centrosomes was cyc B mRNA. Raff and coworkers detected cyc B mRNA associated with spindle poles in a microtubule-dependent manner and further localized cyc B mRNA to anucleated centrosomes within syncytial Drosophila embryos (Raff et al., 1990). Cyc B protein also localizes to centrosomes, where it is required for mitotic progression (Minshull et al., 1989; Murray et al., 1989; Murray and Kirschner, 1989; Hagting et al., 1998; Huang and Raff, 1999; Wakefield et al., 2000). cyc B mRNA also resides at the spindle poles in early Xenopus embryos (Table 1).
The first clue that local mRNA influences centrosome function came from manipulating the RNA-binding protein responsible for targeting cyc B mRNA to spindle poles, CPEB (cytoplasmic polyadenylation element binding), which binds to cognate CPE cites within the cyc B 3′UTR. These experiments revealed that altering cyc B mRNA localization to the centrosome causes spindle morphogenesis defects and mitotic delay (Groisman et al., 2000). Subsequent work revealed the CPE motif is over-represented in datasets of mRNAs enriched on Xenopus and human taxol-stabilized microtubules, raising the possibility that CPEB proteins regulate other centrosome-enriched target mRNAs (Blower et al., 2007). Consistent with this idea, CPEB proteins localize to centrosomes and regulate the expression and localization of the master regulator of centrosome maturation, Plk1 (Groisman et al., 2000; Eliscovich et al., 2008; Pascual et al., 2020). Depletion of CPEB1 in cultured mammalian cells reduces total Plk1 protein expression and recruitment to centrosomes (Pascual et al., 2020). CPEB proteins likely regulate other centrosome-associated genes, including those with mRNAs residing at centrosomes, as suggested by high-throughput datasets obtained using Drosophila and mammalian cell culture systems (Stepien et al., 2016; Pascual et al., 2020).
At Your (Postal) Service: RNA-Binding Proteins and Ribosomes at the Centrosome
Besides CPEB proteins, other RNA-binding proteins, ribosomes, and translation initiation factors also localize to centrosomes (Figure 1A). Further, some RNA-binding proteins are implicated in PCM maintenance or other centrosome functions.
One multifunctional regulator of RNA metabolism is the RNA-binding protein Gle1, best described for its role in mRNA transport (Alcazar-Roman et al., 2006; Weirich et al., 2006), but also implicated in translation initiation and termination through DEAD-box proteins (Bolger et al., 2008). Gle1 localizes to the centrosome and the basal body of cilia, colocalizing with Pericentrin (PCNT) and also required for the recruitment of PCNT and NIN proteins to the centrosome. Further, gle1 depletion impairs microtubule organization and ciliary function (Jao et al., 2017). Given its established role in mRNA transport and translation, Gle1 may promote PCNT and NIN mRNA recruitment to the centrosome, as recent studies note enrichment of these two transcripts at the centrosome through a translation-dependent mechanism (Sepulveda et al., 2018; Chouaib et al., 2020; Kwon et al., 2021; Safieddine et al., 2021).
Although not particularly enriched at centrosomes, Fragile-X Mental Retardation Protein (FMRP) is an RNA-binding protein contributing to centrosome functions and normal mitotic progression. FMRP is encoded by the Fmr1 gene, which, when mutated, is associated with Fragile-X Syndrome (FXS), the most common heritable form of intellectual disability and autism spectrum disorder (Santoro et al., 2012). Loss of FMRP results in disordered microtubules and altered microtubule-dependent intracellular trafficking, which likely influences the pathophysiology of FXS (Yao et al., 2011). Among the putative RNA targets of FMRP, several overlap with CPEB (Costa et al., 2005; Udagawa et al., 2013). FMRP regulates mitotic progression in many tissues, as loss of Fmr1 leads to elevated rates of neural stem cell proliferation, resulting in impaired neurogenesis in Drosophila and, remarkably, in induced pluripotent stem cells (iPSCs) derived from FXS patients (Callan et al., 2010; Raj et al., 2021).
Recent work implicates FMRP as important for the regulation of the centrosome-localized cen mRNA in Drosophila (Ryder et al., 2020). cen mRNA was first localized near spindle poles by a genome-wide RNA localization screen (Lecuyer et al., 2007). Importantly, centrosomal localization of cen mRNA is conserved among various Drosophila species, despite millions of years of evolutionary distance (Table 1; Bergalet et al., 2020). Cen is required for embryonic development, as loss of cen impairs centrosome separation, spindle morphogenesis, and actin cleavage furrow formation, leading to embryonic lethality (Kao and Megraw, 2009). Concurrent work showed cen mRNA assembles into large pericentrosomal RNPs that colocalize with Cen protein during later syncytial embryonic stages (Bergalet et al., 2020; Ryder et al., 2020). Furthermore, puromycylation-proximity ligation assay (puro-PLA) experiments suggest Cen is locally translated near centrosomes (Bergalet et al., 2020). Loss of Fmr1 increases the localization of cen mRNA to Drosophila embryonic centrosomes and enhances translation of Cen protein, suggesting FMRP functions to attenuate cen mRNA localization and translation. Consistently, reduction of cen dosage is sufficient to partially rescue mitotic spindle defects observed in Fmr1 mutants; moreover, cen mRNA and protein associate with FMRP, implicating cen mRNA as an important FMRP target. Finally, mistargeting cen mRNA to the anterior cortex is sufficient to block cen mRNA and protein recruitment to distal centrosomes and recruits excess FMRP (Ryder et al., 2020). Mislocalized cen mRNA disrupts microtubule organization and induces elevated rates of mitotic errors, showcasing local dosage of cen mRNA as a key contributor to centrosome functions (Bergalet et al., 2020; Ryder et al., 2020).
In C. elegans, the RNA-binding protein SZY-20 localizes to the centrosome, suppresses embryonic lethality of the PLK4 ortholog, zyg-1, and restricts centrosome size by impairing recruitment of the PCM components SPD-2, SPD-5, and γ-tubulin; ultimately limiting microtubule-nucleation (Song et al., 2008). Similar findings were observed for Ataxin-2 (ATX-2), a conserved RNA-binding protein associated with spinocerebellar ataxia in humans, which associates with SZY-20 (Stubenvoll et al., 2016). ATX-2 itself localizes to centrosomes and is required for mitotic spindle orientation and successful mitosis (Gnazzo et al., 2016). Further studies are needed to identify relevant mRNA targets of SZY-20 and ATX-2 to understand how they influence centrosome activity.
In response to growth factor stimulation, the RNA-binding protein Hu Antigen R (HuR) is phosphorylated and localizes to centrosomes, relaxing its repression of cyclin A mRNA translation and permitting centrosome amplification characteristic of cancer cells (Filippova et al., 2012; Filippova et al., 2015). The HuR model is one example of how RNA-binding proteins influence centrosomal function by recruiting and stabilizing mRNA at the centrosome until needed for local translation.
Another major protein complex influencing RNA localization and converging on centrosomes is the exon junction complex (EJC). The EJC comprises three protein subunits (Magoh, EIF4A3, and RBM8A (Y14)) and mediates splicing, nonsense-mediated decay, RNA localization, and translation (McMahon et al., 2016). During mouse neurogenesis, loss of Magoh leads to errant mitotic spindle orientation in neuronal progenitors and spindle morphogenesis defects associated with incomplete centrosome separation. Similar phenotypes are observed by depleting other EJC components in cultured mammalian cells. Consequently, Magoh loss results in reduced neural stem cells and precocious neurogenesis, leading to microcephaly (Silver et al., 2010). Further, haploinsufficiency of either Magoh, EIF4A3, or Y14 results in p53-dependent microcephaly in murine models (Mao et al., 2016).
Underscoring this pathophysiology is the localization of EJC components to centrosomes. In cultured mammalian cells, Y14 is significantly enriched at centrosomes (Ishigaki et al., 2014). Likewise, in mouse neural stem cells, EIF4A3 and Y14 localize to the basal body at the base of primary cilia in a microtubule and dynein-dependent manner (Kwon et al., 2021). Although both BICD2 and NIN mRNAs associate with EIF4A3 and Y14 proteins and localize to the ciliary base, only NIN mRNA localization to the basal body is EIF4A3 and Y14-dependent in RPE1 cells. Depletion of EIF4A3 and Y14 also decreases localization of PCNT and γ-tubulin proteins, resulting in impaired microtubule organization and reduced ciliation, consistent with the spindle defects previously observed by Silver and co-workers (Silver et al., 2010; Kwon et al., 2021).
Proteomic analysis from isolated Drosophila centrosomes identified additional translational initiation factors, like EIF4A, and other RNA-binding proteins associated with centrosomes, such as poly(A)-binding protein (Muller et al., 2010). Consistent with a function in centrosome regulation, EIF4A localizes to centrosomes and its depletion impairs recruitment of PCM factors PCNT/PLP, Spd-2, and γ-tubulin, but not centriolar components. These phenotypes are likely unrelated to the role EIF4A plays in translation initiation because disrupting initiation by deleting other members of the EIF4F complex or inhibiting translation elongation by cycloheximide did not diminish the PCM (Muller et al., 2010). Further studies are needed to uncover the mechanism by which EIF4A restricts centrosomal size. As EIF4A promotes expression of oncogenes in pediatric leukemia, understanding how it regulates centrosome activity may inform human disease mechanisms (Wolfe et al., 2014).
Components of the EIF4F initiation complex associate and colocalize with centrosomal OFD1. While OFD1 weakly binds mRNA, the presence of BICC1, an RNA-binding protein also found at the centrosome, allows OFD1 to mediate a stronger association between eIF4F via eIF4E and several centrosomal mRNAs that are implicated in ciliogenesis and renal cyst formation (Iaconis et al., 2017). These studies support the notion that centrosomes serve as hubs for translational control.
Reading the Message: Translation at the Centrosome
The enrichment of ribosomes, mRNA, and translation machinery supports local translation at the centrosome and spindle poles, as evidenced by local puromycylated ribosomes and azidohomoalanine (AHA) to detect nascent peptides (Blower et al., 2007; Bergalet et al., 2020; Pascual et al., 2020). Why is the centrosome a translationally active site? To answer this question, we must first recognize that the centrosome is a center of cellular management. It plays a major role in organizing the microtubule network, nucleating the spindle fibers during cell division, and forming the basal body in ciliated cells. These functions depend on the size and composition of the PCM, which will go through stages of expansion and shedding depending on the cell cycle stage. Altering the composition of the PCM around the centrosome at the mRNA level may be how the centrosome smoothly transitions between its cellular responsibilities. Changing local mRNA levels and translational status is an effective and efficient method of control. When no longer required, the mRNA can then easily be shuttled away in a translationally repressed state or degraded until needed again.
In support of this hypothesis, ribosomes co-purify with microtubules (Goldman and Rebhun, 1969), and ribosomal proteins decorate centrosomes and spindle poles (Blower et al., 2007; Sepulveda et al., 2018; Chouaib et al., 2020; Pascual et al., 2020; Kwon et al., 2021). Persuasive evidence for polyribosomes located near centrosomes comes from ultrastructural analysis; for example, see figure 18 in (Sorokin, 1962) and Plate 10 in (Murray et al., 1965). Similar findings are noted in recent 3D focused ion beam scanning electron microscopy (FIB-SEM) renderings of centrioles and basal bodies (Xu et al., 2017; Muller et al., 2021). Some mRNAs, such as cyc B mRNA, localize to centrosomes independent of translation; indeed, even the localization of ribosomes to centrosomes is translation-independent (Blower et al., 2007). However, many centrosome-enriched transcripts rely upon the presence of intact ribosomes, as determined by puromycin-sensitivity, consistent with a co-translational transport mechanism. These include PCNT, ASPM, NUMA1, HMMR, CEP350, NIN, BICD2, and CCDC88C mRNAs in cultured mammalian cells (Sepulveda et al., 2018; Chouaib et al., 2020; Safieddine et al., 2021) and cen, asp (ASPM), Girdin, mud (NuMA), and BicD mRNAs in Drosophila (Bergalet et al., 2020; Safieddine et al., 2021). Co-imaging nascent peptides using the SunTag system along with endogenous, MS2-aptamer-tagged mRNAs beautifully demonstrates transport of active polysomes translating ASPM and NUMA1 transcripts as they move towards the centrosome (Safieddine et al., 2021). The minus-end directed microtubule motor dynein is implicated in the transport mechanism for some of these mRNAs (e.g., PCNT; Sepulveda et al., 2018). How commonly mRNAs are co-translationally localized by dynein to the centrosomes is still an open question.
Postscript
To date, the rate of discovering RNAs localizing to centrosomes far outpaces their functional characterization, which remains a key bottleneck in the field. Convincing evidence of a direct role for mRNA at centrosomes comes from mistargeting or misexpression analyses of cyc B and cen mRNAs (Groisman et al., 2000; Bergalet et al., 2020; Ryder et al., 2020). These experiments allow experimenters to decipher whether local mRNA or protein affect centrosome functions and should be expanded in future studies. Additional approaches, including the expression of non-translatable transcripts, deletion of identified zipcodes, and mislocalization of aptamer-tagged RNAs will likewise prove informative.
Advances in our ability to detect mRNA at the single-molecule level in vivo, manipulate mRNA localization, and characterize specific protein-mRNA complexes precipitated a recent explosion of research investigating mRNAs at centrosomes, leading to novel insights in a short time. It seems the biological function of mRNA at the centrosome is finally being recognized as a significant regulatory paradigm. However, much work remains to understand which RNAs reside at centrosomes, how they get there, and what, precisely, they are doing.
Acknowledgments
We thank members of the Lerit lab for constructive feedback.
Author Contributions
HZ-S wrote the original draft, and both authors edited the article.
Funding
This work was supported by NIH grants 5K12GM000680 (HZ-S) and R01GM138544 to DL.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Alcázar-Román A. R., Tran E. J., Guo S., Wente S. R. (2006). Inositol Hexakisphosphate and Gle1 Activate the DEAD-Box Protein Dbp5 for Nuclear mRNA export. Nat. Cel Biol 8, 711–716. 10.1038/ncb1427 [DOI] [PubMed] [Google Scholar]
- Alliegro M. C., Alliegro M. A., Palazzo R. E. (2006). Centrosome-associated RNA in Surf Clam Oocytes. Proc. Natl. Acad. Sci. 103, 9034–9038. 10.1073/pnas.0602859103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliegro M. C. (2011). The Centrosome and Spindle as a Ribonucleoprotein Complex. Chromosome Res. 19, 367–376. 10.1007/s10577-011-9186-7 [DOI] [PubMed] [Google Scholar]
- Alon S., Goodwin D. R., Sinha A., Wassie A. T., Chen F., Daugharthy E. R., et al. (2021). Expansion Sequencing: Spatially Precise In Situ Transcriptomics in Intact Biological Systems. Science 371, eaax2656. 10.1126/science.aax2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergalet J., Patel D., Legendre F., Lapointe C., Benoit Bouvrette L. P., Chin A., et al. (2020). Inter-dependent Centrosomal Co-localization of the Cen and Ik2 Cis-Natural Antisense mRNAs in Drosophila. Cel Rep. 30, 3339–3352. 10.1016/j.celrep.2020.02.047 [DOI] [PubMed] [Google Scholar]
- Bertrand E., Chartrand P., Schaefer M., Shenoy S. M., Singer R. H., Long R. M. (1998). Localization of ASH1 mRNA Particles in Living Yeast. Mol. Cel 2, 437–445. 10.1016/s1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- Blower M. D., Feric E., Weis K., Heald R. (2007). Genome-wide Analysis Demonstrates Conserved Localization of Messenger RNAs to Mitotic Microtubules. J. Cel Biol. 179, 1365–1373. 10.1083/jcb.200705163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger T. A., Folkmann A. W., Tran E. J., Wente S. R. (2008). The mRNA export Factor Gle1 and Inositol Hexakisphosphate Regulate Distinct Stages of Translation. Cell 134, 624–633. 10.1016/j.cell.2008.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S. L. (2011). Messengers, Motors and Mysteries: Sorting of Eukaryotic mRNAs by Cytoskeletal Transport. Biochem. Soc. Trans. 39, 1161–1165. 10.1042/bst0391161 [DOI] [PubMed] [Google Scholar]
- Buxbaum A. R., Haimovich G., Singer R. H. (2015). In the Right Place at the Right Time: Visualizing and Understanding mRNA Localization. Nat. Rev. Mol. Cel Biol 16, 95–109. 10.1038/nrm3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan M. A., Cabernard C., Heck J., Luois S., Doe C. Q., Zarnescu D. C. (2010). Fragile X Protein Controls Neural Stem Cell Proliferation in the Drosophila Brain. Hum. Mol. Genet. 19, 3068–3079. 10.1093/hmg/ddq213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouaib R., Safieddine A., Pichon X., Imbert A., Kwon O. S., Samacoits A., et al. (2020). A Dual Protein-mRNA Localization Screen Reveals Compartmentalized Translation and Widespread Co-translational RNA Targeting. Developmental Cel 54, 773–791. 10.1016/j.devcel.2020.07.010 [DOI] [PubMed] [Google Scholar]
- Costa A., Wang Y., Dockendorff T. C., Erdjument-Bromage H., Tempst P., Schedl P., et al. (2005). The Drosophila Fragile X Protein Functions as a Negative Regulator in the Orb Autoregulatory Pathway. Developmental Cel 8, 331–342. 10.1016/j.devcel.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Das S., Vera M., Gandin V., Singer R. H., Tutucci E. (2021). Intracellular mRNA Transport and Localized Translation. Nat. Rev. Mol. Cel Biol 22, 483–504. 10.1038/s41580-021-00356-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D., Parkhurst S. M., Halsell S. R., Lipshitz H. D. (1993). Dynamic Hsp83 RNA Localization during Drosophila Oogenesis and Embryogenesis. Mol. Cel Biol. 13, 3773–3781. 10.1128/mcb.13.6.3773-3781.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliscovich C., Peset I., Vernos I., Méndez R. (2008). Spindle-localized CPE-Mediated Translation Controls Meiotic Chromosome Segregation. Nat. Cel Biol 10, 858–865. 10.1038/ncb1746 [DOI] [PubMed] [Google Scholar]
- Engel K. L., Lo H.-Y. G., Goering R., Li Y., Spitale R. C., Taliaferro J. M. (2021). Analysis of Subcellular Transcriptomes by RNA Proximity Labeling with Halo-Seq. bioRxiv 2021, 447604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal F. M., Han S., Parker K. R., Kaewsapsak P., Xu J., Boettiger A. N., et al. (2019). Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell 178, 473–490. 10.1016/j.cell.2019.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino A. M., Fay F. S., Fogarty K., Singer R. H. (1998). Visualization of Single RNA Transcripts In Situ . Science 280, 585–590. 10.1126/science.280.5363.585 [DOI] [PubMed] [Google Scholar]
- Filippova N., Yang X., King P., Nabors L. B. (2012). Phosphoregulation of the RNA-Binding Protein Hu Antigen R (HuR) by Cdk5 Affects Centrosome Function. J. Biol. Chem. 287, 32277–32287. 10.1074/jbc.m112.353912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova N., Yang X., Nabors L. (2015). Growth Factor Dependent Regulation of Centrosome Function and Genomic Instability by HuR. Biomolecules 5, 263–281. 10.3390/biom5010263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest K. M., Gavis E. R. (2003). Live Imaging of Endogenous RNA Reveals a Diffusion and Entrapment Mechanism for Nanos mRNA Localization in Drosophila . Curr. Biol. 13, 1159–1168. 10.1016/s0960-9822(03)00451-2 [DOI] [PubMed] [Google Scholar]
- Gnazzo M. M., Uhlemann E.-M. E., Villarreal A. R., Shirayama M., Dominguez E. G., Skop A. R. (2016). The RNA-Binding Protein ATX-2 Regulates Cytokinesis through PAR-5 and ZEN-4. MBoC 27, 3052–3064. 10.1091/mbc.e16-04-0219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R. D., Rebhun L. I. (1969). The Structure and Some Properties of the Isolated Mitotic Apparatus. J. Cel Sci 4, 179–209. 10.1242/jcs.4.1.179 [DOI] [PubMed] [Google Scholar]
- Groisman I., Huang Y.-S., Mendez R., Cao Q., Theurkauf W., Richter J. D. (2000). CPEB, Maskin, and Cyclin B1 mRNA at the Mitotic Apparatus. Cell 103, 435–447. 10.1016/s0092-8674(00)00135-5 [DOI] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A. (2004). Splicing of Oskar RNA in the Nucleus Is Coupled to its Cytoplasmic Localization. Nature 428, 959–963. 10.1038/nature02521 [DOI] [PubMed] [Google Scholar]
- Hagting A., Karlsson C., Clute P., Jackman M., Pines J. (1998). MPF Localization Is Controlled by Nuclear export. EMBO J. 17, 4127–4138. 10.1093/emboj/17.14.4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. W., Park J. H., Kim M., Lee J. (1997). mRNAs for Microtubule Proteins Are Specifically Colocalized during the Sequential Formation of Basal Body, Flagella, and Cytoskeletal Microtubules in the Differentiation of Naegleria Gruberi. J. Cel Biol 137, 871–879. 10.1083/jcb.137.4.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C. E., Bullock S. L. (2009). Subcellular mRNA Localization in Animal Cells and Why it Matters. Science 326, 1212–1216. 10.1126/science.1176488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-y., Raff J. W. (1999). The Disappearance of Cyclin B at the End of Mitosis Is Regulated Spatially in Drosophila Cells. EMBO J. 18, 2184–2195. 10.1093/emboj/18.8.2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconis D., Monti M., Renda M., Van Koppen A., Tammaro R., Chiaravalli M., et al. (2017). The Centrosomal OFD1 Protein Interacts with the Translation Machinery and Regulates the Synthesis of Specific Targets. Sci. Rep. 7, 1224. 10.1038/s41598-017-01156-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Nakamura Y., Tatsuno T., Hashimoto M., Iwabuchi K., Tomosugi N. (2014). RNA-binding Protein RBM8A (Y14) and MAGOH Localize to Centrosome in Human A549 Cells. Histochem. Cel Biol 141, 101–109. 10.1007/s00418-013-1135-4 [DOI] [PubMed] [Google Scholar]
- Jan C. H., Williams C. C., Weissman J. S. (2014). Principles of ER Cotranslational Translocation Revealed by Proximity-specific Ribosome Profiling. Science 346, 1257521. 10.1126/science.1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.-P. (2001). mRNA Localization: Message on the Move. Nat. Rev. Mol. Cel Biol 2, 247–256. 10.1038/35067016 [DOI] [PubMed] [Google Scholar]
- Jao L.-E., Akef A., Wente S. R. (2017). A Role for Gle1, a Regulator of DEAD-Box RNA Helicases, at Centrosomes and Basal Bodies. MBoC 28, 120–127. 10.1091/mbc.e16-09-0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery W. R., Tomlinson C. R., Brodeur R. D. (1983). Localization of Actin Messenger RNA during Early Ascidian Development. Developmental Biol. 99, 408–417. 10.1016/0012-1606(83)90290-7 [DOI] [PubMed] [Google Scholar]
- Kao L.-R., Megraw T. L. (2009). Centrocortin Cooperates with Centrosomin to Organize Drosophila Embryonic Cleavage Furrows. Curr. Biol. 19, 937–942. 10.1016/j.cub.2009.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Z. B., Wells A. L., Park H. Y., Wu B., Shenoy S. M., Singer R. H. (2012). -Actin mRNA Compartmentalization Enhances Focal Adhesion Stability and Directs Cell Migration. Genes Development 26, 1885–1890. 10.1101/gad.190413.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L. (1999). The Sudden Recruitment of γ-Tubulin to the Centrosome at the Onset of Mitosis and its Dynamic Exchange throughout the Cell Cycle, Do Not Require Microtubules. J. Cel Biol. 146, 585–596. 10.1083/jcb.146.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley E. P., Chan X. Y., Duan Y., Lambert J. D. (2007). Widespread RNA Segregation in a Spiralian Embryo. Evol. Dev. 9, 527–539. 10.1111/j.1525-142x.2007.00194.x [DOI] [PubMed] [Google Scholar]
- Kislauskis E. H., Zhu X.-c., Singer R. H. (1997). β-Actin Messenger RNA Localization and Protein Synthesis Augment Cell Motility. J. Cel Biol. 136, 1263–1270. 10.1083/jcb.136.6.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O. S., Mishra R., Safieddine A., Coleno E., Alasseur Q., Faucourt M., et al. (2021). Exon junction Complex Dependent mRNA Localization Is Linked to Centrosome Organization during Ciliogenesis. Nat. Commun. 12, 1351. 10.1038/s41467-021-21590-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. D., Nagy L. M. (2002). Asymmetric Inheritance of Centrosomally Localized mRNAs during Embryonic Cleavages. Nature 420, 682–686. 10.1038/nature01241 [DOI] [PubMed] [Google Scholar]
- Lawrence J., Singer R. H. (1986). Intracellular Localization of Messenger RNAs for Cytoskeletal Proteins. Cell 45, 407–415. 10.1016/0092-8674(86)90326-0 [DOI] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., et al. (2007). Global Analysis of mRNA Localization Reveals a Prominent Role in Organizing Cellular Architecture and Function. Cell 131, 174–187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Lerit D. A., Gavis E. R. (2011). Transport of Germ Plasm on Astral Microtubules Directs Germ Cell Development in Drosophila . Curr. Biol. 21, 439–448. 10.1016/j.cub.2011.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Aggarwal M. B., Ke K., Nguyen K., Spitale R. C. (2018). Improved Analysis of RNA Localization by Spatially Restricted Oxidation of RNA-Protein Complexes. Biochemistry 57, 1577–1581. 10.1021/acs.biochem.8b00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K., Jansen R.-P. (1997). Mating Type Switching in Yeast Controlled by Asymmetric Localization of ASH1 mRNA. Science 277, 383–387. 10.1126/science.277.5324.383 [DOI] [PubMed] [Google Scholar]
- Mao H., Mcmahon J. J., Tsai Y.-H., Wang Z., Silver D. L. (2016). Haploinsufficiency for Core Exon Junction Complex Components Disrupts Embryonic Neurogenesis and Causes P53-Mediated Microcephaly. Plos Genet. 12, e1006282. 10.1371/journal.pgen.1006282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F., Rosenbaum J. L. (2000). Are There Nucleic Acids in the Centrosome? Curr. Top. Dev. Biol. 49, 187–205. 10.1016/s0070-2153(99)49009-x [DOI] [PubMed] [Google Scholar]
- Martin K. C., Ephrussi A. (2009). mRNA Localization: Gene Expression in the Spatial Dimension. Cell 136, 719–730. 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcmahon J. J., Miller E. E., Silver D. L. (2016). The Exon junction Complex in Neural Development and Neurodevelopmental Disease. Int. J. Dev. Neurosci. 55, 117–123. 10.1016/j.ijdevneu.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V., Agard D. A., Huang B., Pelletier L. (2014). Amorphous No More: Subdiffraction View of the Pericentriolar Material Architecture. Trends Cel Biol. 24, 188–197. 10.1016/j.tcb.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J., Blow J. J., Hunt T. (1989). Translation of Cyclin mRNA Is Necessary for Extracts of Activated xenopus Eggs to Enter Mitosis. Cell 56, 947–956. 10.1016/0092-8674(89)90628-4 [DOI] [PubMed] [Google Scholar]
- Müller A., Schmidt D., Xu C. S., Pang S., D'costa J. V., Kretschmar S., et al. (2021). 3D FIB-SEM Reconstruction of Microtubule-Organelle Interaction in Whole Primary Mouse Beta Cells. J. Cel Biol 220, e202010039. 10.1083/jcb.202010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H., Schmidt D., Steinbrink S., Mirgorodskaya E., Lehmann V., Habermann K., et al. (2010). Proteomic and Functional Analysis of the Mitotic Drosophila Centrosome. EMBO J. 29, 3344–3357. 10.1038/emboj.2010.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Kirschner M. W. (1989). Cyclin Synthesis Drives the Early Embryonic Cell Cycle. Nature 339, 275–280. 10.1038/339275a0 [DOI] [PubMed] [Google Scholar]
- Murray A. W., Solomon M. J., Kirschner M. W. (1989). The Role of Cyclin Synthesis and Degradation in the Control of Maturation Promoting Factor Activity. Nature 339, 280–286. 10.1038/339280a0 [DOI] [PubMed] [Google Scholar]
- Murray R. G., Murray A. S., Pizzo A. (1965). The fine Structure of Mitosis in Rat Thymic Lymphocytes. J. Cel Biol 26, 601–619. 10.1083/jcb.26.2.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E. A., Raff J. W. (2009). Centrioles, Centrosomes, and Cilia in Health and Disease. Cell 139, 663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Palacios I. M., Gatfield D., St Johnston D., Izaurralde E. (2004). An eIF4AIII-Containing Complex Required for mRNA Localization and Nonsense-Mediated mRNA Decay. Nature 427, 753–757. 10.1038/nature02351 [DOI] [PubMed] [Google Scholar]
- Palazzo R. E., Vogel J. M., Schnackenberg B. J., Hull D. R., Wu X. (2000). Centrosome Maturation. Curr. Top. Dev. Biol. 49, 449–470. 10.1016/s0070-2153(99)49021-0 [DOI] [PubMed] [Google Scholar]
- Pascual R., Segura-Morales C., Omerzu M., Bellora N., Belloc E., Castellazzi C. L., et al. (2020). mRNA Spindle Localization and Mitotic Translational Regulation by CPEB1 and CPEB4. RNA 27, 291–302. 10.1261/rna.077552.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff J. W., Whitfield W. G., Glover D. M. (1990). Two Distinct Mechanisms Localise Cyclin B Transcripts in Syncytial Drosophila Embryos. Development 110, 1249–1261. 10.1242/dev.110.4.1249 [DOI] [PubMed] [Google Scholar]
- Raj A., Van Den Bogaard P., Rifkin S. A., Van Oudenaarden A., Tyagi S. (2008). Imaging Individual mRNA Molecules Using Multiple Singly Labeled Probes. Nat. Methods 5, 877–879. 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj N., Mceachin Z. T., Harousseau W., Zhou Y., Zhang F., Merritt-Garza M. E., et al. (2021). Cell-type-specific Profiling of Human Cellular Models of Fragile X Syndrome Reveal PI3K-dependent Defects in Translation and Neurogenesis. Cel Rep. 35, 108991. 10.1016/j.celrep.2021.108991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A., Barkley D., França G. S., Yanai I. (2021). Exploring Tissue Architecture Using Spatial Transcriptomics. Nature 596, 211–220. 10.1038/s41586-021-03634-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder P. V., Fang J., Lerit D. A. (2020). c Entrocortin RNA Localization to Centrosomes Is Regulated by FMRP and Facilitates Error-free Mitosis. J. Cel Biol 219, e202004101. 10.1083/jcb.202004101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder P. V., Lerit D. A. (2020). Quantitative Analysis of Subcellular Distributions with an Open-Source, Object-Based Tool. Biol. Open 9, bio055228. 10.1242/bio.055228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder P. V., Lerit D. A. (2018). RNA Localization Regulates Diverse and Dynamic Cellular Processes. Traffic 19, 496–502. 10.1111/tra.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safieddine A., Coleno E., Salloum S., Imbert A., Traboulsi A.-M., Kwon O. S., et al. (2021). A Choreography of Centrosomal mRNAs Reveals a Conserved Localization Mechanism Involving Active Polysome Transport. Nat. Commun. 12, 1352. 10.1038/s41467-021-21585-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M. R., Bray S. M., Warren S. T. (2012). Molecular Mechanisms of Fragile X Syndrome: a Twenty-Year Perspective. Annu. Rev. Pathol. Mech. Dis. 7, 219–245. 10.1146/annurev-pathol-011811-132457 [DOI] [PubMed] [Google Scholar]
- Sepulveda G., Antkowiak M., Brust-Mascher I., Mahe K., Ou T., Castro N. M., et al. (2018). Co-translational Protein Targeting Facilitates Centrosomal Recruitment of PCNT during Centrosome Maturation in Vertebrates. Elife 7, e34959. 10.7554/eLife.34959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. A., Plant J. J., Ohsumi T. K., Borowsky M., Blower M. D. (2011). Functional Analysis of the Microtubule-Interacting Transcriptome. MBoC 22, 4312–4323. 10.1091/mbc.e11-07-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D. L., Watkins-Chow D. E., Schreck K. C., Pierfelice T. J., Larson D. M., Burnetti A. J., et al. (2010). The Exon junction Complex Component Magoh Controls Brain Size by Regulating Neural Stem Cell Division. Nat. Neurosci. 13, 551–558. 10.1038/nn.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. H., Aravind L., Müller-Reichert T., O'connell K. F. (2008). The Conserved Protein SZY-20 Opposes the Plk4-Related Kinase ZYG-1 to Limit Centrosome Size. Developmental Cel 15, 901–912. 10.1016/j.devcel.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. (1962). Centrioles and the Formation of Rudimentary Cilia by Fibroblasts and Smooth Muscle Cells. J. Cel Biol 15, 363–377. 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien B. K., Oppitz C., Gerlach D., Dag U., Novatchkova M., Krüttner S., et al. (2016). RNA-binding Profiles of Drosophila CPEB Proteins Orb and Orb2. Proc. Natl. Acad. Sci. USA 113, E7030–E7038. 10.1073/pnas.1603715113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubenvoll M. D., Medley J. C., Irwin M., Song M. H. (2016). ATX-2, the C. elegans Ortholog of Human Ataxin-2, Regulates Centrosome Size and Microtubule Dynamics. Plos Genet. 12, e1006370. 10.1371/journal.pgen.1006370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P. A., Sil A., Swedlow J. R., Herskowitz I., Vale R. D. (1997). Actin-dependent Localization of an RNA Encoding a Cell-Fate Determinant in Yeast. Nature 389, 90–93. 10.1038/38015 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M. E., Stern-Ginossar N., Weissman J. S., Vale R. D. (2015). Regulation of mRNA Translation during Mitosis. Elife 4, e07957. 10.7554/eLife.07957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T., Farny N. G., Jakovcevski M., Kaphzan H., Alarcon J. M., Anilkumar S., et al. (2013). Genetic and Acute CPEB1 Depletion Ameliorate Fragile X Pathophysiology. Nat. Med. 19, 1473–1477. 10.1038/nm.3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J. G., Huang J.-y., Raff J. W. (2000). Centrosomes Have a Role in Regulating the Destruction of Cyclin B in Early Drosophila Embryos. Curr. Biol. 10, 1367–1370. 10.1016/s0960-9822(00)00776-4 [DOI] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner J., Weis K. (2006). Activation of the DExD/H-Box Protein Dbp5 by the Nuclear-Pore Protein Gle1 and its Coactivator InsP6 Is Required for mRNA export. Nat. Cel Biol 8, 668–676. 10.1038/ncb1424 [DOI] [PubMed] [Google Scholar]
- Wilk R., Hu J., Blotsky D., Krause H. M. (2016). Diverse and Pervasive Subcellular Distributions for Both Coding and Long Noncoding RNAs. Genes Dev. 30, 594–609. 10.1101/gad.276931.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. L., Singh K., Zhong Y., Drewe P., Rajasekhar V. K., Sanghvi V. R., et al. (2014). RNA G-Quadruplexes Cause eIF4A-dependent Oncogene Translation in Cancer. Nature 513, 65–70. 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J. B., Hyman A. A., Boke E. (2018). Organization and Function of Non-dynamic Biomolecular Condensates. Trends Biochem. Sci. 43, 81–94. 10.1016/j.tibs.2017.11.005 [DOI] [PubMed] [Google Scholar]
- Woodruff J. B., Wueseke O., Hyman A. A. (2014). Pericentriolar Material Structure and Dynamics. Phil. Trans. R. Soc. B 369, 20130459. 10.1098/rstb.2013.0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Akhmanova A. (2017). Microtubule-Organizing Centers. Annu. Rev. Cel Dev. Biol. 33, 51–75. 10.1146/annurev-cellbio-100616-060615 [DOI] [PubMed] [Google Scholar]
- Xu C. S., Hayworth K. J., Lu Z., Grob P., Hassan A. M., García-Cerdán J. G., et al. (2017). Enhanced FIB-SEM Systems for Large-Volume 3D Imaging. Elife 6, e25916. 10.7554/eLife.25916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao A., Jin S., Li X., Liu Z., Ma X., Tang J., et al. (2011). Drosophila FMRP Regulates Microtubule Network Formation and Axonal Transport of Mitochondria. Hum. Mol. Genet. 20, 51–63. 10.1093/hmg/ddq431 [DOI] [PubMed] [Google Scholar]