Abstract
An understanding of the spatio-temporal distribution of several groups of mastitis pathogens can help to inform programs for the successful control and management of mastitis. However, in the absence of an active surveillance program such information is not readily available. In this retrospective study we analyzed passive surveillance data from a diagnostic laboratory with an aim to describe the spatio-temporal trend of major mastitis pathogens between 2008 and 2017 in Ontario dairy cattle. Data for all milk culture samples submitted to the Animal Health Laboratory (AHL) at the University of Guelph between 2008 and 2017 was accessed. Descriptive analyses were conducted to identify the major pathogens and Chi-square goodness-of-fit tests were used to compare between multiple proportions. Likewise, univariable logistic regression analysis was performed to determine if there was a change in the probability of isolating the major mastitis pathogens depending on geography or time. Seasonality was assessed by calculating the seasonal relative risk (RR). Of a total of 85,979 milk samples examined, more than half of the samples (61.07%) showed no growth and the proportion of samples that showed no growth almost halved during the study period. Of the samples (36.21%, n = 31,133) that showed any growth, the major bacterial pathogens were Staphylococcus aureus (15.60%), Non-aureus Staphylococci (NAS) (5.04%), Corynebacterium spp. (2.96%), and Escherichia coli (2.00%). Of the NAS, the major species reported were Staphylococcus chromogenes (69.02%), Staphylococcus simulans (14.45%), Staphylococcus epidermidis (12.99%), and Staphylococcus hyicus (2.13%). A temporal change in the prevalence of contagious pathogens like S. aureus and Corynebacterium spp. was observed with an increasing odds of 1.06 and 1.62, respectively. Likewise, except for Trueperella pyogenes, the prevalence of all the major environmental mastitis pathogens increased during the study period. The isolation of most of the pathogens peaked in summer, except for S. aureus, T. pyogenes, and Streptococcus dysgalactiae which peaked in spring months. Interestingly, a regional pattern of isolation of some bacterial pathogens within Ontario was also observed. This study showed a marked spatio-temporal change in the prevalence of major mastitis pathogens and suggests that a regional and seasonal approach to mastitis control could be of value.
Keywords: spatio-temporal variation, prevalence, mastitis pathogens, Canada, bovine
Introduction
Mastitis, the inflammation of the mammary gland, is one of the most important production limiting diseases in dairy cattle (1). Farmers have prioritized mastitis as the second most important dairy cattle disease based on the National Dairy Survey in Canada (2). Although the case fatality rate for mastitis is low, it adversely affects farm profitability and can result in significant economic losses (1, 3). Costs due to both clinical and sub-clinical forms of the disease include production costs, treatment costs, and prevention costs (3–5). In addition, there are indirect costs to farmers due to the penalty imposed on milk with a high somatic cell count (6). While the relative importance of the cost components may vary according to the clinical forms of the disease, losses associated with both clinical and sub-clinical forms of mastitis can amount to an average of Canadian $662 per milking animal per annum for Canadian dairy farmers (5). Mastitis is also important from an animal welfare point of view as clinical mastitis causes pain in the animals (7).
The relationships between pathogen, animal, and farm environment and management factors play an important role in the causation, and hence management and treatment of mastitis (8, 9). The management of mastitis is particularly challenging as mastitis in dairy cattle is caused by several groups of pathogens like the most frequently isolated Staphylococcus aureus, and opportunistic pathogens like Streptococcus species, Escherichia coli, Klebsiella species, and coagulase-negative Staphylococci (3, 9). The relative frequency of isolation of these pathogens can vary temporally or spatially and can also depend on the production stage of the animal and the clinical form of the disease (2, 10–15). The treatment of mastitis in cows involves the use of antimicrobials via intramammary and/or parenteral routes, which has been associated with antimicrobial resistance of the mastitis pathogens and future treatment failure (16). This is especially important when antibiotics are used in dairy cows as blanket dry cow therapy without the identification of the causative pathogen and its antimicrobial sensitivity profile (17–20). Antimicrobial resistance (if developed against antibiotics that are critically important for human use) will be detrimental to public health as some of the mastitis pathogens are also human pathogens, opportunistic human pathogens, or can transmit antimicrobial resistance genes to human pathogens. Therefore, information on the pathogens isolated from animals with mastitis will (1) improve situational awareness related to mastitis pathogens in the Ontario dairy population, (2) support the design of mastitis control plans, and (3) help clinicians to identify the hazards that are present in the Ontario dairy environment.
While many cross sectional studies have documented the most common mastitis causing pathogens in Ontario (2, 10–15), understanding how the prevalence of these pathogens has changed over time is an important area of focus. In this study, we estimate the prevalence of mastitis pathogens and describe the overall spatio-temporal trend for the major mastitis pathogens between 2008 and 2017 in Ontario dairy cattle.
Methods
This retrospective study examined laboratory data for all milk culture samples submitted to the Animal Health Laboratory (AHL) at the University of Guelph between 2008 and 2017. These milk samples comprise samples that were routinely submitted to the laboratory for only culture or culture and susceptibility testing. Samples were submitted to the AHL by veterinarians as a part of mastitis testing and are expected to represent a sample from a quarter of an animal udder, while some of the samples were composite samples. Additional information on the clinical stage of the disease, and/or treatment history of the animal was not available.
Bacterial Isolation
At the AHL, a standard microbiological isolation technique was followed to isolate bacteria from a milk sample. Briefly, 10 μl of milk were inoculated on Columbia blood agar (BA) and, if <20 milk samples were submitted, also on MacConkey (MAC) agar. Blood agar plates were incubated at 35°C in the presence of 5% CO2 whereas MAC agar plates were incubated at ambient air at 35°C. All plates were checked for the presence of bacterial growth after 24 and 48 h of incubation. In addition, all milk samples were incubated at 35°C aerobically up to 5 days if no bacterial growth was detected on the initial culture. After 5 days of incubation the milk samples were re-plated on BA only and incubated and checked for the presence of bacterial growth as specified above. Before 2011 bacterial identification was done biochemically following standard operating procedures at AHL whereas from 2011 until 2017 bacterial identification was achieved using matrix assisted time-of-flight mass spectroscopy (MALDI-TOF MS). Briefly, individual bacterial colonies were smeared on stainless steel plate and covered by matrix [α-cyano-4-hydroxycinnamic acid (HCCA)]. The results were read using the Bruker MALDI Biotyper. All major mastitis pathogens were reported. If no bacterial growth was detected the results were reported as no bacterial growth. If bacterial growth was detected but there were no major mastitis pathogens present the results were reported as no bacterial pathogens. Overgrowth with mold and/or Proteus spp. and numerous bacterial species was reported as overgrowth with contaminants.
Data Management and Statistical Analysis
The milk culture data obtained from AHL was tabulated using a spreadsheet (Microsoft® Excel® for Office 365). A single entry represents a test performed on a milk sample from a single mammary quarter, or composite milk from all or some quarters of an animal. Duplicate entries were removed. It was expected that the milk samples tested at the AHL would be representative of milk samples tested for mastitis in the province of Ontario, Canada as the Ontario Ministry of Agriculture, Food, and Rural Affairs (OMAFRA) subsidizes the laboratory costs for samples submitted to the AHL. Hence, only milk samples submitted from farms with an Ontario address were retained in the dataset. In this study we retained the test results corresponding to general bacteriological examination of the milk samples.
For qualitative analysis, pathogens that constituted at least 1.5% of the total isolates were selected for further analysis except for Streptococcus dysgalactiae which was included because of its known importance as a mastitis pathogen. This included bacteria representing contagious mastitis pathogens (S. aureus and Corynebacterium spp.), environmental mastitis pathogens [E. coli, non-aureus Staphylococci (NAS), Trueperella pyogenes, and S. dysgalactiae], environmental mastitis pathogen that can also act as contagious mastitis pathogen (Streptococcus uberis) (21), fungi (all yeast, mold, and other fungus spp. aggregated), and algae (Prototheca spp.) that are frequently isolated from mastitis milk samples and are important causes of bovine mastitis (2, 10–15). The pathogens of genus Corynebacterium (Corynebacterium bovis, Corynebacterium ulcerans, and all other Corynebacterium spp.) were aggregated as Corynebacterium spp. and treated as contagious mastitis pathogen as the majority of the isolates reported at the species level included the contagious mastitis pathogen Corynebacterium bovis. Likewise, NAS includes all Staphylococci other than S. aureus, like Staphylococcus agnetis, Staphylococcus capitis, Staphylococcus caprae, Staphylococcus chromogenes, Staphylococcus epidermidis, Staphylococcus equorum, Staphylococcus haemolyticus, Staphylococcus hyicus, Staphylococcus sciuri, Staphylococcus simulans, Staphylococcus xylosus, and those not reported at species level. While most of these pathogens that are grouped as NAS are coagulase negative (formerly grouped as coagulase negative staphylococci), some others are coagulase positive and coagulase variable Staphylococci.
Date of sample submission was categorized by both month and season—Winter (Dec 21–March 19), Spring (March 20–June 19), Summer (June 20–Sep 21), and Autumn/Fall (Sep 22–Dec 20). Year was modeled as a continuous variable. Likewise, the location of the submitting clinic was categorized into Eastern Ontario, Central Ontario, Southwestern Ontario, and Northern Ontario regions (22). The clinics in Toronto was included in Central Ontario. We used R (23) to perform all the statistical analysis. Chi squared goodness-of-fit tests were used to compare between multiple proportions. Univariable logistic regression analysis was performed to determine if the probability of isolating the major mastitis pathogens (outcome) changed with year, month, and/or geography (24). Seasonal relative risk (RR) and the confidence interval (CI) was calculated as described by Brookhart et al. and is interpreted as a RR measure that compares the month with highest incidence to that of the month with the lowest incidence also called Peak-Low Ratio analysis (25). Statistical significance was determined at P < 0.05.
Results
Between 2008 and 2017, a total of 85,979 milk samples were submitted to the AHL for general bacteriological examination resulting in 91,802 test results. The 85,979 samples included 92 composite milk samples.
Frequency of the Major Mastitis-Associated Pathogens
Between January 1, 2008 and December 31, 2017, the AHL performed milk culture on 85,979 milk samples from dairy cattle in the province of Ontario. The number of samples subjected to milk culture are shown in Table 1. Over the years, the average number of milk samples submitted by a clinic was approximately reduced by half. Consequently, the number of samples tested more than halved from 13,339 to 5,891, resulting in a corresponding decrease in the test results.
Table 1.
The final data included a total of 85,979 milk samples that were cultured between 2008 and 2017 in the Animal Health Laboratory, Ontario, Canada.
| Year | Number of sample submissions | Average number of samples per submission | Total number of samples tested | Total milk culture test results | Average number of test results per sample |
|---|---|---|---|---|---|
| 2008 | 1,929 | 6.91 | 13,339 | 13,473 | 1.01 |
| 2009 | 1,786 | 5.18 | 9,249 | 9,375 | 1.01 |
| 2010 | 1,711 | 5.36 | 9,169 | 9,281 | 1.01 |
| 2011 | 1,905 | 5.40 | 10,289 | 10,373 | 1.01 |
| 2012 | 2,376 | 4.21 | 9,992 | 10,161 | 1.02 |
| 2013 | 1,964 | 4.45 | 8,747 | 8,943 | 1.02 |
| 2014 | 1,637 | 4.05 | 6,625 | 7,988 | 1.21 |
| 2015 | 1,883 | 3.35 | 6,312 | 7,365 | 1.17 |
| 2016 | 1,866 | 3.41 | 6,366 | 7,459 | 1.17 |
| 2017 | 1,769 | 3.33 | 5,891 | 7,384 | 1.25 |
| Total | 18,826 | 4.57 | 85,979 | 91,802 | 1.07 |
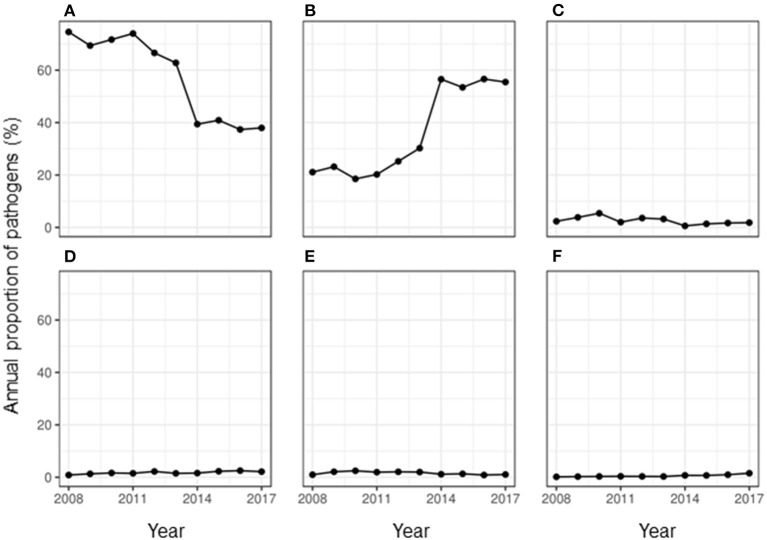
More than half of the milk samples (61.07%, n = 85,979) showed no growth of any group of pathogens. The peak season for observing no growth in milk culture was summer with a RR of 1.21 (95% CI: 1.12–1.31) compared to that in winter. During the study period, the proportion of samples that showed no growth almost halved in the year 2017 when compared to the year 2008 (Figure 1). This overall decreasing trend with each year was found to be significant with an odds ratio (OR) of 0.82 (95% CI: 0.81–0.82).
Figure 1.
Annual proportion of the major classes of pathogens isolated from milk samples submitted to the University of Guelph, Animal Health Laboratory, Ontario, Canada between 2008 and 2017. (A) No growth, (B) bacteria, (C) contamination, (D) algae, (E) fungus, and (F) mixed growth.
Less than half of the samples (36.21%, n = 85,979) showed any growth, while 2.73% of the samples showed contamination. The overall RR of observing contamination was significantly higher during summer compared to that in winter (RR: 21.69, 95% CI: 2.65–177.68). While the proportion of samples showing contamination fluctuated during the study period, an overall decrease in proportion per year was observed (OR = 0.93, 95% CI: 0.91–0.94) compared to the baseline year of 2008. During the study period, the proportion of isolation of bacteria and algae from a milk sample doubled (Figure 1). Of the uncontaminated milk culture results, 89.60% of the samples (n = 31,133) yielded bacterial isolates only, while 1.31% showed mixed cultures of bacteria, fungi and/or algae. Among the major pathogens isolated from the milk samples, most of them were bacterial pathogens (Figure 2) and the top five bacterial species constituted more than two-thirds (78.20%) of the total isolates. Among the fungi, most of the isolates were yeasts (99.56%, n = 1,607). Among the Corynebacterium spp., 14.02% (359) of the isolates were reported as Corynebacterium bovis, three as Corynebacterium ulcerans while rest of the isolates were not reported at the species level. Likewise, of the 5.04% of NAS, less than a half of the NAS (44.36%, n = 4,337) were reported at species level and of those identified at species level, most of them comprised S. chromogenes (69.02%), S. simulans (14.45%), S. epidermidis (12.99%), and S. hyicus (2.13%).
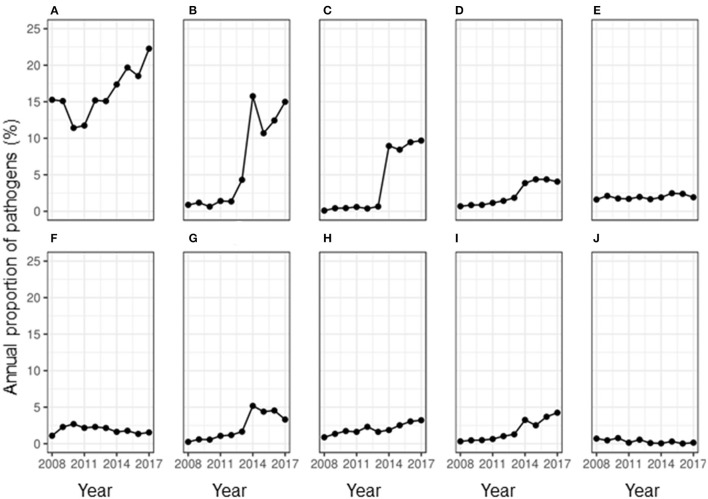
Figure 2.
Annual proportion of major mastitis pathogens isolated from 85,988 milk samples between 2008 and 2017 in Ontario, Canada. (A) Staph. aureus, (B) non-aureus Staphylococci, (C) Corynebacterium spp., (D) E. coli, (E) Trueperella pyogenes, (F) Algae, (G) Streptococcus uberis, (H) Fungus, (I) Strep. dysgalactiae, and (J) Streptococcus agalactiae.
Temporal Change in Frequency of Isolation of Major Mastitis Pathogens in Milk Samples in Ontario
Contagious Mastitis Pathogens
During the study period, there was a fluctuating trend in the proportion of contagious mastitis pathogens isolated from milk. When the number of samples positive for a pathogen was assessed, there was a significant overall increase in the proportion of samples positive for the major contagious mastitis pathogens during the study period (Figure 2). In the year 2008, 15.27% of all the milk samples that were cultured isolated S. aureus. While there was a decrease in the proportion of samples isolating S. aureus in the year 2010 and 2011, an overall increase in prevalence of S. aureus was observed during the study period, such that in 2017, S. aureus was isolated from 22.27% of the milk samples. While the prevalence of Corynebacterium spp. remarkably increased in the milk samples. during the study period, the positivity rate of Streptococcus agalactiae almost halved (Figure 2). Likewise, the prevalence of S. uberis, some strains of which are known to act as a contagious pathogen, also remarkably increased in the milk samples. during the study period (Figure 2). The annual odds of increase were 1.06 for S. aureus and 1.62 for Corynebacterium spp. (Table 2).
Table 2.
Result of univariable regression analysis of the proportion of samples positive for major mastitis pathogens by year.
| Pathogens | Odds ratio | P-value |
|---|---|---|
| (95% Confidence interval) | (Chi square) | |
| Staphylococcus aureus | 1.06 (1.05, 1.07) | <0.01 |
| Non-aureus Staphylococci | 1.47 (1.45, 1.49) | <0.01 |
| Corynebacterium spp. | 1.62 (1.59, 1.65) | <0.01 |
| Escherichia coli | 1.27 (1.25, 1.29) | <0.01 |
| Trueperella pyogenes | 1.03 (1.01, 1.05) | <0.01 |
| Streptococcus dysgalactiae | 1.36 (1.33, 1.39) | <0.01 |
| Algae (Prototheca spp.) | 0.99 (0.97, 1.01) | 0.22 |
| Fungi | 1.13 (1.11, 1.15) | <0.01 |
| Streptococcus uberis | 1.33 (1.31, 1.36) | <0.01 |
| Contamination | 0.93 (0.91, 0.94) | <0.01 |
| No growth | 0.82 (0.81, 0.82) | <0.01 |
P, Probability.
Environmental Mastitis Pathogens
Figure 2 show that there was an increase in the proportion of each major environmental pathogen in the milk samples from the year 2008 to 2017. There was a corresponding 17-fold increase in the prevalence of NAS, an environmental pathogen (Figure 2) during the study period. Likewise, the proportion of samples positive for E. coli increased more than 5-fold from 0.69 to 4.06% and the samples positive for S. dysgalactiae increased more than 13-fold during this time. On contrary, the proportion of samples positive for T. pyogenes remained similar throughout the study period. The annual odds of increase of prevalence per year were significant for all the environmental mastitis pathogens (Table 2).
Minor Mastitis Pathogens
Figure 2 shows that the proportion of samples positive for Algae (Prototheca spp.) changed from 1.09% in the year 2008 to 1.54% in the year 2017 without any significant increase or decrease per year from the base year of 2008 (OR = 0.99; 95% CI: 0.97–1.01, P-value = 0.22). Likewise, the proportion of samples positive for fungi increased from <1 to 3.21% during the study period with an annual odds of increase of 1.13 (95% CI: 1.11–1.15, P-value < 0.01) from the year 2008.
Seasonality
Tables 3–5 show variation in the monthly frequency of isolation of the mastitis pathogens. The odds of increase or decrease of the prevalence of the pathogens during the months of year compared to that in January showed that there was an increasing odds of the prevalence of pathogens like S. aureus during March and April and decreasing odds of isolation during the summer months of August to September (Tables 6–8). Likewise, a higher odds of isolating NAS and E. coli from milk was observed in the spring and summer months compared to that in January. In contrast, the odds of isolating S. dysgalactiae from a milk sample was lower during summer, although this observation was not statistically significant. While compared to January the odd of isolating fungus was higher for both the summer months (July–September) and autumn months (October and November), the highest odds of isolating algae from a milk sample was observed in winter (February) when compared to that in January.
Table 3.
Temporal (monthly) and spatial change in the proportion of bovine milk samples positive for various mastitis pathogens between 2008 and 2017 in Ontario, Canada.
| Parameters | Percentage of samples showing growth of different mastitis pathogens | Total samples per category | |||
|---|---|---|---|---|---|
| Category | Staphylococcus aureus | Corynebacterium spp. | Streptococcus uberis | ||
| Month | January | 7.90 | 4.32 | 5.94 | 6,728 |
| February | 7.85 | 6.99 | 7.49 | 7,001 | |
| March | 10.48 | 7.81 | 7.93 | 7,172 | |
| April | 9.85 | 10.76 | 7.68 | 6,648 | |
| May | 7.78 | 8.09 | 0.85 | 6,461 | |
| June | 8.90 | 7.15 | 8.42 | 7,539 | |
| July | 10.00 | 10.80 | 11.64 | 9,151 | |
| August | 9.11 | 11.54 | 12.94 | 9,595 | |
| September | 7.96 | 7.54 | 8.79 | 7,439 | |
| October | 7.64 | 7.62 | 7.86 | 7,009 | |
| November | 6.73 | 11.58 | 7.06 | 6,073 | |
| December | 5.80 | 5.81 | 5.39 | 5,163 | |
| Location | Central Ontario | 8.35 | 8.13 | 8.85 | 7,165 |
| Eastern Ontario | 28.00 | 31.92 | 20.25 | 22,220 | |
| Northern Ontario | 2.93 | 2.87 | 2.04 | 1,952 | |
| Southwestern Ontario | 60.72 | 57.09 | 68.85 | 54,642 | |
| Total (N) | 13,411 | 2,547 | 1,615 | 85,979 | |
Table 5.
Temporal (monthly) and spatial change in the proportion of samples positive for algae and fungi in bovine milk samples between 2008 and 2017 in Ontario, Canada.
| Parameters | Category | Number of samples showing growth of different minor mastitis pathogens (% of total samples per year) | Total samples per category | |
|---|---|---|---|---|
| Algae (Prototheca spp.) | Fungi (several spp.) | |||
| Month | January | 8.47 | 5.97 | 6,728 |
| February | 12.00 | 6.34 | 7,001 | |
| March | 10.05 | 7.15 | 7,172 | |
| April | 5.48 | 6.22 | 6,648 | |
| May | 7.92 | 5.22 | 6,461 | |
| June | 9.20 | 7.34 | 7,539 | |
| July | 8.47 | 12.06 | 9,151 | |
| August | 9.26 | 13.18 | 9,595 | |
| September | 8.17 | 12.44 | 7,439 | |
| October | 6.89 | 10.20 | 7,009 | |
| November | 7.19 | 7.84 | 6,073 | |
| December | 6.89 | 6.03 | 5,163 | |
| Location | Central Ontario | 9.45 | 10.07 | 7,165 |
| Eastern Ontario | 20.72 | 28.54 | 22,220 | |
| Northern Ontario | 1.89 | 0.56 | 1,952 | |
| Southwestern Ontario | 67.95 | 60.82 | 54,642 | |
| Total (n) | 1,641 | 1,608 | 85,979 | |
Table 6.
Regression analysis of the proportion of samples positive for various mastitis pathogens by month.
| Categories of explanatory variable month | Outcome as proportion of samples positive for each of the mastitis pathogens | ||
|---|---|---|---|
| Staphylococcus aureus | Corynebacterium spp. | Streptococcus uberis | |
| Odds ratio (95% confidence interval) | Odds ratio (95% confidence interval) | Odds ratio (95% confidence interval) | |
| Overall P-value ≤ 0.01 | Overall P-value ≤ 0.01 | Overall P-value ≤ 0.01 | |
| January | 1.00 | 1.00 | 1.00 |
| February | 0.95 (0.86, 1.04) | 1.57 (1.24, 2.00)* | 1.21 (0.93, 1.59) |
| March | 1.30 (1.19, 1.42)* | 1.72 (1.36, 2.18)* | 1.25 (0.96, 1.64) |
| April | 1.33 (1.21, 1.45)* | 2.59 (2.07, 3.25)* | 1.31 (1.00, 1.72) |
| May | 1.03 (0.94, 1.13) | 1.98 (1.57, 2.51)* | 1.56 (1.21, 2.03)* |
| June | 1.01 (0.92, 1.10) | 1.49 (1.17, 1.90)* | 1.27 (0.98, 1.66) |
| July | 0.92 (0.84, 1.00) | 1.86 (1.5, 2.34)* | 1.46 (1.14, 1.87)* |
| August | 0.78 (0.71, 0.85)* | 1.9 (1.53, 2.38)* | 1.54 (1.21, 1.97)* |
| September | 0.9 (0.82, 0.98)* | 1.59 (1.26, 2.03)* | 1.34 (1.04, 1.75)* |
| October | 0.92 (0.83, 1.01) | 1.71 (1.36, 2.18)* | 1.27 (0.98, 1.67) |
| November | 0.93 (0.85, 1.03) | 3.07 (2.47, 3.85)* | 1.32 (1.01, 1.74)* |
| December | 0.95 (0.86, 1.05) | 1.78 (1.39, 2.28)* | 1.18 (0.88, 1.59) |
Represent significant odds ratios.
Table 8.
Regression analysis of the proportion of samples positive for mastitis pathogens, no growth, and contamination by month.
| Categories of explanatory variable month | Outcome as proportion of samples positive for mastitis pathogens, contamination, or no growth | |||
|---|---|---|---|---|
| Algae (Prototheca spp.) | Fungi (several genus) | Contamination | No growth | |
| Odds ratio (95% confidence interval) overall P-value ≤ 0.01 | Odds ratio (95% confidence interval) overall P-value ≤ 0.01 | Odds ratio (95% confidence interval) overall P-value ≤ 0.01 | Odds ratio (95% confidence interval) overall P-value ≤ 0.01 | |
| January | 1.00 | 1.00 | 1.00 | 1.00 |
| February | 1.37 (1.10, 1.71)* | 1.02 (0.77, 1.35) | 0.67 (0.50, 0.91)* | 1.02 (0.95, 1.09) |
| March | 1.12 (0.89, 1.40) | 1.13 (0.86, 1.48) | 0.79 (0.60, 1.05) | 0.75 (0.70, 0.81)* |
| April | 0.65 (0.50, 0.85)* | 1.06 (0.80, 1.40) | 0.80 (0.60, 1.06) | 0.76 (0.71, 0.82)* |
| May | 0.97 (0.76, 1.24) | 0.91 (0.68, 1.22) | 2.01 (1.59, 2.57)* | 0.81 (0.75, 0.86)* |
| June | 0.97 (0.77, 1.22) | 1.10 (0.84, 1.44) | 2.96 (2.38, 3.71)* | 0.72 (0.67, 0.77)* |
| July | 0.73 (0.58, 0.93)* | 1.50 (1.17, 1.92)* | 3.55 (2.88, 4.41)* | 0.68 (0.64, 0.73)* |
| August | 0.76 (0.60, 0.96)* | 1.56 (1.23, 2.00)* | 3.31 (2.69, 4.12)* | 0.79 (0.74, 0.85)* |
| September | 0.87 (0.68, 1.11) | 1.91 (1.50, 2.45)* | 2.83 (2.27, 3.55)* | 0.80 (0.75, 0.86)* |
| October | 0.78 (0.60, 1.00) | 1.66 (1.29, 2.14)* | 1.49 (1.17, 1.92)* | 0.90 (0.84, 0.97)* |
| November | 0.94 (0.73, 1.20) | 1.46 (1.12, 1.92)* | 0.67 (0.49, 0.91)* | 0.87 (0.81, 0.94)* |
| December | 1.06 (0.82, 1.36) | 1.32 (0.99, 1.76) | 1.14 (0.86, 1.52) | 0.93 (0.86, 1.00) |
Represent significant odds ratios.
Table 4.
Temporal (monthly) and spatial change in the proportion of bovine milk samples positive for various environmental mastitis pathogens between 2008 and 2017 in Ontario, Canada.
| Parameters | Number of samples showing growth of different environmental mastitis pathogens (% of total samples) | Total samples per category | ||||
|---|---|---|---|---|---|---|
| Category | Non-aureus Staphylococci | E. coli | Trueperella pyogenes | Streptococcus dysgalactiae | ||
| Month | January | 5.42 | 5.86 | 8.82 | 7.47 | 6,728 |
| February | 5.26 | 4.06 | 18.76 | 5.11 | 7,001 | |
| March | 9.25 | 5.63 | 9.12 | 13.22 | 7,172 | |
| April | 10.72 | 4.76 | 9.74 | 10.23 | 6,648 | |
| May | 7.06 | 8.53 | 8.33 | 9.05 | 6,461 | |
| June | 8.97 | 10.74 | 8.21 | 8.89 | 7,539 | |
| July | 16.16 | 13.23 | 8.70 | 8.42 | 9,151 | |
| August | 13.19 | 14.39 | 8.76 | 9.91 | 9,595 | |
| September | 9.68 | 11.38 | 6.80 | 7.32 | 7,439 | |
| October | 6.11 | 8.59 | 7.96 | 7.00 | 7,009 | |
| November | 4.82 | 7.78 | 8.33 | 7.08 | 6,073 | |
| December | 3.37 | 5.05 | 6.49 | 6.29 | 5,163 | |
| Location | Central Ontario | 5.53 | 2.26 | 7.90 | 4.64 | 7,165 |
| Eastern Ontario | 18.75 | 18.69 | 25.05 | 21.95 | 22,220 | |
| Northern Ontario | 1.38 | 1.74 | 2.57 | 3.54 | 1,952 | |
| Southwestern Ontario | 74.34 | 77.31 | 64.48 | 69.87 | 54,642 | |
| Total (n) | 4,337 | 1,723 | 1,633 | 1,271 | 85,979 | |
Table 7.
Regression analysis of the proportion of samples positive for environmental mastitis pathogens by month.
| Categories of explanatory variable month | Outcome as proportion of samples positive for each of the mastitis pathogens | |||
|---|---|---|---|---|
| Non-aureus Staphylococci | Escherichia coli | Trueperella pyogenes | Streptococcus dysgalactiae | |
| Odds ratio (95% confidence interval) | Odds ratio (95% confidence interval) | Odds ratio (95% confidence interval) | Odds ratio (95% confidence interval) | |
| Overall P-value ≤ 0.01 | Overall P-value ≤ 0.01 | Overall P-value ≤ 0.01 | Overall P-value ≤ 0.01 | |
| January | 1.00 | 1.00 | 1.00 | 1.00 |
| February | 0.93 (0.77, 1.12) | 0.66 (0.49, 0.90)* | 0.95 (0.75, 1.2) | 0.65 (0.47, 0.90)* |
| March | 1.64 (1.39, 1.93)* | 0.9 (0.68, 1.19) | 0.97 (0.77, 1.22) | 1.67 (1.30, 2.17)* |
| April | 2.08 (1.77, 2.44)* | 0.82 (0.61, 1.10) | 1.12 (0.89, 1.41) | 1.39 (1.07, 1.82)* |
| May | 1.37 (1.16, 1.64)* | 1.53 (1.18, 1.98)* | 0.98 (0.78, 1.25) | 1.27 (0.96, 1.67) |
| June | 1.5 (1.28, 1.78)* | 1.65 (1.30, 2.11)* | 0.83 (0.65, 1.05) | 1.06 (0.81, 1.40) |
| July | 2.29 (1.97, 2.67)* | 1.68 (1.33, 2.13)* | 0.72 (0.57, 0.91)* | 0.83 (0.63, 1.09) |
| August | 1.75 (1.5, 2.05)* | 1.74 (1.38, 2.21)* | 0.69 (0.55, 0.87)* | 0.93 (0.71, 1.22) |
| September | 1.65 (1.41, 1.95)* | 1.78 (1.40, 2.27)* | 0.69 (0.54, 0.89)* | 0.88 (0.66, 1.18) |
| October | 1.09 (0.91, 1.3) | 1.42 (1.10, 1.83)* | 0.86 (0.68, 1.1) | 0.9 (0.67, 1.20) |
| November | 0.98 (0.81, 1.19) | 1.48 (1.14, 1.92)* | 1.05 (0.83, 1.33) | 1.05 (0.78, 1.40) |
| December | 0.8 (0.65, 0.99)* | 1.12 (0.84, 1.50) | 0.96 (0.74, 1.23) | 1.1 (0.81, 1.48) |
Represent significant odds ratios.
Likewise, Peak-Low Ratio analysis (Table 9) showed that during a year the isolation of most of the pathogens peaked in summer, except for S. aureus, T. pyogenes, and S. dysgalactiae which peaked in spring.
Table 9.
Seasonal relative risk of isolation of major mastitis pathogens between 2008 and 2017 in Ontario, Canada.
| Isolates | Peak/Low ratio | Peak season |
|---|---|---|
| (95% confidence interval) | ||
| Staphylococcus aureus | 1.36 (1.16, 1.59) | Spring |
| Corynebacterium spp. | 1.35 (1, 1.93) | Summer |
| Non-aureus Staphylococci | 2.76 (1.96, 3.89) | Summer |
| Escherichia coli | 3.04 (1.72, 5.38) | Summer |
| Trueperella pyogenes | 1.12 (1, 1.73) | Spring |
| Streptococcus dysgalactiae | 1.38 (1, 2.29) | Spring |
| Fungi | 1.08 (1, 1.68) | Spring |
| Algae (Prototheca spp.) | 2.11 (1.27, 3.49) | Summer |
| Contamination | 21.69 (2.65, 177.68) | Summer |
| No growth | 1.21 (1.12, 1.31) | Summer |
Spatial Change in Frequency of Isolation of Major Mastitis Pathogens in Milk Samples at Ontario
Tables 3–5 show that frequency of isolation of the major pathogens varied by geographical regions, however, such variation was only significant for some of the pathogens (Table 10). Compared to Central Ontario, in Eastern Ontario, there was a significantly increased odds of samples being positive for S. aureus, and Corynebacterium spp. while there were decreased odds of a sample being positive for S. uberis. In all regions of Ontario, there was an increased odds of isolation of E. coli, and S. dysgalactiae compared to Central Ontario. In contrast, compared to that of Central Ontario, a significant decreasing odds of isolation of algae and fungi was noted in Eastern Ontario (Table 10).
Table 10.
Univariable regression analysis of the proportion of samples showing contamination or no growth and those positive for major contagious and environmental mastitis pathogens by location.
| Pathogens | Locations | Odds ratio (95% confidence interval) | P-value |
|---|---|---|---|
| Staphylococcus aureus | Eastern Ontario | 1.10 (1.02, 1.18)* | <0.01 |
| Northern Ontario | 1.36 (1.20, 1.54)* | ||
| Southwestern Ontario | 0.94 (0.88, 1.01) | ||
| Central Ontario | 1.00 | ||
| Streptococcus uberis | Eastern Ontario | 0.73 (0.60, 0.90)* | <0.01 |
| Northern Ontario | 0.84 (0.57, 1.22) | ||
| Southwestern Ontario | 1.02 (0.86, 1.22) | ||
| Central Ontario | 1.00 | ||
| Corynebacterium spp. | Eastern Ontario | 1.28 (1.10, 1.49)* | <0.01 |
| Northern Ontario | 1.31 (0.99, 1.71) | ||
| Southwestern Ontario | 0.92 (0.79, 1.07) | ||
| Central Ontario | 1.00 | ||
| Non-aureus Staphylococcus | Eastern Ontario | 1.1 (0.95, 1.27) | <0.01 |
| Northern Ontario | 0.92 (0.68, 1.21) | ||
| Southwestern Ontario | 1.81 (1.59, 2.07)* | ||
| Central Ontario | 1.00 | ||
| Escherichia coli | Eastern Ontario | 2.69 (1.95, 3.81)* | <0.01 |
| Northern Ontario | 2.85 (1.75, 4.59)* | ||
| Southwestern Ontario | 4.57 (3.37, 6.39)* | ||
| Central Ontario | 1.00 | ||
| Trueperella pyogenes | Eastern Ontario | 1.02 (0.84, 1.25) | 0.64 |
| Northern Ontario | 1.2 (0.83, 1.69) | ||
| Southwestern Ontario | 1.07 (0.89, 1.29) | ||
| Central Ontario | 1.00 | ||
| Streptococcus dysgalactiae | Eastern Ontario | 1.53 (1.16, 2.05)* | <0.01 |
| Northern Ontario | 2.84 (1.91, 4.19)* | ||
| Southwestern Ontario | 1.99 (1.54, 2.62)* | ||
| Central Ontario | 1.00 | ||
| Prototheca spp. (Algae) | Eastern Ontario | 0.70 (0.58, 0.85)* | <0.01 |
| Northern Ontario | 0.73 (0.49, 1.06) | ||
| Southwestern Ontario | 0.94 (0.80, 1.12) | ||
| Central Ontario | 1.00 | ||
| Fungus | Eastern Ontario | 0.91 (0.76, 1.10) | <0.01 |
| Northern Ontario | 0.20 (0.09, 0.37)* | ||
| Southwestern Ontario | 0.79 (0.67, 0.94)* | ||
| Central Ontario | 1.00 | ||
| Contamination | Eastern Ontario | 1.09 (0.95, 1.26) | <0.01 |
| Northern Ontario | 0.76 (0.56, 1.02) | ||
| Southwestern Ontario | 0.70 (0.61, 0.81)* | ||
| Central Ontario | 1.00 | ||
| No growth | Eastern Ontario | 0.87 (0.82, 0.92)* | <0.01 |
| Northern Ontario | 0.87 (0.78, 0.96)* | ||
| Southwestern Ontario | 0.85 (0.81, 0.90)* | ||
| Central Ontario | 1.00 |
Represent significant odds ratios.
Discussion and Conclusions
In this retrospective study we have described the frequently isolated pathogens from milk samples in Ontario, Canada and evaluated the major trends for nine major mastitis pathogens. This information will facilitate spatial and temporal comparisons with other studies. The study result is not intended to be a diagnostic guide for veterinarians managing the treatment of bovine mastitis as such decisions should be based on testing of individual samples as far as practically possible. However, information on the most frequently isolated pathogens from milk samples and the general trend in their occurrence will be useful for veterinarians as a reference for decision making in the therapy of mastitis.
The major pathogens responsible for mastitis in Ontario have changed significantly over the years examined in this study. The previously important pathogens have been replaced by pathogens which were considered minor pathogens in the past. While this trend is like that in other countries and other parts in Canada, the findings from this study highlight some differences from other studies. This decreasing trend could be attributed to control efforts toward the causative agent of communicable mastitis. However, control efforts should be continued to reduce not only the major contagious pathogens in Ontario but also environmental, minor, and emerging mastitis pathogens.
While S. aureus, S. uberis, and E. coli were more frequently isolated from mastitis milk samples in the Atlantic Provinces of Canada between 1994 and 2013 (26), this study shows that in Ontario NAS were more frequently isolated than E. coli. In contrast to our study finding, in a Canada wide study Enterobacteriaceae (E. coli and Klebsiella spp.) accounted for more than a quarter of isolates from mastitis milk (10). This study also showed that over the years NAS have increasingly been isolated from the milk samples. At the species level, S. chromogenes was the dominant NAS species similar to what was noted in a Canada wide study, the US study and some European studies, however regional variation in the type of NAS species prevalence was noted (27–29). Understanding of the prevalence of NAS at species level is necessary from a disease control and management point of view as infection by some of these species of NAS has been known to be persistent and hence a source of infection to other animals in the herd. Whereas, other species like S. simulans are more of an environmental pathogen (29, 30).
This study shows the low prevalence of some important emerging mastitis pathogens. There was a consistent presence of a potential pathogen Aerococcus spp. in Ontario milk samples since 2014. While no confirmatory evidence is available, Aerococcus viridans has been implicated in subclinical bovine mastitis (31). Likewise, in this study at least 1% of the isolates were identified as Prototheca spp., an algae responsible for incurable mastitis. Prototheca spp. is an emerging pathogen in Canada with it not been reported between 2003 and 2005 (6). A case-control study in 2011 however determined mean within-herd prevalence of Prototheca spp of 5.1% (32). The present study also reported a low prevalence of an important contagious mastitis pathogen Mycoplasma in milk. In a Canada wide study conducted between 2003 and 2005, no Mycoplasma was detected in milk samples from clinically infected animals (6). A low prevalence of Mycoplasma in bulk milk samples was reported in a study from Prince Edward Island, Canada (33). In the current study, no Mycoplasma was detected in milk samples from 2008 to 2012, however, between 2012 and 2018 a total of 98 isolates were identified. While Canada has historically maintained a low prevalence of Mycoplasma compared to other countries (34), it may represent an emerging concern to be monitored.
This study reports interesting seasonal trends of isolation of major mastitis pathogen and clustering of these pathogens by the location of the clinic submitting the samples indicating the possible necessity of seasonal and regional mastitis control approaches. However, caution should be taken while interpreting this as the clustering could be due to stochastic effect of sample submission by the clinics in the geography as some of the clinics conduct milk culture themselves and some sample submission could be to identify the pathogens that they are not capable of doing at their laboratories. In contrast to our study, a Danish study reported that the incidence rate of S. aureus, E. coli (pastured herd), and S. dysgalactiae clinical mastitis was found to be the highest in the winter months (35). Dairy herds in Ontario are predominantly of tie stall types (36) which could be one reason for this unique observation as it was found in a previous study that in case of confined herds, incidence rate of E. coli clinical mastitis was higher in summer than in winter (35). The frequency of isolation of S. uberis was higher not only in summer, similar to what was found in the Danish study where the highest incidence rate of S. uberis clinical mastitis was observed in summer (August), which was associated with pasture (35), but also in autumn.
The spatio-temporal change in the test results could also be a result of a decrease in sample submissions over the years possibly as a result of greater use of in-clinic identification techniques, a decrease in farms and/or animals over the years (37), improvement in the culture techniques, adoption of improved sample collection and transportation, and implementation of mastitis control measures on farm.
This study is based on passively collected data which have limitations. It is a non-systematic study and since laboratory submission requires a veterinarian, the decision to submit a sample will be influenced by the farmer. Likewise, information on farm type, age of animal, their lactational status, clinical form of the disease, and/or treatment history is not known. The availability of this information would have improved our understanding of seasonal and regional variations that were observed in this study. Additionally, some of the submissions may have been a result of a research study on mastitis or on-farm surveillance. However, information was not available to identify such submissions. In this study, we have considered that all the samples were from cattle in either clinical or sub-clinical stages of mastitis. A subset of samples may have originated from animals with a history of treatment failure. Likewise, the samples could be subject to selection bias and likely underrepresent milk samples from mastitis cattle in Ontario as many clinics and farms have their own regular mastitis testing programs in place. Likewise, some findings can also be attributed to the improvement in diagnostic techniques at the AHL as since 2011 spectrometric techniques are being used to identify bacterial species. While the use of an in-house clinic lab can be attractive for improved timeliness, AHL is one of the major laboratories in Ontario, and Ontario laboratory samples are subsidized making submission more economically attractive. Therefore, the milk samples are expected to be representative of mastitis milk samples in Ontario. In addition, the representativeness of these samples could also have been affected by farm economics and disease outbreaks as they have been shown to influence laboratory sample submissions (38).
It is a valid expectation that the culture of milk originating from a mastitis cow would focus on all the pathogens that are known to cause mastitis. Likewise, the pathogens that were isolated and hence reported might come from the milk, or the environment of the animal (skin, farm, milking machine, human handlers, etc.). Nevertheless, knowledge of this is important as it is known that some of the environmental pathogens can persist in the farm environment and be a constant source of infection for the animals. Despite all these limitations, there is a value of the information obtained from these passive surveillance data.
Bulk milk samples are used to estimate the prevalence of mastitis pathogens in a region (33, 39). However, the information they provide can be of limited value as the probability of detecting a pathogen will be reduced many fold by the dilution effect of the bulk milk and the inherent limit of detection of culture techniques especially if only a small fraction of animals are infected in a herd. Testing milk samples from individual quarters is valuable but is more costly making prevalence studies challenging. In this context, information on the prevalence of mastitis pathogens can be obtained using passive surveillance data generated by a diagnostic laboratory, as has been attempted in this study.
A significant number of samples had a culture negative test result, and this reduced significantly during the study period. Reduction in no growth is possibly due to the improvement in sampling of milk and culture techniques. However, it is also possible that the clinics submitted the higher proportion of culture positive samples to the AHL for confirmation. Likewise, the diversification of culturing techniques would have enabled the capacity to identify the presence of pathogens like Mycoplasma which require special media. Given the low prevalence of Mycoplasma in Canada and particularly Ontario, this however looks less likely. Often culture negative test results are attributed to infection by pathogens which are short lived such as E. coli and pathogens which are shed in a cyclical manner such as S. aureus if a single sample has been tested (40, 41). The simultaneous reduction of the proportion of samples positive for E. coli and S. aureus could explain the reduction in culture negative test results. Likewise, as previously mentioned, some of the data in this study may come from research studies like a case-control study conducted in 2011, which might have overestimated the culture negative test result (32).
The present study confirms that the prevalence of mastitis pathogens that do not respond to antibiotic treatment is substantial in the milk samples obtained from Ontario cattle between 2008 and 2017 similar to what was observed Canada wide, in the United States, and in the European countries (2, 3, 42, 43). Likewise, we observed that a large proportion of the mastitis milk was negative on culture as reported previously (24). Thus, the use of antibiotics in most instances of mastitis without the identification of the pathogen and their antimicrobial sensitivity result can be unnecessary. The knowledge of the major pathogens prevalent in Ontario farms will aid veterinarians in evaluating their mastitis treatment protocols and management decision making thereby reducing the unnecessary use of antibiotics, as mastitis is considered one of the main reasons for antibiotic use on a dairy farm (44, 45), and hence improving the future prognosis of mastitis treatment. Similarly, this approach can be adopted in other geographies for estimating the prevalence of major mastitis pathogens by utilizing the passive surveillance data originating from diagnostic laboratories.
Data Availability Statement
The data analyzed in this study is subject to the following licenses/restrictions: On request, the de-identified dataset used in this study can be made available for solely scholarly purposes. Requests to access these datasets should be directed to agreer@uoguelph.ca.
Author Contributions
KA, GB, DS, and AG contributed to the concept of the study and data analysis. KA conducted the data analysis and wrote the initial draft of the manuscript. All the authors contributed to and approved the final manuscript.
Funding
This work was supported by funding from the Canadian Institutes of Health Research (CIHR), under the European Commission's Joint Programming Initiative on Antimicrobial Resistance (5th Joint Call).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Jamali H, Barkema HW, Jacques M, Lavallée-Bourget EM, Malouin F, Saini V, et al. Invited review: incidence, risk factors, and effects of clinical mastitis recurrence in dairy cows. J Dairy Sci. (2018) 101:4729–46. 10.3168/jds.2017-13730 [DOI] [PubMed] [Google Scholar]
- 2.Reyher KK, Dufour S, Barkema HW, Des Côteaux L, DeVries TJ, Dohoo IR, et al. The National Cohort of Dairy Farms—a data collection platform for mastitis research in Canada. J Dairy Sci. (2011) 94:1616–26. 10.3168/jds.2010-3180 [DOI] [PubMed] [Google Scholar]
- 3.Cobirka M, Tancin V, Slama P. Epidemiology and classification of mastitis. Animals. (2020) 10:2212. 10.3390/ANI10122212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogeveen H, Van Der Voort M. Assessing the economic impact of an endemic disease: the case of mastitis. Rev Sci Tech Off Int Epiz. (2017) 36:217–26. 10.20506/rst.36.1.2623 [DOI] [PubMed] [Google Scholar]
- 5.Aghamohammadi M, Haine D, Kelton DF, Barkema HW, Hogeveen H, Keefe GP, et al. Herd-level mastitis-associated costs on Canadian dairy farms. Front Vet Sci. (2018) 5:100. 10.3389/fvets.2018.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blowey R, Edmondson P. Mastitis Control in Dairy Herds. Oxfordshire: CABI Publishing; (2010). 10.1079/9781845935504.0000 [DOI] [Google Scholar]
- 7.Leslie KE, Petersson-Wolfe CS. Assessment and management of pain in dairy cows with clinical mastitis. Vet Clin North Am Food Anim Pract. (2012) 28:289–305. 10.1016/j.cvfa.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 8.Barkema HHW, Schukken YYH, Zadoks RN. Invited Review: The role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J Dairy Sci. (2006) 89:1877–95. 10.3168/jds.S0022-0302(06)72256-1 [DOI] [PubMed] [Google Scholar]
- 9.Zigo F. Vasil' M, Ondrašovičová S, Výrostková J, Bujok J, Pecka-Kielb E. Maintaining optimal mammary gland health and prevention of mastitis. Front Vet Sci. (2021) 8:607311. 10.3389/FVETS.2021.607311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riekerink RGMO, Barkema HW, Kelton DF, Scholl DT. Incidence rate of clinical mastitis on Canadian dairy farms. J Dairy Sci. (2008) 91:1366–77. 10.3168/jds.2007-0757 [DOI] [PubMed] [Google Scholar]
- 11.Sargeant J. Qualitative research part II: participants, analysis, and quality assurance. J Grad Med Educ. (2012) 4:1–3. 10.4300/JGME-D-11-00307.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sargeant JM, Scott HM, Leslie KE, Ireland MJ, Bashiri A. Clinical mastitis in dairy cattle in Ontario: frequency of occurrence and bacteriological isolates. Can Vet J. (1998) 39:33–8. [PMC free article] [PubMed] [Google Scholar]
- 13.Saini V, McClure JT, Léger D, Keefe GP, Scholl DT, Morck DW, et al. Antimicrobial resistance profiles of common mastitis pathogens on Canadian dairy farms. J Dairy Sci. (2012) 95:4319–32. 10.3168/jds.2012-5373 [DOI] [PubMed] [Google Scholar]
- 14.Saini V, McClure JT, Scholl DT, DeVries TJ, Barkema HW. Herd-level relationship between antimicrobial use and presence or absence of antimicrobial resistance in gram-negative bovine mastitis pathogens on Canadian dairy farms. J Dairy Sci. (2013) 96:4965–76. 10.3168/jds.2012-5713 [DOI] [PubMed] [Google Scholar]
- 15.Naqvi SA, De Buck J, Dufour S, Barkema HW. Udder health in Canadian dairy heifers during early lactation. J Dairy Sci. (2018) 101:3233–47. 10.3168/JDS.2017-13579 [DOI] [PubMed] [Google Scholar]
- 16.Saini V, McClure JT, Scholl DT, DeVries TJ, Barkema HW. Herd-level relationship between antimicrobial use and presence or absence of antimicrobial resistance in gram-negative bovine mastitis pathogens on Canadian dairy farms. J Dairy Sci. (2013) 96:4965–76. 10.3168/JDS.2012-5713 [DOI] [PubMed] [Google Scholar]
- 17.Oliveira L, Ruegg PL. Treatments of clinical mastitis occurring in cows on 51 large dairy herds in Wisconsin. J Dairy Sci. (2014) 97:5426–36. 10.3168/jds.2013-7756 [DOI] [PubMed] [Google Scholar]
- 18.Anonymous . Prescription Drug List. Available online at: https://www.canada.ca/en/health-canada/services/drugs-health-products/drug-products/prescription-drug-list/list.html#a2 (accessed March 29, 2019).
- 19.Saini V, McClure JT, Léger D, Dufour S, Sheldon AG, Scholl DT, et al. Antimicrobial use on Canadian dairy farms. J Dairy Sci. (2012) 95:1209–21. 10.3168/JDS.2011-4527 [DOI] [PubMed] [Google Scholar]
- 20.WHO . WHO List of Critically Important Antimicrobials for Human Medicine (WHO CIA List). (2019). Available online at: https://www.who.int/foodsafety/publications/antimicrobials-sixth/en/ (accessed May 21, 2019).
- 21.Zadoks RN, Gillespie BE, Barkema HW, Sampimon OC, Oliver SP, Schukken YH. Clinical, epidemiological and molecular characteristics of Streptococcus uberis infections in dairy herds. Epidemiol Infect. (2003) 130:335–49. 10.1017/S0950268802008221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.First Character of the Postal Code and Corresponding Province Territory or Region . Available online at: https://www150.statcan.gc.ca/n1/pub/92-195-x/2011001/other-autre/pc-cp/tbl/tbl9-eng.htm (accessed January 13, 2021).
- 23.R Core Team . R: A Language and Environment for Statistical Computing. (2019). Available online at: https://www.r-project.org/
- 24.Makovec JA, Ruegg PL. Results of milk samples submitted for microbiological examination in Wisconsin from 1994 to 2001. J Dairy Sci. (2003) 86:3466–72. 10.3168/jds.S0022-0302(03)73951-4 [DOI] [PubMed] [Google Scholar]
- 25.Brookhart MA, Rothman KJ. Simple estimators of the intensity of seasonal occurrence. BMC Med Res Methodol. (2008) 8:67. 10.1186/1471-2288-8-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Awosile BB, Heider LC, Saab ME, McClure JT. Antimicrobial resistance in mastitis, respiratory and enteric bacteria isolated from ruminant animals from the Atlantic Provinces of Canada from 1994–2013. Can Vet J. (2018) 59:1099. [PMC free article] [PubMed] [Google Scholar]
- 27.Condas LAZ, De Buck J, Nobrega DB, Carson DA, Naushad S, De Vliegher S, et al. Prevalence of non-aureus Staphylococci species causing intramammary infections in Canadian dairy herds. J Dairy Sci. (2017) 100:5592–612. 10.3168/JDS.2016-12478 [DOI] [PubMed] [Google Scholar]
- 28.Jenkins SN, Okello E, Rossitto P V, Lehenbauer TW, Champagne J, Penedo MCT, et al. Molecular epidemiology of coagulase-negative Staphylococcus species isolated at different lactation stages from dairy cattle in the United States. PeerJ. (2019) 2019:e6749. 10.7717/peerj.6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Buck J, Ha V, Naushad S, Nobrega DB, Luby C, Middleton JR, et al. Non-aureus Staphylococci and bovine udder health: current understanding and knowledge gaps. Front Vet Sci. (2021) 8:658031. 10.3389/fvets.2021.658031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piessens V, Van Coillie E, Verbist B, Supré K, Braem G, Van Nuffel A, et al. Distribution of coagulase-negative Staphylococcus species from milk and environment of dairy cows differs between herds. J Dairy Sci. (2011) 94:2933–44. 10.3168/JDS.2010-3956 [DOI] [PubMed] [Google Scholar]
- 31.Sun M, Gao J, Ali T, Yu D, Zhang S, Khan SU, et al. Characteristics of Aerococcus viridans isolated from bovine subclinical mastitis and its effect on milk SCC, yield, and composition. Trop Anim Health Prod. (2017) 49:843–9. 10.1007/s11250-017-1271-2 [DOI] [PubMed] [Google Scholar]
- 32.Pieper L, Godkin A, Roesler U, Polleichtner A, Slavic D, Leslie KE, et al. Herd characteristics and cow-level factors associated with Prototheca mastitis on dairy farms in Ontario, Canada. J Dairy Sci. (2012) 95:5635–44. 10.3168/jds.2011-5106 [DOI] [PubMed] [Google Scholar]
- 33.Olde Riekerink RGM, Barkema HW, Veenstra S, Poole DE, Dingwell RT, Keefe GP. Prevalence of contagious mastitis pathogens in bulk tank milk in Prince Edward Island. Can Vet J. (2006) 47:567–72. [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson DJ, Goodell G, Justice-Allen A, Smith ST. Herd-level prevalence of Mycoplasma spp mastitis and characteristics of infected dairy herds in Utah as determined by a statewide survey. J Am Vet Med Assoc. (2009) 235:749–54. 10.2460/javma.235.6.749 [DOI] [PubMed] [Google Scholar]
- 35.Riekerink RGMO, Barkema HW, Stryhn H. The effect of season on somatic cell count and the incidence of clinical mastitis. J Dairy Sci. (2007) 90:1704–15. 10.3168/jds.2006-567 [DOI] [PubMed] [Google Scholar]
- 36.Dairy Barns by Type - Canadian Dairy Information Centre (CDIC) . Available online at: https://www.dairyinfo.gc.ca/eng/dairy-statistics-and-market-information/farm-statistics/dairy-barns-by-type/?id=1502467060775 (accessed January 13, 2021).
- 37.Number of Dairy Cows Heifers by Province - Agricultural Industry Market Information System (AIMIS) - Agriculture Agri-Food Canada (AAFC) . Available online at: https://aimis-simia-cdic-ccil.agr.gc.ca/rp/index-eng.cfm?action=pR&r=219&pdctc= (accessed January 13, 2021).
- 38.O'Sullivan T, Friendship R, Pearl DL, McEwen B, Ker A, Dewey C. The association between submission counts to a veterinary diagnostic laboratory and the economic and disease challenges of the Ontario swine industry from 1998 to 2009. Prev Vet Med. (2012) 106:275–83. 10.1016/j.prevetmed.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 39.Bauman CA, Barkema HW, Dubuc J, Keefe GP, Kelton DF. Canadian National Dairy Study: herd-level milk quality. J Dairy Sci. (2018) 101:2679–91. 10.3168/jds.2017-13336 [DOI] [PubMed] [Google Scholar]
- 40.Sears PM, Smith BS, English PB, Herer PS, Gonzalez RN. Shedding pattern of Staphylococcus aureus from bovine intramammary infections. J Dairy Sci. (1990) 73:2785–9. 10.3168/jds.S0022-0302(90)78964-3 [DOI] [PubMed] [Google Scholar]
- 41.Smith KL, Hogan JS. Environmental mastitis. Vet Clin North Am Food Anim Pract. (1993) 9:489–98. 10.1016/S0749-0720(15)30616-2 [DOI] [PubMed] [Google Scholar]
- 42.Boireau C, Cazeau G, Jarrige N, Calavas D, Madec JY, Leblond A, et al. Antimicrobial resistance in bacteria isolated from mastitis in dairy cattle in France, 2006–2016. J Dairy Sci. (2018) 101:9451–62. 10.3168/JDS.2018-14835 [DOI] [PubMed] [Google Scholar]
- 43.Fernández G, Barreal ML, Pombo MB, Ginzo-Villamayor MJ, González-Manteiga W, Prieto A, et al. Comparison of the epidemiological behavior of mastitis pathogens by applying time-series analysis in results of milk samples submitted for microbiological examination. Vet Res Commun. (2013) 37:259–267. 10.1007/S11259-013-9570-1 [DOI] [PubMed] [Google Scholar]
- 44.APHIS . Antibiotic Use on U.S. Dairy Operations, 2002 and 2007. (2008). [Google Scholar]
- 45.Saini V, McClure JT, Léger D, Dufour S, Sheldon AG, Scholl DT, et al. Antimicrobial use on Canadian dairy farms. J Dairy Sci. (2012) 95:1209–21. 10.3168/jds.2011-4527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data analyzed in this study is subject to the following licenses/restrictions: On request, the de-identified dataset used in this study can be made available for solely scholarly purposes. Requests to access these datasets should be directed to agreer@uoguelph.ca.