Abstract
Unintended DNA rearrangements in a differentiating lymphocyte can have severe, oncogenic consequences, but the mechanisms for avoiding pathogenic outcomes in V(D)J recombination are not well understood. The first level at which fidelity is instituted is in discrimination by the recombination proteins between authentic and inauthentic recombination signal sequences. Nevertheless, this discrimination is not absolute and cannot fully eliminate targeting errors. To learn more about the basis of specificity during V(D)J recombination, we have investigated whether it is possible for the recombination machinery to detect an inaccurately targeted sequence subsequent to cleavage. These studies indicate that even postcleavage steps in V(D)J recombination are sequence specific and that noncanonical sequences will not efficiently support the resolution of recombination intermediates in vivo. Accordingly, interventions after a mistargeting event conceivably occur at a late stage in the joining process and the likelihood may well be crucial to enforcing fidelity during antigen receptor gene rearrangement.
A critical process in B- and T-lymphocyte development is the assembly of antigen receptor genes. In this developmentally regulated DNA rearrangement, variable (V), diversity (D), and joining (J) gene segments become connected to one another to create the variable exon of an immunoglobulin (Ig) or T-cell receptor (TCR) gene. V(D)J recombination entails the site-specific recombination of specific DNA motifs termed recombination signal sequences (RSSs) in a process that can be conceptually as well as biochemically divided into two stages: stage 1, where RSS recognition, synapsis, and cleavage takes place, and stage 2, where DNA ends are modified by nucleotide addition and subtraction and become rejoined. Necessary DNA transactions are accomplished through a collaboration between DNA sequence-specific proteins, RAG-1 and RAG-2, and non-sequence-specific nucleases, polymerases, ligases, and structural components (the list includes terminal deoxynucleotidyltransferase, DNA ligase IV, DNA-PKcs, Ku70, Ku80, and XRCC4; reviewed in reference 15). Ultimately, this multicomponent recombination machinery accomplishes the precisely localized cut-and-paste operations that can convert dispersed V, D, and J gene segments into a functional antigen receptor gene.
Central to stage 1 is the site-specific recognition of two RSSs, each comprised of a heptamer (CACAGTG), a spacer of 12 or 23 bp, and a nonamer (ACAAAAACC). Although a consensus RSS is evident from examination of natural joining signals (30, 44), typically only a small minority of RSSs at an Ig or TCR locus exactly match this canonical sequence (for example, see reference 27). The V(D)J recombination machinery therefore is constrained in two opposing ways: it must have sufficient flexibility to recognize naturally occurring RSS variations, but at the same time it must be able to avoid recombination of inappropriate DNA sequences. Such sequences, termed cryptic RSSs, happen to resemble real RSSs, but if joined will promote unintended genome rearrangement.
The recombination machinery is not able to discriminate between target authentic RSSs and cryptic RSSs with absolute success. This is illustrated by the fact that DNA sequences that match a canonical RSS at only about 50% of the heptamer- or nonamer-equivalent positions will still recombine when tested in an extrachromosomal V(D)J recombination assay (24). The number of cryptic RSSs that can be documented within artificial recombination substrates suggests that a site with functionally relevant similarities to an RSS can be expected to occur at least once every 600 bp in the mammalian genome (24). Even though the intrinsic joining proficiency of most such cryptic RSSs is several orders of magnitude lower than that of an authentic RSS, the number of cryptic sites is overwhelming—fidelity must certainly involve biological strategies that go beyond target site discrimination alone.
Accuracy in V(D)J recombination can be envisioned to rely upon the regulation of the accessibility of genomic sequences to recombination proteins (reviewed in references 35 and 40). Features of chromatin structure are known to allow recombination of appropriate Ig and TCR loci during B- and T-cell differentiation (for recent discussions, see references 8, 13, 45, and 46), and it is reasonable to suppose, by extension, that an inaccessible chromatin configuration may protect every other site in the genome from illegitimate rearrangement. However, the molecular basis of accessibility is only beginning to be elucidated, and how much genome alteration is actually prevented by such a mechanism has not been investigated. Here we have examined an additional, complementary possibility, which is that the recombination enzymes can detect the participation of a cryptic RSS even after cleavage of the mistargeted sequence has already taken place. Postcleavage sequence specificity may provide an important means of error avoidance by allowing for late-stage interventions when mistargeting occurs.
The need for a midcourse correction mechanism is underscored by the observation that site-specific cryptic RSS rearrangement can occur between cryptic signal-like sequences at locations completely removed from any Ig or TCR locus. The threat this type of event poses, and the necessity for safeguards is illustrated for T-cell acute lymphocytic leukemia (T-ALL). In a subset of T-ALL cases, a pair of cryptic RSSs at the scl/tal locus have recombined, resulting in the unscheduled expression of the tal gene and contributing to leukemogenesis (3, 7, 9). Cryptic RSSs have also been shown to mediate a less-disastrous site-specific deletion at the hprt locus, detectable in T cells sampled from healthy adults (11, 34). A key feature illustrated by these examples is that the site-specific cryptic rearrangements can take place without interfering with the concomitant recombination of the Ig or TCR loci in the affected cell. Thus, this type of aberrant outcome is unlikely to be screened out by any of the highly specialized cellular selective forces operating in T- and B-cell development, all of which focus on the products of antigen receptor gene rearrangement. A cell that harbors a potentially harmful rearrangement would be just as likely as any other to undergo proper recombination at its antigen receptor loci and thereby to receive further proliferative signals.
In this regard, clearly a mechanism that can detect mistargeting even though the joining process has already proceeded as far as RSS cleavage, would help to maintain genome stability during V(D)J recombination. To discover whether such a mechanism exists, we have refined an in vivo plasmid assay so that the capacity of a particular RSS to support postcleavage operations can be distinguished from its function in precleavage and cleavage phases of the V(D)J recombination process. By applying this assay to various cryptic RSSs, our results indicate that the events that occur after cleavage in V(D)J recombination are indeed sequence specific. These observations provide in vivo support for suggestions based upon in vitro evidence that RAG1 and RAG2 (RAG1/2) and its maintenance within a site-specific protein-DNA complex has an important role to play in the later steps of Ig and TCR gene recombination (for example, see references 5, 18, 20, 33, and 37). In the context of fidelity and genome stability, the postcleavage sequence specificity observed here raises the possibility that an interruption of V(D)J recombination and/or reduced cellular survival results as a late-stage consequence of target misrecognition.
MATERIALS AND METHODS
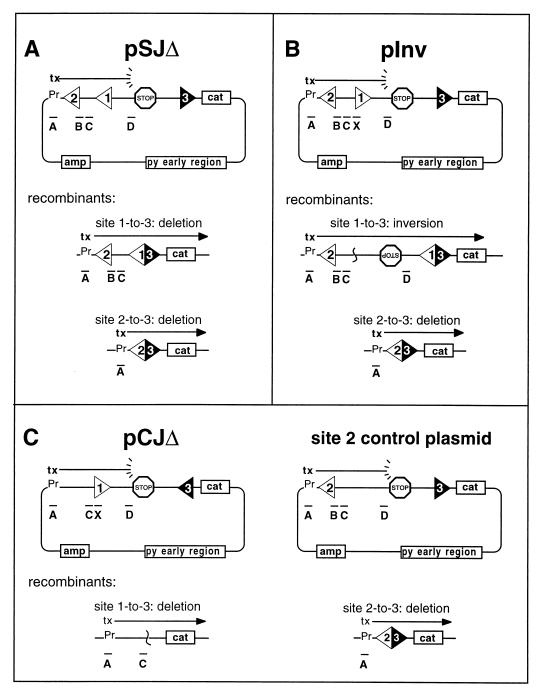
Substrates pWT-SJΔ, pWT-Inv (designated p12x23 in reference 25), p6131-SJΔ, p6131-Inv, pCA-SJΔ, pCA-Inv, pT4Anon-SJΔ, pT4Anon-Inv, p6131hWTnon-SJΔ, p6131hWTnon-Inv, pNOnon-SJΔ, and pNOnon-Inv were derived from p23 (25) and differ from one another only in the identity and orientation of the RSS occupying site 1. Twelve-spacer RSSs (12-RSS) were inserted in the form of a double-stranded oligonucleotide cassette with SalI compatible ends at a unique SalI site in the common p23 plasmid backbone. pWT-CJΔ was derived from p12 (25) after introducing a canonical 23-spacer RSS (23-RSS) that is identical to the 23-RSS in p23 and its derivatives, at a unique BamHI site. An intermediate plasmid, pEA23flip, was constructed from pJH298 (26) after removal of its 23-RSS and replacement with our 23-RSS used in the desired orientation. p6131-CJΔ and pT4Anon-CJΔ were then constructed by insertion of the appropriate 12-RSS oligonucleotide cassette at the SalI site of pEA23flip. All modifications were confirmed by dideoxy sequencing (Sequenase). p6131-Invno2 was constructed from pJH298 and p6131-Inv. The two plasmids were double digested with SalI and AatII, and the small fragment of the former was ligated to the large fragment of the latter. p6131-Inv(506) was constructed from p6131-Invno2 by duplication of the 227-bp ClaI fragment between the two RSSs; p6131-CJΔ(546) was similarly constructed from p6131CJΔ. The control site 2 plasmid is p6912 described elsewhere (24) and is identical to the SJΔ constructs except that it contains a nonfunctional sequence at site 1. In summary, all of the plasmids used in the present study are identical to one another save only for (i) the identity and orientation of the sequence in site 1, and/or (ii) the orientation of the sequence in site 3, and/or (iii) the presence or absence of a functional RSS at site 2.
Cell culture.
A pre-B-like cell line, 204-1-8, derived by Abelson virus transformation was used for all experiments (32). Cells were maintained in RPMI–10% heat-inactivated fetal bovine serum–50 μM β-mercaptoethanol.
Extrachromosomal recombination assay.
Transfections of 204-1-8 cells were performed with 150 ng of substrate DNA and included 1 mM caffeine as described previously (24). Transfected DNA was harvested, and recombinant colonies were selected as described earlier. Every construct was transfected 4 to 10 times, and at minimum 500 colonies were picked in grid arrays to nitrocellulose filters. No more than 20% of a given transfection was sampled. For experiments involving cotransfection with the site 2 control plasmid, 75 ng of each plasmid was used.
Screening of recombinants.
Chloramphenicol-resistant colonies were picked to replicate grids and transferred to nitrocellulose filters for hybridization to 32P-end-labeled oligonucleotides. The diagnostic hybridization patterns are given in Fig. 2. As shown in the figure, probe A is LACPRIME (CTCATTAGGCACCCCAGGCT), probe B is LACMER (TATGTTGTGTGGAATTGT), probe C is PLAC20 (TCCTAACAGCTATGACCATG), and probe D is TER-2 (TCCAAAGTTCTCAATGCTGC). Probe X represents the RSS-specific oligonucleotides used in the screening of CJΔ and Inv substrates: PD12MER (AGGTCGACACAGTGG) was used for WT, T4Anon, and NOnon; O/6131INV (AGGTCGACACAACAT) was used for 6131 and 6131hWTnon; and O/CAINV (AGGTCGACACACACA) was used for CA. Any colony that gave an ambiguous signal by hybridization was reanalyzed. The accuracy of the recombinant designations for all experiments was tested by subjecting DNA samples from random, typed colonies to diagnostic restriction enzyme analysis; in all cases there was 100% concordance.
FIG. 2.
Recombination substrates. In all substrates, 12-RSSs occur at sites 1 or 2, and the canonical 23-RSS is inserted at site 3. The frequency with which a given test RSS positioned at site 1 recombines is tested relative to the frequency with which the invariant sequence at site 2 is rearranged. In each section of the figure, labeled bars represent the oligonucleotide probes that are used in the typing of recombinants (see Materials and Methods). (A) The SJΔ substrate and two possible recombinant deletion products, involving either the site 1 or site 2 RSS. (B) Inv substrate with the inversional recombinant resulting from site-1-to-site-3 rearrangement and site 2 deletion product. (C) The CJΔ substrate is shown with the deleted coding joint product. This plasmid is cotransfected with the site 2 control plasmid, which gives rise to the site 2 deletion product.
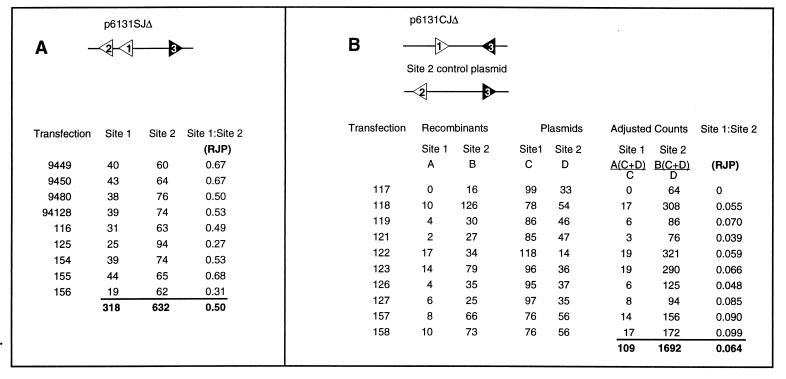
In experiments with a cotransfected control site 2 plasmid, the ratio of plasmids within the transfected cells was determined by treating an aliquot of the recovered plasmid DNA with DpnI prior to transformation of Escherichia coli. Nonrecombinant colonies were selected on media without chloramphenicol and were randomly picked and typed according to whether or not they hybridized to the oligonucleotide B probe (see Fig. 2). This gives the ratio of successfully transfected (replicated) molecules in the cells, and the number was used to normalize the score obtained for each class of recombinant according to the sample calculation shown in Fig. 4B.
FIG. 4.
Reproducibility of the measured RJP. (A) Nine transfections of p6131-SJΔ were conducted and screened. The top four transfections were performed 4 years earlier than the bottom five. These numbers correspond to the pooled data reported in Fig. 3. (B) Ten transfections of p6131-CJΔ cointroduced with the site 2 control plasmid were analyzed. Colonies selected on chloramphenicol were typed to determine the number of site 1 and site 2 recombinants as usual (columns A and B). Colonies isolated on ampicillin after DpnI digestion of the harvested DNA were typed to determine the numbers of each parental plasmid successfully transfected as described in Materials and Methods (columns C and D). Colony counts were normalized as shown (two subsequent columns) to give the RJP (rightmost column).
RESULTS
Experimental design.
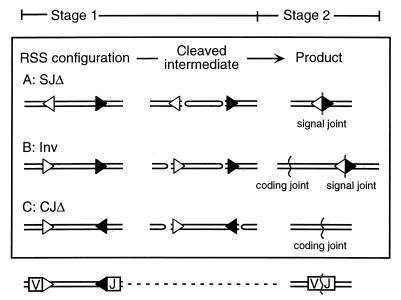
Stage 2 sequence discrimination is detected by the configuration-specific test illustrated in Fig. 1. This test measures the number of complete recombinants recovered when the orientation of the RSSs (and only this) is varied in an artificial recombination substrate. The configuration-specific test (described in more detail below) avoids complications associated with attempts to quantify cut recombination intermediates in vivo. Though seemingly more direct, any measurement of cleaved intermediates must be related in some fashion to rates of formation, degradation, and conversion to product before being meaningfully interpreted, and this is not possible at present for any in vivo V(D)J recombination system.
FIG. 1.
Configuration-specific differences in recombination outcome. RSS orientation dictates the type of junction that will be recovered after recombination. Three different outcomes, a signal joint via deletion, a signal and coding joint via inversion, and a coding joint via deletion via represent the recovered products resulting from the configurations shown in sections A, B, and C, respectively. Regardless of configuration, stage 1 operations involve cleavage at the heptamers of each RSS (shown as an open triangle for the 12-RSS and as a solid triangle for the 23-RSS). At the end of stage 1, as shown in the middle column (‘Cleaved intermediate’), two blunt signal ends and two hairpin-terminated coding ends are formed in every case. Subsequently, during stage 2, there are different operations required for resolution of the coding and/or signal ends. The identity of stage 1 operations and the divergence of stage 2 operations with different configurations forms the basis for distinguishing the postcleavage effects of RSS sequence in vivo.
In brief, a pair of 12- and 23-RSSs will recombine when present in any of three possible configurations, here designated SJΔ, Inv, and CJΔ. Regardless of configuration, identical stage 1 operations must take place; that is, the engagement of two RSSs and the cleavage of four DNA strands. In all cases, the products of RAG-mediated cleavage are one pair of blunt signal ends and one pair of hairpin-terminated coding ends (Fig. 1). However, after stage 1, each of the three configurations results in a different product (reviewed in references 15 and 22). For the SJΔ configuration, a signal joint is created by simple ligation of two signal ends. For CJΔ, DNA ends with hairpin termini must be opened and variably modified in the process of becoming connected to form a coding joint. For the Inv configuration, coding and signal ends are both joined, and the coding and signal joint products must be formed in a temporally correlated fashion (Fig. 1). If all steps in stage 1 are the same and all products in stage 2 are different, it follows by simple logic that configuration-related differences in the joining proficiency of an RSS should be attributed to the processes that take place after stage 1 and during stage 2.
We have employed an extrachromosomal V(D)J recombination assay to detect configuration-related differences in cryptic RSS recombination, and one caveat to the line of reasoning outlined above is that topological differences in our recombination substrates may create configuration-specific stage 1 effects. By altering the relative orientation of the RSSs, it is possible that physical limitations to DNA flexibility introduce different degrees of strain during stage 1 at the time of synapsis (discussed, for example, in reference 36). For the sake of clarity, a detailed description of the experiments performed to address this possibility has been deferred to the end of the Results section.
The stage 2 competence of a given RSS was determined by inserting the sequence at a position termed site 1 within the plasmid construct. The RSS was tested for joining with a canonical 23-RSS positioned at site 3 (Fig. 2) in up to three different configurations (SJΔ, Inv, and CJΔ). For standardization, the recombination of an invariant control 12-RSS in site 2 with the 23-RSS in site 3 was also measured in parallel. The ratio of site 1 and site 2 recombinants gave the relative joining proficiency (RJP) for a given construct.
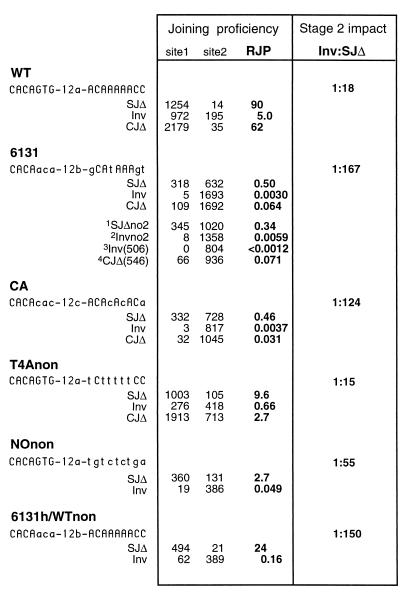
To establish the RJP values for the canonical sequence, three substrates, pWT-SJΔ, pWT-Inv, and pWT-CJΔ, were created bearing a canonical sequence, called WT, in site 1. Each construct was transfected multiple times into the 204-1-8 cell line, which is an Abelson murine leukemia virus-transformed pre-B-like cell active for V(D)J recombination (see Materials and Methods). The RJP of the WT sequence when measured in the SJΔ plasmid was 90 (units are arbitrary and defined by the site 2 control RSS, which is a noncanonical sequence [24, 25]). With pWT-CJΔ, a similar RJP of 62 was obtained. Finally, the RJP for WT in the pWT-Inv plasmid was 5.0, a value much lower than the RJP for WT in either deletional construct (Fig. 3).
FIG. 3.
Variant RSSs and configuration-specific RJP. The sequence of each tested RSS is given, indicating matches to the canonical RSS in uppercase letters. Spacer sequences were changed according to the dictates of a given experiment. The 12a spacer is an arbitrary sequence as given previously (25). The 12b spacer is identical to the spacer of the RSS appearing in site 2, and the 12c spacer is a continuation of the CA repeat. The total number of recombinants typed as either site 1 or site 2 rearrangements is given for each of the SJΔ and Inv constructs. For experiments where the site 2 control was introduced on a separate plasmid, the values shown are calculated as described in Materials and Methods (a sample calculation is given in Fig. 4). The RJP is defined as the ratio of site 1 to site 2 recombinants. The ratio of inversion to SJΔ provides a means by which to compare the stage 2 impact of a different RSS variants. Superscript numbers: 1, cotransfection of p6131-SJΔno2 and the site 2 control plasmid (see Fig. 2C); 2, cotransfection of p6131-Invno2 and the site 2 control plasmid; 3, cotransfection of p6131-Inv(506) (in which the distance between sites 1 and 3 was increased from 279 to 506 bp) and the site 2 control plasmid; and 4, cotransfection of p6131-CJΔ(546) (in which the distance between sites 1 and 3 was increased from 319 to 546 bp) and the site 2 control plasmid.
The fact that the RJP of a consensus RSS in an inversional configuration is not on par with the values measured for the deletional configuration (SJΔ or CJΔ) demonstrates, as observed by others, that the joining proficiency of an RSS is configuration sensitive when measured in the extrachromosomal assay (12). Configuration-specific differences in joining proficiency have been attributed to the fact that inversion requires the coordinate formation of two joints, whereas for deletion only one junction need be created. Having established a canonical standard in each configuration, we can compare the behavior of other, noncanonical DNA sequences tested similarly. That is, with a noncanonical RSS, the RJP for all constructs may be proportionally depressed, in which case there would be no evidence for sequence discrimination during stage 2. Alternatively, the RJP for certain configurations might be disproportionately reduced. The latter situation would indicate that sequence specificity pertains even to the postcleavage phase of V(D)J recombination and would additionally provide clues as to the affected postcleavage process.
Cryptic RSSs show configuration-specific deficits in RJP.
Two cryptic signals were tested for joining proficiency in each of three configurations (Fig. 2). One cryptic RSS, termed 6131 (24), differs from the WT RSS at multiple positions within both the heptamer and nonamer. In all, 7 of 16 nucleotides in the 6131 sequence are noncanonical (Fig. 3). (The 6131 sequence also served as the control against which the RJP of each test RSS was determined.) A second cryptic RSS, termed CA, was comprised of a CA dinucleotide repeated 14 times. CA repeats are highly represented in both mouse and human genomes, occurring once every 30 kb according to one estimate (41), and are known to possess cryptic signal function in an endogenous chromosomal context as well as by the extrachromosomal assay (11, 24). Compared to a consensus RSS, the pure CA repeat diverges at six positions (Fig. 3), and again these differences are distributed between both the heptamer and the nonamer.
Six constructs (representing each of three configurations for the two cryptic RSSs) were tested in 204-1-8 cells. The numbers of site 1 and site 2 recombinants were determined (details are given in Materials and Methods). As expected, both 6131 and CA were much weaker than the consensus WT sequence, regardless of orientation. For example, in an SJΔ-type construct, 6131 and CA both exhibited RJPs of about 0.5, well below the value of 90 obtained for the canonical RSS (pWT-SJΔ; see Fig. 3).
The key result however was that both 6131 and CA were considerably more compromised for inversion than they were for deletional signal joint formation (Fig. 3). For comparison, the ratios of the RJPs obtained with pInv-type plasmids versus the RJPs obtained with SJΔ-type plasmids are presented in Fig. 3. Thus, as mentioned above, the WT sequence supports inversion less readily than deletional signal joint formation, giving an Inv/SJΔ ratio of 1:18. If stage 2 operations were unaffected by substitution of a cryptic RSS, we would observe the same Inv/SJΔ ratio for the cryptic sequences despite an overall depression in recombination proficiency. However, the cryptic RSS 6131 gave a much lower ratio of 1:167, and CA gave a ratio of 1:124. Thus, 6130 and CA are excessively impaired in their ability to support inversion.
The diminished Inv/SJΔ ratio for CA or 6131 does not of itself reveal whether it is the nature of the joints resulting from inversion (i.e., a coding joint and a signal joint) or the number of joints that must form (i.e., two instead of one) that becomes problematic after cryptic RSS cleavage (Fig. 1). This question can be explored through testing CJΔ constructs, where coding joints are formed in a one-joint (deletional) rather than the two-joint (inversional) context. An impediment that relates specifically to coding end joining at stage 2 will be revealed if joining proficiency for a cryptic RSS is excessively reduced for the CJΔ configuration.
It is necessary to reverse the orientation of the 23-RSS at site 3 in the CJΔ configuration (Fig. 1C). Consequently, the 23-RSS is in the wrong orientation with respect to the control RSS at site 2 to measure site 2 deletion formation as before (see Fig. 2C). CJΔ plasmids were therefore cotransfected with a separate control plasmid (Fig. 2C). This control plasmid was identical to an SJΔ plasmids except that at site 1 the RSS was replaced by an inert sequence. For transfection, the CJΔ plasmid was mixed in a 1:1 ratio with the control site 2 plasmid. The effective ratio (of molecules that successfully reached the nucleus) was determined by assessing the representation of each plasmid among replicated DNA molecules after transfection, as described in Materials and Methods. This result was used to normalize the numbers of site 1 or site 2 recombinants measured in the usual manner by filter hybridization of chloramphenicol-resistant transformants (see Fig. 4B).
Whereas the canonical RSS had given similar RJPs when tested in either the SJΔ or the CJΔ configuration (90 and 62, respectively; Fig. 3), this was not the case for either 6131 or CA. The RJP for p6131-SJΔ was 0.50, while the RJP for p6131-CJΔ was only 0.064 (Fig. 3). Similarly, pCA-SJΔ gave an RJP of 0.46, while pCA-CJΔ gave an RJP of only 0.031. Thus, it was clear that both of the cryptic signals, 6131 and CA, were particularly ineffective in supporting coding-joint formation.
The RJP for a given construct is reproducible.
The reproducibility of RJP measurements was good, as shown in Fig. 4A. The construct p6131-SJΔ contains the same sequence (i.e., 6131) at both sites 1 and 2, each being oriented to promote a deletional signal joint formation. Nine separate transfections were performed several years apart by different authors. After analysis by transformation of E. coli with DNA from each of nine transfections, it can be seen that the RJP for any single determination is close to the RJP calculated from the pooled numbers (0.50), and the mean for the nine trials was 0.52 ± 0.15. Based upon the fact that there was little variation in the RJP from one transfection to the next, we have pooled the total number of site 1 and site 2 recombinants to give the RJP for all constructs, as shown in Fig. 3. To assure representative samplings, no more than one-fifth of a given transfection was transformed into E. coli. DNA sequence analysis of coding-joint recombinants indicated that the methods employed were not susceptible to “jackpot” effects (that is, multiple reisolation of a single recombinant due to vector replication in the transfected 204-1-8 cells), even in cases where screening one-fifth of a transfection yielded only a few, rare site 1 recombinants.
The fine-structure features of the products of cryptic RSS recombination are normal.
Our data suggested that engagement of a cryptic RSS by the V(D)J joining machinery affects the efficiency of coding-joint formation. It was therefore of interest to investigate whether the fine structure of coding joints resulting from cryptic RSS recombination was altered. Abnormal loss of DNA sequence up to a limit of 111 bp from the site 1-associated coding end and up to 23 nucleotides from the canonical site 3-associated coding end is detected by the present approach. In addition, it is also informative to score P nucleotide insertion, which reflects details of the hairpin opening step (6, 23, 28).
Analysis of 44 recombinant coding joints, incorporating all variant RSSs, is presented in Fig. 5. There were no gross anomalies. Nucleotide loss as well as insertion at the joint were within normal ranges and fully consistent with prior analyses of joints formed in the same cell line with canonical extrachromosomal recombination substrates (21, 24, 25, 28). Coding joints formed from the CA RSS included bits of the CA repeat itself at the junction, but this was not unexpected given that a CA repeat can be targeted in multiple ways, by shifting the recognized sequence by 2-bp increments (see Fig. 5B for examples). DNA sequence analysis of signal joints recovered from pCA-Inv and pCA-SJΔ were consistent with this interpretation and exhibited recombination sites at the CAC delimiting the 12th or 13th repeat in addition to that appearing at the 14th repeat (data not shown).
FIG. 5.
Coding joints. (A) Sequences of coding joints from randomly selected inversions (Inv) and deletions (CJΔ). Underlined nucleotides can be assigned to either end. (B) Variant target recognition of a CA repeat. As seen in analysis of coding joints in panel A, as well as of signal joints (not shown), a CA repeat is recognized in alternative frames displaced by 2 bp. Heptamer and nonamer elements are in boldface, and capital letters indicate matches to the canonical sequence.
Additionally, at least two signal joints were sequenced for each of the tested RSSs. Dozens more from various experiments were assessed by digestion with ApaLI, (a restriction enzyme specific for the GTGCAC sequence formed at a precise signal joint). No irregularity with respect to signal joint structure was revealed.
Our analysis indicated that a cryptic RSS causes a depression in coding joint recovery without affecting the properties of the junctions that are observed. The apparent interruption of recombination events involving a cryptic RSS is not circumvented in any obvious way by alternative resolution functions.
Both the nonamer and heptamer elements of the RSS are recognized during rejoining of cleaved ends.
To explore the importance of the nonamer motif of the RSS in stage 2 operations, we determined configuration-specific RJP values for the NOnon RSS. As its name implies, NOnon possessed a noncanonical substitution at each position of the nonamer. Otherwise, the heptamer (as well as spacer) sequence of NOnon was identical to that of WT. The result, as shown in Fig. 3, was that inversion was excessively depressed for NOnon, (the Inv/SJΔ ratio was 1:55), indicating that the nonamer portion of the RSS motif is recognized during postcleavage, stage 2, joining operations.
To determine whether stage 2 operations require site-specific interactions with the heptamer as well, we tested an RSS variant, termed 6131h/WTnon (Fig. 3). Here only the three nonamer-proximal nucleotides of the heptamer deviate from the canonical sequence (the heptamer being CACAaca instead of CACAGTG). We found that whereas signal joint formation was only mildly affected by the variant RSS (the RJP was 24 as opposed to the RJP of 90 for a canonical RSS), there was nevertheless a significant stage 2 impact. The inversional construct gave 0.16 for p6131h/WTnon-Inv as opposed to 5.0 for pWT-Inv. In other words, the Inv/SJΔ ratio was 1:150 for the RSS with a mutant heptamer compared to the 1:18 value for pWT (Fig. 3).
The 6131h/WTnon result demonstrates that a configuration-related depression of joining proficiency is seen upon modification of the heptamer only. The apparent stage 2 sequence specificity is revealed upon introduction of a minimal change in the RSS, for which, as seen for the SJΔ construct, joining function has not been greatly compromised. Thus, there is no obvious correlation between the strength of a cryptic signal and its stage 2 impact.
Taken together, the results obtained with 6131h/WTnon and NOnon highlight differences between RSS features that have been predicted to have an impact upon stage 1 binding and cleavage and those that are of more consequence in postcleavage, stage 2 operations. Based upon studies that measure cleavage or binding of RSSs by RAG1/2 (2, 10, 19, 31), the nonamer is more critical than the heptamer for binding and, in particular, the nonamer-proximal nucleotides of the heptamer are relatively unimportant for either precleavage binding or cleavage itself.
Differences between stage 1 and stage 2 sequence specificity are further established with T4Anon.
Previous studies have shown that A-to-T transversions within the A tract of the nonamer result in an RSS that is bound no better than a nonamerless (or randomized-nonamer) RSS by RAG1/2 (2, 10). To determine if A-to-T transversions similarly affect RSS function during stage 2, a final variant, T4Anon, was tested. This sequence was identical to WT except that every A nucleotide within the nonamer had been replaced by a T (i.e., TCTTTTTCC). Overall, the T4Anon RSS is noncanonical at a total of 6 of 16 positions.
Oddly enough, the results with T4Anon in the configuration-sensitive assay contrasted with those of every other variant described above. For T4Anon, the Inv/SJΔ ratio gave no indication of an impairment of stage 2 functions. The ratio was 1:15, a value actually somewhat higher than the Inv/SJΔ ratio for a canonical RSS and much higher than that for the nonamerless RSS (NOnon). T4Anon was also tested in the CJΔ configuration, where the RJPs for signal joints and coding joints were 9.6 versus 2.7. By these tests, the T4A sequence was found to have only a mild, if any, effect on stage 2 joining operations. This is exceptional and indicates that T-to-A transversions in the nonamer, which as noted above have a large impact on binding and cleavage by RAG1/2 in vitro, do not adversely affect stage 2 joining.
The configuration-specific assay is not sensitive to topological and topographical differences between test plasmids.
In the present study experimental variation is strictly limited to RSS orientation, stage 2 effects being deduced on the basis of configuration-sensitive effects on joining proficiency (Fig. 1). The distance between the interacting RSSs had been chosen according to the structure of the original plasmid upon which the present substrates are based (17) and is within a range (roughly 250 to 300 bp) for which it is conceivable that synapsis of the two RSSs might be impeded by the difficulty of bending the intervening DNA in certain configurations but not others. Should this be the case, then configuration-sensitive changes in RJP could reflect a need for strong RSS interactions during synapsis (i.e., during stage 1) and not later, as outlined in Fig. 1. Additionally, a related concern is that although the distance between the midpoint of the RSSs in site 1 and that of site 3 are maintained in all substrates without exception, the distances between the points of DNA cleavage (located at the heptamer edge of each RSS) must differ with orientation. Lastly, a feature that might confound the analysis is that the CJΔ recombinants have to be normalized against a cointroduced control plasmid.
To determine which, if any, of the above factors should be taken into account in the interpretation of the results, several additional experiments were performed. We again measured the RJP for the 6131 RSS in both the SJΔ and the Inv configurations, having first removed site 2 from the test constructs. The new constructs (p6131-SJΔno2 and p6131-Invno2) were then cotransfected with the site 2 control plasmid, and results were normalized as described for the CJΔ series of constructs (Fig. 4). The values obtained whether or not the site 2 RSS was located on the same substrate molecule as the test RSS were quite close: where the RJP of p6131-Inv was 0.0030, that of p6131-Invno2 was 0.0059. Similarly, the RJP of p6131-SJΔ was 0.50, while that of p6131-SJΔno2 was 0.34 (Fig. 3). These results indicated that the relative joining proficiency is roughly the same even if the site 2 control is measured on a separate, cotransfected substrate. Thus, for example, the possibility of secondary rearrangements (between the rearranged 23-RSS in an inversional site 1 recombinant and the site 2 RSS) can be discounted, and it is valid to compare the CJΔ results with results obtained for other configurations.
Concerns about how the varied inter-RSS distances and the different topologies might affect synapsis were also addressed. If the difficulty of bending covalently closed DNA to bring the two RSSs together is a significant factor in these experiments, configuration-specific joining deficits may actually reflect poor RSS binding during stage 1 and not stage 2. Although differences in DNA flexibility do not cause marked differences in ring closure for DNA fragments in the size range separating the cleavage and joining sites in the present substrates (251 bp for SJΔ, 319 bp for CJΔ, and 279 bp for Inv) (38, 39), shortening the intersignal distance still further in V(D)J recombination substrates has been shown to reduce recombination frequencies (36). To determine whether configuration-specific joining deficits are a manifestation of a hindered synapsis at stage 1 caused by close spacing between the RSSs, we looked for an amelioration of the deficits upon increasing the intersignal distance. In particular, for the 6131 RSS, we tested whether nearly doubling the distance between the 12 and 23 signals could improve the RJP for this sequence in either the Inv or the CJΔ configuration.
The constructs p6131-Inv(506) and p6131-CJΔ(546) were each cotransfected with the control site 2 plasmid, and the RJPs were determined. Results for both of the larger substrates were fully consistent with those obtained using the original, standard versions (Fig. 3). No site 1 recombinants were found among the 805 recombinants screened for p6131-Inv(506), which compares well to, and is certainly not better than, the 8 site 1 recombinants per 1,358 obtained with the construct for which the intersignal distance was 279 bp (Fig. 3). Reconstruction experiments confirmed that the recombinant form of p6131-Inv should have been detected (data not shown). The plasmid p6131-CJΔ(546) also had an RJP value like its standard counterpart: 0.071 compared to 0.064. Thus, there was no evidence that precleavage strain due to the different topologies of the variously configured substrates could account for the observed configuration-sensitive depressions in RJP.
DISCUSSION
Cells in which cryptic RSSs have been recognized and cleaved can threaten survival of an animal, even when the breaks are generated at a very low frequency. This is because the proliferative and highly selective nature of the lymphoid tissue is such that a cell with a preleukemic rearrangement caused by illegitimate V(D)J recombination is especially at risk of becoming clonally expanded. We know that the stringency of target site recognition is not so refined as to eliminate mistargeting altogether (discussed in reference 24), and it is of interest to learn what biological mechanism might exist to recognize and rectify recombination errors once the process has proceeded as far as cleavage.
Two cryptic RSSs, 6131 and CA, were chosen for a detailed analysis. Although neither sequence closely matches a canonical RSS, both sequences are able to function at a low level in V(D)J recombination (24). 6131 is a sequence discovered to contribute to an unwanted background in the extrachromosomal assay (17). The other cryptic RSS, CA, corresponds to an abundant dinucleotide repeat in the mouse and human genomes (41) and has been seen to function as a cryptic RSS in the human hprt locus (11). 6131 and CA are representative of millions of naturally occurring RSS-like sequences in the genome with respect to their resemblance to the canonical RSS, being similarly divergent in both the heptamer and the nonamer (24).
To learn whether sequence discrimination occurs not only in the initial binding and cleavage of a V(D)J recombination target (stage 1) but also during the second, rejoining phase of the gene assembly process (stage 2), we tested 6131 and CA for evidence of configuration-sensitive differences in V(D)J joining proficiency. In both cases, the results reveal a defect in carrying out stage 2 operations, indicating that there is indeed sequence recognition late in the joining reaction. These findings extend previous studies that implicate RAG1/2 in postcleavage operations (5, 20, 29, 33, 37, 42) by providing evidence that the protein-DNA interactions are both sequence specific and relevant in vivo.
Significantly, despite the fact that by the second stage of V(D)J recombination the coding ends have become unlinked from their RSSs, the CA and 6130 sequences quite clearly exhibited a deficit in coding-joint formation. This raises the logical question as to why operations that effect coding-end joining should depend upon site-specific DNA recognition between recombination proteins and the signal ends.
One likely explanation for postcleavage sequence specificity is a requirement for RAG1/2 to be retained within a protein-DNA complex in order to carry out an enzymatic role in the postcleavage processing of coding ends. The notion that later steps in V(D)J recombination take place in the context of a specific postcleavage complex has had experimental support from several quarters (1, 14, 18, 20) and, although the complex itself (along with all associated components) is not yet biochemically defined, several stable postcleavage complexes containing RAG1/2 and cleaved DNA ends have been described. The various complexes contain RAG1/2, HMG-1 (or -2) and all four ends (18), RAG1/2 and the cut coding and signal end derived from a single RSS (4), and complexes comprised of RAG1/2 together with two cut signal ends (1). The present results are consistent with an in vivo role for a postcleavage complex minimally involving RAG protein and both types of cut end (signal end as well as coding end) in the resolution of recombination intermediates.
A more specific suggestion that fits well with our data is that RAG1/2 itself is responsible for opening hairpin coding-end intermediates and/or for further processing of the coding ends during their ligation (5, 33, 37). If the signal ends help to anchor RAG1/2 in the vicinity of the hairpin coding ends through RSS-specific interactions and if RAG1/2 provides essential hairpin DNA processing functions, then it follows that sequence-specific interactions involving the RSS should influence the joining of coding ends even though coding ends have, by this time, become unlinked from the RAG recognition motifs.
Further, characterization of stage 2 sequence specificity has revealed the importance of both heptamer and nonamer elements in postcleavage joint formation. Of particular interest, a marked postcleavage effect was revealed with the RSS-like sequence, 6131H/WTnon, in which the only noncanonical alteration was in the three nonamer-proximal nucleotides of the heptamer (i.e., CACAGTG to CACAaga). Mutational analyses of the nonamer-proximal nucleotides in the heptamer have shown relatively small effects on the early binding and/or cleavage of RSS-containing DNA by purified RAG1/2 in vitro (2, 31). In contrast, these three nucleotides have a clear in vivo function during stage 2 of V(D)J joining. It is possible that the interaction in question is fairly weak and/or transient, and it is of particular interest that nonamer-proximal nucleotides of the heptamer have been highlighted as a possible contact site for RAG1/2 in one modification interference study (43).
We suggest that the postcleavage complex serves as a focal point in the control of fidelity in V(D)J recombination. After RSS cleavage, coding ends are excluded from ligation events until the hairpin structure at the DNA ends is opened. For a cryptic RSS, a failure to anchor RAG1/2 in a postcleavage complex through its interactions with the RSS could interfere with efficient hairpin removal. Such a delay in hairpin end opening, perhaps interpreted by cellular control mechanisms as irreparable DNA damage, may trigger an apoptotic response. Given the fact that mistakes in RSS target recognition are not fully avoidable, sequence specificity during stage 2 of V(D)J recombination may provide an essential safeguard against pathogenic rearrangements during pre-B- and pre-T-cell differentiation.
ACKNOWLEDGMENTS
We thank Howard Lipshitz and anonymous reviewers for comments on the manuscript.
This work was supported by a grant from the National Cancer Institute of Canada (NCIC). E.A.A. is a recipient of a Doctoral Research Award from the Medical Research Council of Canada. S.M.L. is a Scientist of the NCIC.
REFERENCES
- 1.Agrawal A, Schatz D B. RAG1 and RAG2 form a stable post-cleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu Y, Oettinger M A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aplan P D, Lombardi D P, Ginsberg A M, Cossman J, Bertness V L, Kirsch I R. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250:1426–1429. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 4.Bailin T, Mo X, Sadofsky M. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol Cell Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besmer E, Mansilla-Soto J, Cassard S, Sawchuk D J, Brown G, Sadofsky M, Lewis S M, Nussenzweig M C, Cortes P. Hairpin coding end opening is mediated by the recombination activating genes RAG1 and RAG2. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 6.Binnie A, Olson S, Wu G E, Lewis S M. Gamma-irradiation directly affects the formation of coding joints in SCID cell lines. J Immunol. 1999;163:5418–5426. [PubMed] [Google Scholar]
- 7.Brown L, Cheng J-T, Chen Q, Siciliano M J, Crist W, Buchanan G, Baer R. Site-specific recombination of the tal-1 gene is a common occurrence in human T cell leukemia. EMBO J. 1990;9:3343–3351. doi: 10.1002/j.1460-2075.1990.tb07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherry S R, Baltimore D. Chromatin remodeling directly activates V(D)J recombination. Proc Natl Acad Sci USA. 1999;96:10788–10793. doi: 10.1073/pnas.96.19.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chervinsky D S, Zhao X F, Lam D H, Ellsworth M, Gross K W, Aplan P D. Disordered T-cell development and T-cell malignancies in SCL LMO1 double-transgenic mice: parallels with E2A-deficient mice. Mol Cell Biol. 1999;19:5025–5035. doi: 10.1128/mcb.19.7.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 11.Fuscoe J C, Zimmerman L J, Lippert M J, Nicklas J A, O'Neill J P, Albertini R J. V(D)J recombinase-like activity mediates hprt gene deletion in human fetal T-lymphocytes. Cancer Res. 1991;51:6001–6005. [PubMed] [Google Scholar]
- 12.Gauss G H, Lieber M R. The basis for the mechanistic bias for deletional over inversional V(D)J recombination. Genes Dev. 1992;6:1553–1561. doi: 10.1101/gad.6.8.1553. [DOI] [PubMed] [Google Scholar]
- 13.Golding A, Chandler S, Ballestar E, Wolffe A P, Schlissel M S. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grawunder U, Lieber M R. A complex of RAG-1 and RAG-2 proteins persists on DNA after single-strand cleavage at V(D)J recombination signal sequences. Nucleic Acids Res. 1997;7:1375–1382. doi: 10.1093/nar/25.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grawunder U, West R B, Lieber M R. Antigen receptor gene rearrangement. Curr Opin Immunol. 1998;10:172–180. doi: 10.1016/s0952-7915(98)80246-x. [DOI] [PubMed] [Google Scholar]
- 16.Hammarsten O, DeFazio L G, Chu G. Activation of DNA-dependent protein kinase by single-stranded DNA ends. J Biol Chem. 2000;275:1541–1550. doi: 10.1074/jbc.275.3.1541. [DOI] [PubMed] [Google Scholar]
- 17.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 18.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 19.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 20.Leu R M J, Eastman Q M, Schatz D G. Coding joint formation in a cell-free V(D)J recombination system. Immunity. 1997;7:303–314. doi: 10.1016/s1074-7613(00)80532-4. [DOI] [PubMed] [Google Scholar]
- 21.Lewis S, Hesse J E, Mizuuchi K, Gellert M. Novel strand exchanges in V(D)J recombination. Cell. 1988;55:1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
- 22.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 23.Lewis S M. P nucleotide insertions and the resolution of hairpin DNA structures in mammalian cells. Proc Natl Acad Sci USA. 1994;91:1332–1336. doi: 10.1073/pnas.91.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis S M, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol Cell Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis S M, Hesse J E. Cutting and closing without recombination in V(D)J joining. EMBO J. 1991;10:3631–3639. doi: 10.1002/j.1460-2075.1991.tb04929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieber M R, Hesse J E, Lewis S, Bosma G C, Rosenberg N R, Mizuuchi K, Bosma M J, Gellert M. The defect in murine severe combined immune deficiency: Joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier J T, Lewis S M. P nucleotides in V(D)J recombination: a fine-structure analysis. Mol Cell Biol. 1993;13:1078–1092. doi: 10.1128/mcb.13.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramdsen D A, Paull T T, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 30.Ramsden D A, Baetz K, Wu G E. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsden D A, McBlane J F, Gent D C V, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg N, Baltimore D. A quantitative assay for transformation of bone marrow cells by Abelson murine leukemia virus. J Exp Med. 1976;143:1453–1463. doi: 10.1084/jem.143.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santagata S, Besmer E, Villa A, Bozzi F, Allingham J, Sobacchi C, Haniford D B, Vezzoni P, Nussenzweig M C, Pan Z-Q, Cortes P. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol Cell. 1999;4:935–947. doi: 10.1016/s1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- 34.Scheerer J B, Xi L, Knapp G W, Setzer R W, Bigbee W L, Fuscoe J C. Quantification of illegitimate V(D)J recombinase-mediated mutations in lymphocytes of newborns and adults. Mutat Res. 1999;431:291–303. doi: 10.1016/s0027-5107(99)00173-6. [DOI] [PubMed] [Google Scholar]
- 35.Schlissel M S, Stanhope-Baker P. Accessibility and the developmental regulation of V(D)J recombination. Semin Immunol. 1997;9:161–170. doi: 10.1006/smim.1997.0066. [DOI] [PubMed] [Google Scholar]
- 36.Sheehan K M, Lieber M R. V(D)J recombination: signal and coding joint resolution are uncoupled and depend on parallel synapsis of the sites. Mol Cell Biol. 1993;13:1363–1370. doi: 10.1128/mcb.13.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shockett P E, Schatz D G. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol Cell Biol. 1999;19:4159–4166. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shore D, Baldwin R L. Energetics of DNA twisting. II. Topoisomer analysis. J Mol Biol. 1983;170:983–1007. doi: 10.1016/s0022-2836(83)80199-5. [DOI] [PubMed] [Google Scholar]
- 39.Shore D, Langowski J, Baldwin R L. DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci USA. 1981;78:4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sleckman B P, Gorman J R, Alt F W. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 41.Smider V, Rathmell W K, Brown G, Lewis S, Chu G. Failure of hairpin-ended and nicked DNA to activate DNA-dependent protein kinase: implications for V(D)J recombination. Mol Cell Biol. 1998;18:6853–6858. doi: 10.1128/mcb.18.11.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stallings R L, Ford A F, Nelson D, Torney D C, Hildebrand C E, Moyzis R K. Evolution and distribution of (GT)n repetitive sequences in mammalian genomes. Genomics. 1991;10:807–815. doi: 10.1016/0888-7543(91)90467-s. [DOI] [PubMed] [Google Scholar]
- 43.Steen S B, Han J O, Mundy C, Oettinger M A, Roth D B. Roles of the “dispensable” portions of RAG-1 and RAG-2 in V(D)J recombination. Mol Cell Biol. 1999;19:3010–3017. doi: 10.1128/mcb.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanson P C, Desiderio S. V(D)J recombination signal recognition: Distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 45.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 46.Whitehurst C E, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- 47.Zhong X P, Carabana J, Krangel M S. Flanking nuclear matrix attachment regions synergize with the T cell receptor delta enhancer to promote V(D)J recombination. Proc Natl Acad Sci USA. 1999;96:11970–11975. doi: 10.1073/pnas.96.21.11970. [DOI] [PMC free article] [PubMed] [Google Scholar]