Abstract
The RNA dependent RNA polymerase (RdRp) plays crucial role in virus life cycle by replicating the viral genome. The SARS-CoV-2 is an RNA virus that rapidly spread worldwide and acquired mutations. This study was carried out to identify mutations in RdRp as the SARS-CoV-2 spread in India. We compared 50217 RdRp sequences reported from India with the first reported RdRp sequence from Wuhan, China to identify 223 mutations acquired among Indian isolates. Our protein modelling study revealed that several mutants can potentially alter stability and flexibility of RdRp. We predicted the potential B cell epitopes contributed by RdRp and identified thirty-six linear continuous and twenty-five discontinuous epitopes. Among 223 RdRp mutants, 44% of them localises in the B cell epitopes region. Altogether, this study highlights the need to identify and characterize the variations in RdRp to understand the impact of these mutations on SARS-CoV-2.
Keywords: COVID-19, SARS-CoV-2, Mutations, B cell epitopes, RNA dependent RNA polymerase (RdRp), India
1. Introduction
SARS-CoV-2 genome encodes 29 protein molecules which are categorised into three groups including structural, non-structural and accessory proteins. SARS-CoV-2 has four structural proteins namely Spike glycoprotein, Membrane protein, Envelope protein and Nucleocapsid Phosphoprotein [1]. It also encodes sixteen non-structural proteins (Nsp1-16) and nine accessory proteins. The 16 non-structural proteins are synthesised as a single polypeptide molecule of 7096 amino acids known as Orf1ab that is subsequently cleaved into 16 separate proteins [2]. The RNA dependent RNA polymerase (RdRp), also known as Nsp12, is a non-structural protein that replicates SARS-CoV-2 RNA genome [1]. It associates with Nsp7 and Nsp8 and exist as a trimeric complex inside the viral envelope structure [3]. By itself, RdRp has a very weak polymerase activity; however, the complex of RdRp with Nsp7 and Nsp8 significantly increases RdRp processivity and template affinity [4].
RdRp of SARS-CoV-2 is 932 residues in length and contains distinct polymerase and nucleotide binding domains with a central connecting domain. Structurally, RdRp is comprised of an N-terminal β-hairpin (residues 31–50) followed by an extended nidovirus RdRp-associated nucleotidyl-transferase domain (NiRAN, residues 115–250) [5]. Following the NiRAN domain is an interface domain (residues 251–365) connected to the RdRp domain (residues 366–920). Further, the domains of RdRp arranges in such a way that it forms a canonical right-handed cup configuration [6], with the finger subdomain (resides 397–581 and residues 621–679) forming a closed circle with the thumb subdomain (residues 819–920) [5].
Bioinformatics has enabled researchers to study large number of epitopes and their properties without the risk of growing pathogens. It has drastically reduced the cost of study and faster output over the conventional methods of vaccine study. Further, the amalgamation of various genome-wide studies with the immunoinformatics has revolutionised the identification of epitopes contributed by a protein or virus and accelerated our understanding of vaccine design and action [7,8]. Several studies show that SARS-CoV-2 RdRp participates in host immune response and thus provides insights into viral pathogenesis [[9], [10], [11], [12]]. RdRp stimulates a considerable amount of immunogenicity due to its lower glycosylation density as compared to other structural proteins and several studies have revealed that RdRp induces both innate and adaptive immune response of host [[9], [10], [11]]. Another study has revealed that RdRp, suppresses host antiviral responses by inhibiting IRF3 nuclear translocation [12]. Furthermore, RdRp is one of the most conserved enzyme across several viral species, such as influenza virus, hepatitis C virus (HCV), ZIKA virus (ZIKV), and coronavirus (CoV), suggesting that its function and mechanism of action might be well conserved [13,14]. As the SARS-CoV-2 spread to new geographical areas, it started to mutate [14]. The mutations acquired by the SARS-CoV-2 are retained as a consequence of natural selection, if the variants are more adaptable. In order to understand the variations occurring in RdRp among Indian geographical area, we analysed 50217 RdRp sequences reported from India to identified 223 mutations. The B cell epitopes contributed by RdRp were predicted in silico and the mutations were also mapped.
2. Material and methods
2.1. Protein sequences retrieval and analysis
The sequences used in this study are available on publicly accessible CoVal database (https://coval.ccpem.ac.uk/). A total of 50217 SARS-CoV-2 sequences reported between Jan 2020 till Sept 2021 from different geographical locations within India were used in this study. The mutations occurring in SARS-CoV-2 were obtained from CoVal webserver. The CoVal Webserver uses sequences from the GISAID repository and updates its information at frequent intervals.
2.2. Identification of RdRp mutants by multiple sequence alignments (MSAs)
In order to identify the variations present in the RdRp sequences among Indian isolates of SARS-CoV-2, the MSAs were conducted by Clustal omega programe [15] as described earlier [16].
2.3. B cell epitope prediction
The prediction of linear continuous B cell epitopes were conducted by IEDB [17]. The IEDB webserver provides training set for evaluation of existing epitope prediction methods and constitute platform for development of novel and better algorithm for prediction. IEDB webserver also provides tools for the prediction of linear B-cell epitopes from protein sequence including amino acid scales and HMMs, DiscoTope, ElliPro, Paratome, and PIGS. The IEDB contains epitopes derived from the peer-reviewed literature, patent applications, direct submission, and other publicly available databases, for example, FIMM, HLA Ligand database, and MHC binding database. The IEDB prediction method known as ‘Bepipred linear epitope prediction method 2.0’ was used in this study. For this prediction the threshold value of 0.500 was used during the evaluation. The prediction of discontinuous B cell epitopes was performed by an online tool ‘DiscoTope 2.0’. For this prediction the threshold value was set at −3.7.
2.4. Protein modelling studies
We performed protein modelling studies by DynaMut programe [18] as described earlier [19]. The DynaMut web server was used for analysing thermodynamic stability of different RdRp mutations observed in this study. The DynaMut webserver introduces dynamics component for mutational analysis to predict difference in free energy (ΔΔG) and vibrational entropy (ΔΔS). This webserver implements Normal mode analysis (NMA) through two different approaches, Bio3D and ENCoM, that provide rapid and simplified access to analyse protein dynamics and stability resulting from vibrational entropy changes [18]. It also enables to assess the effects of missense mutations on protein stability and provide comprehensive suite for protein motion and flexibility analysis and visualization (http://biosig.unimelb.edu.au/dynamut/). For this study, we used recently reported structure of RdRp (PDB ID: 7BV1) [5]. The effect of mutations on protein is shown in terms of difference in free energy (ΔΔG). DynaMut provides difference in vibrational entropy (ΔΔSvib ENCOM) between the wild type and mutant protein. We ran DynaMut webserver to calculate the ΔΔG and ΔΔSvib ENCOM that provides the impact of mutation on protein structure and stability.
3. Results
3.1. RdRp is frequently mutated among Indian isolates of SARS-CoV-2
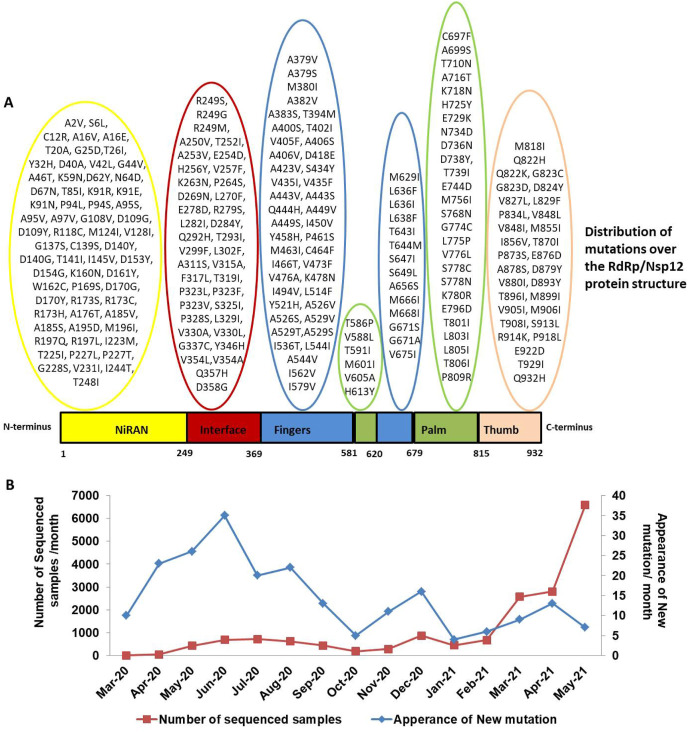
In order to identify the mutations in RdRp, we used CoVal webserver that compares the first reported sequence of RdRp from Wuhan, China with the sequences reported from India. Till Sept 2021, a total of 50217 sequences has been analysed by CoVal webserver. The data revealed 223 mutations present among the Indian sequences of RdRp as shown in Table 1 . The mutations are also demonstrated on the schematic representation of RdRp as shown in Fig. 1 A. Our result show that the mutations are spreading all over the RdRp polypeptide sequence. The distribution of mutations in different domains of RdRp has been highlighted in Fig. 1A. This data strongly indicates that RdRp is one of the most frequently mutated proteins of SARS-CoV-2 because we observed 223 mutations till Sept 2021. Furthermore, we looked at the time course of the samples used for mutational study by CoVal webserver. This webserver shows the monthly appearance of new mutations from India. Based on the mutational analysis of RdRp by CoVAL webserver, we observe that during initial phase of COVID19 pandemic, the rate of occurrence of new mutations were high but it slowed down as the time progresses (Fig. 1B), suggesting that the virus is attaining mutational stability over time.
Table 1.
The list demonstrates the location and details of mutations of RdRp identified by CoVal webserver. The RdRp sequence reported from Wuhan, China was used as wild type sequence for this analysis. The 50217 sequences of RdRp reported from India (till Sept 2021) were used for identifying mutations and their frequency. The ΔΔG and ΔΔSvib ENCOM values were obtained by protein modelling using DynaMut programe. The positive and negative ΔΔG represents increase and decrease in protein stability upon mutation. Similarly, the positive and negative ΔΔSvib ENCOM represents the increase in flexibility and rigidity upon mutations. The mutation that localises in the unmodeled region of RdRp was not used in the analysis of ΔΔG and ΔΔSvib ENCOM and they were left blank (denotes by -) in table. P stands for Polar, NP stands for Non-Polar.
| Serial Number |
Nsp12 mutations | Polarity changes | Charge changes | Frequency of mutation | ΔΔG (kcal/mol) | ΔΔSVib ENCoM (kcal.mol-1.K-1) |
|---|---|---|---|---|---|---|
| 1 | A2V | NP to NP | Neutral to Neutral | 3 | – | – |
| 2 | S6L | P to NP | Neutral to Neutral | 12 | – | – |
| 3 | C12R | P to P | Neutral to Basic | 2 | – | – |
| 4 | A16V | NP to NP | Neutral to Neutral | 8 | – | – |
| 5 | A16E | NP to P | Neutral to Acidic | 2 | – | – |
| 6 | T20A | P to NP | Neutral to Neutral | 11 | – | – |
| 7 | G25D | NP to P | Neutral to Acidic | 4 | – | – |
| 8 | T26I | P to NP | Neutral to Neutral | 13 | – | – |
| 9 | Y32H | P to P | Neutral to Basic | 2 | −0.349 | 0.090 |
| 10 | D40A | P to NP | Acidic to Neutral | 4 | −0.139 | 0.021 |
| 11 | V42L | NP to NP | Neutral to Neutral | 2 | 0.347 | −0.342 |
| 12 | G44V | NP to NP | Neutral to Neutral | 2 | −1.011 | −0.103 |
| 13 | A46T | NP to P | Neutral to Neutral | 2 | −1.031 | −0.174 |
| 14 | K59 N | P to P | Basic to Neutral | 12 | – | – |
| 15 | D62Y | P to P | Acidic to Neutral | 3 | – | – |
| 16 | N64D | P to P | Neutral to Acidic | 2 | – | – |
| 17 | D67 N | P to P | Acidic to Neutral | 4 | – | – |
| 18 | T85I | P to NP | Neutral to Neutral | 14 | −0.707 | −0.074 |
| 19 | K91R | P to P | Basic to Basic | 11 | −0.143 | −0.066 |
| 20 | K91E | P to P | Basic to Acidic | 2 | −0.187 | 0.416 |
| 21 | K91 N | P to P | Basic to Neutral | 2 | −0.311 | 0.295 |
| 22 | P94L | NP to NP | Neutral to Neutral | 6 | 0.858 | −0.242 |
| 23 | P94S | NP to P | Neutral to Neutral | 3 | −0.035 | −0.131 |
| 24 | A95S | NP to P | Neutral to Neutral | 5 | −1.317 | −0.172 |
| 25 | A95V | NP to NP | Neutral to Neutral | 4 | −0.669 | −0.627 |
| 26 | A97V | NP to NP | Neutral to Neutral | 438 | 0.469 | −1.020 |
| 27 | G108V | NP to NP | Neutral to Neutral | 8 | – | – |
| 28 | D109G | P to NP | Acidic to Neutral | 3 | – | – |
| 29 | D109Y | P to P | Acidic to Neutral | 2 | – | – |
| 30 | R118C | P to P | Basic to Neutral | 4 | 0.005 | 0.055 |
| 31 | M124I | NP to NP | Neutral to Neutral | 6 | −0.088 | 0.250 |
| 32 | V128I | NP to NP | Neutral to Neutral | 5 | 1.237 | −0.268 |
| 33 | G137S | NP to P | Neutral to Neutral | 2 | −1.389 | 0.029 |
| 34 | C139S | P to P | Neutral to Neutral | 2 | −0.460 | −0.227 |
| 35 | D140Y | P to P | Acidic to Neutral | 6 | −0.105 | 0.035 |
| 36 | D140G | P to NP | Acidic to Neutral | 3 | −0.180 | 0.035 |
| 37 | T141I | P to NP | Neutral to Neutral | 11 | 1.041 | −0.231 |
| 38 | I145V | NP to NP | Neutral to Neutral | 4 | −0.190 | 0.408 |
| 39 | D153Y | P to P | Acidic to Neutral | 10 | 0.961 | 0.049 |
| 40 | D154G | P to NP | Acidic to Neutral | 2 | −0.771 | 0.111 |
| 41 | K160 N | P to P | Basic to Neutral | 6 | −0.089 | −0.029 |
| 42 | D161Y | P to P | Acidic to Neutral | 2 | 1.107 | −0.621 |
| 43 | W162C | NP to P | Neutral to Neutral | 3 | −1.169 | 1.004 |
| 44 | P169S | NP to P | Neutral to Neutral | 3 | 0.478 | −0.232 |
| 45 | D170G | P to NP | Acidic to Neutral | 3 | −0.527 | 0.156 |
| 46 | D170Y | P to P | Acidic to Neutral | 3 | −0.089 | 0.154 |
| 47 | R173S | P to P | Basic to Neutral | 4 | −0.412 | 0.203 |
| 48 | R173C | P to P | Basic to Neutral | 2 | −0.283 | 0.168 |
| 49 | R173H | P to P | Basic (strongly) to Basic (weakly) | 2 | −0.284 | 0.010 |
| 50 | A176T | NP to P | Neutral to Neutral | 2 | 0.032 | −0.721 |
| 51 | A185V | NP to NP | Neutral to Neutral | 11 | −0.179 | −0.369 |
| 52 | A185S | NP to P | Neutral to Neutral | 2 | – | – |
| 53 | A195D | NP to P | Neutral to Acidic | 2 | −0.478 | −0.397 |
| 54 | M196I | NP to NP | Neutral to Neutral | 7 | −0.554 | 0.094 |
| 55 | R197Q | P to P | Basic to Neutral | 3 | −0.483 | 0.096 |
| 56 | R197L | P to NP | Basic to Neutral | 2 | −0.109 | 0.109 |
| 57 | I223 M | NP to NP | Neutral to Neutral | 13 | −0.051 | −0.084 |
| 58 | T225I | P to NP | Neutral to Neutral | 2 | 0.090 | −0.076 |
| 59 | P227L | NP to NP | Neutral to Neutral | 245 | 0.360 | −0.332 |
| 60 | P227T | NP to P | Neutral to Neutral | 2 | 0.459 | −0.375 |
| 61 | G228S | NP to P | Neutral to Neutral | 18 | −1.113 | −0.158 |
| 62 | V231I | NP to NP | Neutral to Neutral | 3 | −0.529 | 0.229 |
| 63 | I244T | NP to P | Neutral to Neutral | 85 | −3.236 | 0.746 |
| 64 | T248I | P to NP | Neutral to Neutral | 7 | 0.452 | −0.080 |
| 65 | R249S | P to P | Basic to Neutral | 7 | −0.672 | 0.320 |
| 66 | R249G | P to NP | Basic to Neutral | 2 | −0.382 | 0.504 |
| 67 | R249 M | P to NP | Basic to Neutral | 2 | 0.825 | −0.111 |
| 68 | A250V | NP to NP | Neutral to Neutral | 5 | −0.237 | −0.482 |
| 69 | T252I | P to NP | Neutral to Neutral | 4 | 0.265 | −0.060 |
| 70 | A253V | NP to NP | Neutral to Neutral | 2 | −0.415 | −0.586 |
| 71 | E254D | P to P | Acidic to Acidic | 2 | −1.136 | 0.395 |
| 72 | H256Y | P to P | Basic to Neutral | 3 | −0.411 | −0.074 |
| 73 | V257F | NP to NP | Neutral to Neutral | 5 | −0.485 | 0.162 |
| 74 | K263 N | P to P | Basic to Neutral | 5 | −0.336 | 0.316 |
| 75 | P264S | NP to P | Neutral to Neutral | 4 | −0.715 | 0.098 |
| 76 | D269 N | P to P | Acidic to Neutral | 34 | −0.258 | 0.080 |
| 77 | L270F | NP to NP | Neutral to Neutral | 3 | 0.390 | −0.209 |
| 78 | E278D | P to P | Acidic to Acidic | 3 | −0.714 | 0.103 |
| 79 | R279S | P to P | Basic to Neutral | 3 | −2.849 | 1.063 |
| 80 | L282I | NP to NP | Neutral to Neutral | 3 | −0.287 | 0.096 |
| 81 | D284Y | P to P | Acidic to Neutral | 7 | 0.346 | −0.459 |
| 82 | Q292H | P to P | Neutral to Basic | 2 | 0.171 | 0.070 |
| 83 | T293I | P to NP | Neutral to Neutral | 5 | 0.264 | −0.016 |
| 84 | V299F | NP to NP | Neutral to Neutral | 4 | 0.240 | −0.143 |
| 85 | L302F | NP to NP | Neutral to Neutral | 3 | −0.363 | 0.045 |
| 86 | A311S | NP to P | Neutral to Neutral | 8 | −0.229 | −0.014 |
| 87 | V315A | NP to NP | Neutral to Neutral | 3 | −1.724 | 0.711 |
| 88 | F317L | NP to NP | Neutral to Neutral | 2 | 0.045 | 0.379 |
| 89 | T319I | P to NP | Neutral to Neutral | 35 | 1.052 | −0.244 |
| 90 | P323L | NP to NP | Neutral to Neutral | 11094 | 0.530 | −0.252 |
| 91 | P323F | NP to NP | Neutral to Neutral | 24 | 0.297 | −0.199 |
| 92 | P323V | NP to NP | Neutral to Neutral | 3 | 0.578 | −0.313 |
| 93 | S325I | P to NP | Neutral to Neutral | 2 | 1.763 | −0.574 |
| 94 | P328S | NP to P | Neutral to Neutral | 2 | −0.425 | 0.065 |
| 95 | L329I | NP to NP | Neutral to Neutral | 15 | 0.081 | 0.231 |
| 96 | V330A | NP to NP | Neutral to Neutral | 38 | – | – |
| 97 | V330L | NP to NP | Neutral to Neutral | 5 | 0.025 | −0.103 |
| 98 | G337C | NP to P | Neutral to Neutral | 2 | −1.187 | 0.219 |
| 99 | Y346H | P to P | Neutral to Basic | 2 | −0.021 | 0.619 |
| 100 | V354L | NP to NP | Neutral to Neutral | 21 | −0.162 | −0.387 |
| 101 | V354A | NP to NP | Neutral to Neutral | 4 | −1.811 | 0.277 |
| 102 | Q357H | P to P | Neutral to Basic | 7 | 1.218 | −0.094 |
| 103 | D358G | P to NP | Acidic to Neutral | 3 | −0.096 | 0.357 |
| 104 | A379V | NP to NP | Neutral to Neutral | 7 | 0.624 | −0.529 |
| 105 | A379S | NP to P | Neutral to Neutral | 2 | 1.586 | −0.510 |
| 106 | M380I | NP to NP | Neutral to Neutral | 6 | −0.487 | 0.551 |
| 107 | A382V | NP to NP | Neutral to Neutral | 3 | 0.596 | −0.438 |
| 108 | A383S | NP to P | Neutral to Neutral | 2 | −0.131 | −0.021 |
| 109 | T394 M | P to NP | Neutral to Neutral | 3 | 0.947 | −0.648 |
| 110 | A400S | NP to P | Neutral to Neutral | 52 | 0.437 | −0.127 |
| 111 | T402I | P to NP | Neutral to Neutral | 6 | 0.301 | 0.039 |
| 112 | V405F | NP to NP | Neutral to Neutral | 3 | 0.955 | −0.639 |
| 113 | A406S | NP to P | Neutral to Neutral | 7 | 0.497 | −0.246 |
| 114 | A406V | NP to NP | Neutral to Neutral | 2 | −0.148 | −0.047 |
| 115 | D418E | P to P | Acidic to Acidic | 2 | 0.915 | −0.258 |
| 116 | A423V | NP to NP | Neutral to Neutral | 12 | 0.732 | −0.338 |
| 117 | S434Y | P to P | Neutral to Neutral | 2 | 0.277 | −0.606 |
| 118 | V435I | NP to NP | Neutral to Neutral | 4 | 0.150 | −0.391 |
| 119 | V435F | NP to NP | Neutral to Neutral | 2 | −0.063 | −1.096 |
| 120 | A443V | NP to NP | Neutral to Neutral | 4 | 0.937 | −0.250 |
| 121 | A443S | NP to P | Neutral to Neutral | 2 | −0.013 | −0.135 |
| 122 | Q444H | P to P | Neutral to Basic | 5 | 1.310 | −0.346 |
| 123 | A449V | NP to NP | Neutral to Neutral | 7 | 0.871 | −0.560 |
| 124 | A449S | NP to P | Neutral to Neutral | 2 | −0.265 | 0.016 |
| 125 | I450V | NP to NP | Neutral to Neutral | 2 | −1.121 | 0.580 |
| 126 | Y458H | P to P | Neutral to Basic | 3 | −0.169 | 0.293 |
| 127 | P461S | NP to P | Neutral to Neutral | 8 | −0.475 | 0.055 |
| 128 | M463I | NP to NP | Neutral to Neutral | 19 | 0.483 | 0.250 |
| 129 | C464F | P to NP | Neutral to Neutral | 2 | 1.251 | −1.092 |
| 130 | I466T | NP to P | Neutral to Neutral | 2 | −2.428 | 0.355 |
| 131 | V473F | NP to NP | Neutral to Neutral | 24 | −0.802 | −0.941 |
| 132 | V476A | NP to NP | Neutral to Neutral | 3 | −0.477 | 0.578 |
| 133 | K478 N | P to P | Basic to Neutral | 60 | −1.105 | 0.533 |
| 134 | I494V | NP to NP | Neutral to Neutral | 2 | −0.044 | 0.027 |
| 135 | L514F | NP to NP | Neutral to Neutral | 2 | −0.300 | −0.066 |
| 136 | Y521H | P to P | Neutral to Basic | 4 | −0.048 | 0.764 |
| 137 | A526V | NP to NP | Neutral to Neutral | 4 | 0.305 | −0.109 |
| 138 | A526S | NP to P | Neutral to Neutral | 3 | −0.315 | −0.121 |
| 139 | A529V | NP to NP | Neutral to Neutral | 16 | −0.273 | −0.068 |
| 140 | A529T | NP to P | Neutral to Neutral | 3 | −0.874 | 0.002 |
| 141 | A529S | NP to P | Neutral to Neutral | 2 | −0.884 | −0.059 |
| 142 | I536T | NP to P | Neutral to Neutral | 3 | – | – |
| 143 | L544I | NP to NP | Neutral to Neutral | 7 | 0.224 | 0.060 |
| 144 | A544V | NP to NP | Neutral to Neutral | 2 | 0.325 | −0.547 |
| 145 | I562V | NP to NP | Neutral to Neutral | 2 | −0.801 | 0.215 |
| 146 | I579V | NP to NP | Neutral to Neutral | 3 | −0.573 | 0.414 |
| 147 | T586P | P to NP | Neutral to Neutral | 2 | −0.213 | 0.662 |
| 148 | V588L | NP to NP | Neutral to Neutral | 4 | 0.599 | −0.117 |
| 149 | T591I | P to NP | Neutral to Neutral | 4 | 1.218 | −0.416 |
| 150 | M601I | NP to NP | Neutral to Neutral | 18 | −0.362 | −0.033 |
| 151 | V605A | NP to NP | Neutral to Neutral | 18 | −1.528 | 0.596 |
| 152 | H613Y | P to P | Basic to Neutral | 20 | 0.836 | −0.213 |
| 153 | M629I | NP to NP | Neutral to Neutral | 6 | −0.428 | 0.170 |
| 154 | L636F | NP to NP | Neutral to Neutral | 4 | −0.115 | −0.435 |
| 155 | L636I | NP to NP | Neutral to Neutral | 2 | 0.569 | 0.041 |
| 156 | L638F | NP to NP | Neutral to Neutral | 6 | −0.167 | −0.297 |
| 157 | T643I | P to NP | Neutral to Neutral | 10 | −0.383 | −0.074 |
| 158 | T644 M | P to NP | Neutral to Neutral | 3 | −0.084 | −0.037 |
| 159 | S647I | P to NP | Neutral to Neutral | 29 | −0.272 | 0.166 |
| 160 | S649L | P to NP | Neutral to Neutral | 2 | 0.684 | −0.422 |
| 161 | A656S | NP to P | Neutral to Neutral | 20 | −1.416 | 0.063 |
| 162 | M666I | NP to NP | Neutral to Neutral | 6 | −0.229 | 0.365 |
| 163 | M668I | NP to NP | Neutral to Neutral | 11 | 0.104 | 0.066 |
| 164 | G671S | NP to P | Neutral to Neutral | 2665 | 0.786 | −0.246 |
| 165 | G671A | NP to NP | Neutral to Neutral | 2 | 1.315 | −0.906 |
| 166 | V675I | NP to NP | Neutral to Neutral | 49 | 0.050 | −0.115 |
| 167 | C697F | P to NP | Neutral to Neutral | 3 | −1.055 | −0.979 |
| 168 | A699S | NP to P | Neutral to Neutral | 4 | −2.233 | −0.185 |
| 169 | T710 N | P to P | Neutral to Neutral | 2 | 0.703 | −0.185 |
| 170 | A716T | NP to P | Neutral to Neutral | 2 | 0.285 | −0.035 |
| 171 | K718 N | P to P | Basic to Neutral | 5 | −0.208 | 0.051 |
| 172 | H725Y | P to P | Basic to Neutral | 6 | 0.240 | 0.166 |
| 173 | E729K | P to P | Acidic to Basic | 2 | 0.825 | −0.139 |
| 174 | N734D | P to P | Neutral to Acidic | 4 | −0.338 | 0.227 |
| 175 | D736 N | P to P | Acidic to Neutral | 3 | 0.006 | −0.049 |
| 176 | D738Y | P to P | Acidic to Neutral | 5 | 0.861 | −0.137 |
| 177 | T739I | P to NP | Neutral to Neutral | 4 | 0.272 | 0.008 |
| 178 | E744D | P to P | Acidic to Acidic | 11 | −0.566 | 0.229 |
| 179 | M756I | NP to NP | Neutral to Neutral | 11 | 1.019 | 0.205 |
| 180 | S768 N | P to P | Neutral to Neutral | 2 | −0.152 | −0.012 |
| 181 | G774C | NP to P | Neutral to Neutral | 4 | 0.107 | −0.086 |
| 182 | L775P | NP to NP | Neutral to Neutral | 2 | −0.600 | 0.754 |
| 183 | V776L | NP to NP | Neutral to Neutral | 3 | −0.017 | −0.160 |
| 184 | S778C | P to P | Neutral to Neutral | 3 | 0.409 | −0.305 |
| 185 | S778 N | P to P | Neutral to Neutral | 2 | 0.687 | −0.195 |
| 186 | K780R | P to P | Basic to Basic | 5 | 1.115 | −0.590 |
| 187 | E796D | P to P | Acidic to Acidic | 4 | −0.269 | 0.244 |
| 188 | T801I | P to NP | Neutral to Neutral | 5 | 0.114 | −0.047 |
| 189 | L803I | NP to NP | Neutral to Neutral | 2 | 0.397 | −0.105 |
| 190 | L805I | NP to NP | Neutral to Neutral | 3 | −0.189 | 0.217 |
| 191 | T806I | P to NP | Neutral to Neutral | 29 | 0.151 | −0.065 |
| 192 | P809R | NP to P | Neutral to Basic | 5 | −0.615 | −0.234 |
| 193 | M818I | NP to NP | Neutral to Neutral | 2 | 0.129 | 0.221 |
| 194 | Q822H | P to P | Neutral to Basic | 60 | 0.006 | 0.514 |
| 195 | Q822K | P to P | Neutral to Basic | 2 | 1.801 | −0.467 |
| 196 | G823C | NP to P | Neutral to Neutral | 4 | 0.442 | −0.644 |
| 197 | G823D | NP to P | Neutral to Acidic | 2 | 0.467 | −0.400 |
| 198 | D824Y | P to P | Acidic to Neutral | 6 | −0.451 | 0.111 |
| 199 | V827L | NP to NP | Neutral to Neutral | 3 | 0.471 | −0.340 |
| 200 | L829F | NP to NP | Neutral to Neutral | 4 | −1.408 | 0.045 |
| 201 | P834L | NP to NP | Neutral to Neutral | 3 | 1.266 | −0.943 |
| 202 | V848L | NP to NP | Neutral to Neutral | 12 | 0.194 | −0.201 |
| 203 | V848I | NP to NP | Neutral to Neutral | 2 | −0.732 | 0.119 |
| 204 | M855I | NP to NP | Neutral to Neutral | 6 | −0.312 | 0.223 |
| 205 | I856V | NP to NP | Neutral to Neutral | 2 | −0.635 | 0.307 |
| 206 | T870I | P to NP | Neutral to Neutral | 10 | 0.654 | −0.440 |
| 207 | P873S | NP to P | Neutral to Neutral | 6 | −0.054 | 0.023 |
| 208 | E876D | P to P | Acidic to Acidic | 2 | −1.338 | 1.031 |
| 209 | A878S | NP to P | Neutral to Neutral | 2 | 1.053 | −0.488 |
| 210 | D879Y | P to P | Acidic to Neutral | 6 | 1.218 | −0.559 |
| 211 | V880I | NP to NP | Neutral to Neutral | 34 | −0.087 | −0.146 |
| 212 | D893Y | P to P | Acidic to Neutral | 7 | 0.088 | −0.168 |
| 213 | T896I | P to NP | Neutral to Neutral | 2 | – | – |
| 214 | M899I | NP to NP | Neutral to Neutral | 4 | – | – |
| 215 | V905I | NP to NP | Neutral to Neutral | 38 | – | – |
| 216 | M906I | NP to NP | Neutral to Neutral | 2 | – | – |
| 217 | T908I | P to NP | Neutral to Neutral | 5 | – | – |
| 218 | S913L | P to NP | Neutral to Neutral | 19 | −0.116 | 0.012 |
| 219 | R914K | P to P | Basic to Basic | 9 | −0.240 | 0.168 |
| 220 | P918L | NP to NP | Neutral to Neutral | 4 | 0.199 | −0.711 |
| 221 | E922D | P to P | Acidic to Acidic | 8 | −1.106 | 0.193 |
| 222 | T929I | P to NP | Neutral to Neutral | 2 | 0.075 | 4.551 |
| 223 | Q932H | P to P | Neutral to Basic | 2 | 1.077 | −4.934 |
Fig. 1.
The details of the mutation identified in RdRp among Indian SARS-CoV-2 isolates. A) Schematic diagram of the domain architecture of RdRp. Each domain of RdRp is represented by a unique color. The interdomain borders are labeled with residue numbers.The mutations present in RdRp among the Indian isolates of SARS-CoV-2 are demonstrated in the schematics. B) The monthly data of ‘new sequences’ of SARS-CoV-2 and ‘appearance of new mutations’ were obtained from CoVal webserver. These data provided the time course of the SARS-CoV-2 samples reported from India. Based on the mutational analysis of RdRp by CoVal webserver, we observe that during initial phase COVID19 pandemic, the rate occurrence of new mutations were high but it slowed down as the time progresses. The X-axis shows months/year and Y-axis present on the left side of the graph represents number of SARS-CoV-2 samples reported per month while Y –axis on the right side shows the appearance of new mutation in RdRp per month. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Mutations affect RdRp protein dynamic stability and flexibility
We performed protein modelling studies using DynaMut programe to understand, if the mutation observed in RdRp can alter protein structural integrity. Our data revealed that mutations at 89 positions cause stabilisation in protein structure (positive ΔΔG) as shown in Table 1, maximum positive ΔΔG was obtained for Q822K (1.801 kcal/mol). Similarly, the mutations at 111 positions cause destabilisation (negative ΔΔG) in protein structure upon mutation (Table 1), maximum negative ΔΔG was obtained for the mutant I244T (-2.233 kcal/mol).
Subsequently, we measured the changes in vibrational entropy energy (ΔΔSVibENCoM) between the wild type and the mutant. Our data revealed that mutation at 89 positions causes increase in flexibility of mutant protein (positive ΔΔSVibENCoM). The maximum positive ΔΔSVibENCoM was obtained for T929I (4.55 kcal.mol-1.K-1) mutant. Similarly, the mutations at rest of the 111 positions cause rigidification of protein structure (negative ΔΔSVibENCoM) in protein structure upon mutation (Table 1). The maximum negative ΔΔSVibENCoM was obtained for Q932H (-4.93 kcal.mol-1.K-1) mutant. Altogether, our data revealed that the mutation observed in RdRp affects both protein dynamicity and flexibility.
3.3. Identification of B cell epitopes of RdRp
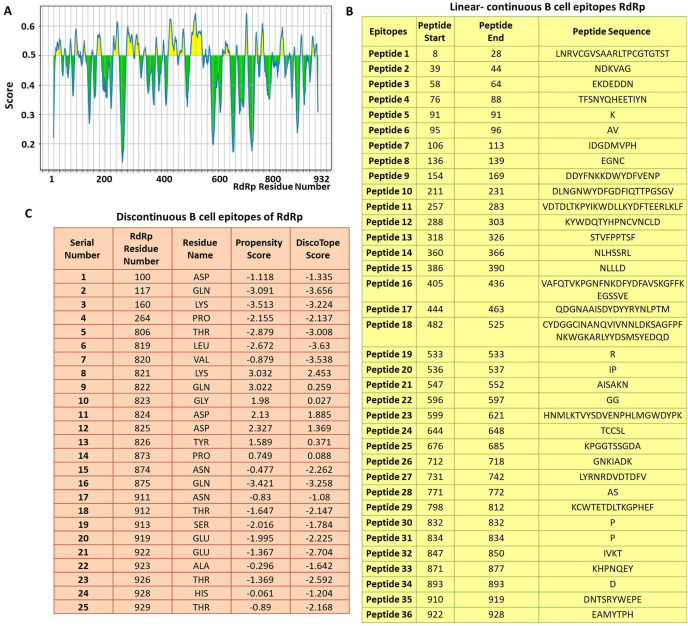
The continuous B-cell epitopes of RdRp were predicted by IEDB webserver tool and the epitopes are shown in Fig. 2 A. The yellow area of the graph corresponds to those regions of the RdRp that can potentially contribute to the B cell epitopes. Our data demonstrated thirty-six epitopes of varying lengths that could potentially act as B cell epitopes (Fig. 2B). Among those peptides, the ‘peptide 18’ is the largest epitope of 44 amino acids (from RdRp residue 482 to 525). Similarly, peptide 5, 19, 30, 31 and 34 are comprised of single amino acid only (Fig. 2B).
Fig. 2.
Prediction of B-cell epitopes of RdRp. A) Linear continuous B-cell epitopes contributed by RdRp, the Y-axis of the graph corresponds to BepiPred score, while the X-axis depicts the RdRp residue positions in the sequence. The data was generated by IEDB webserver using ‘Bepipred Linear Epitope Prediction 2.0’ method. The chart is divided into two parts, yellow and green. The RdRp residues present in the yellow area have higher probability to be part of the linear continuous B cell epitope. B) The details of the linear continuous B cell epitopes are listed. The sequence of each peptide along with its start and end point in the RdRp polypeptide sequence is also mentioned. C) Prediction of discontinuous B-cell epitopes of RdRp by DiscoTope 2.0 web tool. The position of each predicted epitope is mentioned along with its propensity and DiscoTope score. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Subsequently, we predicted the B cell epitopes of RdRp based on its three dimensional structure using DiscoTope 2.0 webserver tool [20]. Our analysis revealed twenty-five discontinuous epitopes of RdRp having high score. The locations of these epitopes are listed in Fig. 2C along with its propensity and DiscoTope score. Among discontinuous epitopes, approximately 80% of them (20 out of 25) reside towards the C-terminal end of RdRp (from residue 800 to 932) as shown in Fig. 2C. Altogether, our data revealed B cell epitopes contributed by RdRp.
3.4. RdRp mutants preferentially localises in the B cell epitopes region
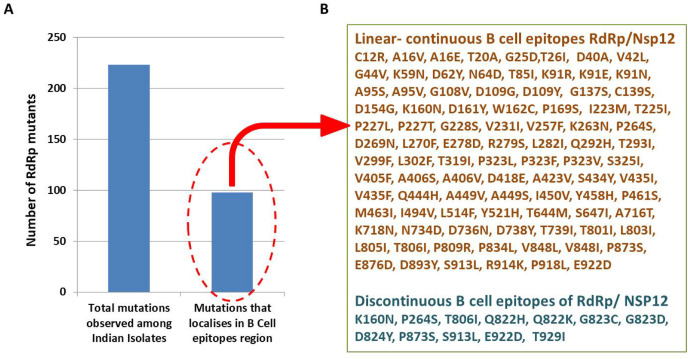
Next, we analysed and compared the RdRp mutations that reside in the linear-continuous and discontinuous B cell epitopes. Our data revealed that out of 223 mutants observed in this study, 98 resides in the B cell epitope region of RdRp (Fig. 3 A). These 98 mutants correspond to 44% of the total mutants observed among Indian isolates. The details of all 98 mutants that localises in B cell epitope region are shown in Fig. 3B. Altogether, our data strongly suggest that several RdRp mutations localises in the B cell epitope region.
Fig. 3.
Correlation of RdRp mutants and B cell epitopes. A) The graph shows the distribution of RdRp mutants observed among Indian SARS-CoV-2 isolates. Out of 223, 98 mutants localises in B cell epitope region of RdRp. B) Detail of the RdRp mutants that localises in B cell epitope region.
4. Discussion
The coronaviruses belongs to RNA viruses that exhibits high rate of mutations in their genome [21]. As these viruses spread to new locations they keep on acquiring mutations and few of them are naturally selected because of their beneficial effect on the virus. The investigation on the genomic variation acquired by SARS-CoV-2 is indispensable for understanding the epidemiology, pathogenesis; devise preventive measures and treatment strategies against COVID-19. The earlier variation studies on SARS-CoV-2 revealed that RdRp is among the mutational hotspot protein [14]. In the similar directions, this study was conducted with an aim to identify mutations in RdRp from Indian isolates. Our earlier study revealed seven crucial mutations in RdRp of SARS-CoV-2 [22] that can have potential impact on this protein function. The present study identifies and characterises B cell epitope contributed by RdRp and correlate them with the observed mutants. In this study, we analysed 50217 RdRp sequences reported from India till Sept 2021 and identified 223 mutations in RdRp, which indicates that RdRp is one of the mutational hotspot protein of SARS-CoV-2. Furthermore, our data revealed that there are thirty-six high rank linear-continuous B cell epitope as well as twenty-five discontinuous B cell epitopes. Moreover, we also identified that out of 223 mutants identified among Indian isolates, 98 resides (44%) in these B cell epitope region.
We used bioinformatics approach to identify probable epitopes that offer various advantages over conventional approaches. However, despite advantages of immunoinformatics, there are certain limitations. Such as the final selection of epitopes from the probable epitopes identified using bioinformatics is still a challenging task. The RdRp epitopes revealed in this study requires validation using in vivo experiments, which is slow and herculean task. Furthermore, the algorithms used for predicting epitopes are liable to alter if the criteria are changed during the tool selection. Therefore, the algorithms are constantly improved to get better output and more reliable data [7,8]. The variations in RdRp or any other protein of SARS-CoV-2 will possibly tell us how the virus is evolving. Earlier studies with RNA viruses have also shown that these viruses keep on mutating to better adapt and survive in the host [23]. Here, in this study, we have reported RdRp mutations, its correlation with B cell epitopes. However, it warrants future studies to understand the possible effect of these mutations on virus infectivity and life cycle.
Acknowledgements
We would like to acknowledge Patna University, Patna, Bihar (India) for providing infrastructural support for this study. This work has been partly funded by (Science and Engineering Board, Department of Science and Technology, Government of India) a project awarded to GKA (Project number: SRG/2020/000808).
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microb Infect. 2020 doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., et al. Structural and biochemical characterization of nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020 doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Te Velthuis A.J.W., Van Den Worm S.H.E., Snijder E.J. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 doi: 10.1126/science.abc1560. 80- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mcdonald S.M. Wiley Interdiscip Rev RNA; 2013. RNA synthetic mechanisms employed by diverse families of RNA viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenbaum J.A., Andersen P.H., Blythe M., Bui H.H., Cachau R.E., Crowe J., et al. Towards a consensus on datasets and evaluation metrics for developing B-cell epitope prediction tools. J Mol Recogn. 2007 doi: 10.1002/jmr.815. [DOI] [PubMed] [Google Scholar]
- 8.Soria-Guerra R.E., Nieto-Gomez R., Govea-Alonso D.O., Rosales-Mendoza S. An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J Biomed Inf. 2015 doi: 10.1016/j.jbi.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Azkur A.K., Akdis M., Azkur D., Sokolowska M., van de Veen W., Brüggen M.C., et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy Eur J Allergy Clin Immunol. 2020 doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yashvardhini N., Kumar A., Jha D.K. Immunoinformatics identification of B-and T-cell epitopes in the RNA-dependent RNA polymerase of SARS-CoV-2. Can J Infect Dis Med Microbiol. 2021 doi: 10.1155/2021/6627141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aftab S.O., Ghouri M.Z., Masood M.U., Haider Z., Khan Z., Ahmad A., et al. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J Transl Med. 2020 doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W., Zhou Z., Xiao X., Tian Z., Dong X., Wang C., et al. SARS-CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell Mol Immunol. 2021 doi: 10.1038/s41423-020-00619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Anirudhan V., Du R., Cui Q., Rong L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J Med Virol. 2021 doi: 10.1002/jmv.26264. [DOI] [PubMed] [Google Scholar]
- 14.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020 doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azad G.K. The molecular assessment of SARS-CoV-2 Nucleocapsid Phosphoprotein variants among Indian isolates. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019 doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigues C.H.M., Pires D.E.V., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azad G.K. Identification and molecular characterization of mutations in nucleocapsid phosphoprotein of SARS-CoV-2. PeerJ. 2021 doi: 10.7717/peerj.10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kringelum J.V., Lundegaard C., Lund O., Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol. 2012 doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., Ciccozzi M. The 2019-new coronavirus epidemic: evidence for virus evolution. J Med Virol. 2020 doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chand G.B., Banerjee A., Azad G.K. Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ. 2020;8 doi: 10.7717/peerj.9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanjuán R., Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]