Abstract
The performance of immune checkpoint inhibitors, which benefit only a subset of patients and can cause serious immune-related adverse events, underscores the need for strategies that induce T-cell immunity with minimal toxicity. The gut microbiota has been implicated in the outcomes of patients following cancer immunotherapy, yet manipulating the gut microbiome to achieve systemic antitumour immunity is challenging. Here, we show, in multiple murine tumour models, that inulin — a widely consumed dietary fibre — formulated as a colon-retentive orally administered gel can effectively modulate the gut microbiome in situ, induce systemic memory-T-cell responses, and amplify the antitumour activity of the checkpoint inhibitor anti-programmed-cell-death-protein-1 (anti-PD-1). Orally delivered inulin-gel treatments increased the relative abundances of key commensal microbes and their short-chain-fatty-acid metabolites, and led to enhanced recall responses for interferon-γ+ CD8+ T cells as well as to the establishment of stem-like T-cell factor-1+ PD-1+ CD8+ T cells within the tumour microenvironment. Gels for the in situ modulation of the gut microbiome may be applicable more broadly to treat pathologies associated with a dysregulated gut microbiome.
One-sentence editorial summary:
An orally administered gel that is retained in the colon modulates the gut microbiome of mice with murine tumours, inducing systemic memory-T-cell responses, and amplifying the antitumour activity of a checkpoint inhibitor.
While immune checkpoint blockers (ICB) have drastically changed the landscape of cancer therapy, only a small subset of patients responds to ICB therapy1,2. Recent studies have highlighted the vast potential of targeting the gut microbiome for the treatment of inflammatory, metabolic, and neurodegenerative diseases as well as cancer3-6. In particular, dysregulated gut microbiota have been implicated in the lack of response to ICB therapy7-13, and clinical investigations are underway to restore healthy gut microbiota in cancer patients by either inoculation of pre-defined probiotics or fecal microbiota transplantation (FMT). However, a favourable consortium of probiotics for cancer therapy remains ill-defined, and the scale-up manufacturing and quality control of FMT products are challenging. In fact, the Food and Drug Administration (FDA) has recently issued an urgent safety warning on FMT procedures due to fatalities resulting from inadvertent transfers of antibiotic-resistant microbes from a doner to recipients14. Microbiome-derived metabolites, including short-chain fatty acids (SCFAs), may serve as the key messengers between the gut microbiota and the host’s immune system15, but it remains unclear how microbial metabolites impact T cell immunity and how to exploit this knowledge for cancer therapy. Here, we report a dietary fiber-based engineering strategy for in situ modulation of the gut microbiome as a safe and straightforward approach for amplifying systemic memory T cell responses and potentiating the therapeutic efficacy of ICB.
Briefly, we screened the FDA’s list of ingredients generally recognized as safe (GRAS) and found that oral administration of inulin, a polysaccharide dietary fiber found in chicory root and Jerusalem artichoke, improved the anti-tumor efficacy of anti-programmed cell death-1 (α-PD-1) ICB therapy. Because “beneficial” commensal microbes (e.g. Akkermansia, Lactobacillus, Roseburia) prevalent in ICB-responsive patients primarily reside in the colon9-11, we engineered “colon-retentive” inulin by formulating it into an oral gel. Oral administration of inulin gel effectively modulated the gut microbiota in situ and promoted the proliferation of commensal microbes with “beneficial” roles in T cell immunity. This triggered microbial metabolite-mediated differentiation of CD8+ T cells into stem-like memory CD8+ T cells that synergized with α-PD-1 therapy (Fig. 1). Given the long history of inulin as a food ingredient with excellent safety profiles and the facile production process for inulin gel, our approach may open new opportunities for improving the anti-tumor effects of ICB.
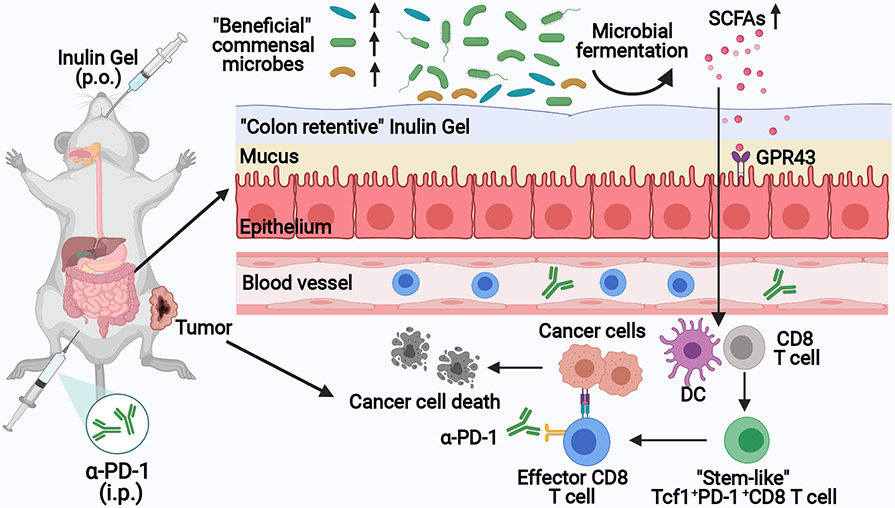
Fig. 1 ∣. In situ modulation of the gut microbiome with “colon-retentive” inulin gel for improving cancer immunotherapy.
Orally administered, inulin gel increases the levels of “beneficial” commensal microbes and their metabolites, including short-chain fatty acids (SCFAs) that induce stem-like T-cell factor-1(Tcf1)+PD-1+CD8+ T cells. Inulin gel synergizes with α-PD-1 immunoglobulin G (IgG) immunotherapy, exerting potent anti-tumor efficacy in multiple tumor models.
Results
Inulin improves the efficacy of α-PD-1 therapy.
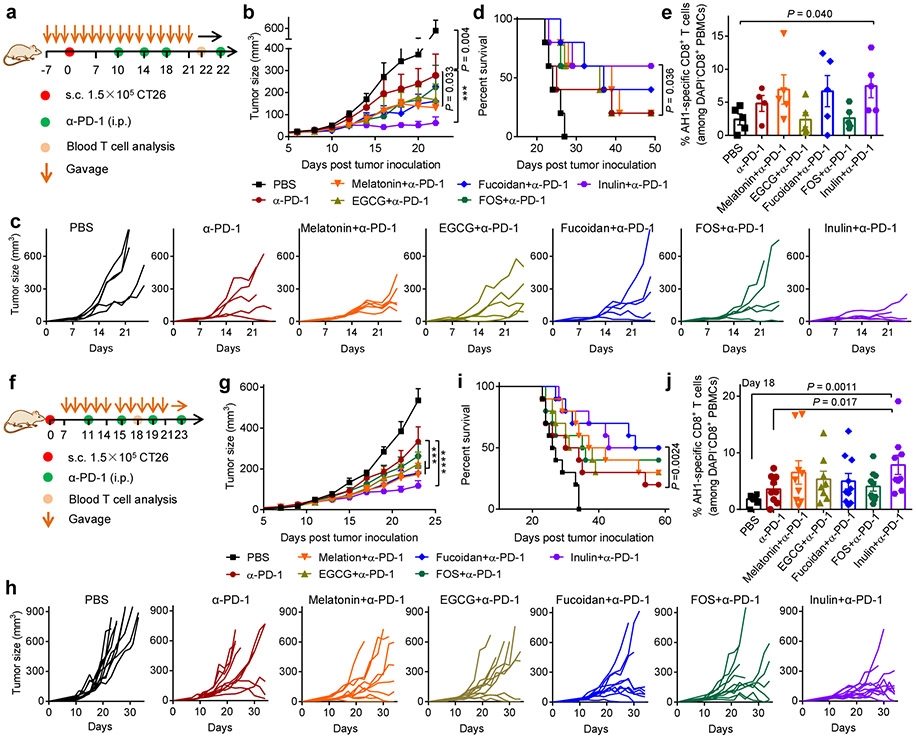
From the FDA GRAS list, we chose candidates known to modulate the gut microbiome and examined whether they could improve the therapeutic efficacy of α-PD-1 ICB. (−)-Epigallocatechin gallate (EGCG), a natural antioxidant found in green tea and red wine, has been shown to alter the gut microbiome16, while fucoidan, fructooligosaccharides (FOS), and inulin are dietary fibers known to increase the richness and diversity of the gut microbiota17. We also examined melatonin, which regulates the sleep cycle and circadian rhythm as well as the gut microbiome18. We first examined these agents in a prophylactic setting. BALB/c mice were pre-treated with these agents via oral gavage for one week, followed by inoculation of CT26 colon carcinoma cells at subcutaneous (s.c.) flank on day 0 (Fig. 2a). On days 10, 14, 18, and 22 the animals were co-treated by systemic intraperitoneal (i.p.) administration of α-PD-1 IgG. Mice were kept on the normal mouse chow diet throughout the study, and all treatments were stopped on day 26. Interestingly, oral gavage with inulin significantly improved the systemic anti-tumor efficacy of α-PD-1 treatment and effectively slowed the growth of CT26 tumors located at s.c. flank (Fig. 2b,c). In fact, inulin plus α-PD-1 treatment group had 60% of mice remaining tumor-free throughout the study (Fig. 2b-d), compared with 20% tumor-free rate for the α-PD-1 monotherapy group. Oral gavage with fucoidan also moderately improved the efficacy of α-PD-1 treatment. Inulin as well as other agents did not significantly alter the body weight of mice (Supplementary Fig. S1). Compared with α-PD-1 alone, inulin plus α-PD-1 significantly increased the frequency of circulating CD8+ T cells specific to an immunodominant epitope from CT26 gp70 (AH1, H-2Ld–restricted SPSYVYHQF) (Fig. 2e, Supplementary Fig. S2a).
Fig. 2 ∣. Improving the anti-tumor efficacy of α-PD-1 therapy with oral candidate agents.
a, Prophylactic treatment regimen. BALB/c mice kept on the normal diet were orally gavaged with the candidate agents (melatonin: 100 mg/Kg dose/bodyweight; EGCG: 100 mg/Kg dose/bodyweight; fucoidan: 200 mg/Kg dose/bodyweight; FOS: 120 mg/dose; inulin:120 mg/dose) starting one week prior to subcutaneous (s.c.) inoculation of 1.5 × 105 CT26 colon carcinoma cells. Mice received the candidate agents until day 26, and 100 μg of α-PD-1 was injected i.p. on days 10, 14, 18 and 22. Shown are (b) the average tumor growth curves, (c) individual tumor growth curves, (d) overall Kaplan-Meier survival curves, and (e) frequency of AH1-specific CD8+ T cells in the peripheral blood on day 22. f, Therapeutic treatment regimen. BALB/c mice were inoculated s.c. at flank with 1.5 × 105 CT26 cells and then treated as shown starting on day 7. The doses were the same as in (a). Shown are (g) the average tumor growth curves, (h) individual tumor growth curves, (i) overall Kaplan-Meier survival curves, and (j) frequency of AH1-specific CD8+ T cells on day 18. Data represent mean ± SEM, from a representative experiment (n = 5 (a-e), or n = 9-10 (f-j) biologically independent samples) from two independent experiments. ***P < 0.001, ****P < 0.0001. Data were analyzed by (b,g) two-way ANOVA or (e,j) one-way ANOVA with Bonferroni's multiple comparisons test, or (d,i) log-rank (Mantel-Cox) test.
We next evaluated these combo-therapies in a therapeutic setting in tumor-bearing mice. BALB/c mice were inoculated at s.c. flank with CT26 cells on day 0, and the treatments were initiated on day 7 (Fig. 2f). Similar as above, inulin plus α-PD-1 significantly slowed the tumor growth (Fig. 2g,h) and increased the survival rate, compared with α-PD-1 alone (Fig. 2i). Inulin plus α-PD-1 induced 2.2-fold higher frequency of AH1-specific CD8+ T cells among the peripheral blood mononuclear cells (PBMCs), compared with α-PD-1 alone (Fig. 2j, Supplementary Fig. S2b). Notably, oral administration of inulin alone exhibited comparable anti-tumor effect and AH1-specific CD8+ T cell response as α-PD-1 IgG monotherapy (Supplementary Fig. S3), showing the crucial combination effects of inulin plus α-PD-1 therapy.
Inulin alters the gut microbiome.
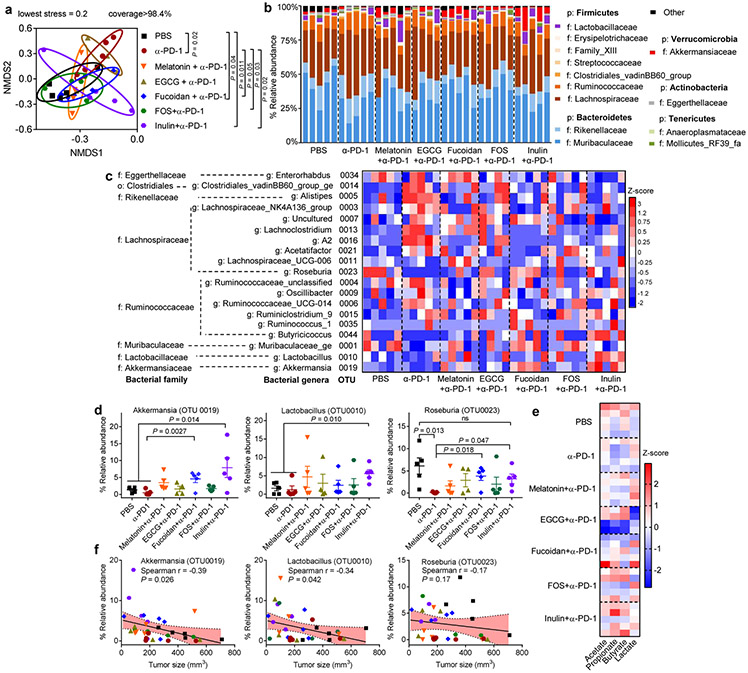
We next examined changes in the gut microbiome by analyzing the fecal samples with 16S rRNA gene sequencing. While all the treatment groups exhibited similar richness and diversity of the gut microbiota from operational taxonomic unit (OTU) and inverse Simpson diversity values (Supplementary Fig. S4), α-PD-1 therapy combined with either inulin, FOS, or fucoidan induced distinct clustering of microbial community structure, as shown by the nonmetric multidimensional scaling analysis (NMDS, known as β-diversity) (Fig. 3a). Specifically, compared with α-PD-1 monotherapy, inulin plus α-PD-1 treatment significantly increased the relative abundances of some bacterial genera, and Akkermansia, Lactobacillus, and Roseburia (Fig. 3b-d), which have been clinically associated with ICB-responsive patients7-9,11. In particular, α-PD-1 treatment alone caused the depletion of Roseburia, which was normalized with inulin co-treatment (Fig. 3d). Lactobacillus and Roseburia are known as the major producers of SCFAs, such as acetate, propionate, and butyrate19-21, and accordingly, inulin plus α-PD-1 treatment increased the levels of SCFAs and lowered the pH of the fecal samples, compared with α-PD-1 monotherapy (Fig. 3e, Supplementary Fig. S5). Spearman’s correlation coefficient analyses suggested that tumor sizes were inversely correlated with the relative abundances of Akkermansia and Lactobacillus (Fig. 3f), and there was also a trend for inverse correlation between the tumor size and the levels of Roseburia, propionate, and butyrate (Supplementary Fig. S6). These results suggest a potential link between the commensal microbes, their SCFA metabolites, and the treatment outcomes of ICB therapy. Based on the potent modulation effect of gut microbiome, anti-tumor effects, and anti-tumor CD8+ T cell response, we focused on improving inulin plus α-PD-1 combo-therapy for the remainder of the study.
Fig. 3 ∣. Distinct gut microbial communities promoted by inulin plus α-PD-1 therapy.
CT26 tumor-bearing BALB/c mice were treated as in Fig. 2f, and the fecal pellets collected on day 21 were analyzed by 16S rRNA gene sequencing. Shown are the (a) NMDS score plot (based on Bray-Curtis); (b) relative abundances of the gut commensal microbes at the phylum and family levels; (c) heatmap showing Z-score normalized values of microbe abundances at family and genus levels, p: phylum level, o: order level, f: family level, g: genus level; (d) relative abundances of Akkermansia, Lactobacillus, and Roseburia; (e) heatmap showing Z-score normalized values of acetate, propionate, butyrate, and lactate in fecal pellets. f, Spearman’s correlation between the tumor sizes and relative abundances of Akkermansia, Lactobacillus, and Roseburia. Shaded band indicates 95% confidence interval of the values fitted by linear regression. Data represent mean ± SEM, from a representative experiment (n = 5 biologically independent samples) from two independent experiments. Data were analyzed by (a) analysis of molecular variance (AMOVA) test, (d) one-way ANOVA with Bonferroni's multiple comparisons test, or (f) Spearman’s rank correlation test.
Oral inulin gel for improved modulation of the gut microbiome.
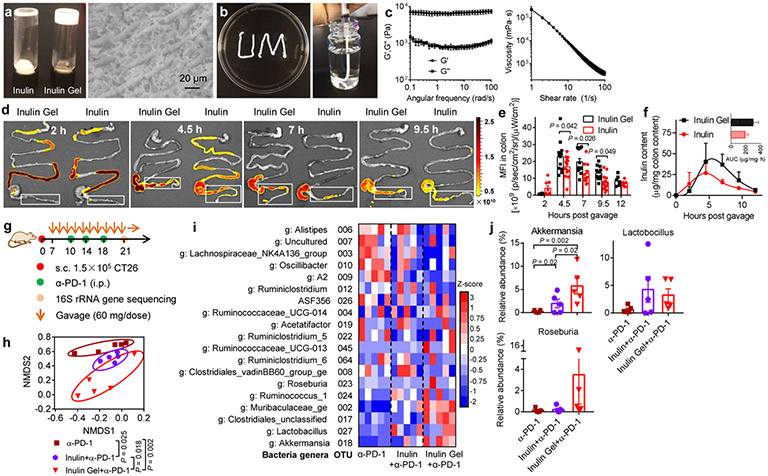
As these commensal microbes primarily reside in the colon, we examined whether the impact of inulin on anti-tumor responses could be further improved using inulin formed into an oral hydrogel with “colon-retentive” properties. This was motivated by prior studies showing that polysaccharide fibers in a gel state can prolong gastric emptying (due to thickening effect), adsorb to the intestinal mucous layer, and prolong the residence time22. To test this idea, we have developed an oral inulin gel formulation with a meso-fibrous morphology via a simple heating-cooling method, which is amenable for a large scale production (Fig. 4a-c, Supplementary Fig. S7). We employed 60 mg dose of inulin all the subsequent studies, presenting 50% reduction from the studies shown above. Compared with fluorescein isothiocyanate (FITC)-inulin, oral gavage with FITC-labeled inulin gel indicated inulin gel was retained longer in the colon (and cecum) in a gel strength-dependent manner (Fig. 4d-e, Supplementary Fig. S8), thereby increasing the cumulative inulin exposure in the colon (Fig. 4f). To examine the impact of inulin gel on the gut microbial community structure, mice were inoculated with CT26 cells on day 0 and treated with oral gavage of inulin or inulin gel starting day 7, followed by α-PD-1 treatment on days 10, 14, and 18 (Fig. 4g). Inulin gel with prolonged colon retention further induced a distinct shift in the gut microbial community structure by day 21, compared with inulin (Fig. 4h, Supplementary Fig. S9a,b). In particular, compared with free inulin plus α-PD-1, inulin gel plus α-PD-1 treatment further increased the relative abundances of Akkermansia (Fig. 4i,j), and there was a trend for increased abundance of Roseburia although this was not statistically significant. Notably, lipopolysaccharide-producing Oscillibater was significantly reduced in mice treated with either inulin or inulin gel combined α-PD-1 (Supplementary Fig. S9c).
Fig. 4 ∣. Colon-retentive inulin gel for modulation of the gut microbial communities.
a,b, Images of inulin gel for colon-targeting after oral administration. c, Dynamic rheological testing of inulin gel: G’ (elastic modulus) and G” (viscous modulus), and viscosity measurement of inulin gel. d,e, Mice were orally gavaged with FITC-labeled inulin gel or inulin (60 mg/dose), followed by visualization of the gastrointestinal tract over 12 h and measurement of the mean fluorescence intensity (MFI) in colon. Shown are representative images from three biologically independent samples in each group. f, Inulin content was quantified in colon digesta using unlabeled inulin or inulin gel. The insert shows the area under the curve (AUC) value. g-j, CT26 tumor-bearing BALB/c mice were treated with inulin or inulin gel (each 60 mg/dose) starting day 7, followed by α-PD-1 treatment as indicated. g, Treatment regimen. h,i, Fecal samples were analyzed on day 21 via 16S rRNA gene sequencing. Shown are the (h) NMDS score plot, (i) heatmap showing Z-score normalized values of microbe abundances at genus level, g: genus level, and (j) relative abundances of Akkermansia, Lactobacillus, and Roseburia. Data represent mean ± SEM, from a representative experiment (n = 3 (a-c), n = 4 (f), or n = 5 (g-j) biologically independent samples) from three (a-c) or two (d-j) independent experiments. Data were analyzed by (e) two-way ANOVA or (j) one-way ANOVA with Bonferroni's multiple comparisons test, or (h) AMOVA test.
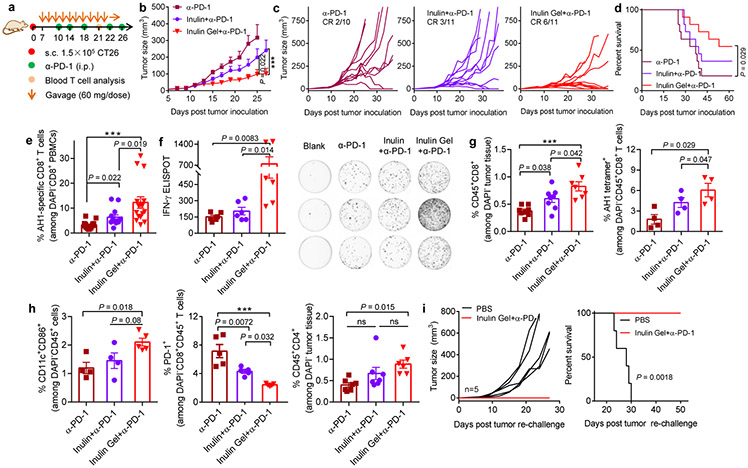
Potent anti-tumor efficacy of oral inulin gel plus α-PD-1 therapy.
Having shown the impact of inulin gel on the gut microbiome, we sought to assess the anti-tumor efficacy of inulin gel plus α-PD-1 therapy. Compared with free inulin plus α-PD-1, inulin gel plus α-PD-1 combo-therapy markedly delayed the tumor growth and increased the rate of complete tumor regression by 2-fold (Fig. 5a-d). The anti-tumor efficacy of combo-therapy was dependent on the dose of inulin gel (Supplementary Fig. S10). On the other hand, oral administration of inulin gel alone exhibited only a modest anti-tumor effect similar to α-PD-1 IgG monotherapy (Supplementary Fig. S11), showing the importance of inulin gel plus α-PD-1 combo-therapy. Interestingly, compared with α-PD-1 given with or without free inulin, inulin gel plus α-PD-1 augmented the systemic anti-tumor CD8+ T cell responses, as evidenced by significantly higher frequencies of AH1-specific CD8+ T cells among PBMCs and interferon (IFN)-γ+CD8+ T cells in spleen (Fig. 5e,f, Supplementary Fig. S12). In addition, inulin gel plus α-PD-1 increased the frequencies of intratumoral CD8+ T cells, AH1-tetramer+CD8+ T cells, CD4+ T cells, and activated CD86+CD11c+ dendritic cells (DCs), while decreasing PD-1+CD8+ T cells (Fig. 5g,h). Moreover, survivor mice in the inulin gel plus α-PD-1 group were completely protected against CT26 tumor re-challenge performed on day 62 (Fig. 5i), indicating long-term memory response.
Fig. 5 ∣. Oral inulin gel potently amplifies the therapeutic efficacy of α-PD-1 therapy.
a, Therapeutic treatment regimen. CT26 tumor-bearing BALB/c mice were treated with inulin or inulin gel (each 60 mg/dose) starting day 7, followed by α-PD-1 treatment as indicated. Shown are (b) the average tumor growth curves, (c) individual tumor growth curves, and (d) overall Kaplan-Meier survival curves. e, The average frequency of AH1-specific CD8+ T cells on day 21 and (f) AH-1-specific IFN-γ enzyme-linked immune absorbent spot (ELISPOT) assay on splenocytes isolated on day 23. Tumor tissues were analyzed for the frequencies of (g) CD8+ T cells, AH1-tetramer+ CD8+ T cells, (h) CD11c+CD86+ DCs, PD-1+CD8+ T cells, and CD4+ T cells. i, Survivors in the inulin gel plus α-PD-1 group were re-challenged with 1.5 × 105 CT26 cells inoculated on the contralateral s.c. flank on day 62, followed by tumor monitoring. Data represent mean ± SEM, from a representative experiment (n = 10-11 (b-e), n = 4-7 (f-h), or n = 5 (i) biologically independent animals) from three (b,c) or two (d-i) independent experiments. ***P < 0.001. Data were analyzed by (e-h) one-way ANOVA or (b) two-way ANOVA with Bonferroni's multiple comparisons test, or (d,i) log-rank (Mantel-Cox) test.
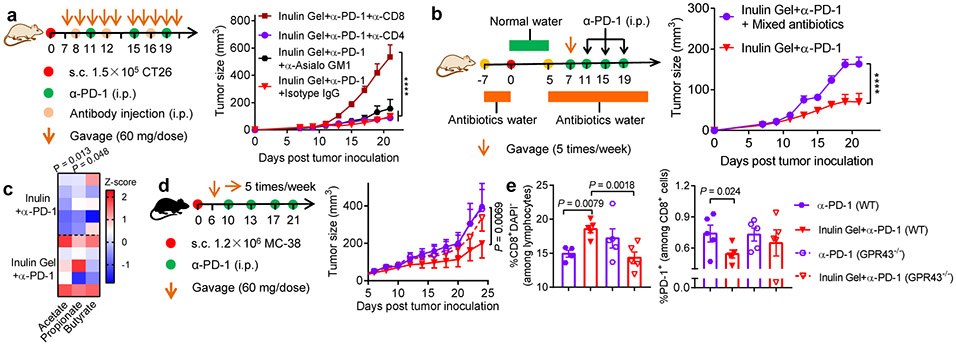
The interplay between the gut microbiota, metabolites, and stem-like memory CD8+ T cells.
We sought to delineate factors contributing to the systemic anti-tumor efficacy of inulin gel plus α-PD-1 combination therapy. Tumor-bearing mice given inulin gel plus α-PD-1 failed to inhibit tumor growth after administration of depletion antibody against CD8+ T cells (Fig. 6a), but not CD4+ T cells or natural killer (NK) cells, indicating CD8+ T cells as the major anti-tumor effector cells. In addition, broad-spectrum antibiotics added in the drinking water abrogated the anti-tumor effects of inulin gel plus α-PD-1 therapy (Fig. 6b), suggesting the gut commensal microbes as a crucial link.
Fig. 6 ∣. The interplay between the gut microbiota, metabolites, and anti-tumor efficacy.
a, CT26 tumor-bearing BALB/c mice were treated as shown and also administered i.p. with 200 μg of antibodies against CD8+ T cells (αCD8), CD4+ T cells (αCD4) or NK cells (αAsialo GM1) or isotype IgG control on days 8, 12, and 16. Shown are the average tumor growth curves. b, Broad-spectrum antibiotics (ampicillin/colistin/streptomycin) treatment regimen in CT26 tumor model during therapy. Shown are the average tumor growth curves. c, CT26 tumor-bearing BALB/c mice were treated as Fig. 5a, and shown are the Z-score normalized values of SCFAs in feces on day 21. d,e, MC-38 tumor-bearing GPR43 knock-out (GPR43−/−) or wide type (WT) C57BL/6N mice were treated as shown and monitored for (d) tumor growth. e, The frequencies of CD8+ T cells and PD-1+CD8+ T cells in mLNs were assessed on day 24. Data represent mean ± SEM, from a representative experiment (n = 5 biologically independent animals) from two (a,b,d,e) or three (c) independent experiments. ****P < 0.0001. Data were analyzed by (e) one-way ANOVA or (a,b,d) two-way ANOVA with Bonferroni's multiple comparisons test, or by (c) two-tailed, unpaired Student’s t-test.
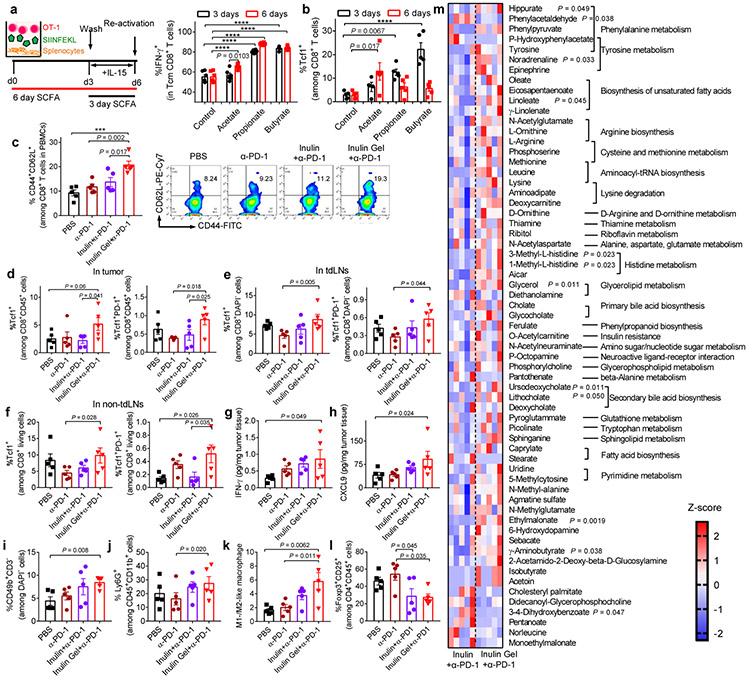
We next examined how the commensal microbes and their metabolites impact CD8+ T cell responses. Inulin gel plus α-PD-1 therapy increased the concentrations of SCFAs in tumor-bearing mice (Fig. 6c). In G protein-coupled receptor 43 (GPR43, known as a key SCFA receptor in the gastrointestinal tract23) knockout mice, inulin gel plus α-PD-1 combo-therapy lost their ability to exert anti-tumor efficacy, increase CD8+ T cell response, and decrease PD-1+CD8+ T cells in mesenteric lymph nodes (mLNs) (Fig. 6d,e), suggesting the importance of microbial fermentation-derived SCFAs. However, oral administration of free SCFAs failed to improve the anti-tumor efficacy of α-PD-1 IgG (Supplementary Fig. S13), probably due to rapid absorption of SCFAs in the small intestine24. Taken together, these results suggest that SCFAs derived from microbial fermentation of inulin gel trigger GPR43-dependent, anti-tumor CD8+ T cell response.
Indeed, SCFAs (acetate, propionate, and butyrate) added to in vitro culture of major histocompatibility complex (MHC) class I-restricted, ovalbumin (OVA)-specific, CD8+ T cells (OT-I CD8+ T cells) for either 3 or 6 days significantly augmented IFN-γ recall response among memory OT-I CD8+ T cells (Fig. 7a)25,26. Importantly, SCFAs induced robust upregulation of T-cell factor-1 (Tcf1, encoded by Tcf7) (Fig. 7b, Supplementary Fig. S14). In agreement with these in vitro results, CT26 tumor-bearing mice treated with inulin gel plus α-PD-1 elicited substantially higher frequencies of CD44+CD62L+CD8+ memory T cells among PBMCs as well as Tcf1+CD8+ T cells and Tcf1+PD-1+CD8+ T cells within the tumor microenvironment, tumor-draining lymph nodes (tdLNs), and non-tumor-draining lymph nodes (non-tdLNs) (Fig. 7c-f). Notably, Tcf1+PD-1+CD8+ T cells, which are a progenitor, stem-like subset of PD-1+CD8+ T cells, are associated with clinical responses to PD-1 ICB therapy in patients27-29. Inulin gel plus α-PD-1 treatment also increased the intratumoral concentrations of IFN-γ, interferon-inducible chemokine (C-X-C motif) ligand 9 (CXCL9), and the frequencies of CD3−CD49b+ NK cells, and Ly6G+CD11b+ neutrophils, while inducing M1-skewed polarization of macrophages and reducing CD4+CD25+Foxp3+ regulatory T cells (Fig. 7g-I, Supplementary Fig. S15). In addition, mice treated with inulin gel plus α-PD-1 therapy maintained the normal complete blood counts and serum biochemistry (Supplementary Fig. S16), indicating safety of the combination therapy. While we mainly focused on the role of SCFAs in our current study, we also detected changes in other metabolites in feces. Among 255 metabolites that we measured, inulin gel plus α-PD-1 therapy substantially increased the levels of hippurate, 1-methyl-L-histine, 3-methyl-L-histine, L-arginine, and oleate (Fig. 7m). Interestingly, these metabolites have been reported to be associated with clinical responders to α-PD-1 IgG therapy30.
Fig. 7 ∣. The interplay between the gut microbiota, metabolites, and induction of stem-like memory CD8+ T cells.
a,b, SCFAs (400 μM) were added during in vitro memory differentiation of OVA-specific OT-I T cells. Shown are the (a) frequencies of IFN-γ+ among CD44+CD62L+ Tcm CD8+ T cells, and the expression levels of (b) Tcf1 among CD8+ T cells after T cell activation. c-m, CT26 tumor-bearing were treated as in Fig. 5a. c, Peripheral blood was analyzed on day 19 for the frequency of CD44+CD62L+ Tcm CD8+ T cells. d, Tumor tissues, (e) tdLNs, and (f) non-tdLNs were analyzed on day 21 for the frequencies of Tcf1+ and Tcf1+PD-1+ T cells among CD45+CD8+ T cells. The levels of (g) IFN-γ and (h) CXCL9; i, CD3-CD49b+ NK cells; j, Ly6G+CD11b+ neutrophils; k, ratio between M1- and M2-like macrophages; and (l) Foxp3+CD25+ Tregs on day 21. m, Metabolites detected in feces. 255 metabolites in feces were measured on day 21. Shown is the heatmap of Z-score normalized value of discriminative metabolites. Data represent mean ± SEM, from a representative experiment (n = 6 (a,b), or n = 5 (c-m) biologically independent samples) from three (a,b) or two (c-m) independent experiments. ***P < 0.001, ****P < 0.0001. Data were analyzed by (c-l) one-way ANOVA or (a,b) two-way ANOVA with Bonferroni's multiple comparisons test, or by (m) two-tailed, unpaired Student’s t-test.
Therapeutic efficacy in multiple murine tumor models.
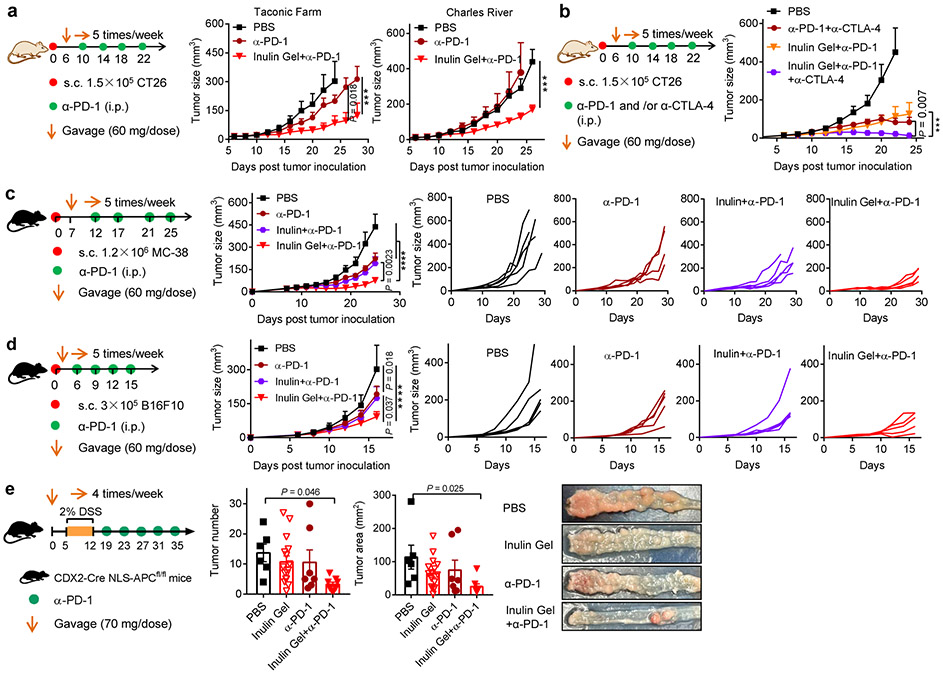
Lastly, we sought to validate our results in mice obtained from different vendors since a recent report suggested that mice from various vendors harbored distinct commensal microbiota and responded differently to α-PD-1 therapy8. In alignment with the above results obtained from CT26 tumor-bearing BALB/c mice (Jackson Laboratory), inulin gel plus α-PD-1 combo-therapy exerted strong anti-tumor efficacy in mice obtained from Charles River and Taconic Farm (Fig. 8a), thus ruling out the vendor-specific effects. Moreover, inulin gel also notably augmented the anti-tumor efficacy of α-PD-1 and anti-cytotoxic T-lymphocyte-associated protein 4 (α-CTLA-4) combo-therapy, leading to regression of CT26 tumors in BALB/c mice (Fig. 8b). Furthermore, inulin gel plus α-PD-1 combo-therapy also generated strong CD8+ T cell responses with robust anti-tumor efficacy in C57BL/6 mice (Jackson Laboratory) bearing either MC-38 colon carcinoma or B16F10 melanoma (Fig. 8c,d), thus showing utility of our strategy in murine models of colorectal carcinoma and melanoma using multiple mouse strains. Finally, in a transgenic mouse model of colorectal cancer31, inulin gel plus α-PD-1 combo-therapy inhibited the growth of dextran sulfate sodium (DSS)-accelerated colon tumor formation in CDX2-Cre NLS-APCfl/fl mice (Fig. 8e).
Fig. 8 ∣. Therapeutic efficacy of inulin gel plus α-PD-1 therapy in multiple tumor models.
a, CT26 tumor-bearing BALB/c mice from Taconic Farm or Charles River were treated as shown (inulin: 60 mg/dose), and tumor growth was monitored. b, CT26 tumor-bearing BALB/c mice from Jackson Laboratory were treated with inulin gel with or without α-PD-1 and α-CTLA-4, and tumor growth was monitored. C57BL/6 mice bearing (c) MC-38 colon carcinoma and (d) B16F10 melanoma were treated as shown (inulin: 60 mg/dose), and tumor growth was monitored. e, CDX2-Cre NLS-APCfl/fl mice were kept on 2%DSS in drinking water for one week to promote the formation of tumors in the colon, followed by the indicated treatments. 3 out of 9 mice in phosphate buffer saline (PBS) group, 2 out of 17 mice in inulin gel group, and 1 out of 8 mice in α-PD-1 group were euthanized due to severe body weight drop or rectal bleeding/prolapse. On day 45, colon tumor number and size were measured among the survivors. Representative colon tissue images are shown. Data represent mean ± SEM, from a representative experiment (n = 5 (a-d), or n = 8-9 (e) biologically independent animals) from two independent experiments. ***P < 0.001, ****P < 0.0001. Data were analyzed by (a-d) two-way ANOVA or (e) one-way ANOVA with Bonferroni's multiple comparisons test.
Discussion
In summary, we have shown that inulin, a widely-consumed dietary fiber with a long history of safety, synergizes with α-PD-1 therapy and elicits memory CD8+ T cell responses with robust anti-tumor efficacy (Fig. 2). We have also demonstrated that inulin formed into a hydrogel improved colon retention (Fig. 4d-f), modulated commensal microbes in situ (Fig. 4g-j), and further amplified anti-tumor efficacy of α-PD-1 therapy (Fig. 5, Fig. 8). Mechanistically, our results suggest that SCFAs produced as metabolites from inulin gel increased the memory potential of antigen-primed CD8+ T cells and triggered their differentiation into stem-like Tcf1+PD-1+CD8+ T cells (Fig. 7a-f), leading to potent, long-lived anti-tumor effects. These results are in line with recent clinical studies showing that concentrations of SCFAs were positively associated with the efficacy of ICB therapy in patients32,33. Interestingly, it was recently reported that mice kept on a high dose, free inulin diet (~450 mg daily dose) for 14 days prior to tumor inoculation could slow down tumor growth, but this did not synergize with α-PD-1 therapy34, potentially due to decreased gut microbiota diversity triggered by the monotonous diet with very high inulin intake35,36. In contrast, we have shown that oral colon-retentive inulin gel given at 60 mg intermittent doses potently amplified the efficacy of α-PD-1 therapy in a therapeutic setting. Thus, in addition to FMT and consortia of probiotics currently being considered for improving cancer immunotherapies37,38, our dietary fiber gel-based approach offers a powerful, safe, and facile strategy for augmenting immune checkpoint therapies.
While biomaterials have been explored extensively for various therapeutic applications, this is, to the best of our knowledge, the first demonstration of biomaterials engineered to target and manipulate the gut microbiota in situ for promoting systemic immunity. The oral hydrogel strategy outlined here represents a straightforward pathway to improve ICBs with minimal dose and favorable safety profiles. Notably, the recommended daily intake of dietary fibers is 25-38 g for adults, and the mean dietary fiber intake is only 15 g in the United States39, thus leaving much room for dietary fiber-based therapeutics. As dysregulated gut microbiota and their metabolites have now been implicated in inflammatory, metabolic, and neurodegenerative diseases, our strategy may provide a general framework for safely modulating the gut microbiome for treating various diseases.
Methods
Preparation and characterization of inulin gel.
To prepare inulin gel, 300 mg of inulin from chicory (Sigma-Aldrich) was dissolved in 1.0 mL deionized (DI) water. Then inulin solution was heated at 70°C for 5 min and kept at room temperature for 12 h. For scanning electron microscope (SEM) observation, inulin gel was plunged into liquid nitrogen and the frozen sample was freeze-dried. Afterwards, the sample was sputter coated with gold for 30s. Samples were visualized using MIRA3 TESCAN (voltage 15 kV) via Michigan Center for Materials Characterization at the University of Michigan. The rheology study of inulin gel was conducted in 25 mm sandblasted parallel plates with a Peltier temperature-controlled plate and hood (Anton Parr MCR702 Rheometer).
In vivo cancer immunotherapy.
Animals were cared for following the federal, state, and local guidelines. The University of Michigan, Ann Arbor is an AAALAC international accredited institution, and all work conducted on animals was in accordance with and approved by the Institutional Animal Care and Use Committee (IACUC). Female (6-8-week-old) BALB/c mice from Jackson Laboratory, Charles Rivers, or Taconic Farm maintained on the normal mouse chow diet were inoculated with 1.5 × 105 CT26 cells per mouse, while C57BL/6 mice (Jackson Laboratory) were inoculated with 1.2 × 106 MC-38 cells or 3 × 105 B16F10 cells per mouse on the right flank by subcutaneous injection. The normal chow included 24.1% protein, 11.4% fat, 5.2% crude fiber, 48.7% nitrogen-free extract, 6.9% ash and less than 12.0% moisture (PicoLab® Laboratory Rodent Diet 5L0D*, Lab Supply). Tumor-bearing mice were randomly assigned to different treatment groups. For prophylactic anti-tumor studies in CT26 tumor model, mice were gavaged with melatonin (100 mg/Kg dose/bodyweight, Alfa Aesar), EGCG (100 mg/Kg dose/bodyweight, Sigma-Aldrich), fucoidan from Fucus vesiculosus (200 mg/Kg dose/bodyweight, Sigma-Aldrich), FOS from Chicory (120 mg/dose, Sigma-Aldrich), inulin (120 mg/dose) three times in one week prior to tumor inoculation. After tumor inoculation, mice were still gavaged three times in the first week, followed by five times per week. Mice were i.p. injected with α-PD-1 antibody (100 μg/dose, clone: RMP1-14, Bioxcell) on days 10, 14, 18 and 22 post tumor inoculation. For combination therapies in CT26 tumor model, mice were gavaged with the indicated samples from day 7 post tumor inoculation. Mice were gavaged with the indicated samples (same dose as in the prophylactic anti-tumor studies) five times per week, and α-PD-1 antibody (100 μg/dose) was injected i.p. on days 11, 15, 19, and 23. For inulin gel involved combination therapy in CT26 tumor model, inulin gel or inulin was 30 mg/dose, 60 mg/dose, or 120 mg/dose, while α-PD-1 antibody (100 μg/dose) was injected i.p. on days 10, 14, 18, 22, and 26. For α-CTLA-4 involved combination therapy in CT26 tumor model, inulin gel or inulin was 60 mg/dose, while α-PD-1 antibody (100 μg/dose) and α-CTLA-4 antibody (100 μg/dose, clone: 9D9, Bioxcell) were i.p. injected on days 10, 14, 18, and 22. For GPR43−/− mice involved study, GPR43−/− or WT C57BL/6N male mice were inoculated with 1.2 × 106 MC-38 cells per mouse on the right flank by subcutaneous injection. Mice were gavaged with inulin gel (60 mg/dose). α-PD-1 antibody (100 μg/dose) was i.p. injected on days 10, 13, 17 and 21. For spontaneous colorectal cancer therapy, 6-8-week-old CDX2-Cre NLS-APCfl/fl mice were orally gavaged with inulin gel (70 mg/dose) 4 times per week. Mice were given 2% DSS (Mw: 4 kDa, Alfa Aesar) in drinking water from days 5 to 12, α-PD-1 antibody (100 μg/dose) was i.p. injected on days 19, 23, 27, 31 and 35. Mice were euthanized on day 45 and colon tumors were counted and measured under a magnifier. For SCFAs plus α-PD-1 study in CT26 tumor model, drinking water was added with 120 mM sodium chloride or the mixture of 65 mM sodium acetate, 25 mM sodium propionate, and 30 mM sodium butyrate. α-PD-1 (100 μg/dose) injection was performed on days 11, 14, 17, and 20. Tumor size was measured, and tumor volume was calculated as ½ (length × width2). Tumor-bearing mice were euthanized when the tumor size reached 1.5 cm in any diameter or when animals became moribund with severe weight loss (> 20%) or tumor ulceration. For the immune cell depletion studies, depletion antibodies (200 μg/dose) against anti-CD8 (Bioxcell, clone 2.43, #BP0061), anti-CD4 (Bioxcell, clone GK1.5, #BP0003-1), or anti-AsialoGM1 (Wako chemicals USA, Inc, #986-10001) were administered i.p. on days 8, 12 and 16 post tumor inoculation and treated as above.
In vivo immunological analyses.
PBMCs were analyzed for the tumor antigen-specific CD8+ T cells. Red blood cells were lysed, discarded, and the remaining cells were blocked with CD16/32 antibody for 10 min and then stained with PE-tagged peptide-MHC tetramer (H-2Ld-restricted SPSYVYHQF). Tetramers were provided by NIH Tetramer Core Facility. Cells were further stained with APC-CD8 Rat Anti-mouse (Clone: 53-6.7(RUO), BD Biosciences). To further analyze the memory phenotype of tetramer+ CD8+ T cells, cells were additionally stained with FITC-CD44 Rat anti-Human/Mouse (Clone: IM7, eBioscience), PE-Cy7-CD62L Monoclonal Antibody (Clone: MEL-14, eBioscience). Stained cells were incubated with 4’,6-diamidino-2-phenylindole (DAPI) prior to flow cytometry analysis. For lymph node tissue analysis, tdLNs, non-tdLNs and mLNs were isolated, ground, and filtered through a 70-μm strainer. Cells were stained with designated antibodies as well as fixable viability dye eFluor™ 450 (eBioscience) for flow cytometry analysis. Tumor tissues were excised at preset time points for analysis of tumor-infiltrating T cells. Tumor tissues were weighed and cut into small pieces and incubated with collagenase type IV (1 mg/ml) and DNase I (0.1 mg/ml) under gentle shaking. After 30 min, cell suspension was filtered through a 70-μm strainer. The filtrate was diluted with PBS to 3 mL and chemokines/cytokines were detected via enzyme-linked immunosorbent assay (ELISA) by Cancer Center Immunology Core of the University of Michigan. The cells were washed with fluorescence-activated cell sorting (FACS) buffer and blocked with CD16/32 antibody. Cells were then stained with designated antibodies: PE-CD4 Monoclonal Antibody (Clone: GK1.5, eBioscience), APC-Anti-CD45 Rat Monoclonal Antibody (Clone: 30-F11, BioLegend), FITC-Anti-CD45 Rat Monoclonal Antibody (Clone: 30-F11, BioLegend), FITC-Rat Anti-mouse CD8 (Clone: 53-6.7(RUO), BD Biosciences), PE labeled H-2Ld-restricted SPSYVYHQF, FITC-Anti-CD86 Rat Monoclonal Antibody (Clone: GL-1, BioLegend), PE-CD11c Armenian Hamster Anti-mouse (Clone: N418, eBioscience), PE-Cy7-Anti-mouse PD-1 (Clone: 29F.1A12, BioLegend), PE-Tcf1/Tcf7 Rabbit mAb (Clone: C63D9, Cell Signaling Technology), PE-Cy7-Anti-mouse CD86 (Clone: GL1, BD Bioscience), FITC-Anti-mouse/human CD11b (Clone: M1/70, BioLegend), PE-Anti-mouse F4/80 (Clone: BM8, BioLegend), APC-Anti-mouse CD206 (MMR, Clone: C068C2, BioLegend), PE-Foxp3 Monoclonal Antibody (Clone: FJK-16s, eBioscience), APC-CD25 Rat Anti-mouse (Clone: PC61, BD Biosciences, FITC-Anti-CD49b Rat Monoclonal Antibody (clone: DX5, BioLegend), Brilliant Violet 786-CD3 Molecular Complex Rat anti Mouse (Clone: 17A2, BD Biosciences), FITC-CD279 (PD-1) Rat anti-mouse (Clone: RMP1 30, eBioscience), PE-Ly6G Monoclonal Antibody (Clone: 1A8-Ly6g, eBioscience)). The cells were washed and stained with DAPI, or 7-aminoactinomycin D (7-AAD), or fixable viability dye eFluor™ 450 (eBioscience) for flow cytometric analysis. For the IFN-γ ELISPOT assay, ELISPOT plate was coated with IFN-γ capture antibody for 24 h and blocked with dulbecco's modified eagle medium (DMEM) + 10% foetal bovine serum (FBS) for 2 h. Splenocytes obtained from pre-treated mice were added to 96-well plate with a density of 2 × 105 live cells/well. SPSYVYHQF peptide (20 μg/mL) was added to stimulate splenocytes. Ionomycin and phorbol myristate acetate (PMA) were employed as the positive control. After 18 h, IFN-γ spots were detected with biotinylated detection antibody, followed by streptavidin-HRP and AEC substrate kit. The IFN-γ spot number and size were measured in the Cancer Center Immunology Core at the University of Michigan.
Microbial community analyses.
CT26 tumor-bearing BALB/c mice were treated as the above, and on day 21, fecal pellets were collected and stored at −80°C before microbial analysis. Microbiome DNA isolation and 16S rRNA gene sequencing were completed by the Microbial Systems Molecular Biology Laboratory in the University of Michigan. Isolation of microbial DNA from mice feces was performed via Qiagen MagAttract Power Microbiome kit. The V4 region of the 16S rRNA-encoding gene was amplified from extracted DNA using the barcoded dual-index primers. Briefly, barcoded dual-index primers specific to the V4 region of the 16S rRNA gene amplified the DNA. Polymerase chain reaction (PCR) was conducted in the following order: 2 min at 95°C, 30 cycles of 95°C for 20 s, 55°C for 15 s, 72°C for 5 min and 72°C for 10 min. The size of the amplicon library (~399 bp) was confirmed by Agilent Bioanalyzer. The pooled amplicon library was then sequenced on the Illumina MiSeq platform using the 500 cycle MiSeq V2 Reagent kit (catalog no. MS-102-2003) according to the manufacturer's instructions with modifications of the primer set with custom read 1/read 2 and index primers added to the reagent cartridge. The “Preparing Libraries for Sequencing on the MiSeq” (part 15039740, Rev. D) protocol was used to prepare libraries with a final load concentration of 5.5 pM, spiked with 15% PhiX to create diversity within the run. FASTQ files are distributed to the client when the 2 × 250 bp sequencing completes. The raw microbial 16S rRNA gene sequencing data were analyzed by Mothur (mothur v.1.40.5, Running 64Bit Version). Silva reference files (release 132) were used to align sequences and the open reference OTU picking protocol was used at 97% sequence identity. The data processing steps based on the MiSeq standard operating procedure40 included reducing sequencing and PCR errors, processing improved sequences (align and filter the sequences to remove the overhangs at both ends, de-noise sequences, remove chimeric sequences, undesirables, etc.), assessing error rates, preparing for analysis, analysis, and OTU-based analysis. The pcr.seqs command aligns sequences to the reference (silva.nr_v132.align) alignment. For classifying sequences, Bayesian classifier with the classify.seqs command was used with cutoff of 80, silva.nr_v132.align and silva.nr_v132.tax were used as the reference and taxonomy, respectively.
Quantification of fecal pH, SCFAs, and other metabolites.
Fresh fecal pellets of mice were collected and weighed on day 21 post tumor inoculation and treatment. The fecal pellets were homogenized in DI water and centrifuged at 3000xg for 5 min. The pH of the supernatant was detected by pH meter. For SCFAs measurement, SCFAs were extracted from fecal pellets using Milli-Q water. The solution was centrifuged for 5 min at 10000xg (4°C) to pellet bacteria and other solids. The supernatant was collected and SCFAs were analyzed using a Dionex Integrion high performance ion chromatography (HPIC) system outfitted with an AS-11HC column (250 mm × 4 mm ID; 4 micron packing) by the University of Michigan Biological Station. The separation was accomplished by using a hydroxide gradient (1-5 mM over the course of 20 minutes, followed by a rapid ramp to 60 mM to clear the column of anions), the detection was conducted under suppressed conductivity. For measuring other metabolites, chilled methanol/water solution (80% MeOH vol/vol) was added to feces (1 g of feces to 10 mL of solution). The mixture was vortexed in cold room for 20 minutes, followed by centrifugation at 17000xg for 20 minutes at 4°C. Supernatant was collected in fresh tubes, dried down in a vacuum evaporator and stored in −80°C. The dried supernatant was reconstituted in 300 μL of water, sonicated for 20 minutes, and then filtered. 100 μL of filtrate was transferred to the liquid chromatography (LC) vials and analyzed on an Agilent 6520 QTOF LC/mass spectrum (MS) machine using ACQUITY UPLC BEH C18 Column, 130Å, 1.7 μm, 2.1 mm X 150 mm coupled with Van-Guard Pre-Columns. The column compartment was set at 40°C and the analysis was performed in both positive and negative modes. Mobile phase A was water with 0.1% formic acid, while mobile phase B was acetonitrile with 0.1% formic acid. The gradient method was set as follows: 0 min: 1% B; 1 min: 1% B; 8 min: 99% B, 13 min: 85% B; 13.1 min: 1% B; 16 min: 1% B. The metabolite peaks were extracted using Agilent Masshunter Profinder based on our in-house library. Any metabolites with RSD greater than 30% in the quality control measurements were removed from further analysis. The peak areas were normalized by the weight of the fecal matters. Pathway analysis was performed with Metaboanalyst®.
Retention of inulin gel in gastrointestinal system.
To prepare FITC-tagged inulin gel, 47.5 mg inulin and 12.5 mg FITC-inulin (Sigma-Aldrich) were mixed in 200 μL DI water, followed by inulin gel formation as described above. A mixture of 47.5 mg inulin and 12.5 mg FITC-inulin was used as the soluble FITC-inulin control. For in vivo imaging, mice were gavaged with FITC-tagged inulin or inulin gel and euthanized at preset time points (2, 4.5, 7, 9.5, 12 h). The stomach, small intestine, cecum, colon and rectum were harvested and washed briefly with PBS and visualized using the IVIS optical imaging system. To detect inulin content in colon, mice were gavaged with inulin or inulin gel (60 mg/dose) and euthanized at preset times (2, 4.5, 7, 9.5, 12 h) post gavage. The digesta in colon was harvested, weighed, and added to 1.5 mL DI water. The samples were homogenized and centrifuged, and the supernatants were diluted 20 times for quantifying the inulin content using the PicoProbe™ Inulin Assay Kit (BioVision) according to the manufacturer’s instructions.
Antibiotics treatment.
BALB/c mice were pre-treated with sterile drinking water containing antibiotics (0.3 mg/mL ampicillin, 2.5 mg/mL streptomycin, and 0.3 mg/mL colistin) for 7 days. After tumor inoculation on day 0, mice were provided with sterile drinking water for 5 days to avoid any potential effects of antibiotics on tumor engraftment and formation. Starting day 5, antibiotics were added in the drinking water throughout the remainder of the study. The antibiotic drinking water was replaced twice weekly. The dilatation of cecum was observed to confirm the antibiotic activity. Gavage of inulin gel (60 mg/dose) began from day 7 and α-PD-1 was injected on day 11, 15 and 19.
In vitro stimulation of OT-I T cells with SCFAs.
Splenocytes from OT-I mice were obtained aseptically. Cells were cultured in 96-well plate (105 cells/well) and activated with SIINFEKL peptide (4 × 10−9 M) in the presence or absence of SCFAs (400 μM) for 3 days. Then cells were washed 3 times with T cell medium and further cultured with interleukin-15 (IL-15, 10 ng/mL, from PeproTech, Rocky Hill, NJ) in the presence or absence of SCFAs for 3 days to generate of OVA-specific OT-I memory CD8+ T cells. Thereafter, cells were washed twice and re-activated with SIINFEKL peptide (10 μM) for 2 h. Subsequently, brefeldin A was added (1 : 1000 dilution) and incubated for another 2 h. The suspension cells were collected, blocked with CD16/32 antibody, and stained with fixable viability dye eFluor™ 450, FITC-CD44 Rat anti-Human/Mouse, PE-Cy7-CD62L Monoclonal Antibody. Intracellular IFN-γ (PE-anti-mouse IFN-γ, clone: XMG1.2, Biolegend) and Tcf1 (PE-Tcf1/Tcf7 Rabbit mAb) positive T cells were detected via flow cytometry.
Safety evaluation.
CT26 tumor-bearing mice were treated as in Fig. 5a. On day 29, the blood was collected for complete blood count (CBC) (including eosinophils, lymphocytes, monocytes, platelet count, red blood cell count, hemoglobin, mean platelet volume, red cell distribution width (RDW), and mean corpuscular hemoglobin (MCH)). Meanwhile, some blood was collected, solidified, and centrifuged to obtain the serum for biochemistry analysis: glutamic pyruvate transaminase (ALT); aspartate aminotransferase (AST); blood urea nitrogen (BUN); glucose; cholesterol; creatine phosphokinase (CPK). Besides, mice were euthanized on day 29. The lung, liver, spleen, heart, and kidney were collected for hematoxylin and eosin (H&E) staining. All these tests were performed via In-Vivo Animal Core of University of Michigan.
Statistical analysis.
All experiments were performed at least twice with duplicate repeated measures. The results are expressed as means ± SEM. A one-way or two-way analysis of variance (ANOVA), followed by Bonferroni’s multiple comparisons post hoc test was used for testing differences among groups. Data were approximately normally distributed, and variance was similar between the groups. Experiments were repeated multiple times as independent experiments as indicated in the figure captions. Shown in the figure is a complete dataset from one representative, independent experiment. No samples were excluded from analysis. Statistical significance is indicated as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA) was used for statistical analyses.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that data supporting the findings of this study are available within the article and its Supplementary Information files. The raw and analysed datasets generated during the study are too large to be publicly shared, thus, they are available for research purposes from the corresponding author on reasonable request. Bacterial 16S rRNA sequencing data has been deposited in the NCBI Sequence Read Archive (accession number: PRJNA715170).
Supplementary Material
Acknowledgements
This work was supported by NIH (R01AI127070, R01EB022563, R01CA210273, U01CA210152, R01DK125087, P30CA046592). J.J.M. is supported by NSF CAREER Award (1553831). D.N. is supported by grants from NCI R01CA227622, R01CA222251, R01CA204969, Rogel Cancer Center grant, and Forbes Scholar Award. We acknowledge the NIH Tetramer Core Facility (contract HHSN272201300006C) for provision of MHC-I tetramers and Dr. Chang H. Kim for provison of GPR43−/− mice. Opinions interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. We thank Marisa Aikins for critical review of the manuscript, Yao Xu for ELISPOT tests, Dr. Keara Saud for rheology tests, Dr, Junnan Lu for the help in microbiota data analysis, and Dr. Hongliang He for helping with animal studies. Some figures were created by the authors with BioRender.com.
Footnotes
Competing interests
A patent application for in situ modulation of the gut microbiome has been filed, with J.J.M., K.H., and J.X. as inventors.
Supplementary information is available for this paper.
Peer review information Nature Biomedical Engineering thanks Reviewer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Topalian SL, Taube JM, Anders RA & Pardoll DM Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 16, 275–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribas A & Wolchok JD Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch SV & Pedersen O The human intestinal microbiome in health and disease. N. Engl. J. Med 375, 2369–2379 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Gilbert JA et al. Current understanding of the human microbiome. Nat. Med 24, 392–400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt TSB, Raes J & Bork P The human gut microbiome: from association to modulation. Cell 172, 1198–1215 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Cryan JF, O'Riordan KJ, Sandhu K, Peterson V & Dinan TG The gut microbiome in neurological disorders. Lancet Neurol. 19, 179–194 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Vetizou M et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sivan A et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matson V et al. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Routy B et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Gopalakrishnan V et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zitvogel L, Ma Y, Raoult D, Kroemer G & Gajewski TF The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 359, 1366–1370 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Helmink BA, Khan MW, Hermann A, Gopalakrishnan V & Wargo JA The microbiome, cancer, and cancer therapy. Nat. Med 25, 377–388 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Giles EM, D’Adamo GL & Forster SC The future of faecal transplants. Nat. Rev. Microbiol 17, 719–719 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Skelly AN, Sato Y, Kearney S & Honda K Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol 19, 305–323 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Man GCW, Chan TH, Kwong J & Wang CC A prodrug of green tea polyphenol (−)-epigallocatechin-3-gallate (Pro-EGCG) serves as a novel angiogenesis inhibitor in endometrial cancer. Cancer Lett. 412, 10–20 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Makki K, Deehan EC, Walter J & Bäckhed F The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23, 705–715 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Paulose JK, Wright JM, Patel AG & Cassone VM Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS One 11, e0146643, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart ML, Savarino V & Slavin JL Assessment of dietary fiber fermentation: Effect of Lactobacillus reuteri and reproducibility of short - chain fatty acid concentrations. Mol. Nutr. Food Res 53, S114–S120 (2009). [DOI] [PubMed] [Google Scholar]
- 20.La Rosa SL et al. The human gut Firmicute Roseburia intestinalis is a primary degrader of dietary β-mannans. Nat. Commun 10, 905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J et al. Fiber-rich foods affected gut bacterial community and short-chain fatty acids production in pig model. J. Funct. Foods 57, 266–274 (2019). [Google Scholar]
- 22.Cao Y & Mezzenga R Design principles of food gels. Nat. Food 1, 106–118 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Sun M et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun 9, 3555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Groot PF et al. Oral butyrate does not affect innate immunity and islet autoimmunity in individuals with longstanding type 1 diabetes: a randomised controlled trial. Diabetologia 63, 597–610, (2020). [DOI] [PubMed] [Google Scholar]
- 25.Balmer ML et al. Memory CD8+ T cells require increased concentrations of acetate induced by stress for optimal function. Immunity 44, 1312–1324 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Bachem A et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 51, 285–297 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Siddiqui I et al. Intratumoral Tcf1+ PD-1+ CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195–211 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Jansen CS et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature, 576, 465–470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sade-Feldman M et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatae R et al. Combination of host immune metabolic biomarkers for the PD-1 blockade cancer immunotherapy. JCI Insight 5, e133501–e133518 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinoi T et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 67, 9721–9730 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Nomura M et al. Association of short-chain fatty acids in the gut microbiome with clinical response to treatment with nivolumab or pembrolizumab in patients with solid cancer tumors. JAMA Netw. 3, e202895–e202895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Botticelli A et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J. Transl. Med 18, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y et al. Prebiotic-induced anti-tumor immunity attenuates tumor growth. Cell Rep. 30, 1753–1766 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senghor B, Sokhna C, Ruimy R & Lagier J-C Gut microbiota diversity according to dietary habits and geographical provenance. Hum. Microbiome J 7, 1–9 (2018). [Google Scholar]
- 36.Salgia NJ et al. Stool microbiome profiling of patients with metastatic renal cell carcinoma receiving anti-PD-1 immune checkpoint inhibitors. Eur. Urol 78, 498–502 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Derosa L, Routy B, Kroemer G & Zitvogel L The intestinal microbiota determines the clinical efficacy of immune checkpoint blockers targeting PD-1/PD-L1. Oncoimmunology 7, e1434468–e1434470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan MAW, Ologun G, Arora R, McQuade JL & Wargo JA Gut microbiome modulates response to cancer immunotherapy. Dig. Dis. Sci 65, 885–896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King DE, Mainous AG 3rd & Lambourne CA Trends in dietary fiber intake in the United States, 1999–2008. J. Acad. Nutr. Diet 112, 642–648 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Kozich JJ, Westcott SL, Baxter NT, Highlander SK & Schloss PD Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that data supporting the findings of this study are available within the article and its Supplementary Information files. The raw and analysed datasets generated during the study are too large to be publicly shared, thus, they are available for research purposes from the corresponding author on reasonable request. Bacterial 16S rRNA sequencing data has been deposited in the NCBI Sequence Read Archive (accession number: PRJNA715170).