Abstract
During HIV-1 assembly, the viral Gag polyprotein specifically selects the dimeric RNA genome for packaging into new virions. The 5′ untranslated region (5′UTR) of the dimeric genome may adopt a conformation that is optimal for recognition by Gag. Further conformational rearrangement of the 5′UTR, promoted by the nucleocapsid (NC) domain of Gag, is predicted during virus maturation. Two 5′UTR dimer conformations, the kissing dimer (KD) and the extended dimer (ED), have been identified in vitro, which differ in the extent of intermolecular basepairing. Whether 5′UTRs from different HIV-1 strains with distinct sequences have access to the same dimer conformations has not been determined. Here, we applied fluorescence cross-correlation spectroscopy and single-molecule Förster resonance energy transfer imaging to demonstrate that 5′UTRs from two different HIV-1 subtypes form (KDs) with divergent stabilities. We further show that both 5′UTRs convert to a stable dimer in the presence of the viral NC protein, adopting a conformation consistent with extensive intermolecular contacts. These results support a unified model in which the genomes of diverse HIV-1 strains adopt an ED conformation.
Significance
During virion assembly, HIV-1 packages dimerized copies of its single-stranded RNA genome. This process may be regulated by the conformation of the genomic 5′UTR. Conformational changes in the 5′ untranslated region (UTR) have also been implicated during additional steps of the virus life cycle, including reverse transcription and virion maturation. Thus far, two conformations of the 5′UTR dimer have been described, which differ in the extent of intermolecular basepairing, although whether genomic RNA from different HIV-1 strains that have distinct 5′UTR sequences form similar dimer conformations has not been fully determined. Here, we demonstrate that 5′UTRs from distinct HIV-1 strains form similar conformations with divergent intrinsic stabilities. These results support a unified model of genome dimerization for different HIV-1 strains.
Introduction
HIV-1 packages two copies of its unspliced single-stranded RNA genome into assembling virions (1,2). Incorporation of two genomes occurs when the viral Gag polyprotein preferentially selects dimerized genomes from an excess of monomeric genomes and subgenomic viral and cellular messenger RNAs. This selection occurs by way of interactions between the nucleocapsid (NC) domain of Gag and an RNA packaging signal within the 5′ untranslated region (5′UTR) of the viral genome (2,3). The optimal substrate for recognition by Gag may be a 5′UTR conformation that is unique to the dimerized genome. After assembly and release of virions from infected cells, viral maturation is required for subsequent rounds of infection. Maturation occurs when the viral protease cleaves the Gag polyprotein into its constituent parts, which induces morphological changes in the viral capsid (4). This reorganization of the virion interior coincides with stabilization of the dimeric genome, which may reflect further conformational changes in the 5′UTR (5). Roles for 5′UTR conformation have also been suggested during reverse transcription (6,7) and splicing and translation of viral messenger RNAs (8,9).
Biochemical studies of the 5′UTR led to the hypothesis that the conformation of the 5′UTR determines the HIV-1 genome’s ability to dimerize (10). The nucleation of intermolecular basepairing between two genomic RNA molecules is localized to the six-nucleotide palindromic dimerization-initiation sequence (DIS) within the 5′UTR (11,12). An NMR structure of the 155-nucleotide monomeric core-encapsidation signal, which contains all the structural elements necessary for dimeric genome packaging into virions, demonstrated that the DIS resides in a hairpin loop that is not engaged in intramolecular basepairing (13), which had previously been suggested by the structure of the isolated DIS hairpin (14). In this dimer-competent monomer (DCM) conformation, the DIS is exposed and available to engage in basepairing with the DIS of another genome molecule, and the U5 and AUG regions are engaged in intramolecular basepairing (Fig. 1 A). Alternatively, the 5′UTR can adopt a conformation in which the DIS is sequestered through basepairing with the upstream U5 region, displacing AUG and forming the dimer-incompetent monomer (DIM) conformation. Although initially, this conformational change was envisioned to occur spontaneously (15, 16, 17), subsequent studies implicated heterogeneity in the number of guanosine nucleotides at the 5′ terminus as determining the conformation adopted by the monomeric 5′UTR (18,19).
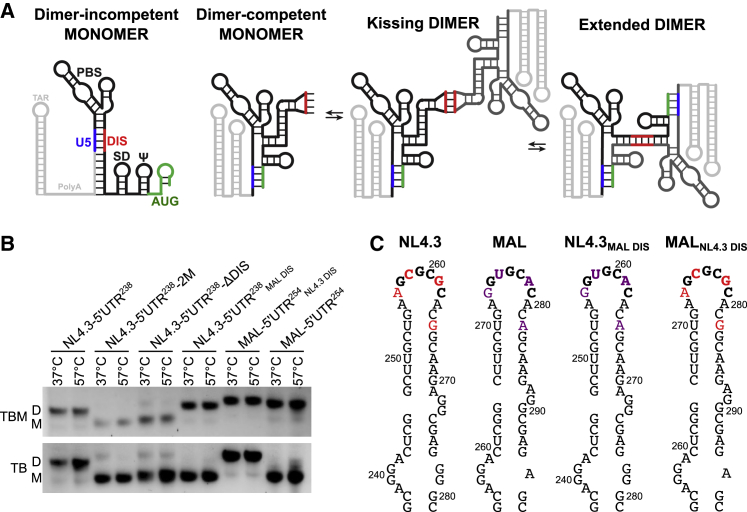
Figure 1.
The DIS sequence mediates 5′UTR dimer stability. (A) Model of monomeric and dimeric 5′UTR conformations. Key structural elements of the 5′UTR are labeled. The TAR and PolyA hairpins, shown here in light gray, are not present in the NL4.3-5′UTR238 and MAL-5′UTR254 constructs. In the DIM conformation, the DIS loop (red) is sequestered through basepairing with U5 (blue). In the DCM conformation, the AUG hairpin (green) unfolds and basepairs with U5, displacing DIS. The DIS is then exposed and capable of intermolecular basepairing with the DIS of another 5′UTR, forming the KD. Unfolding of the DIS hairpins and extension of the intermolecular interface gives rise to the extended-dimer conformation. (B) Analysis of 5′UTR dimer stability with nondenaturing RNA gel electrophoresis. The indicated RNA constructs were dimerized in PI buffer (10 mM Tris-HCl, 140 mM KCl, 10 mM NaCl, 1 mM MgCl2 (pH 7.0)) for 1 h at 37 or 57°C, followed by evaluation on 1% agarose gels in TB (44 mM Tris-borate (pH 8.3)) and TBM (44 mM Tris-borate (pH 8.3), 0.2 mM MgCl2) buffers. Monomer (M) and dimer (D) species are indicated. In the NL4.3-5′UTR238 construct, the ΔDIS mutant contains a GAGA tetraloop in place of the DIS loop; 2M contains the G108C mutation in U5 and the compensatory C258G in DIS (20). (C) Secondary structures of the DIS hairpins and mutants thereof evaluated in (B). Nucleotides highlighted in red are specific to the NL4.3-5′UTR; nucleotides highlighted in purple are specific to the MAL-5′UTR. The chimeric constructs, NL4.3-5′UTR238MAL DIS and MAL-5′UTR254NL4.3 DIS, were formed by swapping the highlighted nucleotides, as indicated. The complete sequences and secondary structural models of NL4.3-5′UTR238 and MAL-5′UTR254 are shown in Fig. S1. To see this figure in color, go online.
Genome dimerization is initiated through intermolecular DIS-DIS basepairing, forming a loop-loop “kissing-dimer” (KD) conformation (Fig. 1 A) (11,21, 22, 23, 24, 25, 26, 27). A conformational change in the 5′UTR can lead to extension of the intermolecular basepairing and conversion from the KD to the stable “extended-dimer” (ED) conformation (Fig. 1 A) (22,28,29). Of particular note, an NMR study of the dimerized 344-nucleotide 5′UTR from the NL4.3 strain of HIV-1 demonstrated the existence of intermolecular basepairing consistent with an ED (30). The relevance of these interactions during genome dimerization in vivo is supported by the observation that genomes with complementary DIS sequences are co-packaged into assembling virions more frequently than random assortment (31, 32, 33). Furthermore, a minimal portion of the 5′UTR that includes the DIS is sufficient to facilitate competitive packaging of heterologous RNAs against authentic viral genomes (34,35). Finally, constructs that omit the 5′UTR but contain the rest of the viral genome are outcompeted by constructs that contain the 5′UTR (34). Taken together, these observations support the notion that intermolecular DIS-DIS interaction initiates in vivo genome dimerization, which is critical to genome packaging, and that further conformational changes in the dimerized 5′UTR may occur.
During virion assembly, the NC domain of Gag selectively recruits dimeric genomes through a combination of low- and high-affinity binding sites within the 5′UTR (36, 37, 38). The ψ hairpin was identified as a key element recognized by NC (39). Later work specified that the ψ hairpin is part of a complex tertiary structure that forms the core packaging signal (13,40). Unpaired guanosines in the 5′UTR are specifically implicated in high-affinity interactions with NC (36,37). High-affinity NC binding to the 5′UTR may function to nucleate initial formation of Gag dimers or multimers, in which subsequent assembly is driven by Gag-Gag interactions and lower-affinity binding of NC to the rest of the viral genome (41). During virion maturation, the Gag polyprotein is cleaved into component parts, allowing the isolated NC protein to potentially reorganize the conformation of the genome. In support of this idea, binding of the NC protein alone promotes dimerization of the 5′UTR (26) and specifically facilitates transition to a thermostable dimer of the 5′UTR from HIV-1 and other retroviruses (42, 43, 44, 45, 46). Single-molecule Förster resonance energy transfer (smFRET) data suggested an NC-induced conversion of the dimeric 5′UTR to a conformation consistent with the ED conformation characterized by NMR, thus suggesting that the ED may correspond to the thermostable form previously observed (30,20).
Importantly, the existence of the ED conformation has not been conclusively demonstrated for diverse 5′UTR sequences, thus leaving the relevance of the ED during HIV-1 replication in question. Different HIV-1 subtypes have distinct DIS sequences, and RNAs containing those sequences differ in their dimerization properties (19,47, 48, 49). The predominant DIS variants are 5′-GCGCGC-3′, found in subtypes B and D, and 5′-GUGCAC-3′, found in other subtypes including A, C, and F. Subtype-specific DIS sequences also have highly conserved characteristic flanking purine nucleotides. These differences in DIS sequences lead to RNA dimers with distinct stabilities. RNAs that include the subtype B DIS sequence form stable dimers that resist dissociation during nondenaturing gel electrophoresis in the absence of Mg2+ (22,27,42,50,51). These stable dimer species form spontaneously and are promoted by NC and elevated temperature. As noted above, structural and single-molecule studies have suggested a correlation of this stable species with the ED conformation of the 5′UTR (30,20). Collectively, these studies form the foundation of hypotheses regarding the importance of the ED in packaging or maturation. In contrast, RNAs that include the subtype A DIS sequence form dimers that are less stable during nondenaturing gel electrophoresis in the absence of Mg2+ (19,21,50,51). ED formation has not been demonstrated for subtype A 5′UTR sequences. An NMR study of a 359-nucleotide subtype A 5′UTR, dimerized in the absence of NC, found no evidence of the ED and confirmed basepairing consistent with the KD (19). To date, only studies of short oligonucleotides corresponding to subtype A DIS hairpins provide evidence of ED formation, which is enhanced by NC binding (28,43,44).
In our study, we sought to draw a direct correspondence between dimer stability and conformation. We sought to determine whether distinct 5′UTRs, with documented differences in dimer stabilities, have access to equivalent dimer conformations. Specifically, we aimed to determine whether the ED conformation is specific to 5′UTRs from subtype B strains or whether it is a general feature of diverse subtypes. To this end, we developed complementary biophysical assays that report on the stability and conformation of 5′UTR dimers from HIV-1 strains NL4.3 and MAL, which typify subtypes B and A, respectively. Using fluorescence cross-correlation spectroscopy (FCCS), we monitored the dissociation of 5′UTR dimers in solution under native conditions. We then applied two orthogonal smFRET imaging assays with alternating laser excitation to visualize the conformation of 5′UTR constructs from NL4.3 and MAL. Using these assays, we explicitly determined the stability of 5′UTR dimers in distinct conformations. Our results conclusively indicate that the NL4.3 5′UTR forms dimers in the absence of NC that remain in the KD conformation and are stable under physiological ion conditions. In contrast, MAL dimers in the KD conformation are comparatively labile. We further show that 5′UTR dimers from both subtypes transition to the ED conformation in the presence of isolated NC protein and that the dimer linkages in NL4.3 genome-containing virions display similar but slightly elevated thermostability relative to those with MAL genomic RNAs. These data thus provide support for the generality of the current mechanistic model of 5′UTR dimerization and indicate that energetic differences between dimeric species are imposed by distinct DIS sequences.
Materials and methods
RNA production
Sequences for NL4.3 and MAL were downloaded from GenBank (accession numbers M19921.2 and X04415.1, respectively), and constructs were designed to include nucleotides −1 to 356 for NL4.3 and the corresponding −1 to 370 for MAL (Fig. S1). The sequences were purchased from Genscript (Piscataway, NJ) and inserted into pUC57. Mutations to swap the DIS sequences were also purchased from Genscript. The template for in vitro transcription was generated by polymerase chain reaction (PCR) amplifying the regions of the NL4.3 and MAL 5′UTR plasmids corresponding to nucleotides 106–343 and 105–358, respectively, using primers that encoded a T7 promoter. For use in generation of RNA for smFRET imaging, a primer encoding a 5′ extension was used. As described below, this extension facilitated immobilization of 5′UTR RNA onto quartz microscope slides for total internal reflection fluorescence (TIRF) microscopy. The PCR amplicons were purified by excision from 1% agarose gels and concentrated by ethanol precipitation. The purified template DNA was resuspended in nuclease-free water and stored at −20°C. The templates were verified by Sanger sequencing before use in in vitro transcription.
NL4.3-5′UTR238 and MAL-5′UTR254 RNAs were synthesized by in vitro transcription using the T7 RiboMax Express Large-Scale RNA Production system (Promega, Madison, WI). Reaction mixtures were prepared according to the manufacturer’s protocol at a scale of 5 μg DNA template in a 100 μL total volume. The reactions were incubated for 2.5 h at 37°C before addition of 5 U of RQ1 RNase-Free DNase (1 U per μg of template DNA; Promega) followed by an additional 15-min incubation at 37°C. 300 μL of cold (4°C) low-ionic-strength buffer (LI buffer; 10 mM Tris-HCl (pH 7.5), 10 mM NaCl) was added, and the reaction mixtures were purified by size-exclusion chromatography.
RNA samples were purified from the reaction mix and short abortive transcripts with a Superdex 200 Increase 10/300 GL column (Cytiva, Marlborough, MA) at a flow rate of 0.65 mL/min on an Akta liquid chromatography system (Cytiva) in LI buffer. Peak fractions were identified by absorbance at 260 nm. Pooled fractions (∼400 μL) were loaded into a 50-kDa molecular weight cut-off Vivaspin 6 centrifugal concentrator (Millipore Sigma, Burlington, MA) and diluted to 6 mL total volume with nuclease-free water. Samples were spun at 4000 × g for 5–8 min. This dilution and concentration procedure was repeated three times to desalt the sample. The RNA samples were aliquoted and stored at −20°C until use.
Fluorescent labeling of RNA
For use in FCCS or smFRET experiments, 5′UTR RNAs were 3′ labeled with Alexa Fluor 647- or Alexa Fluor 488-hydrazide (Thermo Fisher Scientific, Waltham, MA) using periodate chemistry (52). 2 nmol of RNA was mixed with 38 μL 25 mM NaIO4 (freshly prepared) and 38 μL 1 M NaOAc (pH 5.2) and raised to a final volume of 200 μL with nuclease-free water. The RNA was incubated on ice for 2 h, followed by concentration by ethanol precipitation. The resulting RNA pellet was dissolved in 20 μL 1 M NaOAc (pH 5.2) 9 μL 1 M NaCl, and 11 μL nuclease-free water. 1 mg of Alexa Fluor 647- or 488-hydrazide dye was resuspended to 10 mM in dimethyl sulfoxide. 5 μL 10 mM fluorophore was added to the RNA mixture. The labeling reaction was protected from light and incubated for 2 h at room temperature with gentle mixing. The labeling reactions were then diluted to 400 μL in LI buffer and purified by size-exclusion chromatography, desalted, and stored as described above.
Fluorescent labeling and purification of DNA
The DNA handle (5′-TTTTGCTCCATAACCTCCAACC-3′) used to immobilize 5′UTR RNA for smFRET imaging with TIRF microscopy was purchased from IDT (Coralville, IA) with a 5′ C6 amino modifier and 3′ biotin-triethyleneglycol modifier. The DNA handle was labeled at the 5′ end with LD550-NHS (Lumidyne Technologies, New York, NY) by standard protocols (20,52). The labeled DNA handle was purified from unattached dye by size-exclusion chromatography as described above for RNA samples. Peak fractions were pooled and ethanol precipitated. The precipitated handle was resuspended in 0.5 mL 0.1 M triethylammonium acetate (pH 6.5). Labeled handle was purified away from unlabeled handle by C-18 reverse phase chromatography on an Agilent Infinity high performance liquid chromatography system (Santa Clara, CA) in 0.1 M triethylammonium acetate (pH 6.5) using a linear gradient of 0–75% acetonitrile. Peak fractions were pooled, ethanol precipitated, resuspended in nuclease-free water, and stored at −20°C until use.
Purification of NC protein
The 55-amino acid NCNL4.3 protein (MQKGN FRNQR KTVKC FNCGK EGHIA KNCRA PRKKG CWKCG KEGHQ MKDCT ERQAN, 6369 Da) was overexpressed and purified as previously described (37). The codon-optimized gene sequence encoding the 54-amino acid NCMAL protein (MQRGN FKGQK RIKCF NCGKE GHLAR NCRAP RKKGC WKCGK EGHQM KDCTE RQAN) was ordered from Genewiz (Morrisville, NC) and inserted into the pLATE11 plasmid, using the aLICator LIC cloning system (Thermo Fisher Scientific). The pLATE11 plasmid was transformed into BL21(DE3) expression cells. The protein was purified under nondenaturing conditions. Briefly, cells were lysed by microfluidization, followed by purification with ion exchange chromatography using MonoQ and MonoS columns (Cytiva). After elution from the SP column, concentrated fractions were furthered purified and buffer exchanged with size-exclusion chromatography on the Superdex 30 prep grade column (Cytiva). The molecular weight of NCMAL was verified by mass spectrometry to be 6521.
Dimerization and gel electrophoresis of RNA
RNA samples were assessed for homogeneity by denaturing gel electrophoresis on a 1% agarose gel in 1% bleach (0.0525% sodium hypochlorite) and TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) (Fig. S2). Samples were heated to 60°C in nuclease-free water and mixed with heated loading buffer before loading. Gels were run at 30°C and stained with ethidium bromide.
Before dimerization, RNAs were thawed on ice and diluted in cold 50 mM HEPES (pH 7.5) to 10 μM. The RNAs were then heated to 80°C for 2 min, 60°C for 4 min, and 37°C for 6 min, cooled to 4°C, and then placed on ice for 10 min. The RNAs were then dimerized by dilution to 0.5 μM in physiological ion (PI) buffer (10 mM Tris-HCl, 140 mM KCl, 10 mM NaCl, 1 mM MgCl2 (pH 7.0)). PI buffer with either 0 or 5 mM MgCl2 was also used when indicated in the Results. Samples of RNA in PI buffer were then dimerized by incubation at either 37 or 57°C in a thermocycler for 1 h and placed on ice until evaluation. When indicated, neomycin was included in the dimerization mixture at the indicated concentration.
20 pmol of dimerized RNA (in PI buffer or as indicated) was mixed with 6× RNA loading buffer (20% sucrose w/v, without dye) and loaded onto a 1% agarose gel in either TBM (44 mM Tris-borate (pH 8.3), 0.2 mM MgCl2) or TB (44 mM Tris-borate (pH 8.3)) buffers. Gels were pre-equilibrated to 4°C and run for 90 min at 120 V. Gels were prepared with ethidium bromide as indicated.
FCCS experiments
5′UTR RNA samples used for smFRET imaging were dimerized as described above with minor modifications. Included in the dimerization reaction was 0.4 μM 5′UTR RNA labeled at the 3′ terminus with Alexa Fluor 647 and 0.4 μM 5′UTR RNA labeled at the 3′ terminus with Alexa Fluor 488. The dimerization mixtures were incubated as described above. When indicated, NC was included in the dimerization reaction at a 40:1 molar ratio of NC/RNA, which equates to ∼6 nucleotides per NC. After the 1-h incubation to allow dimerization, NC was removed by addition of proteinase K (Millipore Sigma) to a final concentration of 1.0 μg/μL and incubated for an additional 30 min at 37°C. Bulk fluorescence anisotropy measurements were performed in a QuantaMaster 400 fluorimeter (Horiba, Kyoto, Japan) to confirm NC binding (Fig. S3 A) (53). Removal of NC was required to alleviate the fluorescence anisotropy induced by NC binding, which is detrimental to the smFRET experiments described below. Bulk fluorescence anisotropy measurements confirmed the reduction of anisotropy after removal of NC with proteinase K (Millipore Sigma) (Fig. S3 B). Dimerized RNA samples were placed on ice until use. For FCCS measurements, the dimerized 5′UTR RNAs were diluted to 5 nM with PI buffer with the indicated MgCl2 concentration (0–5 mM). The diluted sample was placed on a pegylated No. 1.5 coverslip (ThorLabs, Newton, NJ) and mounted on a CorTector SX100 (LightEdge Technologies, Beijing, China) equipped with 488- and 647-nm lasers. FCCS data were acquired at room temperature in 30-s intervals for 60 min. The cross-correlation (CC) curves were then averaged over 6-min intervals, yielding 10 CC curves per experiment. Each CC curve was fitted to a model of a single diffusing species,
where G(0) is the CC amplitude at 0 lag time (τ), τD is the correlation time, and s is the structure factor (54). G(0) and τD were determined by fitting the experimental CC curve to G(τ) with a least-squares algorithm in MATLAB (The MathWorks, Waltham, MA). The structure factor was determined to be ∼2.9 by fitting the correlation data for a sample of known diffusion coefficient. For each 60-min experiment, the 10 G(0)-values were normalized to the maximal observed value and reported in Figs. 3 and 8.
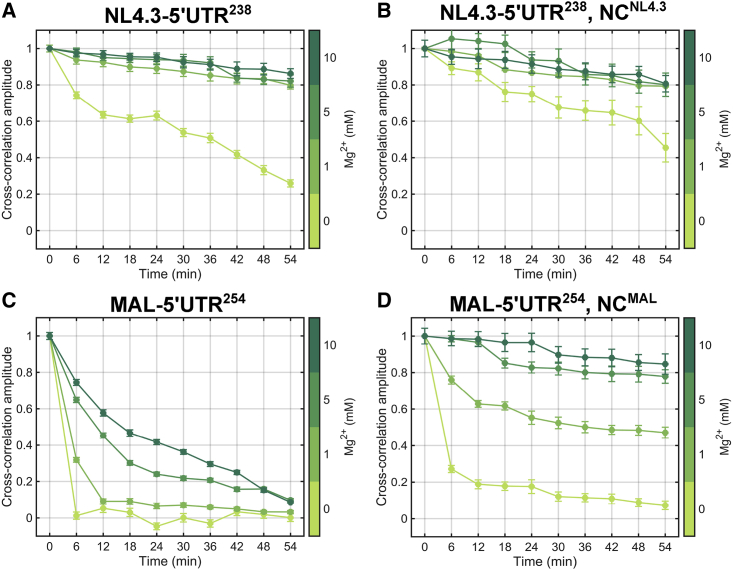
Figure 3.
Stability of 5′UTR dimers from NL4.3 and MAL strains of HIV-1. The amplitude of the CC (at 0-ms lag time) observed at 6-min intervals over a 1-h FCCS acquisition is shown. RNAs were dimerized in PI buffer (with 1 mM Mg2+) for 1 h at 37°C, followed by dilution into PI buffer at the indicated Mg2+ concentration (0–10 mM). All FCCS acquisitions were conducted at room temperature. The CC amplitudes were extracted by fitting the CC functions to a model for a single diffusing species (see Materials and methods). The CC amplitudes at 0-ms lag time were then normalized to the maximal observed value. (A) Normalized CC amplitudes from NL4.3-5′UTR238 dimerized in the absence of NCNL4.3. (B) The same data from NL4.3-5′UTR238 dimerized in the presence of cognate NCNL4.3. Similarly, normalized CC amplitudes from the dimerized MAL-5′UTR254 in the (C) absence and (D) presence of cognate NCMAL are given. The data reflect the mean from three independent measurements, with error bars representing the standard error of the mean. NCNL4.3 and NCMAL binding to NL4.3-5′UTR238 and MAL-5′UTR254, respectively, was verified by bulk fluorescence anisotropy (Fig. S3). To see this figure in color, go online.
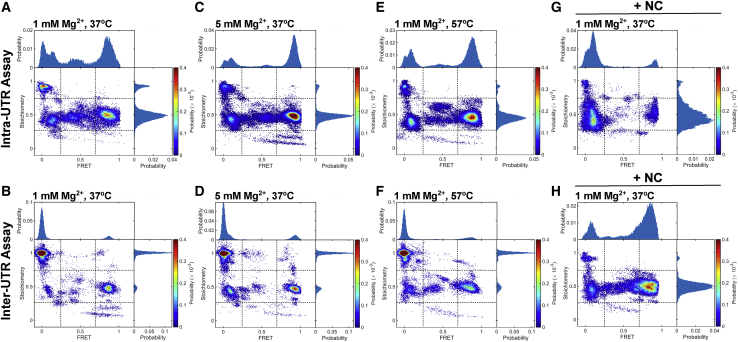
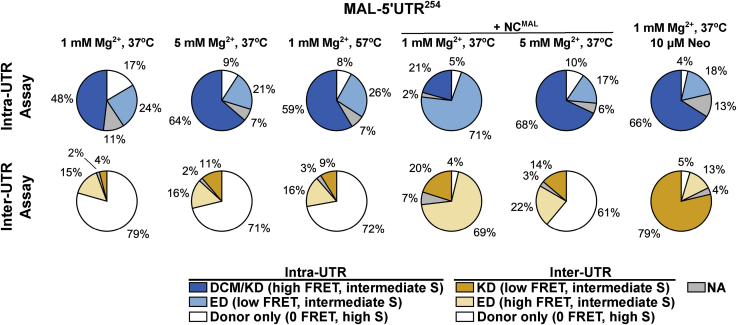
Figure 8.
smFRET analysis of the 5′UTR dimer from the MAL strain. (A) ES plot for the MAL-5′UTR254 dimer, formed and imaged in PI buffer at 1 mM Mg2+ and acquired with the intra-UTR FRET assay. The histograms are normalized to the total number of data points included in each data set. The probability per histogram bin is displayed according to the indicated colormap, with red indicating the highest probability. Also shown are univariate histograms displaying the distributions of FRET efficiency (plotted along the top horizontal axis) and stoichiometry (plotted along the right vertical axis). (B) Intra-UTR FRET data for MAL-5′UTR254 dimerized and imaged in PI buffer with 5 mM Mg2+. (C) Intra-UTR FRET data for MAL-5′UTR254 dimerized in the presence of NCMAL in PI buffer. (D) ES plot for the MAL-5′UTR254 dimer, formed and imaged in PI buffer at 1 mM Mg2+ and acquired with the inter-UTR FRET assay. (E) Inter-UTR FRET data for MAL-5′UTR254 dimerized and imaged in PI buffer with 5 mM Mg2+. (F) Inter-UTR FRET data for MAL-5′UTR254 dimerized in the presence of NCMAL in PI buffer. (G) Intra-UTR FRET data for MAL-5’UTR254 dimerized in the presence of cognate NCMAL in PI buffer at 1 mM Mg2+ for 1 hr at 37°C. (H) The same data obtained with the Inter-UTR FRET Assay. To see this figure in color, go online.
Virus production and genomic RNA isolation
HIV-1 NL4.3 vector L688-RRE-puro-LTR (LTR, long-terminal repeat), containing the intact 688-base HIV-1 5′ leader, Rev-response element (RRE), and puromycin resistance gene was described previously (35). The corresponding MAL sequence was generated by cloning a PCR fragment templated by MAL-GPP-pA (19) into the NL4.3 vector, replacing the first 368 bases with the corresponding bases of the HIV-1 MAL leader. Ψ-HIV-1 helper pCMVΔR8.2 was described previously (55).
293T cells were grown in 10-cm plates containing Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 50 μg/mL gentamicin at 37°C with 5% CO2 before transient transfection at 70% confluence. To produce viral particles, cells were co-transfected using polyethylenimine as described previously (56), with 4 μg of plasmid encoding either NL4.3 or MAL leader vectors and 4 μg of Ψ-helper pCMVΔR8.2. Virus-containing media were harvested 48 and 72 h after transfection. RNA was extracted from virus-containing media using a nondenaturing protocol as described previously (57) and resuspended in 10 mM Tris, 1 mM EDTA, 1% sodium dodecyl sulfate, and 50 mM NaCl.
Nondenaturing Northern blot
Nondenaturing Northern blotting was performed as described (57) with minor changes; the RNA was denatured in an agarose gel after electrophoresis with 10% formaldehyde, transferred to a Zeta-Probe membrane (Bio-Rad, Hercules, CA), and prehybridized at 55°C for 6 h in 6× SSC buffer (900 mM NaCl, 90 mM trisodium citrate (pH 7)), 4× Denhardt’s solution, 20 mM mono and dibasic Na3PO4, 1% SDS, and 250 μg of salmon sperm DNA. RNA immobilized on the membrane was hybridized with a DNA probe (5'-AGCACGACGGCGCTGCCCAGACCCTTGCCCTGG-3') specific to the puromycin resistance gene, labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) according to the manufacturer’s protocol. Bands were quantified using a phosphorimager and ImageQuant software.
smFRET-alternating laser excitation imaging
5′UTR RNA samples used for smFRET imaging were dimerized as described above with minor modifications. Included in the dimerization reaction was 0.4 μM LD550-labeled DNA handle, 0.2 μM 5′UTR RNA that contained the 5′ handle-annealing sequence, and 1.0 μM 5′UTR RNA that lacked the handle-annealing sequence. For use in the intra-UTR assay, the 5′UTR RNA that anneals to the handle was labeled with Alexa 647 at the 3′ terminus, and the 5′UTR RNA that lacks the handle-annealing sequence was unlabeled. For use in the inter-UTR assay, the 5′UTR RNA that anneals to the handle was unlabeled, and the 5′UTR RNA that lacks the handle-annealing sequence was Alexa-647-labeled at the 3′ terminus. In all cases, the dimerization mixtures were incubated as described above. The presence of the fluorophores and the DNA handle did not affect the ability of the RNAs to dimerize (Fig. S4). When indicated, NC was included in the dimerization reaction and removed through proteolysis with proteinase K as described above.
Dimerized RNA samples were placed on ice before imaging. To remove unannealed DNA handle, immediately before imaging, the dimerized RNA complexes (20–40 μL) were applied to a 0.5 mL 50,000-kDa MWCO Amicon spin filter (Thermo Fisher), brought to 0.5 mL in PI buffer, and spun at 13,000 × g at 4°C for 8 min. The dilution and spin were repeated once. The recovered retentate was then diluted to 75 pM in PI buffer and immobilized on a passivated, streptavidin-coated quartz microfluidic chamber for evaluation with TIRF microscopy. Removal of the unannealed handle was required because it competes with the RNA-handle duplex for binding to the coated quartz, dramatically reducing the experimental throughput. The RNA samples were imaged in PI buffer supplemented with 1 mM trolox, 1 mM cyclooctatetraene, 1 mM nitrobenzyl alcohol, 2 mM protocatechuic acid, and 8 nM protocatechuate 3,4-deoygenase (Millipore Sigma), which stabilize fluorescence and remove molecular oxygen (58,59).
smFRET imaging was performed on a wide-field prism-based TIRF microscope centered on an Olympus IX-73 stand equipped with a 60× 1.2-NA water-immersion objective lens (Olympus, Tokyo, Japan). The donor fluorophore was excited with a 532-nm laser (Coherent, Santa Clara, CA) at 1.6 kW/cm2, and the acceptor fluorophore was excited with a 647-nm laser (Coherent) at 1.2 kW/cm2. Fluorescence was passed through an Di02-R532 long-pass filter (Semrock, Rochester, NY) to remove excitation light, separated into donor and acceptor channels by an LP02-FF640 dichroic filter (Semrock) mounted in an Optisplit III (Cairn Research, Faversham, UK), and passed through FF01-590/4 (Semrock) and LP02-RU647 (Semrock) emission filters, respectively, before being detected in parallel halves of a Prime 95B scientific complementary metal–oxide–semiconductor camera (Photometrics, Tucson, AZ). Videos were acquired in Micromanager consisting of 800 frames at 300-ms exposure time. Video acquisition parameters were designed to maximize the duration of fluorescence before photobleaching to capture any dynamics occurring on the timescale of minutes. Synchronously with each exposure, the lasers were alternated through direct modulation with the voltage pulse output from the camera. All smFRET imaging was performed at room temperature.
Time-resolved fluorescence intensity traces were extracted from the videos using the SPARTAN software package in MATLAB (The MathWorks) (60) with custom modifications to allow processing of alternating laser excitation (ALEX) data sets. FRET efficiency was calculated according to FRET = IA/(ID + IA), where ID is the fluorescence intensity of the donor and IA is that of the acceptor. Stoichiometry was calculated according to S = (IDDex + IADex)/(IDDex + IADex + IAAex), where IDDex and IADex are the donor and acceptor fluorophore intensities, respectively, under donor excitation, and IAAex is the acceptor fluorophore intensity under direct excitation of the acceptor. Fluorescence traces were selected for analysis according to the following criteria: signal/noise > 8, background noise < 70, number of acceptor blinks < 4, traces indicating multiple photobleaching events were removed, donor lifetime > 10 frames, stoichiometry lifetime > 10 frames, and average FRET efficiency < 1. Automatically selected traces were confirmed by manual inspection.
Results
The DIS sequence mediates 5′UTR dimer stability
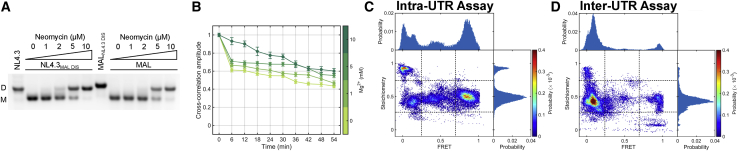
We first sought to evaluate the stability of 5′UTR dimers from the NL4.3 and MAL strains of HIV-1 using all RNA elements necessary for dimerization and packaging. We therefore in vitro transcribed RNA corresponding to nucleotides 106–343 of the NL4.3 genome (NL4.3-5′UTR238), which contain the entire 5′UTR except for the TAR and PolyA hairpins (Fig. S1). We also in vitro transcribed the homologous region of the MAL genome, which consisted of nucleotides 105–358 (MAL-5′UTR254). To preserve co-transcriptional folding, RNA was purified without denaturation using size-exclusion chromatography in low-ionic strength buffer (LI buffer: 10 mM Tris-HCl (pH 7.0)) (61). Previous studies of HIV-1 genomic 5′UTR dimer stability have relied upon the sensitivity of dimers in the absence or presence of Mg2+ in the running buffer of nondenaturing RNA gels. Therefore, to verify the function of our transcripts in dimerization, we incubated the 5′UTR constructs for 1 h at 37°C in PI buffer (10 mM Tris-HCl, 140 mM KCl, 10 mM NaCl, 1 mM MgCl2 (pH 7.0)) (16), followed by evaluation with nondenaturing gel electrophoresis. Consistent with previous reports, NL4.3-5′UTR238 formed a dimer that was stable irrespective of whether or not Mg2+ was included in the running buffer (Fig. 1 B) (62,63). In contrast, MAL-5′UTR254 formed a dimer that was stable only when Mg2+ was present in the running buffer; in the absence of Mg2+, the MAL-5′UTR254 dimers dissociated into monomers. Replacement of the DIS sequences with GAGA tetraloops (ΔDIS), which prevents the formation of intermolecular basepairing in the KD conformation (20), confirmed the identification of the monomer species (Fig. 1 B). Similarly, the 2 M mutation, which destabilizes U5-AUG basepairing and thus favors the DIM conformation (20), failed to dimerize (Fig. 1 B). Formation of 5′UTR dimers at elevated temperature (57°C) did not affect the stabilities of either of the 5′UTR dimers (Fig. 1 B).
We next tested the role of the DIS sequence in formation of the stable dimer species that is resistant to dissociation in the absence of Mg2+. To this end, we swapped the DIS sequences in NL4.3-5′UTR238 and MAL-5′UTR254, forming NL4.3-5′UTR238MAL DIS and MAL-5′UTR254NL4.3 DIS (Fig. 1 C). The chimeric 5′UTR constructs displayed stability phenotypes that were determined by the identity of their DIS sequences. That is, NL4.3-5′UTR238MAL DIS formed a dimer that was stable in the presence of Mg2+ but dissociated in the absence of Mg2+ (Fig. 1 B). Likewise, MAL-5′UTR254NL4.3 DIS formed a dimer that was stable regardless of the presence of Mg2+. Here again, dimerization at 57°C did not affect the stability of the dimers formed (Fig. 1 B). These data demonstrate that the stability of the 5′UTR dimer and its resistance to dissociation in the absence of Mg2+ is due to the DIS sequence and is not affected by the other sequence differences between the NL4.3 and MAL strains.
Characterization of 5′UTR dimer stability
We then characterized the stability of 5′UTR dimers under native conditions in solution and specified their sensitivity to Mg2+ using FCCS (64). The in vitro-transcribed 5′UTR RNA constructs were labeled using periodate oxidation of the 3′ ribose, followed by coupling to either Alexa Fluor 488- (Alexa488) or Alexa Fluor 647-hydrazide (Alexa647; see Materials and methods) (52). The labeled species were mixed at a 1:1 ratio and dimerized through incubation in PI buffer for 1 h at 37°C. The complexes were then diluted down to 5 nM total RNA concentration. Using FCCS, we then monitored the diffusion of fluorescent 5′UTR dimers through a confocal detection volume (Fig. 2 A). Dimers containing both fluorophores generated nonzero CC of the two fluorescence signals (Fig. 2 B). The CC data were then fitted to a model of a single diffusing species, which allowed quantification of the CC amplitude. When the mixture of labeled 5′UTR molecules was analyzed by FCCS before dimerization, zero CC was observed (Fig. S5). Thus, the amplitude of CC reports specifically on the concentration of dimers containing both fluorophores.
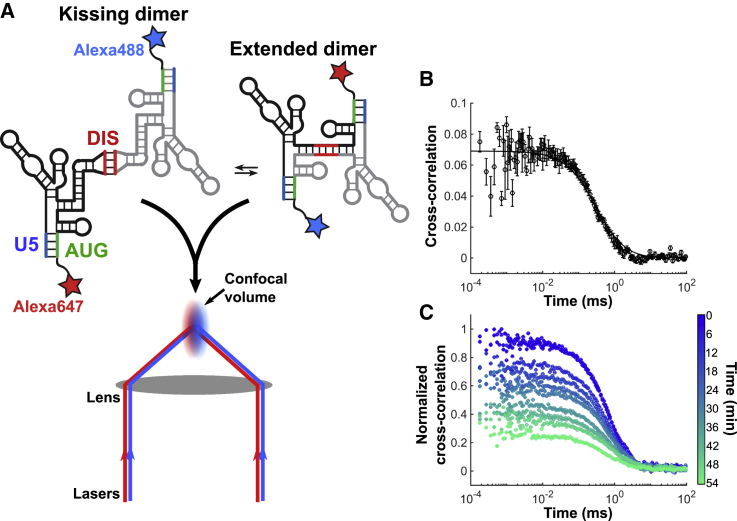
Figure 2.
An FCCS assay to monitor 5′UTR dimer stability in solution under native conditions. (A) Experimental setup of FCCS assay. 5′UTR RNA was labeled with either Alexa488 (blue star) or Alexa647 (red star) at the 3′ terminus (see Materials and methods). The RNAs were dimerized at 800 nM total RNA concentration at 37°C in PI buffer, followed by dilution to 5 nM. The sample was excited with simultaneous illumination at 488 and 647 nm as the RNAs diffuse through a confocal detection volume. (B) The CC of the emitted fluorescence from the Alexa488 and Alexa647 fluorophores. Error bars represent the standard deviation from three independent measurements. The CC was fitted to a model for a single diffusing species, which allowed determination of the CC at 0-ms lag time (solid line; see Materials and methods). The CC amplitude at 0 ms provides a measure of the concentration of dimerized RNAs with both fluorophores. (C) CC data acquired over 1 h and averaged over 6-min intervals. The decay in CC amplitude reflects the dissociation of 5′UTR dimers. To see this figure in color, go online.
To monitor the dissociation of 5′UTR dimers, the complexes were diluted into buffer of varying Mg2+ concentrations, and FCCS data were collected over 1 h. When dimers of NL4.3-5′UTR238 were diluted into water, the CC amplitude gradually decayed over the 1-h acquisition, reflecting the dissociation of the 5′UTR dimers (Fig. 2 C). These data demonstrate that after dilution into water, the NL4.3-5′UTR238 dimer was ∼75% dissociated after 1 h (Fig. 3 A). In contrast, the presence of physiological or elevated Mg2+ concentration rendered the NL4.3-5′UTR238 dimer stable over 1 h, with the modest decrease of CC amplitude reflecting photobleaching of the fluorophores. When dimerization of NL4.3-5′UTR238 was performed in PI buffer in the presence of cognate NCNL4.3, the stability of the dimers after dilution into water was greater than that seen without NCNL4.3, such that the CC amplitude decreased by only 55% after 1 h (Fig. 3 B). Dimer stability was maintained after dilution of buffer containing physiological or elevated Mg2+. Taken together, these data demonstrate that the NL4.3 5′UTR forms a stable dimer through incubation in PI buffer and that Mg2+ and NC further stabilize the dimer.
We next applied FCCS to evaluate the stability of MAL 5′UTR dimers. Here again, dimers were formed through incubation in PI buffer, followed by dilution into water or buffer at various Mg2+ concentrations. After dilution of MAL-5′UTR254 dimers into water, the CC amplitude rapidly decreased, reaching zero amplitude within 6 min (Fig. 3 C). Dilution into buffer containing 1 mM Mg2+ reduced the rate of CC amplitude decay. Increasing the Mg2+ concentration to 5–10 mM further decreased the rate of CC amplitude decay. But, across all Mg2+ concentrations tested, the CC lost at least 90% amplitude after 1 h. These data demonstrate that the MAL 5′UTR dimerizes through incubation in PI buffer, but the dimer is unstable and sensitive to Mg2+ concentration as compared to the NL4.3 5′UTR. We next asked whether NCMAL promotes stabilization of the MAL-5′UTR254 dimer. When the dimers were diluted into water, the CC amplitude decayed rapidly, although somewhat slower than in the absence of NCMAL (Fig. 3 D). The presence of 1 mM Mg2+ reduced the rate of CC decay such that ∼50% amplitude remained after 1 h (Fig. 3 D). Elevated Mg2+ (5–10 mM) stabilized the MAL-5′UTR254 dimers such that the modest decay in CC amplitude was comparable to that seen for NL4.3. These data demonstrate that NC promotes stabilization of the MAL 5′UTR dimer. But the MAL 5′UTR dimer, even when formed in the presence of NC, maintains its sensitivity to Mg2+ concentration.
Stability of genomic RNA dimers isolated from virions
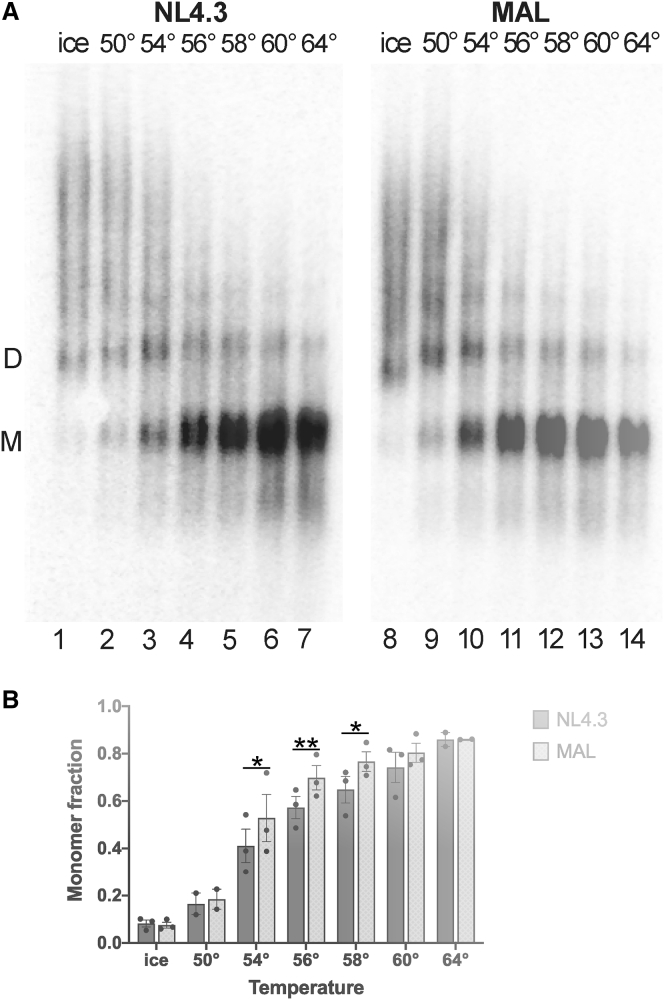
We next evaluated the stability of NL4.3 and MAL genomic RNA dimers isolated from mature HIV-1 virions. To this end, genomic RNA was extracted from virions under nondenaturing conditions and incubated at varying temperatures for 5 min. The fraction of monomer and dimer RNAs was evaluated using nondenaturing Northern blot analysis. Both NL4.3 and MAL genomic RNA showed predominant dimers, with minimal dissociation into monomers (<10%), after incubation on ice (Fig. 4). After 50°C incubation, the fraction of monomers ranged from ∼10 to 20% for both strains. For both strains, the fraction of monomeric RNA increased with increasing incubation temperature, reaching nearly 90% monomeric at temperatures above 62°C. After incubation at temperatures between 54 and 58°C, MAL showed a significantly higher fraction of monomeric RNA, consistent with greater dissociation of the dimeric genome (Fig. 4 B). Thus, both genomic RNAs form dimers in the mature virion that dissociate upon incubation at elevated temperature, with the NL4.3 RNA forming a dimer with greater stability.
Figure 4.
Stability of NL4.3 and MAL genomic RNA dimers isolated from virions. (A) Nondenaturing Northern blot of viral RNA extracted from media of cells transfected with HIV-1 Ψ-helper and NL4.3 (lanes 1–7) or MAL (lanes 8–14) leader RNAs. RNA was incubated at indicated temperature for 5 min before electrophoresis. Migration of viral genomic RNA dimers (D) and monomers (M) are indicated. (B) Percentage of viral genomic RNA monomers at different incubation temperatures as quantified by phosphorimager. Quantifications of three independent experiments are presented, except temperatures 52 and 62°C, which show data from one blot only. Bars reflect the average and standard deviation. Statistical significance was determined using two-way analysis of variance and indicated with asterisk notation.
Evaluation of 5′UTR dimer conformation with smFRET imaging
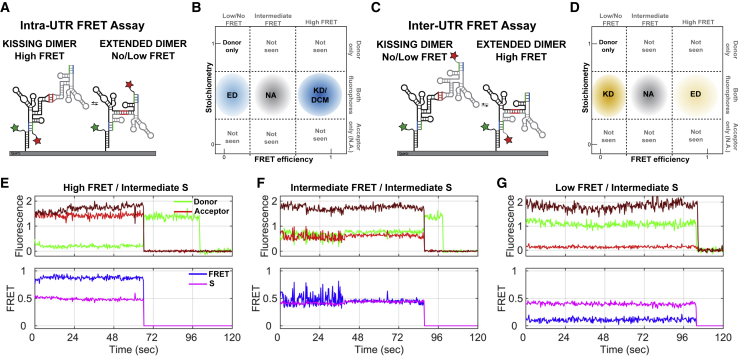
The different stability phenotypes displayed by the NL4.3- and MAL-5′UTR dimers prompted us to ask whether the two strains form dimers with distinct conformations. We previously established two orthogonal smFRET imaging assays that report on the conformation of 5′UTR dimers (20). Both assays required the insertion of a 5′ extension into the 5′UTR constructs that facilitates annealing to a DNA oligonucleotide. The DNA handle contained a 3′ biotin to enable immobilization on a streptavidin-coated quartz surface for imaging with TIRF microscopy. In the intra-UTR assay, the DNA handle was labeled at the 5′ terminus with the donor fluorophore (LD550), and the hybridized 5′UTR RNA was labeled at the 3′ terminus with the acceptor fluorophore (Alexa647). The intra-UTR assay reports on the conversion from the KD to the ED conformation (Fig. 5 A). This assay does not distinguish between the DCM conformation and the KD. In contrast, the inter-UTR assay, in which the acceptor fluorophore was moved to the 3′ terminus of the 5′UTR molecule that is not directly attached to the quartz surface, reports only on the conversion from the KD to the ED conformation and does not detect the presence of monomers (Fig. 5 C). Our previous study demonstrated that the majority of the NL4.3-5′UTR238 dimers visualized in the inter-UTR assay yielded an FRET value that was not detectable above background (20). This observation is consistent with the interfluorophore distance being beyond the range over which FRET can be reliably detected (30–80 Å). This limitation prevented us from making an accurate accounting of the dimer conformations adopted by NL4.3-5′UTR238. Similarly, in the intra-UTR assay, the ED conformation may place the fluorophores farther apart than can be clearly detected, further limiting our determination of the relative occupancies of the dimer species.
Figure 5.
smFRET imaging assays for determining the conformation of 5′UTR dimers. (A) In the intra-UTR FRET assay, 5′UTR RNA was transcribed with a 5′ handle-annealing sequence. The 5′UTR was then labeled at the 3′ end with the Alexa647 fluorophore. The labeled 5′UTR was hybridized to the DNA handle, which was labeled at the 5′ end with the LD550 fluorophore and the 3′ end with biotin. The RNA-DNA duplex was incubated with unlabeled 5′UTR RNA for 1 h at 37°C in PI buffer. The presence of the DNA handle and the 3′ fluorophore on the 5′UTR did not affect dimerization, as determined by nondenaturing gel electrophoresis (Fig. S4). (B) Schematic of bivariate histograms (ES plots), which display the distribution of FRET efficiency and S-values observed over a population of individual 5′UTR molecules. Identification of subpopulations of complexes, based on FRET and stoichiometry, is indicated and colored as in Figs. 7 and 9. Molecules in intermediate-FRET states were not assigned (NA) to a conformation. Molecules with only a donor fluorophore could arise from unannealed DNA handle or incomplete labeling of the RNA. (C) In the inter-UTR FRET assay, unlabeled 5′UTR RNA with the handle-annealing sequence was hybridized to the same LD550-labeled and biotinylated DNA handle. The RNA-DNA duplex was incubated with 5′UTR RNA labeled at the 3′ end with Alexa647 for 1 h at 37°C in PI buffer. In both assays, the dimerized complexes were immobilized on streptavidin-coated quartz microscope slides and imaged with prism-based TIRF microscopy at room temperature. The anticipated FRET values are shown for the KD and ED conformations. (D) Schematic of ES plots for the inter-UTR assay. As in (B), the locations of subpopulations are indicated. Here again, molecules in intermediate-FRET states were not assigned (NA) to a conformation. Molecules with only a donor fluorophore could arise from 5′UTR monomers, unannealed DNA handle, or incomplete labeling of the RNA. (E, top) Example fluorescence traces (donor, green; acceptor intensity during excitation by way of FRET, red; acceptor intensity during direct excitation at 647 nm, maroon) and (bottom) the calculated high-FRET efficiency (blue) and intermediate S-value (pink). (F) Example traces displayed as in (E) with intermediate-FRET efficiency (blue) and intermediate S-value and (G) intermediate-FRET efficiency (blue) and intermediate S-value. To see this figure in color, go online.
To address the limitations of our previous smFRET imaging approach and improve our accounting of 5′UTR dimer conformations, we incorporated ALEX into our imaging approach (65). In conventional smFRET imaging applications, only the donor fluorophore is excited by a laser. The FRET efficiency is then calculated according to FRET = IA/(ID + IA), where ID is the fluorescence intensity of the donor and IA is that of the acceptor. The presence of the acceptor fluorophore is verified only if the efficiency of FRET is high enough to generate acceptor fluorescence that is detectable above the background. The ALEX approach involves alternating the illumination of the sample with lasers that excite either the donor or the acceptor fluorophore synchronously with camera exposures. Excitation of the donor fluorophore permits calculation of FRET efficiency as in standard smFRET imaging. Excitation of the acceptor fluorophore allows calculation of the stoichiometry, defined as S = (IDDex + IADex)/(IDDex + IADex + IAAex), where IDDex and IADex are the donor and acceptor fluorophore intensities, respectively, under donor excitation, and IAAex is the acceptor fluorophore intensity under direct excitation of the acceptor. For 5′UTR molecules with a single fluorophore pair, S = 0.5. The observed FRET and S-values are compiled into a bivariate histogram (ES plots), which permits identification of subpopulations of molecules based on conformation and presence of one or both fluorophores (Fig. 5, B and D). This technical advance facilitated the differentiation of 5′UTR monomers from dimer conformations in which the distance between the fluorophores was greater than we can reliably detect in a conventional smFRET assay. This resulted in obtaining a better accounting of the conformations of individual 5′UTR dimers.
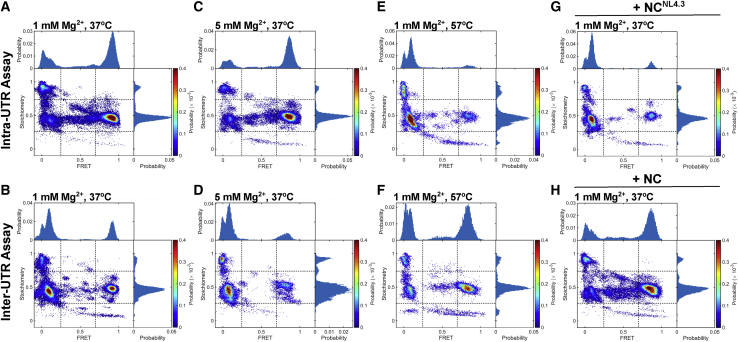
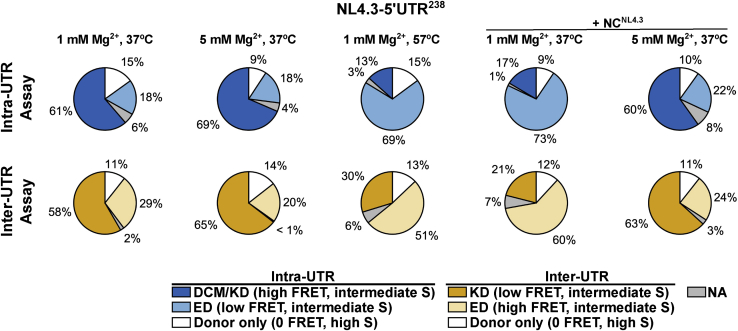
We first evaluated the conformation of the NL4.3-5′UTR238 dimer. The RNA was dimerized in PI buffer at 37°C, followed by imaging at room temperature in the same buffer. Example traces acquired from individual RNA complexes are shown in Fig. 5, E–G. The intra-UTR assay indicated 61% occupancy in a high-FRET state, whereas the inter-UTR assay indicated 58% occupancy in a low-FRET state (Fig. 6, A and B). Both results are consistent with the KD conformation predominating. Taken together with the FCCS results, these data indicate that the NL4.3-5′UTR238 KD is stable under PI buffer conditions on the timescale probed here. The intra- and inter-UTR assays also showed 18 and 29% occupancy, respectively, in FRET states consistent with the ED, suggesting that the ED can form under the PI buffer condition. These occupancies are summarized in Fig. 7 and Table S1. A minority of molecules were found in intermediate-FRET states (6 and 2% for the intra- and inter-UTR assays, respectively), which occasionally showed evidence of dynamics (Fig. 5 F). These may reflect molecules that are misfolded or have adopted unidentified intermediate conformations. Dimerization of NL4.3-5′UTR238 in PI buffer at elevated Mg2+ (5 mM) led to an increase in high FRET occupancy in the intra-UTR assay (69%) and a corresponding increase in low FRET occupancy in the inter-UTR assay (65%) (Figs. 6, C and D and 7). These data are consistent with elevated Mg2+ modestly promoting transition to the ED conformation. Given the anticipated extent of refolding necessary to transition to the ED conformation, we next asked whether dimerization at elevated temperature (57°C) in PI buffer would induce transition to the ED. The intra-UTR assay indicated 69% occupancy in the low-FRET state, whereas the inter-UTR assay indicated 51% occupancy in the high-FRET state (Figs. 6, E and F and 7). Both assays are consistent with elevated temperature promoting transition to the ED conformation. The 30% occupancy in the low-FRET state seen in the inter-UTR assay may indicate some heat-induced refolding into an off-pathway dimer conformation. Finally, given the modest stabilization of the NL4.3-5′UTR238 dimer seen after dimerization in the presence of cognate NCNL4.3 in the FCCS assay, we asked whether NCNL4.3 would similarly promote transition to the ED conformation. As expected, dimerization in the presence of NCNL4.3 in PI buffer at 37°C led to an increase in the low FRET occupancy in the intra-UTR assay (73%) and an increase in high FRET occupancy in the inter-UTR assay (60%; Figs. 6, G and H and 7). Both observations are consistent with NCNL4.3 promoting transition to the ED conformation. In contrast, NL4.3-5′UTR238 dimerization with NCNL4.3 at 5 mM Mg2+ failed to induce transition to the ED (Figs. 7 and S6, A and B). Taken together, these data demonstrate that under PI buffer conditions, NL4.3-5′UTR238 forms a stable dimer in the KD conformation, and transition to the ED conformation is promoted by NCNL4.3 and elevated temperature. Both assays consistently showed evidence of minor populations of ∼0-FRET and high-S molecules indicative of unannealed DNA handle, incompletely labeled RNA, or 5′UTR monomers in the case of the inter-UTR assay (Fig. 7; Table S1).
Figure 6.
smFRET analysis of the 5′UTR dimer from the NL4.3 strain. (A) Bivariate histogram (ES plot) for the NL4.3-5′UTR238 dimer, formed in PI buffer at 1 mM Mg2+ for 1 h at 37°C and acquired at room temperature with the intra-UTR FRET assay. The histograms are normalized to the total number of data points included in each data set. The probability per histogram bin is displayed according to the indicated colormap, with red indicating the highest probability. Dotted lines indicate the thresholds used for identification of subpopulations of molecules, as defined in Fig. 5. The sizes of the subpopulations are summarized in Fig. 7. Also shown are univariate histograms displaying the distributions of FRET efficiency (plotted along the top horizontal axis) and stoichiometry (plotted along the right vertical axis). (B) Equivalent data, displayed in the same manner, obtained with the inter-UTR assay. (C) ES plot for NL4.3-5′UTR238 dimerized and imaged in PI buffer with 5 mM Mg2+ with the intra-UTR assay and (D) the same data obtained with the inter-UTR assay. (E) ES plot for NL4.3-5′UTR238 dimerized and PI buffer at 1 mM Mg2+ for 1 h at 57°C, followed by imaging at room temperature with the intra-UTR assay and (F) the same data obtained with the inter-UTR assay. (G) Intra-UTR FRET data for NL4.3-5′UTR238 dimerized in the presence of cognate NCNL4.3 in PI buffer at 1 mM Mg2+ for 1 h at 37°C. (H) The same data obtained with the inter-UTR FRET assay. To see this figure in color, go online.
Figure 7.
Summary of the NL4.3-5′UTR238 conformation occupancies. The occupancies in the observed NL4.3-5′UTR238 conformations are displayed in pie graphs. The results of all smFRET experiments displayed in Fig. 6, performed with both the intra-UTR and inter-UTR assays, are shown. Conformations were identified according to FRET efficiency and S-value and are abbreviated as follows: KD, kissing dimer; ED, extended dimer; DCM, dimer-competent monomer. The occupancies were calculated as the number of FRET observations in high or low FRET, as indicated, divided by the total number of FRET observations and displayed as a percentage. In the intra-UTR assay, “Donor only” molecules arise from incomplete labeling of the RNA and from unannealed DNA handle. In the inter-UTR assay, the “Donor only” population arises additionally from 5′UTR monomers. smFRET trajectories that displayed intermediate-FRET values that are inconsistent with the identified conformations were not assigned (NA). These data are also presented in Table S1. To see this figure in color, go online.
Stabilization of the MAL-5′UTR dimer by NC
We next evaluated the conformation formed by MAL-5′UTR254 using our smFRET assays. When MAL-5′UTR254 was dimerized in PI buffer at 37°C, the intra-UTR assay yielded 48% occupancy in the high-FRET state (Fig. 8 A). The FCCS assay indicated that the MAL-5′UTR254 dimer dissociated rapidly under PI buffer conditions (Fig. 3 C, 1 mM Mg2+). Therefore, the high-FRET state predominantly reflects the DCM conformation, which is indistinguishable from the KD in the intra-UTR FRET assay. The inter-UTR assay yielded 79% occupancy in an ∼0-FRET high-S state, which indicates molecules with only a donor fluorophore (Fig. 8 B). Consistent with the intra-UTR assay result, this subpopulation likely consists mainly of 5′UTR monomers. However, a portion may also arise because of dissociation of the 5′UTR from the LD550-labeled DNA handle, which readily binds the surface. This likely explains why higher occupancy in the 0-FRET high-S state was seen in the inter-UTR assay as compared to high-FRET intermediate-S occupancy in the intra-UTR assay. Both assays showed a minor population of molecules in FRET states consistent with the ED, suggesting that, like NL4.3-5′UTR238, MAL-5′UTR254 can form the ED during incubation in PI buffer. Additionally, some of the 24% occupancy in low-FRET intermediate S seen in the intra-UTR assay may reflect a DIM, as previously reported (20), which would further contribute to the 79% 0-FRET high-S occupancy in the inter-UTR assay. These occupancies are summarized in Fig. 9 and Table S1. We next asked whether dimerization at elevated Mg2+ would stabilize the dimers in the KD conformation or promote further transition to the ED. Consistent with the FCCS data that indicated modest stabilization of the MAL-5′UTR254 dimer at 5 mM Mg2+ (Fig. 3 C), both smFRET assays indicated an increase in the occupancy in the KD conformation (Figs. 8, C and D and 9). When dimerization of MAL-5′UTR254 was performed in PI buffer at 57°C, a similar effect was seen (Figs. 8, E and F and 9). When viewed alongside the FCCS data, the smFRET experiments demonstrate that MAL-5′UTR254 can form an ED but that it reflects a minor proportion of the overall population of 5′UTR molecules. In contrast to the NL4.3-5′UTR238 dimer, elevated temperature did not promote transition of MAL-5′UTR254 to the ED conformation.
Figure 9.
Summary of the MAL-5′UTR254 conformation occupancies. The occupancies in the observed MAL-5′UTR254 conformations are displayed in pie charts. The results of all smFRET experiments shown in Figs. 8 and 10, performed with both the intra-UTR and inter-UTR assays, are shown. Conformations were identified according to FRET efficiency and S-value and are abbreviated as follows: KD, kissing dimer; ED, extended dimer; DCM, dimer-competent monomer. The occupancies were calculated as the number of FRET observations in high or low FRET, as indicated, divided by the total number of FRET observations and displayed as a percentage. In the intra-UTR assay, “Donor only” molecules arise from incomplete labeling of the RNA and from unannealed DNA handle. In the inter-UTR assay, the “Donor only” population arises additionally from 5′UTR monomers. smFRET trajectories that displayed intermediate-FRET values that are inconsistent with the identified conformations were not assigned (NA). These data are also presented in Table S1. To see this figure in color, go online.
Given the FCCS results demonstrating stabilization of the MAL-5′UTR254 by NCMAL, we asked whether NCMAL stabilized the KD conformation or promoted transition to the ED conformation as seen for NL4.3-5′UTR238. We therefore evaluated the conformation of MAL-5′UTR254 dimers formed in PI buffer in the presence of NCMAL. In this case, the intra-UTR assay yielded 71% occupancy in the low-FRET state (Figs. 8 G and 9). The inter-UTR assay yielded 69% occupancy in the high-FRET state (Figs. 8 F and 9). Thus, both assays indicated formation of the MAL-5′UTR254 ED conformation in the presence of NCMAL. Like NL4.3-5′UTR238, NCMAL failed to promote transition to the MAL-5′UTR254 ED conformation in the presence of elevated Mg2+ (Figs. 9 and S6, C and D). When viewed alongside the FCCS data indicating some persistent dissociation in 1 mM Mg2+ of MAL-5′UTR254 dimers formed in the presence of NCMAL, these observations demonstrate that the MAL-5′UTR254 dimer in the ED conformation fails to achieve the same level of stability seen for NL4.3-5′UTR238.
Neomycin stabilizes the MAL-5′UTR KD
Previous studies demonstrated that aminoglycosides bind at the intermolecular DIS interface and stabilize the 5′UTR dimer from HIV-1 subtype A strains (48,66). We therefore sought to determine whether neomycin promotes transition to the ED conformation or stabilizes dimers in the KD conformation, as suggested by chemical footprinting experiments (48,66). Analysis of the stability of MAL-5′UTR254 dimers formed in the presence of varying concentrations of neomycin by nondenaturing gel electrophoresis confirmed the formation of a dimer resistant to dissociation in the absence of Mg2+ (Fig. 10 A). Similarly, our FCCS assay indicated that MAL-5′UTR254 dimers formed in PI buffer in the presence of 10 μM neomycin remained stable over the 1-h time course when diluted into water and across a range of Mg2+ concentrations (1–5 mM; Fig. 10 B). We next evaluated the conformation of MAL-5′UTR254 dimers formed in PI buffer in the presence of 10 μM neomycin using our smFRET imaging assays. The intra-UTR assay indicated 66% occupancy in the high-FRET state, whereas the inter-UTR assay yielded 79% occupancy in the low-FRET state (Fig. 10, C and D). That is, both assays indicated efficient formation and stabilization of the KD conformation and minimal transition to the ED. The FCCS data and the agreement between the results generated with the smFRET assays are consistent with the MAL-5′UTR254 dimer remaining stable in the KD conformation over the course of our experiments in the presence of neomycin.
Figure 10.
Stabilization of the MAL-5′UTR dimer by neomycin. (A) Nondenaturing gel electrophoresis of the indicated RNAs in TB buffer. Samples were dimerized in PI buffer for 1 h at 37°C with 0–10 μM neomycin. Dimer (D) and monomer (M) species are indicated. (B) FCCS data displayed as in Fig. 3. MAL-5′UTR254 was dimerized as in (A) in the presence of 10 μM neomycin. The presence of neomycin was maintained during acquisition of FCCS data. (C) ES plot for the MAL-5′UTR254 dimer, formed and imaged in PI buffer at 1 mM Mg2+ with 10 μM neomycin and acquired with the intra-UTR FRET assay. (D) Inter-UTR FRET data for MAL-5′UTR254 dimerized and imaged in PI buffer with 1 mM Mg2+ and 10 μM neomycin. ES plots and univariate histograms are displayed as in Figs. 6 and 8. To see this figure in color, go online.
Discussion
Previous studies support the mechanistic model in which 5′UTR dimerization is initiated when two 5′UTR molecules in the DCM conformation form intermolecular basepairing at the DIS sequence, giving rise to the KD conformation (11,21, 22, 23, 24, 25, 26, 27). The spontaneous extension of the dimer interface to form the stable ED conformation was observed by NMR for the subtype B NL4.3 5′UTR (30). No such extension of the dimer interface was reported in an NMR study of the subtype A MAL 5′UTR (19). To date, only studies of short oligonucleotides reflective of 5′UTR elements from MAL have suggested the existence of an ED conformation (28,43,44). The data summarized in Figs. 7 and 9 and tabulated in Table S1 demonstrate that 5′UTR dimers from subtype B HIV-1 strains adopt a stable KD conformation in the absence of NC, whereas subtype A strains dissociate rapidly from the KD conformation. The differences in stability between the two strains can be explained by their characteristic DIS sequences, rather than by accessing distinct dimer conformations. We find that both 5′UTRs can transition to a stable ED conformation in the presence of NC (Figs. 7 and 9). Thus, the chaperone activity of NC enables refolding of the 5′UTR into a thermodynamically favorable state (5,67,68). Our results promote the generality of the model shown in Fig. 11 A and indicate that 5′UTRs from both subtypes A and B dimerize by a similar mechanism and have access to comparable conformations (11,21, 22, 23, 24, 25, 26, 27).
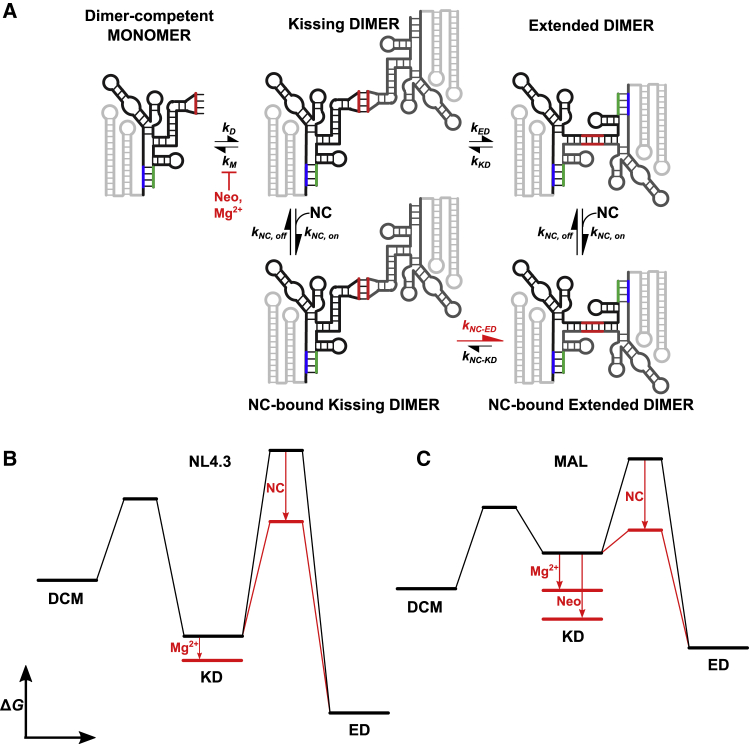
Figure 11.
Kinetic and energetic model of 5′UTR dimer stability and conformation. (A) Kinetic model describing 5′UTR dimer conformational changes. According to this model, the DCM and KD exist in a dynamic equilibrium mediated by forming and breaking intermolecular DIS-DIS interactions. Based on our FCCS experiments, the rate of dimer dissociation (kM) for NL4.3-5′UTR is slower than for MAL-5′UTR. Neomycin and Mg2+ stabilize the MAL-5′UTR dimer, reducing the rate of dimer dissociation (kM) (highlighted in red). Spontaneous exchange between the KD and ED conformations (kKD and kED) is slow in the absence of NC. NC binding increases the rate of transition to the ED (kNC-ED > kED) (highlighted in red). (B) Proposed energy landscape determining NL4.3-5′UTR dimer conformational changes. Physiological Mg2+ has a modest stabilizing effect on the KD conformation. NC facilitates access to the thermodynamically stable ED by reducing the activation energy for the KD-to-ED transition. (C) The equivalent energy landscape governing the MAL-5′UTR dimer conformations. The KD conformation of MAL-5′UTR is less stable than for NL4.3-5′UTR. Neomycin and Mg2+ stabilize the MAL-5′UTR KD conformation. Here again, NC facilitates access to the thermodynamically stable ED conformation. To see this figure in color, go online.
Although we find that the model shown in Fig. 11 A applies equally well to both strains studied here, differences in the thermodynamics of the states and the kinetics of transition are apparent. The data presented are consistent with the NL4.3-5′UTR238 forming a stable KD conformation in the absence of NC, which is insensitive to the presence of elevated Mg2+ (Fig. 7). Our smFRET data indicate minimal occupancy in the ED conformation at physiological Mg2+ and only a modest increase in ED occupancy at elevated Mg2+. We therefore conclude that on the timescale of our incubation and measurement, NL4.3-5′UTR238 remains predominantly in the KD conformation, which is stable under PI buffer conditions. At present, we cannot rule out the possibility that conversion to the ED conformation might occur under these conditions on a longer timescale. The extended timescale of data collection could explain why basepairing consistent with the ED conformation of a 344-nucletide NL4.3-5′UTR construct was detected by NMR (30). Regardless of whether conversion to the ED conformation might require longer incubation, the strength of the intermolecular DIS-DIS basepairing in the KD conformation is sufficient to explain the observed stability of the NL4.3-5′UTR dimer.
Previous studies have demonstrated that the 5′UTR of the subtype A MAL strain forms a labile dimer that requires Mg2+ to remain intact (19,21,50,51). Our study supports these prior observations and specifies that the instability of the MAL-5′UTR dimer is due to the DIS sequence, which is distinct from that of NL4.3. The data presented further demonstrate that the rapid dissociation of the MAL-5′UTR dimer, as compared to the NL4.3-5′UTR, is due to the instability of the KD conformation. The distinct stability phenotypes are not due to differing levels of intrinsic access to the stable ED conformation; neither strain forms the stable ED conformation to a significant extent in the absence of NC under the conditions tested here. Consistent with previous studies, we find that the aminoglycoside antibiotic neomycin stabilizes the MAL-5′UTR dimer (66). Our smFRET data specify that this is due to stabilization of the KD conformation, presumably because of binding at the DIS-DIS interface. Despite the stability of the dimer in the presence of neomycin, no appreciable conversion to the ED conformation was seen. Elevated Mg2+ similarly increases the stability of the KD conformation without promoting conversion to the ED. Thus, stabilization of the MAL-5′UTR dimer in the KD conformation does not induce conversion to the ED conformation. The different rates of dissociation of the 5′UTR dimer in the KD conformation provide a mechanistic explanation for the phenotypic difference in 5′UTR dimer stability seen for the MAL and NL4.3 strains, rather than distinct intrinsic access to the stable ED conformation. In both strains, dimer stability can be achieved by stabilizing the KD conformation without promoting transition to the ED conformation.
The data presented here demonstrate that dimerization in the presence of NC promotes conversion of both NL4.3-5′UTR238 and MAL-5′UTR254 to the ED conformation. The observation that elevated temperature also promotes conversion of the NL4.3-5′UTR238 dimer to the ED conformation indicates that a high activation energy limits access to the ED conformation; elevated temperature increases the probability that the 5′UTR transits the activation energy without the assistance of NC. The failure of elevated temperature to promote ED formation for MAL is consistent with the activation energy for dimer dissociation in the KD conformation being significantly smaller than the activation energy for transition to the ED. The mechanism of NC can be described as reducing the activation energy for transition to the ED conformation (Fig. 11 B). Previous calorimetry measurements showed that NC engages in both high- and low-affinity binding to the 5′UTR (16,34). Elevated Mg2+ prevented low-affinity, nonspecific NC binding to the 5′UTR and unfolding of the RNA (13,16). Consistent with these results, we find here that NC has no effect on the conformation of either 5′UTR dimer when incubated at elevated Mg2+ (5 mM). This observation implicates low-affinity, nonspecific binding of NC to the 5′UTR in promoting the conversion from the KD to the ED conformation. Only minimal dissociation of the NL4.3-5′UTR dimer occurs over the course of our FCCS measurements, whether or not NC is included during dimerization. In contrast, dissociation of the MAL-5′UTR dimer persists even after dimerization in the presence of NC, albeit at a significantly reduced rate as compared to dimers formed in the absence of NC. Our smFRET data show that conversion of the MAL-5′UTR to the ED conformation is efficient in the presence of NC. Taken together, these data show that the ED conformation of MAL-5′UTR is reversible on the timescale of our measurements. That is, the instability of the MAL-5′UTR dimer as compared to NL4.3-5′UTR persists in both the KD and ED conformations. Our analysis of RNA dimers isolated from mature virions demonstrates that the greater stability of the NL4.3 dimer as compared to MAL exists in the context of the viral genome as well as isolated 5′UTRs. Further studies are necessary to determine whether this phenotypic difference has implications during the replication of these distinct strains of HIV-1.
It is well established that dimerization of the HIV-1 genome regulates its recognition by NC during virion assembly (69, 70, 71). Dimerization may facilitate formation of a specific three-dimensional arrangement of NC binding sites that support nucleation of assembly. Our observations here provide the further insight that in vitro, NC initially interacts with the 5′UTR in the KD conformation and subsequently triggers conversion to the ED. In vivo, the ED conformation may then provide the optimal substrate for subsequent NC binding necessary to nucleate virion assembly. Alternatively, the conformational transition from the KD to the ED may reflect an event that occurs during maturation of the HIV-1 virion (5). Genomic RNAs isolated from HIV-1 virions and other retroviruses become more stably dimerized after capsid maturation, indicating a maturation-induced change in the intermolecular interface (72, 73, 74, 75, 76). This could be explained by the conversion of the 5′UTR from the KD to the ED conformation induced after NC has been proteolytically liberated from Gag. Whatever the relevance of the ED conformation is to virus replication, our study reinforces the idea that NC acts as a chaperone to manipulate the conformation of the 5′UTR, thus accelerating the transition to the ED conformation. In this regard, NC destabilizes nucleic acid duplexes to facilitate transition to thermodynamically favorable conformations (5,67,68).
Specifying the role of the KD-to-ED conversion during virus replication will require elucidation of the 5′UTR conformation in intact virions or at virus assembly sites. Further studies of 5′UTR RNA in functional complex with viral or host factors, such as the recent description of the reverse-transcription initiation complex assembled on the primer-binding site determined using cryo-electron microscopy (77), are also likely to be informative in this regard. Analysis of the structure of the 5′UTR in the context of the virion has been confined to ex vivo interrogation of RNA isolated from virions either by chemical footprinting assays (78, 79, 80), which informed subsequent computational modeling (81,82), or observations of low-resolution genome ultrastructure by electron microscopy (83). These studies clearly indicated intermolecular DIS-DIS basepairing, but the nature of the 5′UTR conformation was not resolved and thus remains unknown. Extension of smFRET imaging approaches, such as that presented here, to interrogation of RNA structure inside HIV-1 virions may find utility in elucidating the conformation of the 5′UTR or other regions of the genome in its native context.
Conclusions
Here, we have shown that 5′UTR dimers from HIV-1 subtypes A and B adopt a common KD conformation, although the subtype B 5′UTR achieves greater dimer stability. We further show that both 5′UTRs convert to the ED conformation in the presence of the viral NC protein, although the subtype B 5′UTR dimer still demonstrates greater stability. The phenotypic differences in 5′UTR dimer stability are attributable to sequence variation in the DIS stem loops. These results support a unified model in which the genomes of diverse HIV-1 strains dimerize by a common mechanism and adopt an ED in the presence of NC.
Author contributions
R.J.B. conducted all biochemical and smFRET experiments involving the 5′UTR RNA. J.B.M. conducted the FCCS experiments. C.B. and S.K. conducted the experiments involving HIV-1 virions. C.S. expressed and purified the NC protein. R.J.B. and J.B.M. drafted the manuscript. All authors contributed to editing the manuscript.
Acknowledgments
We gratefully acknowledge Dr. Michael Summers for helpful discussion and critical reading of the manuscript. We also thank Dr. Benjamin Brigham for assistance with initial implementation of the FRET assays.
This work was supported by National Institutes of Health grant R21 AI136711 and the Center for HIV RNA Studies (U54 AI150470).
Editor: Mark C. Williams.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.09.017.
Supporting material
References
- 1.Dubois N., Marquet R., et al. Bernacchi S. Retroviral RNA dimerization: from structure to functions. Front. Microbiol. 2018;9:527. doi: 10.3389/fmicb.2018.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rein A. RNA packaging in HIV. Trends Microbiol. 2019;27:715–723. doi: 10.1016/j.tim.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz P., Telesnitsky A. Multiple, switchable protein:RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu. Rev. Virol. 2018;5:165–183. doi: 10.1146/annurev-virology-092917-043448. [DOI] [PubMed] [Google Scholar]
- 4.Pornillos O., Ganser-Pornillos B.K. Maturation of retroviruses. Curr. Opin. Virol. 2019;36:47–55. doi: 10.1016/j.coviro.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rein A., Henderson L.E., Levin J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 6.Seif E., Niu M., Kleiman L. Annealing to sequences within the primer binding site loop promotes an HIV-1 RNA conformation favoring RNA dimerization and packaging. RNA. 2013;19:1384–1393. doi: 10.1261/rna.038497.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coey A., Larsen K., et al. Viani Puglisi E. Heterogeneous structures formed by conserved RNA sequences within the HIV reverse transcription initiation site. RNA. 2016;22:1689–1698. doi: 10.1261/rna.056804.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeras I., Seufzer B., et al. Boris-Lawrie K. The basal translation rate of authentic HIV-1 RNA is regulated by 5′UTR nt-pairings at junction of R and U5. Sci. Rep. 2017;7:6902. doi: 10.1038/s41598-017-06883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbink T.E.M., Berkhout B. RNA structure modulates splicing efficiency at the human immunodeficiency virus type 1 major splice donor. J. Virol. 2008;82:3090–3098. doi: 10.1128/JVI.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huthoff H., Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skripkin E., Paillart J.C., et al. Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paillart J.C., Marquet R., et al. Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 13.Keane S.C., Heng X., et al. Summers M.F. RNA structure. Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence D.C., Stover C.C., et al. Summers M.F. Structure of the intact stem and bulge of HIV-1 Ψ-RNA stem-loop SL1. J. Mol. Biol. 2003;326:529–542. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 15.Ooms M., Huthoff H., et al. Berkhout B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 2004;78:10814–10819. doi: 10.1128/JVI.78.19.10814-10819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu K., Heng X., et al. Summers M.F. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Bel N., Ghabri A., et al. Berkhout B. The HIV-1 leader RNA is exquisitely sensitive to structural changes. Virology. 2015;483:236–252. doi: 10.1016/j.virol.2015.03.050. [DOI] [PubMed] [Google Scholar]
- 18.Kharytonchyk S., Monti S., et al. Summers M.F. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc. Natl. Acad. Sci. USA. 2016;113:13378–13383. doi: 10.1073/pnas.1616627113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown J.D., Kharytonchyk S., et al. Summers M.F. Structural basis for transcriptional start site control of HIV-1 RNA fate. Science. 2020;368:413–417. doi: 10.1126/science.aaz7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brigham B.S., Kitzrow J.P., et al. Munro J.B. Intrinsic conformational dynamics of the HIV-1 genomic RNA 5′UTR. Proc. Natl. Acad. Sci. USA. 2019;116:10372–10381. doi: 10.1073/pnas.1902271116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paillart J.C., Skripkin E., et al. Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muriaux D., Fossé P., Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 23.Mujeeb A., Clever J.L., et al. Parslow T.G. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 24.Ennifar E., Walter P., et al. Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 25.Zhang K., Keane S.C., et al. Chiu W. Structure of the 30 kDa HIV-1 RNA dimerization signal by a hybrid cryo-EM, NMR, and molecular dynamics approach. Structure. 2018;26:490–498.e3. doi: 10.1016/j.str.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darlix J.L., Gabus C., et al. Barré-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 27.Andersen E.S., Jeeninga R.E., et al. Kjems J. Dimerization and template switching in the 5′ untranslated region between various subtypes of human immunodeficiency virus type 1. J. Virol. 2003;77:3020–3030. doi: 10.1128/JVI.77.5.3020-3030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mundigala H., Michaux J.B., et al. Rueda D. HIV-1 DIS stem loop forms an obligatory bent kissing intermediate in the dimerization pathway. Nucleic Acids Res. 2014;42:7281–7289. doi: 10.1093/nar/gku332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girard F., Barbault F., et al. Lancelot G. Dimer initiation sequence of HIV-1Lai genomic RNA: NMR solution structure of the extended duplex. J. Biomol. Struct. Dyn. 1999;16:1145–1157. doi: 10.1080/07391102.1999.10508323. [DOI] [PubMed] [Google Scholar]
- 30.Keane S.C., Van V., et al. Summers M.F. NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc. Natl. Acad. Sci. USA. 2016;113:13033–13038. doi: 10.1073/pnas.1614785113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J., Nikolaitchik O., et al. Hu W.-S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. USA. 2009;106:13535–13540. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore M.D., Fu W., et al. Hu W.-S. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 2007;81:4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin M.P.S., Rhodes T.D., et al. Hu W.-S. Identification of a major restriction in HIV-1 intersubtype recombination. Proc. Natl. Acad. Sci. USA. 2005;102:9002–9007. doi: 10.1073/pnas.0502522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heng X., Kharytonchyk S., et al. Summers M.F. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 2012;417:224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kharytonchyk S., Brown J.D., et al. Telesnitsky A. Influence of gag and RRE sequences on HIV-1 RNA packaging signal structure and function. J. Mol. Biol. 2018;430:2066–2079. doi: 10.1016/j.jmb.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolaitchik O.A., Somoulay X., et al. Hu W.-S. Unpaired guanosines in the 5′ untranslated region of HIV-1 RNA act synergistically to mediate genome packaging. J. Virol. 2020;94:e00439-20. doi: 10.1128/JVI.00439-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding P., Kharytonchyk S., et al. Summers M.F. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc. Natl. Acad. Sci. USA. 2020;117:17737–17746. doi: 10.1073/pnas.2008519117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb J.A., Jones C.P., et al. Musier-Forsyth K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging. RNA. 2013;19:1078–1088. doi: 10.1261/rna.038869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Guzman R.N., Wu Z.R., et al. Summers M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y., Nikolaitchik O.A., et al. Hu W.-S. HIV-1 sequence necessary and sufficient to package non-viral RNAs into HIV-1 particles. J. Mol. Biol. 2017;429:2542–2555. doi: 10.1016/j.jmb.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comas-Garcia M., Datta S.A., et al. Rein A. Dissection of specific binding of HIV-1 Gag to the ‘packaging signal’ in viral RNA. eLife. 2017;6:4304. doi: 10.7554/eLife.27055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muriaux D., De Rocquigny H., et al. Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 43.Rist M.J., Marino J.P. Mechanism of nucleocapsid protein catalyzed structural isomerization of the dimerization initiation site of HIV-1. Biochemistry. 2002;41:14762–14770. doi: 10.1021/bi0267240. [DOI] [PubMed] [Google Scholar]
- 44.Mihailescu M.-R., Marino J.P. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc. Natl. Acad. Sci. USA. 2004;101:1189–1194. doi: 10.1073/pnas.0307966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Y.X., Copeland T.D., et al. Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baba S., Takahashi K., et al. Kawai G. Role of the zinc fingers of HIV-1 nucleocapsid protein in maturation of genomic RNA. J. Biochem. 2003;134:637–639. doi: 10.1093/jb/mvg200. [DOI] [PubMed] [Google Scholar]
- 47.Ennifar E., Dumas P. Polymorphism of bulged-out residues in HIV-1 RNA DIS kissing complex and structure comparison with solution studies. J. Mol. Biol. 2006;356:771–782. doi: 10.1016/j.jmb.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 48.Ennifar E., Paillart J.-C., et al. Walter P. HIV-1 RNA dimerization initiation site is structurally similar to the ribosomal A site and binds aminoglycoside antibiotics. J. Biol. Chem. 2003;278:2723–2730. doi: 10.1074/jbc.M205726200. [DOI] [PubMed] [Google Scholar]
- 49.Jossinet F., Paillart J.-C., et al. Marquet R. Dimerization of HIV-1 genomic RNA of subtypes A and B: RNA loop structure and magnesium binding. RNA. 1999;5:1222–1234. doi: 10.1017/s1355838299990982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paillart J.C., Westhof E., et al. Marquet R. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J. Mol. Biol. 1997;270:36–49. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- 51.Lodmell J.S., Paillart J.-C., et al. Marquet R. Oligonucleotide-mediated inhibition of genomic RNA dimerization of HIV-1 strains MAL and LAI: a comparative analysis. Antisense Nucleic Acid Drug Dev. 1998;8:517–529. doi: 10.1089/oli.1.1998.8.517. [DOI] [PubMed] [Google Scholar]
- 52.Hermanson G. Second Edition. Elsevier; Amsterdam: 2008. Bioconjugate Techniques. [Google Scholar]
- 53.Jones C.P., Datta S.A.K., et al. Musier-Forsyth K. Matrix domain modulates HIV-1 Gag’s nucleic acid chaperone activity via inositol phosphate binding. J. Virol. 2011;85:1594–1603. doi: 10.1128/JVI.01809-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weidemann T., Schwille P. Dual-color fluorescence cross-correlation spectroscopy with continuous laser excitation in a confocal setup. Methods Enzymol. 2013;518:43–70. doi: 10.1016/B978-0-12-388422-0.00003-0. [DOI] [PubMed] [Google Scholar]
- 55.Naldini L., Blömer U., et al. Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 56.Boussif O., Lezoualc’h F., et al. Behr J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flynn J.A., An W., et al. Telesnitsky A. Nonrandom dimerization of murine leukemia virus genomic RNAs. J. Virol. 2004;78:12129–12139. doi: 10.1128/JVI.78.22.12129-12139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dave R., Terry D.S., et al. Blanchard S.C. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 2009;96:2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aitken C.E., Marshall R.A., Puglisi J.D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Juette M.F., Terry D.S., et al. Blanchard S.C. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods. 2016;13:341–344. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenna S.A., Kim I., et al. Puglisi J.D. Purification and characterization of transcribed RNAs using gel filtration chromatography. Nat. Protoc. 2007;2:3270–3277. doi: 10.1038/nprot.2007.480. [DOI] [PubMed] [Google Scholar]
- 62.Laughrea M., Jetté L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 63.Tran T., Liu Y., et al. Summers M.F. Conserved determinants of lentiviral genome dimerization. Retrovirology. 2015;12:83. doi: 10.1186/s12977-015-0209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwille P., Meyer-Almes F.J., Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kapanidis A.N., Lee N.K., et al. Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ennifar E., Paillart J.-C., et al. Marquet R. Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell. Nucleic Acids Res. 2006;34:2328–2339. doi: 10.1093/nar/gkl317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turner K.B., Hagan N.A., Fabris D. Understanding the isomerization of the HIV-1 dimerization initiation domain by the nucleocapsid protein. J. Mol. Biol. 2007;369:812–828. doi: 10.1016/j.jmb.2007.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cruceanu M., Urbaneja M.A., et al. Williams M.C. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nikolaitchik O.A., Dilley K.A., et al. Hu W.-S. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013;9:e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakuragi J., Shioda T., Panganiban A.T. Duplication of the primary encapsidation and dimer linkage region of human immunodeficiency virus type 1 RNA results in the appearance of monomeric RNA in virions. J. Virol. 2001;75:2557–2565. doi: 10.1128/JVI.75.6.2557-2565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakuragi J., Sakuragi S., Shioda T. Minimal region sufficient for genome dimerization in the human immunodeficiency virus type 1 virion and its potential roles in the early stages of viral replication. J. Virol. 2007;81:7985–7992. doi: 10.1128/JVI.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ohishi M., Nakano T., et al. Sakuragi J. The relationship between HIV-1 genome RNA dimerization, virion maturation and infectivity. Nucleic Acids Res. 2011;39:3404–3417. doi: 10.1093/nar/gkq1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song R., Kafaie J., et al. Laughrea M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 2007;371:1084–1098. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 74.Fu W., Gorelick R.J., Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu W., Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jalalirad M., Laughrea M. Formation of immature and mature genomic RNA dimers in wild-type and protease-inactive HIV-1: differential roles of the Gag polyprotein, nucleocapsid proteins NCp15, NCp9, NCp7, and the dimerization initiation site. Virology. 2010;407:225–236. doi: 10.1016/j.virol.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 77.Larsen K.P., Mathiharan Y.K., et al. Puglisi E.V. Architecture of an HIV-1 reverse transcriptase initiation complex. Nature. 2018;557:118–122. doi: 10.1038/s41586-018-0055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paillart J.-C., Dettenhofer M., et al. Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 79.Watts J.M., Dang K.K., et al. Weeks K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilkinson K.A., Gorelick R.J., et al. Weeks K.M. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lavender C.A., Gorelick R.J., Weeks K.M. Structure-based alignment and consensus secondary structures for three HIV-related RNA genomes. PLoS Comput. Biol. 2015;11:e1004230. doi: 10.1371/journal.pcbi.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sükösd Z., Andersen E.S., et al. Kjems J. Full-length RNA structure prediction of the HIV-1 genome reveals a conserved core domain. Nucleic Acids Res. 2015;43:10168–10179. doi: 10.1093/nar/gkv1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Höglund S., Ohagen A., et al. Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.