Abstract
Purpose:
High-dose-rate (HDR) prostate brachytherapy involves treatment catheter placement, which is currently empirical and physician dependent. The lack of proper catheter placement guidance during the procedure has left the physicians to rely on a heuristic thinking-while-doing technique, which may cause large catheter placement variation and increased plan quality uncertainty. Therefore, the achievable dose distribution could not be quantified prior to the catheter placement. To overcome this challenge, we proposed a learning-based method to provide HDR catheter placement guidance for prostate cancer patients undergoing HDR brachytherapy.
Methods:
The proposed framework consists of deformable registration via registration network (Reg-Net), multi-atlas ranking and catheter regression. To model the global spatial relationship among multiple organs, binary masks of the prostate and organs-at-risk are transformed into distance maps which describe the distance of each local voxel to the organ surfaces. For a new patient, the generated distance map is used as fixed image. Reg-Net is utilized to deformably register the distance maps from multi-atlas set to match this patient’s distance map and then bring catheter maps from multi-atlas to this patient via spatial transformation. Several criteria, namely prostate volume similarity, multi-organ semantic image similarity and catheter positions criteria (far from the urethra and within the partial prostate), are used for multi-atlas ranking. The top-ranked atlas’ deformed catheter positions are selected as the predicted catheter position for this patient. Finally, catheter regression is used to refine the final catheter positions. A retrospective study on 90 patients with a five-fold cross validation scheme was used to evaluate the proposed method’s feasibility. In order to investigate the impact of plan quality from the predicted catheter pattern, we optimized the source dwell position and time for both the clinical catheter pattern and predicted catheter pattern with the same optimization settings. Comparisons of clinically relevant dose volume histogram (DVH) metrics were completed.
Results:
For all patients, on average, both the clinical plan dose and predicted plan dose meet the common dose constraints when prostate dose coverage is kept at V100 = 95%. The plans from predicted catheter pattern have slightly higher hotspot in terms of V150 by 5.0% and V200 by 2.9% on average. For bladder V75, rectum V75 and urethra V125, the average difference is close to zero, and the range of most patients is within ±1 cc.
Conclusion:
We developed a new catheter placement prediction method for HDR prostate brachytherapy based on a deep-learning-based multi-atlas registration algorithm. It has great clinical potential since it can provide catheter location estimation prior to catheter placement, which could reduce the dependence on physicians’ experience in catheter implantation and improve the quality of prostate HDR treatment plans. This approach merits further clinical evaluation and validation as a method of quality control for HDR prostate brachytherapy.
Keywords: HDR, catheter, deep learning, prostate
1. INTRODUCTION
High-dose-rate (HDR) brachytherapy has been widely practiced as a standard treatment option for localized prostate cancer since the 1990s.1,2 In prostate HDR brachytherapy, 10 to 18 catheters are interstitially implanted to cover the prostate gland using transrectal ultrasound (TRUS) guidance.3,4 After catheter implantation and CT simulation, source dwell position and duration are optimized by a treatment planning system, aiming to deliver conformal dose encompassing the whole gland while minimizing hot spots and sparing organs-at-risk. Clinically, prostate HDR is favored for patients with disease confined to the prostate or immediate surrounding tissue, and it serves as either a boost treatment combined with external beam radiation therapy (EBRT) or a standalone therapy (monotherapy), depending on the cancer risk types.5-7
Among the multiple steps of HDR brachytherapy, catheter placement is one of the important steps that directly impact the plan quality and potential treatment outcome. Currently, the catheter positions are selected by the radiation oncologist or urologist based on a standard implantation pattern using the largest prostate axial cross section as reference view.8 However, the standard pattern may not be optimal for all patients due to the large variation in prostate size, shape and distance to organs-at-risk (OARs).9 Brachytherapists have adopted a heuristic method to adapt catheter positions in real-time based on individual patient anatomy. This important step is often rushed due to the time-pressure associated with the operating room schedule and anesthetized patient. This empirical process introduces uncertainty and variation in catheter placement among different operators, which adversely impacting HDR plan quality consistency and standardization. Additionally, it is difficult to assess the plan quality quantitatively without prior knowledge of catheter positions. Thus, it is desirable to have a high quality patient-specific catheter position map available immediately prior to catheter insertion. This type of guidance can help physicians evaluate the achievable dose distribution prior to needle insertion, and minimize catheter placement variation.
The improvement of HDR prostate brachytherapy plans has been an active area of research in recent years. However, most of these studies focused on optimizing the source dwell time on the empirical catheter placement.10-12 Optimal catheter placement prediction combined with the source dwell time optimization is a novel promising strategy to improve the current HDR procedures, which has been rarely investigated in the literature.13 Thus, an efficient, automated and clinically feasible catheter placement prediction method is desirable.14
In this study, we propose a new learning-based method to provide patient specific HDR catheter position pattern. The patient specific catheter pattern is predicted from a multi-atlas dataset using deep learning-based deformable registration and is expected to generate an HDR plan with similar quality of physicians’ empirical catheter placement. The proposed framework consists of three major steps, which are (1) deformable registration15 via registration network (Reg-Net), (2) multi-atlas ranking and (3) catheter regression. To model the global spatial relationship among multiple organs, binary masks of the target and organs at risk (OAR) were transformed into distance maps which describe the distance of each local voxel to the organ surfaces. Then, Reg-Net is utilized to deformably register the distance maps and contours of multi-atlas to match those of an arrival patient. By multi-atlas ranking and spatial transformation, the corresponding catheter locations of top-ranked multiple atlases are registered and fused to generate synthetic catheter locations of an arrival patient. The final catheter positions of this patient are then regressed from the deformed catheter locations. To evaluate our proposed method, we retrospectively investigated 90 prostate cancer patients who were treated with HDR brachytherapy. HDR plans were optimized for both the predicted and clinical catheter placement with same optimization parameters, and were normalized to the same prostate dose coverage for dosimetric comparison.
2. METHODS AND MATERIALS
2.A. Method overview
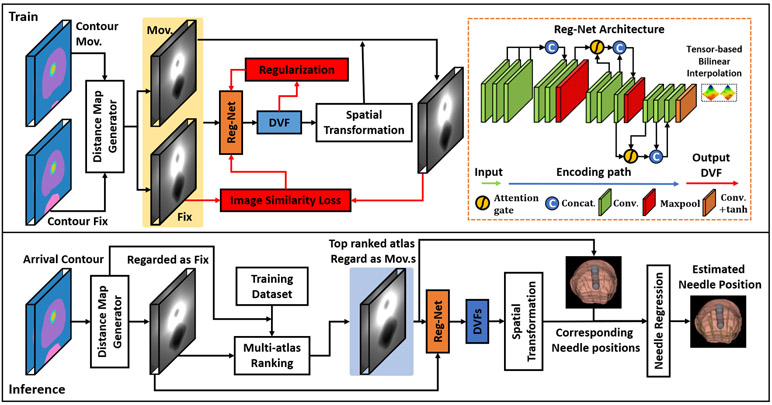
Figure 1 outlines the schematic flow chart of this prediction process. During training, a Reg-Net is trained without the need of ground truth deformation vector field (DVF) under several supervision mechanisms that perform learnable parameters optimization. The Reg-Net is implemented via a 3D CNN. Binary organ masks contain no image gradient within the organ, which cannot be directly used to drive the registration. We thus propose to generate organ distance maps by encoding the binary masks with distance from the organ surface. Voxels within a specific organ were encoded with negative distance values while voxels outside the organ were encoded with positive distance values. Distance maps of the prostate and OARs were concatenated as the input. The organ distance maps of the atlas and the patient are registered using the proposed Reg-Net. During inference, the resultant DVF of the registration is used to deform the atlas catheter locations to predict the patient catheter locations.
Figure 1.
Schematic flow chart of the proposed method for catheter position prediction. The first row shows the training procedure and the network architecture used in this work. The second row shows the inference procedure.
2.B. Multi-atlas ranking
For a new arrival patient whose multi-organ contours and generated distance map are used as fixed data, our goal is to select its multi-atlas data and bring the catheter maps of these data to match the new arrival patient’s data. Situation similarity is used as ranking criterion for multi-atlas ranking. In this work, the situation similarity is composed of three criteria. Firstly, we assume that patients’ data with similar volume size of organs (urethra, prostate and bladder) should have similar number of inserted catheters. Thus, the first criterion is the contour volume. From atlas data, based on contour volume size, we first select top k patients’ data as rough multi-atlas dataset.
Secondly, image similarity between new patient’s semantic image of contours and that from rough multi-atlas dataset are used as second criterion to pick top m ≤ k patients’ data as fine multi-atlas data. In order to assess the similarity between the new patient’s image and the multi-atlas datasets, initial rigid registration was necessary to roughly align the organs in the datasets. A supervised deep learning-based warping method is used to rapidly align the multi-atlas dataset to new patient’s data. This model is a spatial transformer network but implemented via 3D. The details of the network is introduced in 16. The model took multi-atlas dataset and new patient’s dataset as input and output the parameters that can perform affine registration that match the manual contour of multi-atlas data to the new patient’s data. The DSC calculated between deformed contour and new patient’s contour is used to training this network. The aligned multi-atlas datasets were then used to calculate the similarity and choose the top m atlases. In our work, the image distance map of atlas data and the new patient’s data is used as moving and fixed input image, respectively. The warped OAR contours of atlas data and new patient’s data are used for label similarity measurement, which is a loss term to supervise the network. The output of this network is the estimated parameters of rigid registration. After registration, NCC is used to compare the similarity between rough multi-atlas dataset’s semantic images and new patient’s semantic image. Since the number of catheters should be correlated to the volume size of prostate, the prostate volume size similarity is also used as similarity measurement for ranking.
Finally, we perform deformable registration based on the proposed Reg-Net (see details in the next section) to bring semantic image and distance map from fine multi-atlas dataset to match that of new patient. We further use two catheter position criteria for deformed catheters to select the best catheter positions. The catheter positions are expressed as a set of center points of the catheters on each slice. The first aspect is that the catheter position should be far from urethra. The second aspect is that the catheter positions should be within prostate. These two aspects are measured by the distance between center-of-mass of each catheter and surface of urethra and prostate. Based on the distance criterion, we select the deformed catheter location of the top similar atlas as the temporary deformed catheter locations. Note that the temporary deformed catheter locations may be over-bended and not physically achievable due to large deformation. Catheter regression is then performed to restrict the predicted catheter locations on straight lines in 3D volume. Details of this step can be found in section 2.D.
2.C. Reg-Net
Due to the limited number of atlas libraries, it is difficult to find an exact matching atlas to the new patient’s image in terms of prostate and OAR’s size and shape. To enhance the robustness of the proposed method, multiple top-ranking atlases which were selected according to our atlas-selection rules were registered to the new patient. Reg-Net,17 a deep learning-based network that does not need ground truth DVF, is applied to perform deformable registration. Its network architecture is shown in Figure 1. The size of the input image pairs is reduced in the encoding path after several convolutional layers. In order to generate DVFs with the same matrix sizes as the input images, we used a bilinear interpolation to up-sample the DVFs. It is true that transpose-convolution layers with trainable parameters is an alternative for up-sampling. However, we prefer bilinear interpolation with no trainable parameters since it tends to generate smoother DVFs that are more realistic in medical image registration, while transpose-convolution layers often derive unrealistic DVFs even with heavily weighted DVF smoothness regularization term. We also integrate two attention gates, called self-attention strategy, into the generator to highlight informative differencing features between moving and target input. This is achieved by feeding the feature maps of one layer and its previous layer from the encoding path to the attention gate.18 It can facilitate the network to capture structural differences between the moving and fixed images. Our previous studies show that the self-attention strategy allow Reg-Net parameters in shallower layers to be updated based on spatial regions that are most relevant to deformation estimation.17,19
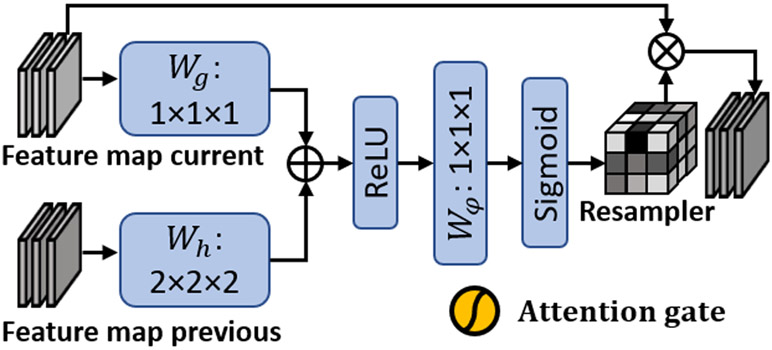
Attention gates have been explored in the context of semantic segmentation in recent studies.20 Previous works demonstrated that the most relevant semantic contextual information can be captured by integrating attention gates into a standard U-Net without a very large reception field 21. In this study, we incorporated attention gates into the design of our Reg-Net. Figure 2 shows the subnetwork architecture of attention gate. The attention gates combined feature maps of adjacent/current layer and previous layer, i.e. the feature maps of different scales. To achieve the same resolution, convolutional layers Wg and Wh with different kernel sizes were used. Then, the two feature maps were added together and passed through an additional convolutional layer Wφ to obtain a matrix, called resampler. The resampler is regarded as a weighted matrix, whose elements correspond to the feature importance of the current feature map. Then, element-wise multiplication between resampler and current feature map is performed to derive the feature map that is weighted via the attention gate.
Figure 2.
Subnetwork architecture of attention gate.
As can be seen from Figure 1, the attention gates operations were performed immediately prior to the concatenation in order to retain only relevant activations and remove irrelevant/noisy responses. Additionally, the attention gates filtered the neuron activations during both the forward pass and the backward pass. Gradients originating from image background regions were down weighted during the backward pass. This allows model parameters in shallower layers to be updated based on spatial regions that are most relevant to a given task, i.e., deformation estimation. Thus, the attention gates could have the ability to highlight the features from previous layers, which can well represent the deformation between the selected atlas patient and test patient.
The accuracy and realism of generated DVF between new patient’s data and multi-atlas patients’ data should rely on the design of supervision, i.e., loss function. In this work, to achieve both accuracy and realism, the loss function consists of two parts which are the data similarity loss and the regularization loss. As discussed above, the moving and fixed data include both semantic image of contours and generated distance map, thus, the data similarity loss include both the similarity of these two kinds of data. For semantic image of contours, Dice loss is used as data similarity loss. For distance map, normalized cross-correlation (NCC) loss is used as data similarity loss. Let denotes a set of the moving semantic image of contours and moving distance map. Let denotes a set of the fixed semantic image of contours and moving distance map, respectively. Let Ψ denotes the Reg-Net, and φ = Ψ(Imov, Ifix) represents the predicted DVF for a moving and target data pair. Then, the data similarity loss Lsim can be represented as follows:
| (1) |
where and denotes the deformed semantic image of contours and deformed distance map, respectively, which were obtained by warping the moving contours and distance map by DVF φ using spatial transformer.22 DSC(∙) denotes the Dice similarity coefficient (DSC) between moving and fixed contours. NCC(∙) denotes the NCC between moving and fixed distance maps. α denotes the balancing parameter between Dice and NCC loss term.
Considering that the network is trained in a completely unsupervised manner, i.e., with no ground truth DVF for supervision,23 we added DVF regularization term to enforce smoothness constraint.. In this work, bending energy, which is a second derivatives of the DVF, is used as DVF regularization.2
| (2) |
In total, the trainable parameters of Reg-Net are optimized via minimizing the similarity and regularization losses:
| (3) |
where β denotes the regularization parameter. Regarding the regularization parameter values, a rule of thumb is that the initial loss terms should be in the same order of magnitude numerically given equal priority. Therefore, we have empirically set the β to be 1.5 and α to be 1.2.
2.D. Catheter Regression
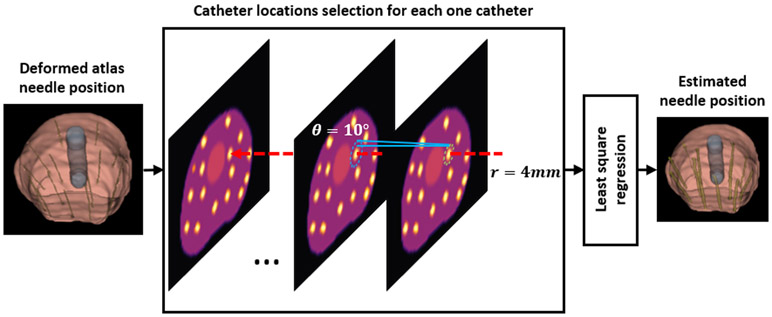
The temporary predicted catheter locations may not be physically achievable due to catheter over-bending and large deformation. Therefore, we need to post-process the predicted catheter locations so that the predicted catheters are straight, separate, and reasonably spaced with respect to each other. To achieve this, circles on the transverse plane with varying radius were drew to group the temporary predicted catheter locations inside these circles as one single catheter. As is shown in Figure 3, for a deformed atlas needle mask image that is derived by Reg-Net, we first set a circle (shown as green dashed circle in Figure 3) that only covers one catheter position on first axial slice and is centered at that catheter position. The radius of this circle is set to r = 4mm. Because we found that the minimum distance between any two catheters’ location on one axial slice is about 8 mm. We then find the most related catheter position on next axial slice. Since the catheter may not be exact perpendicular to the axial slices, we set an angle tolerance θ so that the radius on any two adjacent slices follow r′ = r + d∙tanθ, where θ is set to θ = 10°, d denotes the slice thickness. In addition, these two circles share same center location. By this setting, we can find the catheter location of the next slice if it is located within the blue dashed circle. We can then trace the center locations of the catheter slice by slice. Least square regression method was used to regress the temporary catheter locations on straight lines in 3D volume as the final catheter placement pattern.
Figure 3.
Schematic flow chart of the needle regression step used in the proposed method. The green dashed circle denotes the circle region which centered at one needle position of the most left axial slice of deformed atlas needle mask image. The radius for this circle is set to r = 4mm. Then, for next slice, the searching region is shown by the blue dashed circle. The blue dashed circle has same center with the previous green dashed circle, but with a larger radius r′ = r + d∙tanθ, where θ denotes the angle threshold along z-axis and is set by θ = 10°, d denotes the slice thickness. The red dashed arrow denotes one regressed catheter.
2.E. Evaluation and validation
We retrospectively investigated 90 prostates HDR brachytherapy patients, each of which has clinical delivered treatment plans including catheter positions, CT images and contours of OARs. Twelve to eighteen (depending on prostate size) Nucletron ProGuide Sharp 5F catheters were placed under TRUS guidance. CT images were acquired after catheter implantation by Brilliance Big Bore (Philips, Netherlands) with 140 kVp, 350 mAs, 0.5 mm pixel size and 1 mm slice thickness. On CT images, physicist reconstructed catheters, and radiation oncologist delineated the whole prostate and OARs including bladder, rectum, and urethra. Note that the CT images were not input to our model. The delineated contours were converted to binary maps first, and the binary maps and the distance maps calculated from the binary maps were input to our model. For all patients, both maps were set to the size of 320×320×72 pixels with same pixel resolution as CT images. Both maps were also central cropped and zero-padded with the center set at the center-of-mass of prostate. This step is to align the atlas prostate with the testing patient prostate prior to Reg-Net registration.
A five-fold cross-validation was performed for evaluation. The 90 prostate datasets were first randomly and equally grouped into five subgroups, then, one group was selected as test data and the rest four groups were used as atlas. The experiment was repeated five times to let each subgroup used as test data exactly once. The aim of this five-fold cross-validation is not to determine the number of training epochs to stop the model training. The number of training epochs is determined by how fast the loss curve converged during training. If the loss is converged or reached at maximum training epoch number, the training of Reg-Net is stopped. The aim of the five-fold cross-validation is to let each patient used as test patient such that a Reg-Net model can be trained on the training data with no overlapping with the test patient.
In order to investigate the impact of plan quality from the predicted catheter pattern, we optimized the source dwell position and duration for both the clinical catheter pattern and predicted catheter pattern with the same optimization settings (Prostate: D90>100%, V100>90% of prescribed dose; OAR: Bladder and Rectum V75<1cc, Urethra V125<1cc). The optimization algorithm is inverse planning by simulated annealing (IPSA), which is currently used in Oncentra Brachy (Elekta, Sweden) treatment planning system of our clinic. For a fair comparison, no manual dose adjusting was done after auto optimization, and plan dose for both scenarios was normalized to cover 95% of prostate using prescription dose (V100=95%). The plan dose was quantified by clinically relevant DVH metrics and was compared with each other.
Our algorithm was implemented in Python 3.6.9 and Tensorflow 1.8 with Adam gradient descent optimizer and was trained and tested on a NVIDIA Tesla V100 GPU with 32 GB of memory. The batch size is set to 8. The learning rate is set to 1E-4. The maximum training epoch number is set to 200. We add this information in revised manuscript. We also used several libraries and toolboxes such as numpy, scikit-image, pydicom, h5py, and scipy. In our work, the training of our proposed Reg-Net takes about 7.5 hours. For a new patient, the estimation of catheter placement takes about 5 minutes.
3. RESULTS
To evaluate the registration accuracy of the proposed Reg-Net, we calculate the dice similarity coefficients (DSCs) between the OAR masks of the deformed atlas patient via Reg-net and those of the testing patient. To demonstrate the effectiveness of this deformable registration step, we also calculate the DSCs between the OAR masks of the selected atlas patient before Reg-net and those of the testing patient. These metrics are shown in Table I. As can be seen from the comparison of the row of “atlas vs. test patient” and the row of “deformed atlas vs. test patient (α = 1.2)” (which is the performance of our hyper-parameter setting), before performing Reg-Net, the DSCs between the OAR mask image of the selected atlas patient and test patient are 0.83±0.06 for prostate, 0.43±0.15 for urethra, 0.65±0.18 for bladder, and 0.53±0.18 for rectum. After performing Reg-Net to deform the selected atlas patient to match the testing patient, the DSCs between the OAR mask image of the deformed atlas patient and test patient are improved to 0.95±0.04 for prostate, 0.86±0.07 for urethra, 0.93±0.11 for bladder, and 0.86±0.04 for rectum.
Table I.
Mean±Std of OAR mask DSC between testing patient and atlas patient under different conditions.
| Prostate | Urethra | Bladder | Rectum | |
|---|---|---|---|---|
| Atlas vs. test patient | 0.83±0.06 | 0.43±0.15 | 0.65±0.18 | 0.53±0.18 |
| Deformed atlas vs. test patient (α = 0.6) | 0.94±0.04 | 0.75±0.11 | 0.85±0.14 | 0.83±0.05 |
| Deformed atlas vs. test patient (α = 0.8) | 0.95±0.03 | 0.79±0.1 | 0.89±0.14 | 0.84±0.06 |
| Deformed atlas vs. test patient (α = 1.0) | 0.95±0.03 | 0.82±0.09 | 0.93±0.08 | 0.86±0.04 |
| Deformed atlas vs. test patient (α = 1.2) | 0.95±0.04 | 0.86±0.07 | 0.93±0.11 | 0.86±0.04 |
| Deformed atlas vs. test patient (α = 1.4) | 0.95±0.04 | 0.85±0.06 | 0.92±0.12 | 0.86±0.04 |
In addition, the registration performance of Reg-Net depends on the hyper-parameter setting. To evaluate the influence of different choices of α in Eq. (1), we set the α by different values ranging from 0.6 to 1.4. To evaluate the performance of the registration accuracy, the DSCs between the OAR masks of the deformed atlas patient via Reg-Net under different settings of α and those of the test patient are calculated. By comparing the metrics of the DSC performance under different settings of α, we can evaluate the influence of different choices of α. These metrics is shown in Table I. As can be seen from the comparison of the row of “deformed atlas vs. test patient (α = 0.6 to 1.4)”, we can see that when α is set ranging from 0.8 to 1.4, the DSC of prostate shows little difference. Under the setting of α = [1.0, 1.2], the DSC of bladder shows highest average performance. Under the setting of α = [1.0, 1.4], the DSC of rectum shows highest average performance. In addition, under the setting of α = 1.2, the DSC of urethra reaches at highest average performance. Thus, we set the hyper-parameter α to 1.2 in this work.
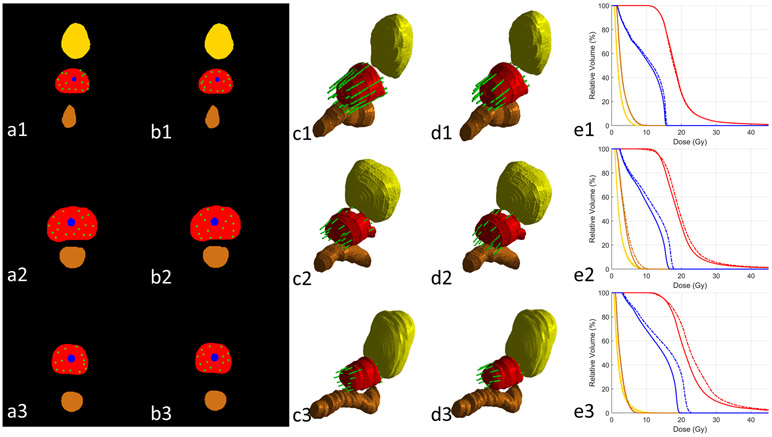
Representative results of the proposed method are compared side-by-side with the clinical ground truth for 3 patients in Figure 4. The results in Fig. 4 (1), (2) and (3) represent cases with above-average, average and under-average performance, respectively. Note that the “average” is regarded as that the overall DVH metrics difference between prediction and clinical dose of that case is close to the average DVH metrics difference among all patients (Table II). For above-average case (Fig. 4 (1)), the predicted catheter pattern appears close to the clinical pattern in sufficient peripheral catheter placements, which results into similar DVHs as clinical plan. The average case (Fig. 4 (2)) keeps good peripheral loading posteriorly but subtopimal laterally. The under-average case (Fig. 4 (3)) has noticeable insufficient catheter coverage at the patient left posteriolateral prostate, which causes higher heterogeneity in the prostate and OAR doses than clinical plan when prostate coverage (V100=95%) is normalized.
Figure 4.
The comparison between clinical and predicted catheter placements and their resultant DVHs of three representative cases (row (1), (2) and (3)). Columns (a) and (b) show the 2D clinical and predicted catheter placements at the central slice of prostate. Columns (c) and (d) show the 3D rendering clinical and predicted catheter placements. Column (e) demonstrate the DVHs of treatment plans from clinical (solid lines) and predicted (dashed lines) catheter placements. Red: prostate; yellow: bladder; brown: rectum; blue: urethra; green: catheter.
Table II.
Mean±Std of DVH metrics on plans based on clinical and predicted catheter placement and their difference.
| Prostate |
Bladder |
Rectum |
Urethra |
|||||
|---|---|---|---|---|---|---|---|---|
| V150 (%) |
V200 (%) | D90 (%) |
D2cc (%) | V75 (cc) |
D2cc (%) |
V75 (cc) |
V125 (cc) | |
| Clinical | 43.4±9.9 | 11.0±4.6 | 109.4±1.6 | 57.6±9.2 | 0.4±0.6 | 57.3±10.8 | 0.5±0.7 | 0.4±0.4 |
| Predicted | 48.4±10.9 | 13.9±6.9 | 110.3±1.9 | 61.1±9.4 | 0.6±0.8 | 58.7±11.0 | 0.6±0.7 | 0.8±0.5 |
| Difference | 5.0±6.5 | 2.9±4.3 | 0.9±1.5 | 3.5±3.4 | 0.2±0.4 | 1.5±4.1 | 0.1±0.4 | 0.3±0.4 |
The difference distribution of plan DVH metrics between the clinical and predicted catheter placement among the 90 patients is shown in Fig. S1 of Supplementary Document as box plot. The statistical results are summarized in Table II. Overall, both scenarios met the common clinical dose constraints on average when prostate dose coverage is kept at V100=95%. The plans from predicted catheter pattern have slightly higher dose heterogeneity in terms of V150 by 5.0% and V200 by 2.9% on average. For bladder V75, rectum V75 and urethra V125, the average difference is close to zero, and the range for most patients is within ±1cc.
4. CONCLUSIONS AND DISCUSSION
This study presented a novel method to predict catheter placement pattern for HDR prostate brachytherapy a priori. In the proposed learning-based method, combined criteria were used to rank multi-atlas data. Then, deep learning-based multi-atlas registration was used to bring the catheter positions from the top-ranked atlases to the new patient. The proposed method provides clinically feasible catheter patterns and plans with similar quality in terms of target and OAR DVHs as clinical catheter placement. This strategy has great potential to improve HDR prostate brachytherapy standardization and plan quality control by reducing reliance on physician experience.
We investigated the dosimetric impact of the predicted catheter pattern by comparing its DVHs with those of plans generated using the clinical catheter pattern. The higher hot spot in prostate and volume dose in OAR indicates that the plans based on the predicted catheter pattern, although very close, are still slightly inferior to the clinical catheter plans. This can be attributed to several limitations. First, the performance and robustness of catheter plan prediction of this proposed method depends on the multi-atlas dataset’s variation. If the OAR contour size, shape and distribution of a new arrival patient are significantly different than any of those in the atlas dataset, the predicted catheter plan of this patient would be substantially affected. Including more patient data to increase the applicability of the multi-atlas dataset will be our future work. Second, the training of the Reg-Net model, that brings catheter position from atlas patient to new arrival patient, is performed by the loss of comparing the binary masks of deformed OAR contours and target OAR contours. No ground truth deformation vector field was used to supervise this model. Thus, learning-based deformable registration is inherently an ill-posed problem that only contours information is available for DVF estimation in our work. Additional constraints are therefore necessary to regularize the DVF to be physically reasonable and physiologically plausible. Although we used regularization term (bending energy) to force the realism of the deformation vector field, the realism of deformation vector field would still be difficulty to guarantee when ground truth deformation vector field is not available. Recently, biomechanically constrained model was integrated into deep learning to enhance the realism of generated deformation vector field,24,25 which may help our method in our future work.
After the deformable registration, we selected the best catheter positions from the deformed catheters based on the distance from the catheters to the urethra and prostate surface. In this study, it is quantified by the distance between the center-of-mass of catheters and the surface of urethra/prostate. Such distance describes the general distance between the catheters and organs, and may not be the optimal metric for all patients. Other metrics such as orthogonal distance that quantifies the shortest distance may apply a more conservative criterion, and are worthy of further investigation.
In addition to the catheter pattern, the proposed method also provides the plan parameters (source dwell time and positions) and dose distribution that are optimized based on the predicted catheter pattern. It should be noted that the final plan and dose to be delivered to the patient may be different. One of the reasons is that the physicians have operating uncertainty in inserting catheters at predicted catheter locations.26 The specific anatomy of prostate and pubic bones of patients may also bend the catheters or even prohibit the placement of certain catheters, which leads to deviations from predicted catheter patterns. On the other hand, manual dose adjustment, which is not considered in this study, is usually applied after automatic optimization in clinical practice. It not only provides a better prostate dose coverage with less OAR dose for most patients, but also enables a more customized plan dose when physicians consider patient specific target/OAR dose tradeoff. Although the plan and dose that are directly optimized from the predicted catheter pattern may be changed during implementation, it still provides the physicians with the knowledge of potential achievable dose.
In this study, the binary maps of prostate and OARs used as input for the proposed method are from manual contouring on CT images. It thus requires a pre-plan CT image scan before OR. However, since the binary maps only provide the location and shapes of the organs, as long as they are accurate, our method is agnostic to the imaging modality used to generate contours. It is actually preferred to be from TRUS images obtained in the OR rather than CT because the patient has the same lithotomy position during TRUS images and catheter placement, which minimizes the difference of patient anatomy between prediction and catheter insertion. Moreover, the contours can also be obtained from automatic segmentation methods which have been developed with promising results on CT or TRUS in the last few years.27-30 Using auto-segmentation on TRUS can further streamline intraoperative catheter prediction.
In this study, we only considered the binary maps of prostate and OAR contours in deformation registration and multi-atlas ranking steps. Other factors such as the aforementioned pubic bone are not included as a factor in catheter prediction, which may cause deviation from predicted catheter pattern during clinical implementation. Future study may include the planning CT images that contains more anatomy information than binary contour maps as one of the registration loss in the step of deformation registration and as one of ranking criterion in the step of multi-atlas ranking. Moreover, dominant intraprostatic lesions have been recently considered as a focal boost target in HDR brachytherapy.31-34 Incorporating the information of dominant intraprostatic lesion with prostate/OAR may enable catheter prediction for HDR prostate focal boost brachytherapy.
Supplementary Material
Boxplot of DVH metrics difference between plans from clinical and predicted catheter placement. The central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers, and the outliers are plotted individually using the ‘+’ symbol.
ACKNOWLEDGEMENTS
This research was supported in part by the National Cancer Institute of the National Institutes of Health Award Number R01CA215718 and the Emory Winship Cancer Institute pilot grant.
Footnotes
Disclosure: None
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Yoshioka Y Current status and perspectives of brachytherapy for prostate cancer [published online ahead of print 2009/02/20]. International journal of clinical oncology. 2009;14(1):31–36. [DOI] [PubMed] [Google Scholar]
- 2.Peach MS, Trifiletti DM, Libby B. Systematic Review of Focal Prostate Brachytherapy and the Future Implementation of Image-Guided Prostate HDR Brachytherapy Using MR-Ultrasound Fusion [published online ahead of print 2016/06/14]. Prostate cancer. 2016;2016:4754031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, Rossi P, Jani A, et al. Improved prostate delineation in prostate HDR brachytherapy with TRUS-CT deformable registration technology: A pilot study with MRI validation [published online ahead of print 2017/03/16]. J Appl Clin Med Phys. 2017;18(1):202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, Rossi P, Jani A, Mao H, Curran WJ, Liu T. 3D Transrectal Ultrasound (TRUS) Prostate Segmentation Based on Optimal Feature Learning Framework [published online ahead of print 2016/02/01]. Proc SPIE Int Soc Opt Eng. 2016;9784:97842F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherr D, Swindle PW, Scardino PT. National Comprehensive Cancer Network guidelines for the management of prostate cancer. Urology. 2003;61(2, Supplement):14–24. [DOI] [PubMed] [Google Scholar]
- 6.Zaorsky NG, Doyle LA, Yamoah K, et al. High dose rate brachytherapy boost for prostate cancer: a systematic review [published online ahead of print 2013/11/16]. Cancer treatment reviews. 2014;40(3):414–425. [DOI] [PubMed] [Google Scholar]
- 7.Yoshioka Y, Yoshida K, Yamazaki H, Nonomura N, Ogawa K. The emerging role of high-dose-rate (HDR) brachytherapy as monotherapy for prostate cancer [published online ahead of print 2013/04/02]. Journal of radiation research. 2013;54(5):781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slessinger ED. Practical considerations for prostate HDR brachytherapy [published online ahead of print 2009/10/27]. Brachytherapy. 2010;9(3):282–287. [DOI] [PubMed] [Google Scholar]
- 9.Lei Y, Wang T, Tian S, et al. Male pelvic CT multi-organ segmentation using synthetic MRI-aided dual pyramid networks [published online ahead of print 2021/03/30]. Phys Med Biol. 2021;doi: 10.1088/1361-6560/abf2f9. [DOI] [PubMed] [Google Scholar]
- 10.Lessard E, Pouliot J. Inverse planning anatomy-based dose optimization for HDR-brachytherapy of the prostate using fast simulated annealing algorithm and dedicated objective function. Med Phys. 2001;28(5):773–779. [DOI] [PubMed] [Google Scholar]
- 11.Luong NH, Alderliesten T, Bel A, Niatsetski Y, Bosman PAN. Application and benchmarking of multi-objective evolutionary algorithms on high-dose-rate brachytherapy planning for prostate cancer treatment. Swarm and Evolutionary Computation. 2018;40:37–52. [Google Scholar]
- 12.Dinkla AM, van der Laarse R, Kaljouw E, et al. A comparison of inverse optimization algorithms for HDR/PDR prostate brachytherapy treatment planning [published online ahead of print 2014/12/03]. Brachytherapy. 2015;14(2):279–288. [DOI] [PubMed] [Google Scholar]
- 13.Edmundson GK, Yan D, Martinez AA. Intraoperative optimization of needle placement and dwell times for conformal prostate brachytherapy. International journal of radiation oncology, biology, physics. 1995;33(5):1257–1263. [DOI] [PubMed] [Google Scholar]
- 14.Sharma N, Aggarwal LM. Automated medical image segmentation techniques [published online ahead of print 2010/02/24]. Journal of medical physics. 2010;35(1):3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X. Deep learning in medical image registration: a review [published online ahead of print 2020/03/29]. Phys Med Biol. 2020;65(20):20TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaderberg M, Simonyan K, Zisserman A, Kavukcuoglu K. Spatial Transformer Networks. NIPS. 2015;arXiv:1506.02025. [Google Scholar]
- 17.Fu Y, Lei Y, Wang T, et al. LungRegNet: An unsupervised deformable image registration method for 4D-CT lung [published online ahead of print 2020/02/06]. Med Phys. 2020;47(4):1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra D, Chaudhury S, Sarkar M, Soin AS. Ultrasound Image Segmentation: A Deeply Supervised Network with Attention to Boundaries. IEEE Trans Biomed Eng. 2018;In press. [DOI] [PubMed] [Google Scholar]
- 19.Lei Y, Fu Y, Wang T, et al. 4D-CT deformable image registration using multiscale unsupervised deep learning [published online ahead of print 2020/02/26]. Phys Med Biol. 2020;65(8):085003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romera-Paredes B, Torr PHS. Recurrent Instance Segmentation. Paper presented at: Computer Vision – ECCV 2016; 2016//, 2016; Cham. [Google Scholar]
- 21.Oktay O, Schlemper J, Folgoc LL, et al. Attention U-Net: Learning Where to Look for the Pancreas. ArXiv. 2018;abs/1804.03999. [Google Scholar]
- 22.Zeng Q, Fu Y, Tian Z, et al. Label-driven magnetic resonance imaging (MRI)-transrectal ultrasound (TRUS) registration using weakly supervised learning for MRI-guided prostate radiotherapy [published online ahead of print 2020/04/25]. Phys Med Biol. 2020;65(13):135002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei Y, Fu Y, Harms J, et al. 4D-CT Deformable Image Registration Using an Unsupervised Deep Convolutional Neural Network. Paper presented at: Artificial Intelligence in Radiation Therapy; 2019//, 2019; Cham. [Google Scholar]
- 24.Fu Y, Lei Y, Wang T, et al. Biomechanically constrained non-rigid MR-TRUS prostate registration using deep learning based 3D point cloud matching. Medical Image Analysis. 2021;67:101845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y, Wang T, Lei Y, et al. Deformable MR-CBCT prostate registration using biomechanically constrained deep learning networks [published online ahead of print 2020/11/10]. Med Phys. 2020;DOI: 10.1002/mp.14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damore SJ, Syed AM, Puthawala AA, Sharma A. Needle displacement during HDR brachytherapy in the treatment of prostate cancer [published online ahead of print 2000/03/22]. International journal of radiation oncology, biology, physics. 2000;46(5):1205–1211. [DOI] [PubMed] [Google Scholar]
- 27.Yang L, Tonghe W, Sibo T, et al. Male pelvic CT multi-organ segmentation using synthetic MRI-aided dual pyramid networks. Phys Med Biol. 2021. [DOI] [PubMed] [Google Scholar]
- 28.Lei Y, Dong X, Tian Z, et al. CT prostate segmentation based on synthetic MRI-aided deep attention fully convolution network. Medical physics. 2020;47(2):530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong X, Lei Y, Tian S, et al. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network [published online ahead of print 2019/10/22]. Radiother Oncol. 2019;141:192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei Y, Tian S, He X, et al. Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net. Medical physics. 2019;46(7):3194–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T, Press RH, Giles M, et al. Multiparametric MRI-guided dose boost to dominant intraprostatic lesions in CT-based High-dose-rate prostate brachytherapy. The British Journal of Radiology. 2019;92(1097):20190089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Zhou J, Tian S, et al. A planning study of focal dose escalations to multiparametric MRI-defined dominant intraprostatic lesions in prostate proton radiation therapy. The British Journal of Radiology. 2020;93(1107):20190845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dearnaley DP, Sydes MR, Graham JD, et al. Escalated-dose versus standard-dose conformal radiotherapy in prostate cancer: first results from the MRC RT01 randomised controlled trial. The Lancet Oncology. 2007;8(6):475–487. [DOI] [PubMed] [Google Scholar]
- 34.Gomez-Iturriaga A, Casquero F, Urresola A, et al. Dose escalation to dominant intraprostatic lesions with MRI-transrectal ultrasound fusion High-Dose-Rate prostate brachytherapy. Prospective phase II trial. Radiother Oncol. 2016;119(1):91–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Boxplot of DVH metrics difference between plans from clinical and predicted catheter placement. The central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers, and the outliers are plotted individually using the ‘+’ symbol.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.